Abstract
Objective:
To explore circulating microRNAs (miRNAs) in cell-free CSF as novel biomarkers for multiple sclerosis (MS).
Methods:
Profiling of miRNAs in CSF of pooled patients with clinically isolated syndrome (CIS), patients with relapsing-remitting MS, and inflammatory and noninflammatory neurologic disease controls was performed using TaqMan miRNA arrays. Two independent patient cohorts (n = 142 and n = 430) were used for validation with real-time PCR.
Results:
We reliably detected 88 CSF miRNAs in the exploratory cohort. Subsequent validation in 2 cohorts demonstrated significantly higher levels of miR-150 in patients with MS. Higher miR-150 levels were also observed in patients with CIS who converted to MS compared to nonconverters, and in patients initiating natalizumab treatment. Levels of miR-150 correlated with immunologic parameters including CSF cell count, immunoglobulin G index, and presence of oligoclonal bands, and with candidate protein biomarkers C-X-C motif chemokine 13, matrix metallopeptidase 9, and osteopontin. Correlation with neurofilament light chain (NFL) was observed only when NFL was adjusted for age using a method that requires further validation. Additionally, miR-150 discriminated MS from controls and CIS converters from nonconverters equally well as the most informative protein biomarkers. Following treatment with natalizumab, but not fingolimod, CSF levels of miR-150 decreased, while plasma levels increased with natalizumab and decreased with fingolimod, suggesting immune cells as a source of miR-150.
Conclusions:
Our findings demonstrate miR-150 as a putative novel biomarker of inflammatory active disease with the potential to be used for early diagnosis of MS.
Classification of evidence:
This study provides Class II evidence that CSF miR-150 distinguishes patients with MS from patients with other neurologic conditions.
Multiple sclerosis (MS) is an immune-mediated demyelinating disease of the CNS that has, during the last decades, shifted from being a disease with very limited treatments to being among the most dynamic fields in clinical neurology regarding disease-modifying drugs (DMDs). While these drugs present new opportunities for personalized treatments, they also increase the need for reliable biomarkers to provide accurate diagnosis and prognosis, and to predict response to treatments.1 A large number of molecules detectable in blood or CSF has been suggested to reflect different disease processes; however, few have been replicated in large and controlled cohorts, which is necessary for potential clinical applications.2
MicroRNAs (miRNAs) are small, ∼22 nucleotide, noncoding RNAs, which regulate gene expression in a post-transcriptional manner.3,4 Besides intracellular functions, cell-free miRNAs have been detected in various biofluids,5 where they are packaged in vesicles (exosomes and microvesicles)6 and thereby protected from degradation. The potential for using circulating miRNAs as biomarkers for diagnostic, prognostic, and predictive applications has been a hot topic of investigation in many different diseases, including MS.7–10 At present, only one study comprehensively investigated miRNAs in CSF from patients with MS and reported differences in the levels of 3 miRNAs between patients with MS and controls.10
In this study, we profiled circulating miRNAs in cell-free CSF in a large cohort of patients with MS and controls identifying miR-150 as a putative novel biomarker for MS.
METHODS
Classification of evidence.
We included patients with relapsing-remitting MS (RRMS) and patients with clinically isolated syndrome (CIS) with a diagnosis according to the 2005 revisions to the McDonald criteria11 and neurologic disease controls who displayed no clinical or neuroradiologic features of MS. Furthermore, noninflammatory neurologic disease controls (NINDC) had no inflammatory lesions on MRI and no signs of intrathecal inflammation as shown by the presence of any of the following: oligoclonal bands (OCBs), increased immunoglobulin G (IgG) index, or pleocytosis (higher than 2 × upper normal limit, i.e., >10,000 cells/mL). Inflammatory neurologic disease controls (INDC) may or may not have inflammatory lesions on MRI together with the presence of intrathecal inflammation as defined above. The inclusion of samples into 4 different diagnostic categories (CIS, RRMS, NINDC, and INDC) was based on obtaining a comparable group size, age, female-to-male ratio, and the period of sampling among the 4 categories. The miR-150 assays were carried out on randomized samples in a blinded fashion and the quantification analysis of miR-150 levels was performed by 2 people, independently, who were blind to the diagnostic status of the samples.
The objective of this study was to profile circulating miRNAs in cell-free CSF in a large cohort of patients with MS and controls to identify a novel biomarker for MS. This study provides Class II evidence.
Patients.
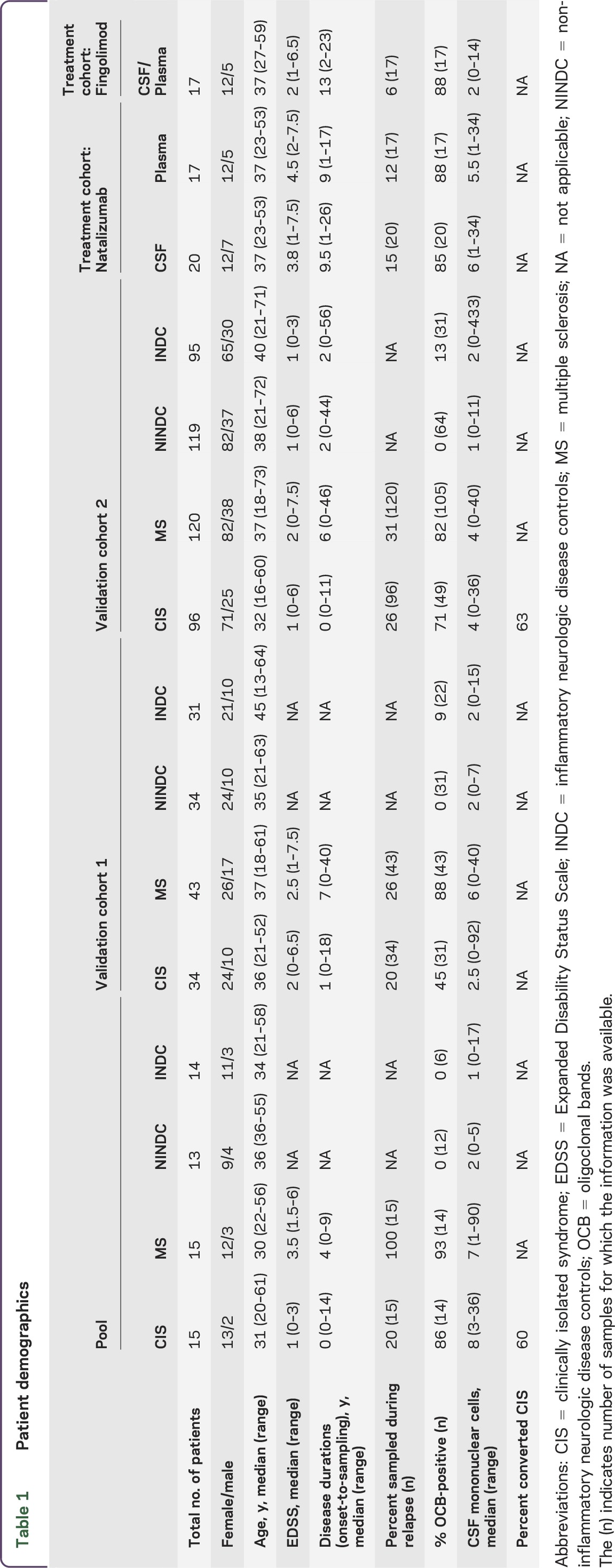
During routine visits to the neurology clinic at Karolinska University Hospital, patients' Expanded Disability Status Scale (EDSS) score was determined by the treating neurologist and CSF and blood samples were collected. Patients were subdivided into RRMS, CIS, INDC, and NINDC. MRI examinations on 1.5T or 3T scanners were done according to clinical routine. Here we collected information on number of T2 and gadolinium-enhancing lesions as judged from the original neuroradiologic evaluation of scans performed within ±6 months of sampling. For detailed patient demographics, see table 1.
Table 1.
Patient demographics
Handling of samples.
The CSF samples were centrifuged immediately after lumbar puncture at 440g for 10 minutes at room temperature to separate cells and larger particles from the CSF supernatant. The blood samples were collected in EDTA tubes and centrifuged at 1,500g for 15 minutes. Both the plasma phase and the CSF supernatant were batched and stored at −80°C until use. Methodologic details regarding measurements of C-X-C motif chemokine 13 (CXCL13), matrix metallopeptidase 9 (MMP-9), osteopontin (OPN), and neurofilament light chain (NFL) in CSF can be found in the original study.12 Levels of miRNA in CSF and plasma of patients with MS were determined at baseline and 12 months after open-label treatment with natalizumab or fingolimod.
Standard protocol approvals, registrations, and patient consents.
CSF and blood samples were handled according to consensus guidelines.13 The study was approved by the regional ethical committee (ethical permit 2009/2107-31/2) and written informed consent was obtained from all patients.
RNA isolation from cell-free CSF.
For miRNA TaqMan low density arrays (TLDA), 4 CSF pools were created by combining equal volumes of CSF from patients with CIS (n = 15), MS (n = 15), NINDC (n = 13), and INDC (n = 14). RNA was isolated from 500 μL using the miRCURY RNA isolation kit for biofluids (Exiqon, Vedbaek, Denmark) according to manufacturer's protocol. For normalization of sample-to-sample variation, 2 fmol synthetic Arabidopsis thaliana miRNA, ath-miR159a, 5′-UUUGGAUUGAAGGGAGCUCUA-3′ (RNA oligonucleotide, Eurofins Genomics, Ebersberg, Germany), was spiked into each CSF pool after addition of Lysis Solution BF.
In addition to TLDA cards where CSF pools were used, individual CSF samples used in the profiling pools (n = 57) and validation cohort 1 (n = 142) were subjected to total RNA isolation using a standard TRIzol protocol, where 700 μL of TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA) was added to 300 μL CSF sample. For normalization of sample-to-sample variation, 2 fmol synthetic Arabidopsis thaliana miRNA ath-miR159a was spiked into each CSF sample after the addition of TRIzol.
RNA isolation for validation cohort 2 (n = 450) and natalizumab- (n = 20) and fingolimod-treated (n = 17) patients was performed on 300 μL CSF using the miRCURY RNA isolation kit for biofluids. For normalization of sample-to-sample variation, synthetic Caenorhabditis elegans miRNAs cel-miR-39, 5′-UCA CCG GGU GUA AAU CAG CUU G-3′, cel-miR-54, 5′-UAC CCG UAA UCU UCA UAA UCC GAG-3′, cel-miR-238, 5′-UUU GUA CUC CGA UGC CAU UCA GA-3′ (RNA oligonucleotides, Integrated DNA Technologies, Coralville, IA), were added as a mixture of 3 fmol per oligonucleotide to each denatured sample. All eluted RNA was immediately stored at −80°C.
RNA isolation from cell-free plasma.
RNA isolation from natalizumab- (n = 17) and fingolimod-treated (n = 17) patients as well as a plasma cohort (n = 156) was performed from 100 μL plasma using the miRCURY RNA isolation kit for biofluids according to manufacturer's protocol. For normalization of sample-to-sample variation, synthetic Caenorhabditis elegans miRNAs cel-miR-39, cel-miR-54, and cel-miR-238 were added as a mixture of 15 fmol per oligonucleotide to each denatured sample. Eluted RNA was immediately stored at −80°C.
MiRNA profiling with TLDA.
MiRNA was reverse transcribed using Megaplex RT Primers, Human Pool Set v3.0, and TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Life Technologies) according to manufacturer's instructions. The reverse transcription (RT) products were preamplified using Megaplex PreAmp Primers, Human Pool Set v3.0, and TaqMan PreAmp Master Mix Kit. The preamplified (PreAmp) products were loaded onto TLDA cards (TaqMan Array Human MicroRNA A+B Cards Set v3.0) and run on the ABI 7900HT real-time PCR system. Raw data were analyzed using the ExpressionSuite Software (Life Technologies). To ensure good quality detection and to avoid false-positives, a stringent cutoff for detection was set where at least 2 samples should have Ct <32, AmpScore >1.1, and minimum 2 samples with adequate Cq Confidence (AmpScore and Cq Confidence were provided by ExpressionSuite software). Cutoff thresholds were set in consultation with technical experts at Applied Biosystems. Raw Ct values were normalized to the spike-in control using the following equation: normalized sample Ct = raw sample Ct − (spike-in Ct value for sample − average of all spike Ct values). A and B cards were normalized separately.
Quantitative real-time PCR.
Multiplexed RT and preamplification products were created using the Protocol for Creating Custom RT and Preamplification Pools using TaqMan MicroRNA Assays (publication 4465407, Applied Biosystems). Preamplified products were diluted 8× in 0.1×TE prior to real-time PCR using individual miRNA assays on the CFX384 real-time PCR detection system (Bio-Rad, Hercules, CA). Individuals with undetectable miRNA levels were excluded from analysis. Raw data were analyzed and Ct values extracted using CFX Manager software. For validation cohort 1, miRNA levels were calculated using the following equation: ΔCt = Ct miRNA − Ct ath-miR159a, ΔΔCt = 2^ −(ΔCt − Avg ΔCt). All ΔΔCt values were normalized to the lowest ΔΔCt. For validation cohort 2, treatment, and plasma cohorts, 3 interplate controls were run for each miRNA assay on each plate to adjust for plate-to-plate variations. MiRNA levels were calculated using the following equation: ΔCt = Ct miRNA − Ct Avg spike-in, ΔΔCt = 2^ −(ΔCt − Avg ΔCt). All ΔΔCt values were normalized to the lowest ΔΔCt.
Statistical analysis.
Qualitative data were analyzed using Kruskal-Wallis one-way analysis of variance combined with the Dunn test of multiple comparisons. For 2-group comparisons, Mann-Whitney U test was used. Wilcoxon signed rank test for matched pairs was used to analyze the treatment cohort. All correlations were performed on ln-transformed values (ln[x + 1] transformation was applied for all parameters apart from NFL adjusted for age-related changes, where ln[x + 281] was used) and analyzed using Spearman rank test. All statistical analyses were performed using GraphPad Prism 5 (San Diego, CA). NFL adjustment for age-related changes was performed by subtracting the expected level for the given age calculated following linear relationship reported in healthy individuals (i.e., 11.8 ng/L × age − 95 ng/L23) from the measured NFL level.
Receiver operating characteristic (ROC) curve analyses, between MS and NINDC as well as CIS and CIS-MS, were performed using pROC and Epi packages in R. Independent contribution of multiple variables was tested using generalized linear model in Rcmdr package in R.
RESULTS
Detection of miRNAs in cell-free CSF.
Profiling of miRNAs using TLDA cards enables quantification of the 754 most common human miRNAs. We profiled miRNAs in pooled samples from CIS (n = 15), MS (n = 15), and controls (NINDC, n = 13, and INDC, n = 14) and we identified 88 miRNAs in cell-free CSF (63 miRNAs passed the most stringent detection cutoff and an additional 25 miRNAs were detected when allowing for reduced amplification quality in maximum 2 samples) (table e-1 at Neurology.org/nn). Subsequently, 15 miRNAs were selected, based on quantifiable levels (detection in at least 75% of the individual samples of the profiling pools) and an indication of a larger difference between MS and NINDC (table e-1), for examination in an independent sample cohort (validation cohort 1, n = 142). Out of the tested miRNAs, only miR-145 and miR-150 displayed significant differences between MS and NINDC (p = 0.0038 and p = 0.0027, respectively) (figure e-1). Together these results demonstrate detectability of circulating miRNAs in cell-free CSF and differential presence of miR-145 and miR-150.
Levels of miR-150 are elevated in patients with MS and associate with markers of inflammation.
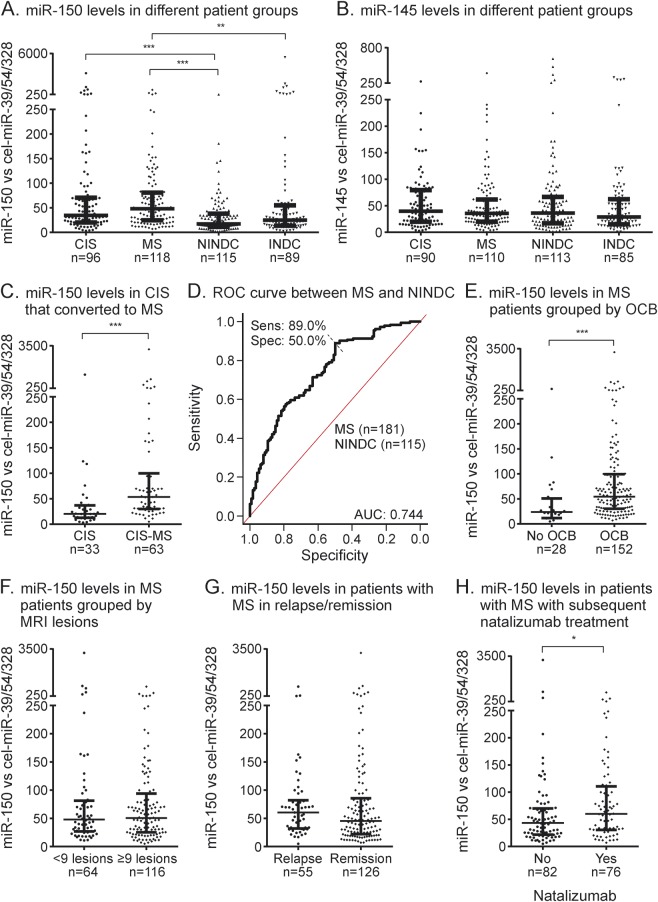
We next sought to determine whether the observed differences could be confirmed in a larger cohort, validation cohort 2 (n = 430). MiR-145 was particularly interesting as it had been reported as a biomarker of MS in plasma and serum,7,14 as well as in peripheral blood mononuclear cells (PBMC).7,15 MiR-150 has not been reported in MS biofluids, but is differentially regulated, albeit in small cohorts, in PBMC and T cells.16,17 In this large cohort, we could replicate significantly higher levels of miR-150 in MS compared to both control groups as well as between CIS and NINDC (p < 0.0001) (figure 1A). Notably, however, we were not able to reproduce the differences observed for miR-145 (p = 0.73) (figure 1B). We also observed higher miR-150 levels in CSF from patients with CIS who subsequently converted to MS (CIS-MS) compared to those who did not convert during follow-up (median period of 52 months) (p < 0.0001) (figure 1C). To evaluate the diagnostic value of miR-150 in differentiating patients with MS and controls, we constructed a ROC curve (figure 1D). Area under the curve (AUC) for miR-150 was 0.744 (50% specificity and 89% sensitivity).
Figure 1. Levels of miR-150 are elevated in patients with multiple sclerosis (MS) and patients with clinically isolated syndrome (CIS) who convert to MS.
Relative levels of mature microRNAs (miRNAs) were measured using multiplexed specific TaqMan miRNA assays and normalized to an average of 3 spike-ins (cel-miR-39, cel-miR-54, and cel-miR-238). Levels of (A) miR-150 are significantly different between disease groups, while (B) there was no difference between levels of miR-145. Levels of miR-150 are also (C) higher in patients with CIS who converted to MS (CIS-MS) compared to those who never converted and (D) can discriminate MS from noninflammatory neurologic disease controls (NINDCs) based on receiver operating characteristic (ROC) curve analysis. Graph intersection indicates the cutoff value for miR-150 that proved the best specificity and sensitivity. Levels of miR-150 in relation to descriptive disease parameters (E) oligoclonal bands (OCB), (F) relapse and remission, (G) number of MRI lesions, and (H) subsequent treatment with natalizumab. Lines in dot plots represent median and interquartile range. *p < 0.05, **p < 0.01, ***p < 0.001. INDC = inflammatory neurologic disease controls.
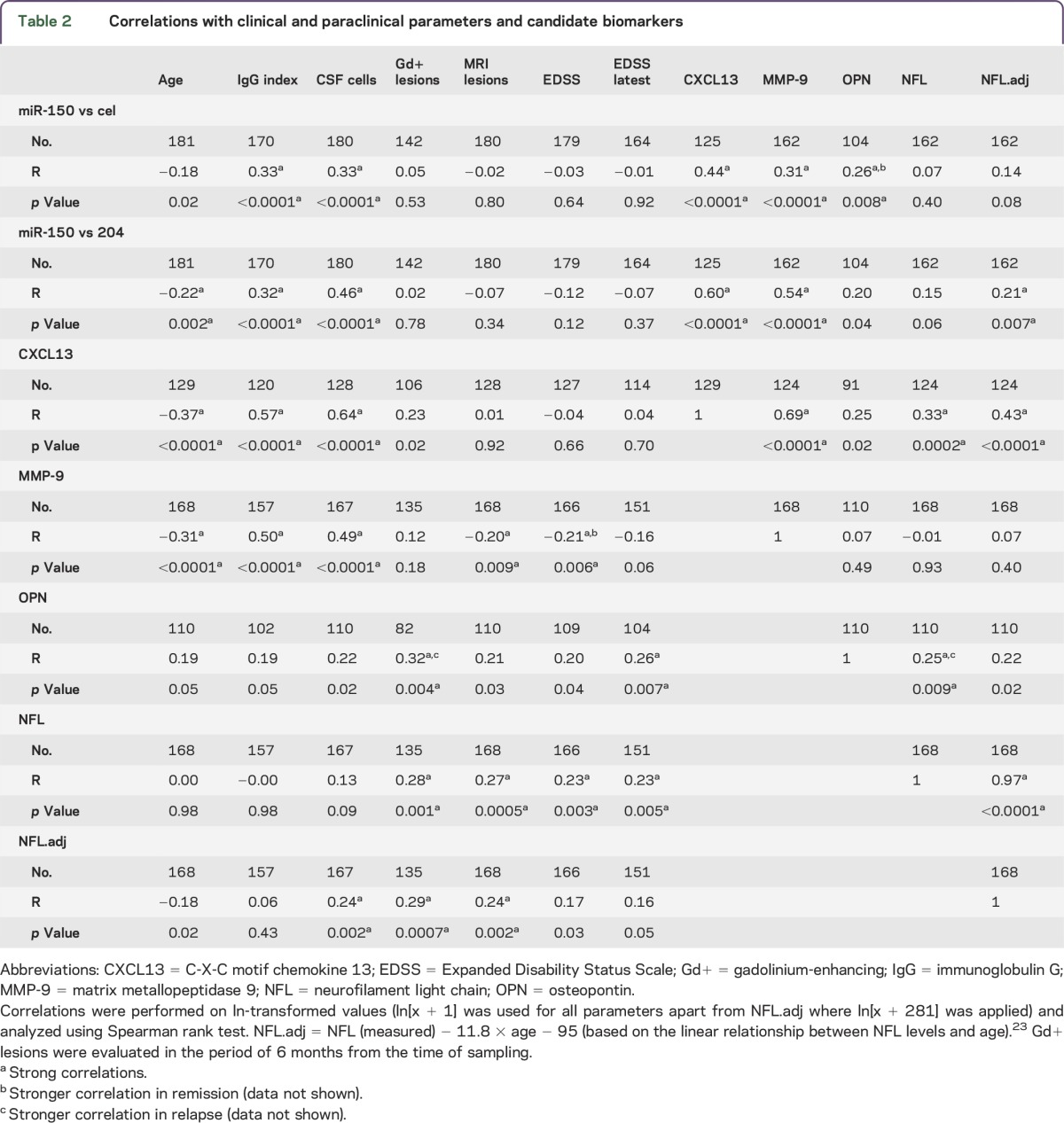
To explore the relation of miR-150 to MS disease processes, we correlated miR-150 with established laboratory markers of disease. We observed significantly higher levels of miR-150 in patients with OCBs compared to OCB-negative patients (p = 0.0003) (figure 1E). High levels of miR-150 correlated with higher CSF cell numbers (p < 0.0001, r = 0.33) and higher IgG index (p < 0.0001, r = 0.33) (table 2), indicating that miR-150 associates with active inflammation. In contrast, miR-150 levels did not correlate with the number of MRI T2 lesions and EDSS score (figure 1F, table 2) and there was only a tendency for higher miR-150 levels in relapse (p = 0.1) (figure 1G). However, patients with RRMS who subsequently initiated treatment with natalizumab displayed higher levels compared to all other patients with RRMS (p = 0.022) (figure 1H) and this effect was independent from the number of MRI T2 lesions (p = 0.038). Finally, we also investigated the relationship between miR-150 levels and candidate CSF biomarkers for MS, CXCL13, MMP-9, OPN, and NFL. We observed that higher levels of miR-150 correlate with higher levels of CXCL13 (p < 0.0001, r = 0.44), MMP-9 (p < 0.0001, r = 0.31), and OPN (p = 0.008, r = 0.26) (table 2). Together these results indicate that miR-150 is a marker of CNS inflammation.
Table 2.
Correlations with clinical and paraclinical parameters and candidate biomarkers
Effect of DMDs on miR-150 levels in CSF and plasma.
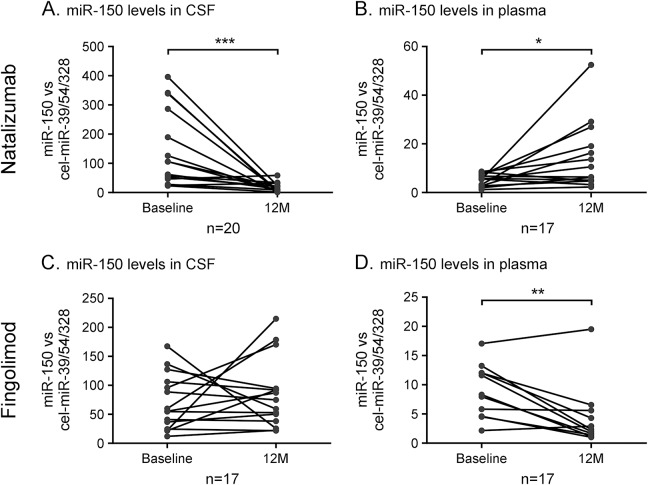
Well-established DMDs with known actions include natalizumab and fingolimod. Patients treated with natalizumab displayed a significant reduction of CSF miR-150 levels after 12 months of treatment (figure 2A). Concomitantly, an increase in plasma levels of miR-150 was found in these patients (figure 2B). A divergent pattern was observed in patients initiating fingolimod, where plasma miR-150 levels decreased following treatment, while CSF levels remained unchanged (figure 2C,D). The findings of regulated miR-150 levels in plasma following treatment raised the question of whether miR-150 could act as a biomarker for MS also in plasma. To investigate this, we analyzed 156 patients and controls from validation cohort 2 from whom plasma samples were available. However, we could not observe differences in miR-150 levels between any of the disease groups (data not shown). This suggests that the treatments affect levels of miR-150 but that plasma levels of miR-150 are unlikely to serve as a marker of MS.
Figure 2. Levels of miR-150 are altered in CSF and plasma following treatment with disease-modifying drugs.
Relative levels of mature miR-150 were measured in CSF and in plasma from patients with multiple sclerosis treated with (A,B) natalizumab and (C,D) fingolimod at baseline and at 12 months. Quantification was performed using multiplexed specific TaqMan microRNA assays and normalized to an average of 3 spike-ins (cel-miR-39, cel-miR-54, and cel-miR-238). *p < 0.05, **p < 0.01, ***p < 0.001.
Ratio of miR-150 and miR-204 as a biomarker in CSF.
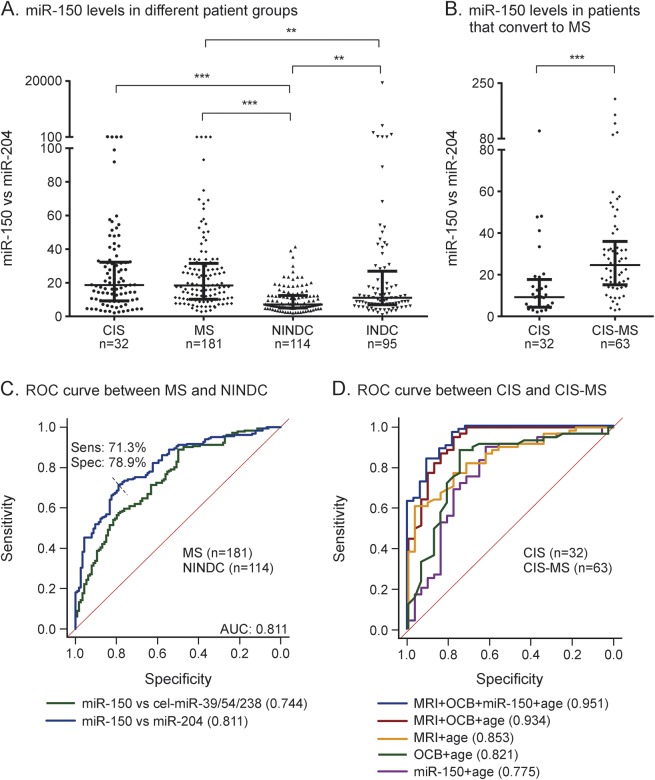
The rationale for spiking CSF samples with synthetic miRNAs is the absence of circulating endogenous miRNA for normalization.18,19 An alternative is to use miRNA pairs,20 where the ratio (ΔCt) of all miRNAs are calculated and the pair that provides the highest accuracy in differentiating patients and controls is selected. We performed this comparison for the miRNAs in validation cohort 1 and established that the pair of miR-150 and miR-204 provides the largest difference between MS and NINDC (table e-2). Levels of miR-150 normalized with miR-204 were significantly higher in MS compared to NINDC and INDC in validation cohort 2 (figure 3A) as well as in patients with CIS who subsequently converted to MS compared to those who did not convert (p < 0.0001) (figure 3B). The miR-150/miR-204 ratio also displayed a stronger correlation compared to miR-150 alone with candidate CSF biomarkers (especially CSF cells, CXCL13 and MMP-9), including significant correlation with age-adjusted NFL levels (p < 0.007, r = 0.21) (table 2). Additionally, AUC identifies miR-150/miR-204 ratio as a better marker in differentiating MS and NINDC (AUC 0.811, 79% specificity and 71% sensitivity) (figure 3C) compared to miR-150 normalized to spike-ins. In this respect, miR150/miR-204 ratio performed as well as the current best protein biomarker, CXCL13 (figure e-2A). Finally, miR-150/miR-204 ratio could differentiate patients with CIS who converted to MS compared to those who did not convert (AUC = 0.775) and had an independent effect (p = 0.017) from the factors known to affect conversion, namely age, OCB status, and MRI lesions (figure 3D). Even in this respect, the miR-150/miR-204 ratio performed similarly to informative protein biomarkers CXCL13 and MMP-9, while NFL had no predictive power (figure e-2B). Together this demonstrates the potential for using the miR-150/miR-204 ratio as a biomarker for discriminating patients with MS from controls and patients with CIS that will convert to MS.
Figure 3. MicroRNA (miRNA) pairs can be used to normalize miRNA levels.
The ratio of miR-150 and miR-204, calculated and normalized using the ΔΔCT method (see Methods), is significantly different (A) between disease groups and (B) between patients with clinically isolated syndrome (CIS) who have converted or not converted to multiple sclerosis (MS). The miR-150/miR-204 ratio improves (C) the area under receiver operating characteristic (ROC) (AUC) curve, indicating improved specificity and sensitivity to discriminate MS from noninflammatory neurologic disease controls (NINDC) compared to miR-150 normalized to spike-ins. The miR-150/204 ratio can furthermore (D) predict conversion from CIS to MS as indicated by the ROC curve generated to compare converters and nonconverters. The miR-150/miR-204 ratio is an independent significant (p = 0.017) predictor of conversion when known risk factors such as age, oligoclonal bands (OCB), and MRI lesions are taken into account, and it can improve their combined predictive value. Lines represent (A, B) median and interquartile range. **p < 0.01, ***p < 0.001. INDC = inflammatory neurologic disease controls.
DISCUSSION
In the last few years, circulating miRNAs have become a new class of biomarkers for various diseases including CNS disorders. In this study using an unbiased exploratory approach with subsequent validation in 2 large cohorts we identified CSF levels of circulating miR-150 as a promising candidate biomarker for MS.
We identified higher levels of circulating miR-150 in CSF of patients with MS compared to both inflammatory and noninflammatory controls in 2 cohorts jointly comprising nearly 600 individuals. The levels of miR-150 correlated with established immunologic parameters, i.e., CSF cell numbers, IgG index, presence of OCB, as well as the candidate immune-associated biomarkers CXCL13, MMP-9, and OPN. The strongest correlation was observed with CXCL13 and MMP-9, suggesting that miR-150 reflects similar qualities of the immune responses. We also observed correlation with NFL, which is released during ongoing axonal injury, e.g., upon inflammatory attack,21 and is a marker of disease activity in MS,22 but only after adjusting levels of NFL for age-related changes.23 However, correlation with NFL should be taken with caution considering the novelty of our approach to age-adjust NFL levels, which requires further validation. Together these results indicate that miR-150 is a marker of CNS inflammation in the context of MS. Additionally, miR-150 was higher in patients with CIS who converted to MS compared to those who did not convert during follow-up, indicating a potential to facilitate earlier diagnosis. The observation that miR-150 is not immediately affected by disease dynamics, i.e., relapse vs remission, suggests that it may serve as a more stable prognostic marker than for instance CXCL13 and NFL, which are both upregulated during relapses (p = 0.01 and 0.02, respectively). Accordingly, we detected higher levels of miR-150 in patients who were subsequently treated with natalizumab, suggesting that higher miR-150 levels associate with more inflammatory active disease.
Previously, 63 miRNAs were reported to be differentially expressed between MS lesions and healthy white matter.24 Nearly 40% of these miRNAs were also detected in our study, indicating that circulating miRNAs in the CSF may mirror events in the target tissue. Surprisingly, we could not detect any of the 3 miRNAs (miR-181c, miR-633, and miR-922)10 previously reported to be differentially regulated in CSF of patients with MS. This lack of overlap between the studies probably reflects differences in separation of cells from the cell-free CSF fraction. Several recent studies report miR-150 among the more abundant miRNAs in cell-free CSF using different methods,25–28 corroborating our method of measuring circulating miRNAs in cell-free CSF, which is handled according to consensus guidelines.13
Although it is challenging to establish the source of circulating miRNAs, our data suggest that immune cells release miR-150. Thus, natalizumab reduced miR-150 levels in the CSF with a concurrent increase of miR-150 levels in plasma, which parallels drug-induced changes in immune cell numbers in the 2 compartments.29 Along this line, we also observed reduced levels of miR-150 in plasma in patients on fingolimod treatment, reflecting sequestration of immune cells within secondary lymphoid organs.30 It has been shown that activated monocytes and T and B cells selectively package and secrete miR-150 in microvesicles.31,32 All these cell types are found in CSF and CNS of patients with MS33 and could thus contribute to miR-150 release, although the exact source remains to be defined.
Functional studies of intracellular miR-150 suggest a role in regulating T- and B-cell development34–37; however, the functionality of circulating miRNAs is far from clarified. They might mediate cell-to-cell communication,38,39 e.g., lymphocyte-derived exosomes are known to transfer miRNAs to CD8+ T cells and antigen-presenting cells.38 Our observation of higher levels of extracellular miR-150 compared to previous reports of reduced levels of miR-150 in PBMCs16 and T cells17 in patients with MS suggests that packaging of miR-150 is likely an active process and that exported miR-150 may have functional roles. It has been shown that infiltrating monocytes actively secrete miR-150, thereby affecting endothelial cells.32,39 However, any functional implications of circulating miR-150 in MS remain to be investigated.
Given the heterogeneity and complexity of MS it is unlikely that a single biomarker will be able to satisfy all needs for disease monitoring. Our data indicate that candidate biomarkers, while largely overlapping, also reflect slightly different qualities of inflammatory responses (table 2) and while some can discriminate MS from controls (CXCL13, MMP-9, and NFL), others cannot (OPN) (figure e-2A). Similarly, while CXCL13 and MMP-9 can discriminate CIS converters from nonconverters, NFL has very little predictive value (figure e-2B). A panel of markers is more likely to achieve sufficient sensitivity and specificity. Indeed, we here show that the difference between miR-150 and miR-204 has improved biomarker characteristics compared to miR-150 alone, which may reflect the regulation of both miRNAs or more accurate normalization compared to spike-ins. Indeed, the miR-150/miR-204 ratio had very similar predictive capacity to discriminate MS from controls compared to the current best protein biomarker that we tested, CXCL13. It also performed similarly to CXCL13 and MMP-9 in discriminating CIS converters from nonconverters and it has an additive independent effect even compared to known risk factors such as OCB and MRI lesions. At present, most candidate biomarkers in MS, both in serum and CSF, are proteins.2 Compared to proteins, assays to study miRNAs are easily multiplexed, providing a possibility to detect a broad spectrum of biomarkers from a single isolation procedure. This together with the extraordinary stability of circulating miRNAs even during unfavorable conditions40 makes miRNAs attractive candidates for biomarker selection.
Using large cohorts and an unbiased detection strategy, we demonstrate the potential of miR-150 as an early marker of inflammatory active disease that warrants further investigation. Future studies employing broad screening methods in large cohorts hold the potential to define novel sensitive and specific biomarkers based on panels of miRNAs.
Supplementary Material
GLOSSARY
- AUC
area under the curve
- CIS
clinically isolated syndrome
- CXCL13
C-X-C motif chemokine 13
- DMD
disease-modifying drug
- EDSS
Expanded Disability Status Scale
- IgG
immunoglobulin G
- INDC
inflammatory neurologic disease controls
- miRNA
microRNA
- MMP-9
matrix metallopeptidase 9
- MS
multiple sclerosis
- NFL
neurofilament light chain
- NINDC
noninflammatory neurologic disease controls
- OCB
oligoclonal band
- OPN
osteopontin
- PBMC
peripheral blood mononuclear cells
- ROC
receiver operating characteristic
- RRMS
relapsing-remitting multiple sclerosis
- RT
reverse transcription
- TLDA
TaqMan low density arrays
Footnotes
Supplemental data at Neurology.org/nn
AUTHOR CONTRIBUTIONS
Dr. Bergman: designed the study, performed experimental work, contributed to statistical analyses, interpreted data, and wrote and edited the manuscript. E. Piket: contributed to experimental work and writing and submission of the manuscript. Dr. Khademi: managed samples. T. James: contributed to statistical analyses. Prof. Brundin: contributed to sample collection and data interpretation. Prof. Olsson: contributed to sample collection and data interpretation. Prof. Piehl: contributed to sample collection and data interpretation and critically revised the manuscript. Dr. Jagodic: designed the study, supervised the study, performed statistical analysis, interpreted data, and wrote and edited the manuscript.
STUDY FUNDING
This work was supported by grants from the Swedish Research Council, the Swedish Association for Persons with Neurological Disabilities, the Swedish Brain Foundation, the Swedish Medical Society, Petrus and Augusta Hedlunds Foundation, the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, and AstraZeneca (AstraZeneca-Science for Life Laboratory collaboration).
DISCLOSURE
P. Bergman reports no disclosures. E. Piket received research support from Neuro Sweden. M. Khademi and T. James report no disclosures. L. Brundin is on the scientific advisory board for Genzyme and Biogen; received speaker honoraria from Biogen and Genzyme; provided written information for the Swedish MPA on MS treatment; receives royalties from the Swedish Brain Fund; participated in an airline commercial; and received research support from Swedish MRC, Karolinska Institutet Stockholm County, and Soderberg Foundation. T. Olsson served on the scientific advisory boards for Merck-Serono, Biogen Idec, Genzyme/Sanofi Aventis, and Novartis; received travel funding and/or speaker honoraria from Novartis, Biogen Idec, Sanofi Aventis, Merck, Genzyme, and Medimmune; was co-editor for Current Opinion in Immunology; and received research support from Merck, Biogen Idec, Sanofi Aventis, Bayer, Novartis, Astra Zeneca, The Swedish Research Council, EU fp7, Euratrans Neuroinox, combiMS, Swedish Brain Foundation, AFA Foundation, Knut and Alice Wallenberg Foundation, Bayer Schering, Genzyme/Sanofi-Aventis, Biogen Idec, and Astra Zeneca. F. Piehl served on the data safety monitoring committee for Parexel/Chugai; and received research support from Biogen, Novartis, Genzyme, and Swedish Medical Research Council. M. Jagodic is on the editorial board for Physiological Genomics; and received research support from Astra Zeneca, The Swedish Research Council, ALF, The Swedish Association for Persons with Neurological Disabilities, The Swedish Medical Society, and Petrus and Augusta Hedlunds Foundation. Go to Neurology.org/nn for full disclosure forms.
REFERENCES
- 1.Piehl F. A changing treatment landscape for multiple sclerosis: challenges and opportunities. J Intern Med 2014;275:364–381. [DOI] [PubMed] [Google Scholar]
- 2.Comabella M, Montalban X. Body fluid biomarkers in multiple sclerosis. Lancet Neurol 2014;13:113–126. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 4.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene 2006;25:6163–6169. [DOI] [PubMed] [Google Scholar]
- 5.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi R, Healy B, Gholipour T, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol 2013;73:729–740. [DOI] [PubMed] [Google Scholar]
- 8.Fenoglio C, Ridolfi E, Cantoni C, et al. Decreased circulating miRNA levels in patients with primary progressive multiple sclerosis. Mult Scler 2013;19:1938–1942. [DOI] [PubMed] [Google Scholar]
- 9.Siegel SR, Mackenzie J, Chaplin G, Jablonski NG, Griffiths L. Circulating microRNAs involved in multiple sclerosis. Mol Biol Rep 2012;39:6219–6225. [DOI] [PubMed] [Google Scholar]
- 10.Haghikia A, Haghikia A, Hellwig K, et al. Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology 2012;79:2166–2170. [DOI] [PubMed] [Google Scholar]
- 11.Polman CH, Reingold SC, Edan G, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria.” Ann Neurol 2005;58:840–846. [DOI] [PubMed] [Google Scholar]
- 12.Khademi M, Dring AM, Gilthorpe JD, et al. Intense inflammation and nerve damage in early multiple sclerosis subsides at older age: a reflection by cerebrospinal fluid biomarkers. PLoS One 2013;8:e63172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teunissen CE, Petzold A, Bennett JL, et al. A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology 2009;73:1914–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sondergaard HB, Hesse D, Krakauer M, Sorensen PS, Sellebjerg F. Differential microRNA expression in blood in multiple sclerosis. Mult Scler 2013;19:1849–1857. [DOI] [PubMed] [Google Scholar]
- 15.Keller A, Leidinger P, Lange J, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One 2009;4:e7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinelli-Boneschi F, Fenoglio C, Brambilla P, et al. MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci Lett 2012;508:4–8. [DOI] [PubMed] [Google Scholar]
- 17.Jernas M, Malmestrom C, Axelsson M, et al. MicroRNA regulate immune pathways in T-cells in multiple sclerosis (MS). BMC Immunol 2013;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts TC, Coenen-Stass AM, Wood MJ. Assessment of RT-qPCR normalization strategies for accurate quantification of extracellular microRNAs in murine serum. PLoS One 2014;9:e89237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010;50:298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA 2011;108:3713–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Povlishock JT. Pathobiology of traumatically induced axonal injury in animals and man. Ann Emerg Med 1993;22:980–986. [DOI] [PubMed] [Google Scholar]
- 22.Lycke JN, Karlsson JE, Andersen O, Rosengren LE. Neurofilament protein in cerebrospinal fluid: a potential marker of activity in multiple sclerosis. J Neurol Neurosurg Psychiatry 1998;64:402–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vågberg M, Norgren N, Dring A, et al. Levels and age dependency of neurofilament light and glial fibrillary acidic protein in healthy individuals and their relation to the brain parenchymal fraction. PLoS One 2015;10:e0135886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junker A, Krumbholz M, Eisele S, et al. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009;132:3342–3352. [DOI] [PubMed] [Google Scholar]
- 25.Bjersing JL, Lundborg C, Bokarewa MI, Mannerkorpi K. Profile of cerebrospinal microRNAs in fibromyalgia. PLoS One 2013;8:e78762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holm A, Bang-Berthelsen CH, Knudsen S, et al. MiRNA profiles in cerebrospinal fluid from patients with central hypersomnias. J Neurol Sci 2014;347:199–204. [DOI] [PubMed] [Google Scholar]
- 27.Denk J, Boelmans K, Siegismund C, Lassner D, Arlt S, Jahn H. MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer's disease. PLoS One 2015;10:e0126423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgos KL, Javaherian A, Bomprezzi R, et al. Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing. RNA 2013;19:712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stuve O, Marra CM, Jerome KR, et al. Immune surveillance in multiple sclerosis patients treated with natalizumab. Ann Neurol 2006;59:743–747. [DOI] [PubMed] [Google Scholar]
- 30.Chun J, Hartung HP. Mechanism of action of oral fingolimod (FTY720) in multiple sclerosis. Clin Neuropharmacol 2010;33:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Candia P, Torri A, Gorletta T, et al. Intracellular modulation, extracellular disposal and serum increase of MiR-150 mark lymphocyte activation. PLoS One 2013;8:e75348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Zhang Y, Liu Y, et al. Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem 2013;288:23586–23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stangel M, Fredrikson S, Meinl E, Petzold A, Stuve O, Tumani H. The utility of cerebrospinal fluid analysis in patients with multiple sclerosis. Nat Rev Neurol 2013;9:267–276. [DOI] [PubMed] [Google Scholar]
- 34.Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007;131:146–159. [DOI] [PubMed] [Google Scholar]
- 35.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA 2007;104:7080–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghisi M, Corradin A, Basso K, et al. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood 2011;117:7053–7062. [DOI] [PubMed] [Google Scholar]
- 37.Trifari S, Pipkin ME, Bandukwala HS, et al. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci USA 2013;110:18608–18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2011;2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 2010;39:133–144. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.