Abstract
Leukemia stem cells (LSCs) are self-renewable leukemia-initiating populations that are often resistant to traditional chemotherapy and tyrosine kinase inhibitors (TKI) currently used for treatment of acute or chronic myeloid leukemia (AML or CML). The persistence and continued acquisition of mutations in resistant LSCs represent a major cause for refractory disease and/or relapse following remission. Understanding the mechanisms regulating LSC growth and survival is critical for devising effective therapies that will improve treatment response and outcome. Several recent studies now indicate that the p53 tumor suppressor pathway is often inactivated in de novo myeloid leukemia through oncogenic specific mechanisms, which converge on aberrant p53 protein deacetylation. Here, we summarize our current understanding of various mechanisms underlying deregulation of histone deacetylases (HDACs), which could be exploited to restore p53 activity and enhance targeting of LSCs in molecularly defined patient subsets.
Introduction
Leukemia stem cells (LSCs), characterized by unlimited self-renewal capacity, are shown to be central to the initiation, growth and relapse of acute and chronic myelogenous leukemia (AML and CML). Studies in recent years have led to the view that the persistence of these clonal LSC subpopulations could be a major driving mechanism contributing to treatment refractory and/or relapse following remission [1-3]. It has also recently been brought to light that after chemotherapy treatment, clonal evolution from preleukemic hematopoietic stem cells (HSCs) could occur and promote development of chemoresistant relapse [4-6]. The heterogeneity and the dynamic nature of malignant disease progression appear increasingly complex. Meanwhile, it is now clear that new therapies more effective in targeting quiescent and chemoresistant LSCs are needed to improve treatment outcome and cure.
The tumor suppressor protein p53 is arguably the most studied molecule due to its central role in coordinating regulatory circuits that sense and respond to a wide variety of stressors including DNA damage and oncogenic events and ultimately control fundamental cell fate decisions such as cell cycle progression, apoptosis, senescence, metabolism, and autophagy [7,8]. The important role of p53 in cancer is underscored by the fact that genetic mutations in TP53 have been detected in approximately half of all human cancers and disruption of other p53 pathway components is prevalent in the remainder [9]. In myeloid leukemias, however, TP53 mutations are relatively infrequent (less than 10%) and mostly associate with complex karyotype and therapy related neoplasms [10-13]. Nevertheless, TP53 mutation is recognized as an adverse risk factor for chemotherapy response and prognosis [14,15]. As a master coordinator of important cellular processes, p53 function is regulated by a wide spectrum of post-translational modifications including phosphorylation, ubiquitination, acetylation, methylation and sumoylation [7,16-19]. It has been suggested that inactivation of non-mutated p53 frequently occurs through binding to its principal regulator MDM2, a E3 ubiquitin ligase that mediates degradation of p53 [20-22]. Compounds that directly interfere with the binding of p53 and MDM2, including Nutlins and MI-series inhibitors, have been developed and evaluated for anti-leukemia efficacy [23-32]. Multiple mechanisms have been observed to influence the efficacy of MDM2 inhibitors, underscoring the need to further dissect the heterogeneity and oncogene-specific mechanisms inhibiting p53 response in various types of leukemia. In particular, LSCs pertinent to refractory disease and relapse could rely heavily on alternative p53-inactivating mechanisms for survival and continued evolution during and following chemotherapy. Understanding these mechanisms presents new opportunities to specifically reactivate p53 and elicit LSC-selective vulnerability.
Histone deacetylases (HDACs) are enzymes that catalyze the removal of acetyl moieties from ε-amino groups of lysine residues in a variety of proteins, including histones and nonhistone proteins [33]. Based on homology to the yeast HDACs and their enzymatic activities, HDAC proteins are categorized into four classes, including class I (HDAC1, 2, 3 and 8), class II (HDAC4, 5, 6, 7, 9 and 10), class III (SIRT1, 2, 3, 4, 5, 6 and 7) and class IV (HDAC11). HDACs are widely recognized as important epigenetic regulators of gene expression via histone modification and chromatin remodeling. Many broad spectrum HDAC inhibitors have potent anticancer activities and are in various stages of clinical trials [33-38]. However, these inhibitors are highly toxic and lack selectivity, which have greatly hampered their clinical application and efficacy. More selective inhibition of mechanistically defined HDAC targets is needed to effectively eliminate cancer cells and minimize toxicity. Several members of the class I (HDAC1, 2 and 3) [39-41] and class III HDACs [42,43] are known to deacetylate the p53 protein. Given that acetylation modification of the p53 protein is essential for stabilization, nuclear localization, and transcriptional activation [44,45], p53 activity can be specifically altered by deregulation of HDACs. Here we focus on recent advances in our understanding of divergent p53-inactivating mechanisms and how deregulation of specific HDAC proteins could be exploited to restore p53 activity and enhance targeting of LSCs.
BCR-ABL activates SIRT1 expression in CML
CML has served as a paradigm for neoplasia evolution and targeted molecular therapy [46]. CML usually presents in a chronic phase and progresses through an accelerated phase followed by a terminal acute leukemia-like blast crisis [47]. It is uniformly associated with a chromosomal translocation t(9;22)(q34; q11) which results in the generation of BCR-ABL fusion gene. A unique feature of CML is that a single genetic lesion encoding the BCR-ABL fusion protein is sufficient to initiate malignant transformation of hematopoietic stem cells (HSCs). The use of tyrosine kinase inhibitor (TKI) to target BCR-ABL signaling has revolutionized the standard of care and greatly improved patient outcome. Treatment with TKI such as imatinib (IM), nilotinib, and dasatinib has been effective in inducing complete cytogenetic remissions and prolonging survival of chronic phase CML patients, but less effective against advanced phases of disease [48]. Even though TKI treatment effectively inhibited BCR-ABL kinase activity and reduced proliferation of primitive CML LSCs, it has been unable to eliminate residue LSC populations that may be potential sources of relapse [49-52]. In addition, mutations in BCR-ABL that confer resistance to TKI are common [53-55].
Sirtuin 1 (SIRT1) is a member of the sirtuin family of nicotinamide adenosine dinucleotide (NAD)-dependent deacetylases that regulate numerous biological processes, including aging, DNA repair, cell cycle, metabolism, and cell survival [56,57]. SIRT1 is shown to play important roles in the maintenance and differentiation of HSCs, especially under conditions of stress [58,59]. In CML, Wang et al. showed that SIRT1 deacetylase promotes acquisition of TKI resistant BCR-ABL mutations [60]. Given that acetylation is indispensible for transcriptional activation of p53 protein [44,45], SIRT1 functions as a negative regulator of p53 by deacetylating several lysine sites [42,43,61,62]. SIRT1 expression can be upregulated by multiple mechanisms including epigenetic silencing of a negative regulator HIC1 [63] or altered miRNA regulation [64]. In a study by Yuan et al., it was shown that BCR-ABL activates SIRT1 through STAT5 signaling and SIRT1 act as a survival pathway, which promotes oncogenic transformation and leukemogenesis [65]. Meanwhile, Li et al. showed that SIRT1 is overexpressed in primitive CML stem and progenitor cells compared to their normal counterparts [66]. Genetic knock-down of SIRT1 or pharmacological inhibition by the small molecule inhibitor tenovin-6 (TV-6) [67] impaired proliferation and induced apoptosis of CML stem and progenitor cells. In addition, combination of TV-6 with IM TKI treatment significantly reduced CML LSC growth and prolonged survival in vivo. Inhibition of SIRT1 led to enhanced p53 acetylation, and p53 activation is required for observed growth inhibitory effects of CML stem/progenitor cells. Another recent study by Wang et al. further demonstrated that genetic loss of SIRT1 depleted maintenance of CML LSCs [68]. Collectively, these studies establish that inhibiting the SIRT1-dependent survival pathway effectively activates p53 response and enhances targeting of CML LSCs. Combination of SIRT1 inhibitors with TKI could be efficacious for treating advanced CML disease and/or eradicating minimal residual disease.
FLT3-ITD induces SIRT1-c-MYC network in AML
AML is a form of highly heterogeneous hematopoietic malignancy with diverse cytogenetic, genetic and molecular abnormalities [69]. Identification of cytogenetic and genetic lesions has revolutionized AML disease classification and prognosis stratification [70-73]. However, treatment outcome in the majority of patients remains poor, with frequent and fatal relapse. Seminal work by Lapidot et al. provided the first proof that the continued growth and propagation of AML depends on a rare population of leukemia-initiating LSCs [74]. With the advent of next generation sequencing technologies, the profound heterogeneity in genomic and epigenetic landscapes in AML is undoubtedly clear [75,76]. It has also allowed detection of stepwise acquisition of AML driving mutations and infer clonal architecture [4-6,77]. In addition, it has led to identification of preleukemic stem cells harboring one or few founding mutations and the ability to acquire additional mutations contributing to relapse. The dynamic clonal and subclonal evolution during or following treatment further contributes to the complexity and heterogeneity of therapy response and outcome in AML. In-depth understanding of molecular alterations and oncogenic mechanisms underlying diverse genetic lesions and LSC resistance is needed to devise effective targeted therapies.
Activating mutations in receptor tyrosine kinases and signaling components constitute one of the classical types of mutations associated with AML. FMS-like tyrosine kinase-3 (FLT3) internal tandem duplication (ITD) is observed in 25–30% of AML patients and predicts poor prognosis [78-84]. The ITD mutation disrupts the negative regulatory function of the juxtamembrane domain, rendering FLT3 receptor constitutively active [85-87]. FLT3-ITD mutation activates canonical receptor tyrosine kinase signaling, most prominently via STAT5, RAS/MAPK, and PI3K [86,88-91]. Expression of FLT3-ITD from the endogenous promoter results in loss of HSC quiescence and a myeloproliferation neoplasm, which is reversible by FLT3-TKI treatment [92]. There are several small molecule FLT3 TKIs including quizartinib (AC220) and sorafenib being evaluated in clinical trials; however, responses have been heterogeneous and transient [93-96]. These results suggest that the leukemia-initiating LSCs may be escaping FLT3 TKI-induced cytotoxicity [96-99].
In an effort to better understand drug resistance mechanisms, Li et al. showed that FLT3-ITD caused increased SIRT1 protein expression via enhanced expression of USP22 deubiquitinase induced by c-MYC [100,101], which is activated by PIM1 as well as SIRT1-c-MYC feed forward loop in FLT3-ITD AML cells [102,103]. Inhibition of SIRT1 by shRNA-mediated knock-down or pharmacological inhibitor TV-6 reciprocally increased c-Myc acetylation and reduced its stability. SIRT1 knock-down or inhibition by TV-6 resulted in enhanced p53 acetylation and p53 target gene expression. Combination of TV6 with AC220 reduced FLT3-ITD+ AML CD34+ cell growth and survival, and enhanced TKI-mediated targeting of AML LSCs in vivo [100]. Meanwhile, Sasca et al. demonstrated that tyrosine kinase signaling including STAT5 and RAS activation likely acts in concert to activate SIRT1 expression [104]. In addition, it is proposed that FLT3-ITD regulates p53 acetylation via the ATM-DBC1-SIRT1 axis, which could also be regulated by irradiation-induced genotoxic stress [104]. In murine AML models driven in combination with MLL-AF9 or RUNX1-ETO, the combination of TV-6 and TKI modestly enhanced inhibition of proliferation [104]. The impact of additional genetic and cytogenetic aberrations on the sensitivity to SIRT1 inhibition remains to be determined. It is noteworthy that this p53 activating effect elicited by SIRT1 inhibition was not seen in FLT3 non-mutated AML cells or normal cells, underscoring the importance of identifying oncogene-specific adaptive response in the face of chemotherapy and other targeted therapy. However, there appears to be some discrepancy regarding whether SIRT1 activation was selective for FLT3-ITD+ AML and not for AML with FLT3-TKD mutations. Further studies are needed to clarify the nature and spectrum of oncogenic stimuli rendering SIRT1 activation and sensitivity to SIRT1 inhibition.
HDAC8 mediates deacetylation of p53 in inv(16) AML
In AML, chromosomal abnormalities frequently result in transcription factor fusion proteins that contribute to the unique etiology and prognosis of distinct cytogenetic subsets [105]. As a master transcriptional regulator of hematopoiesis, the core-binding factor (CBF) complex is a common target of leukemia-associated mutations [106,107]. Among the most common cytogenetic aberrations found in AML patients is chromosome 16 inversion inv(16)(p13.1q22) or translocation t(16;16)(p13.1;q22) [108]. Inv(16) generates a fusion gene Cbfb-MYH11, leading to expression of a fusion protein CBFβ-SMMHC [109,110]. A series of studies revealed that CBFβ-SMMHC dominantly inhibits CBF function, impairs hematopoietic differentiation and predisposes for leukemia transformation [111-115]. Dominant inhibition of RUNX proteins, either through cytoplasmic sequestration [116,117] or constitutive repression [118,119], was considered the main leukemogenic mechanism of CBFβ-SMMHC chimeric protein. However, more recent studies indicate that functional RUNX proteins are in fact needed for CBFβ-SMMHC leukemogenesis and growth of CBF AML cells [120-124]. It was previously reported that p53 response was reduced by CBFβ-SMMHC [125], although the underlying mechanism was not clear. A recent study by Qi et al. revealed that CBFβ-SMMHC gains p53-inhibiting function via aberrant protein-protein interaction with HDAC8 and the p53 protein [126]. HDAC8 is a member of the zinc-dependent class I HDAC enzyme known to deacetylate lysine residues in a variety of proteins, including histones and transcription factors [127-129]. Qi et al. showed that like other members of class I HDAC, HDAC8 is capable of deacetylating the p53 protein. Thus, CBFβ-SMMHC promoted HDAC8-mediated deacetylation of p53 by recruiting HDAC8 and p53 into an aberrant protein complex. Consequently, p53 induction and target gene expression is largely inhibited in the presence of CBFβ-SMMHC. Although CBFβ-SMMHC binds p53 and HDAC8 independently via distinct protein domains, the p53-inhibiting activity is dependent on the presence of both p53 and HDAC8 proteins in the ternary complex. Depleting CBFβ-SMMHC or HDAC8 resulted in restoration of p53 acetylation and activation upon exposure to genotoxic stress such as irradiation. Genetic deletion of Hdac8 in a conditional CBFβ-SMMHC knock-in mouse model dramatically diminished LSC transformation, as evidenced by greatly reduced AML incidence and delayed onset. Qi et al. also found that HDAC8 expression was significantly higher in the primitive CD34+ population and that inv(16)+ AML CD34+ cells express 5-12 fold higher levels of HDAC8 compared to non-inv(16) AML or normal CD34+ cells. In line with the differential HDAC8 expression, pharmacologic inhibition of HDAC8 enzyme using HDAC8 isoform-selective inhibitors (HDAC8i) [130,131] resulted in enhanced p53 acetylation, p53 target gene activation, and p53-dependent apoptosis selectively in inv(16)+ AML CD34+ cells while sparing the normal CD34+ stem/progenitor population. This activity further translated into elimination of AML propagation and leukemia-initiating activity in both murine AML and human AML xenograft models in vivo. Importantly, HDAC8i treatment was capable of enhancing the chemosensitivity of inv(16)+ CD34+ cells. Despite having a relatively favorable prognosis, only approximately half of the patients with inv(16) AML eventually achieve long-term survival with the standard chemotherapy regimens [132,133]. These results highlight the potential efficacy of HDAC8i in overcoming chemotherapy resistance and relapse of inv(16)+ AML.
Conclusion
In recent years, several alternative p53 inactivation mechanisms specific to the underlying oncogenic lesions have been shown for CML, FLT3-ITD+ AML, and inv(16)+ AML. These divergent pathways converge on inhibiting p53 acetylation via deregulation of alternative protein deacetylases (Figure 1). These results partly explain the heterogenous response to other p53 activating agents such as MDM2 inhibitors. Given that TP53 is rarely mutated in de novo myeloid neoplasm, these findings present new opportunities to regain control of p53 activity and enhance response to chemotherapy or other targeted therapies. Importantly, LSC populations relevant to refractory disease and relapse are selectively sensitive to perturbation of the specific protein deacetylase defined by the specific oncogenic mechanism. Thus, selective inhibition of context-specific HDAC isoforms is a promising approach to eradicate residual drug resistant LSCs, prevent further acquisition of mutations and reduce relapse. This highlights the importance to dissect the genetic and molecular heterogeneity, particularly in AML. These studies also demonstrate that by attacking cancer-specific vulnerability, the normal HSC counterpart can largely be spared. Further development of isoform specific HDAC inhibitors is critical to translate these insights into the clinic.
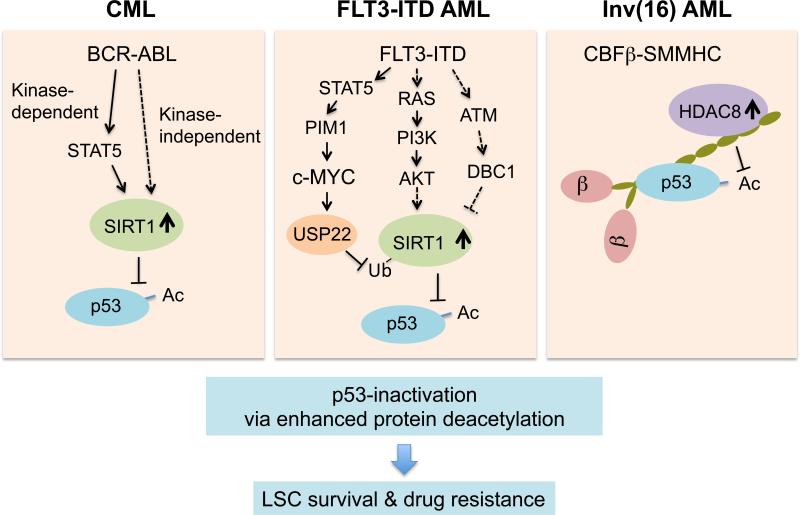
Figure 1. Multiple leukemogenic pathways converge on p53-inactivation through enhanced p53 protein deacetylation.
SIRT1 deacetylase is stabilized and activated through multiple mechanisms downstream of BCR-ABL in CML (left) and FLT3-ITD (center) signaling in FLT3-ITD+ AML. In inv(16) AML (right), CBFβ-SMMHC fusion protein recruits HDAC8 and p53 in a protein complex, thereby promoting deacetylation of p53 by HDAC8. Inhibition of oncogenic context-specific deacetylase is a promising approach to specifically activate p53 pathway and enhance sensitivity of leukemia-initiating LSCs to TKI or chemotherapy.
Acknowledgements
The authors would like to acknowledge the research support from the American Cancer Society (123278-RSG-12-140-01-CSM to Y.-H.K.), and the National Cancer Institute of the National Instituted of Health under award number R01CA178387 (to Y.-H.K.) and P30CA033572 (to COH). The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declared that no conflicts of interest exist.
Reference
- 1.Dick JE. Acute myeloid leukemia stem cells. Ann N Y Acad Sci. 2005 Jun;1044:1–5. doi: 10.1196/annals.1349.001. [DOI] [PubMed] [Google Scholar]
- 2.Chan W-I, Huntly BJP. Leukemia stem cells in acute myeloid leukemia. Semin Oncol. 2008 Aug;35(4):326–35. doi: 10.1053/j.seminoncol.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Reinisch A, Chan SM, Thomas D, Majeti R. Biology and clinical relevance of acute myeloid leukemia stem cells. Semin Hematol. 2015 Jul;52(3):150–64. doi: 10.1053/j.seminhematol.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corces-Zimmerman MR, Majeti R. Pre-leukemic evolution of hematopoietic stem cells: The importance of early mutations in leukemogenesis. Leukemia. 2014 Dec;28(12):2276–82. doi: 10.1038/leu.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014 Feb 20;506(7488):328–33. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corces-Zimmerman MR, Hong W-J, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014 Feb 18;111(7):2548–53. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vousden KH, Lane DP. P53 in health and disease. Nat Rev Mol Cell Biol. 2007 Apr;8(4):275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 8.Kruiswijk F, Labuschagne CF, Vousden KH. P53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015 Jul;16(7):393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 9.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009 Apr 30;458(7242):1127–30. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rücker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012 Mar 1;119(9):2114–21. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- 11.Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015 Feb 26;518(7540):552–5. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008 Aug;22(8):1539–41. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]
- 13.Ok CY, Patel KP, Garcia-Manero G, Routbort MJ, Peng J, Tang G, et al. TP53 mutation characteristics in therapy-related myelodysplastic syndromes and acute myeloid leukemia is similar to de novo diseases. J Hematol Oncol. 2015;8:45. doi: 10.1186/s13045-015-0139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wattel E, Preudhomme C, Hecquet B, Vanrumbeke M, Quesnel B, Dervite I, et al. P53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994 Nov 1;84(9):3148–57. [PubMed] [Google Scholar]
- 15.Nakano Y, Naoe T, Kiyoi H, Kitamura K, Minami S, Miyawaki S, et al. Prognostic value of p53 gene mutations and the product expression in de novo acute myeloid leukemia. Eur J Haematol. 2000 Jul;65(1):23–31. doi: 10.1034/j.1600-0609.2000.90138.x. [DOI] [PubMed] [Google Scholar]
- 16.Brooks CL, Gu W. P53 regulation by ubiquitin. FEBS Lett. 2011 Sep 16;585(18):2803–9. doi: 10.1016/j.febslet.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marouco D, Garabadgiu AV, Melino G, Barlev NA. Lysine-specific modifications of p53: A matter of life and death? Oncotarget. 2013 Oct;4(10):1556–71. doi: 10.18632/oncotarget.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks CL, Gu W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell. 2011 Jun;2(6):456–62. doi: 10.1007/s13238-011-1063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai C, Gu W. P53 post-translational modification: Deregulated in tumorigenesis. Trends Mol Med. 2010 Nov;16(11):528–36. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bueso-Ramos CE, Yang Y, deLeon E, McCown P, Stass SA, Albitar M. The human MDM- 2 oncogene is overexpressed in leukemias. Blood. 1993 Nov 1;82(9):2617–23. [PubMed] [Google Scholar]
- 21.Bueso-Ramos CE, Manshouri T, Haidar MA, Huh YO, Keating MJ, Albitar M. Multiple patterns of MDM-2 deregulation in human leukemias: Implications in leukemogenesis and prognosis. Leuk Lymphoma. 1995 Mar;17(1-2):13–8. doi: 10.3109/10428199509051698. [DOI] [PubMed] [Google Scholar]
- 22.Seliger B, Papadileris S, Vogel D, Hess G, Brendel C, Störkel S, et al. Analysis of the p53 and MDM-2 gene in acute myeloid leukemia. Eur J Haematol. 1996 Sep;57(3):230–40. doi: 10.1111/j.1600-0609.1996.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 23.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004 Feb 6;303(5659):844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 24.Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the mdm2-p53 interaction. J Med Chem. 2006 Jun 15;49(12):3432–5. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 25.Secchiero P, Zerbinati C, Melloni E, Milani D, Campioni D, Fadda R, et al. The MDM-2 antagonist nutlin-3 promotes the maturation of acute myeloid leukemic blasts. Neoplasia. 2007 Oct;9(10):853–61. doi: 10.1593/neo.07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008 Mar 11;105(10):3933–8. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurosu T, Wu N, Oshikawa G, Kagechika H, Miura O. Enhancement of imatinib-induced apoptosis of BCR/abl-expressing cells by nutlin-3 through synergistic activation of the mitochondrial apoptotic pathway. Apoptosis. 2010 May;15(5):608–20. doi: 10.1007/s10495-010-0457-0. [DOI] [PubMed] [Google Scholar]
- 28.Ding Q, Zhang Z, Liu J-J, Jiang N, Zhang J, Ross TM, et al. Discovery of RG7388, a potent and selective p53–MDM2 inhibitor in clinical development. J Med Chem. 2013 Jul 25;56(14):5979–83. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Ding Q, Liu J-J, Zhang J, Jiang N, Chu X-J, et al. Discovery of potent and selective spiroindolinone MDM2 inhibitor, RO8994, for cancer therapy. Bioorg Med Chem. 2014 Aug 1;22(15):4001–9. doi: 10.1016/j.bmc.2014.05.072. [DOI] [PubMed] [Google Scholar]
- 30.Andreeff M, Kelly KR, Yee K, Assouline S, Strair R, Popplewell L, et al. Results of the phase I trial of RG7112, a small-molecule MDM2 antagonist in leukemia. Clin Cancer Res. 2015 Oct 12; doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter BZ, Mak PY, Mak DH, Ruvolo VR, Schober W, McQueen T, et al. Synergistic effects of p53 activation via MDM2 inhibition in combination with inhibition of bcl-2 or bcr-abl in CD34+ proliferating and quiescent chronic myeloid leukemia blast crisis cells. Oncotarget. 2015 Oct 13;6(31):30487–99. doi: 10.18632/oncotarget.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long J, Parkin B, Ouillette P, Bixby D, Shedden K, Erba H, et al. Multiple distinct molecular mechanisms influence sensitivity and resistance to MDM2 inhibitors in adult acute myelogenous leukemia. Blood. 2010 Jul 8;116(1):71–80. doi: 10.1182/blood-2010-01-261628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006 Sep;5(9):769–84. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 34.Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: Mechanisms of cell death and promise in combination cancer therapy. Cancer Lett. 2008 May 5; doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 35.Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009 Aug 8;280(2):125–33. doi: 10.1016/j.canlet.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 36.Ma X, Ezzeldin HH, Diasio RB. Histone deacetylase inhibitors: Current status and overview of recent clinical trials. Drugs. 2009 Oct 1;69(14):1911–34. doi: 10.2165/11315680-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 37.Wanczyk M, Roszczenko K, Marcinkiewicz K, Bojarczuk K, Kowara M, Winiarska M. HDACi--going through the mechanisms. Front Biosci. 2011;16:340–59. doi: 10.2741/3691. [DOI] [PubMed] [Google Scholar]
- 38.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008 Jan 15;409(2):581–9. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 39.Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS, Wu CW. Histone deacetylases specifically down-regulate p53-dependent gene activation. J Biol Chem. 2000 Jul 7;275(27):20436–43. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 40.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000 Nov 16;408(6810):377–81. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 41.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002 Nov 15;21(22):6236–45. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, et al. Negative control of p53 by sir2alpha promotes cell survival under stress. Cell. 2001 Oct 19;107(2):137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 43.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. HSIR2(SIRT1) functions as an nad-dependent p53 deacetylase. Cell. 2001 Oct 19;107(2):149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 44.Prives C, Manley JL. Why is p53 acetylated? Cell. 2001 Dec 28;107(7):815–8. doi: 10.1016/s0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- 45.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008 May 16;133(4):612–26. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007 Jun;7(6):441–53. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 47.Sawyers CL. Chronic myeloid leukemia. N Engl J Med. 1999 Apr 29;340(17):1330–40. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- 48.Eiring AM, Khorashad JS, Morley K, Deininger MW. Advances in the treatment of chronic myeloid leukemia. BMC Med. 2011;9:99. doi: 10.1186/1741-7015-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holtz MS, Forman SJ, Bhatia R. Nonproliferating CML CD34+ progenitors are resistant to apoptosis induced by a wide range of proapoptotic stimuli. Leukemia. 2005 Jun;19(6):1034–41. doi: 10.1038/sj.leu.2403724. [DOI] [PubMed] [Google Scholar]
- 50.Chu S, McDonald T, Lin A, Chakraborty S, Huang Q, Snyder DS, Bhatia R. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood. 2011 Nov 17;118(20):5565–72. doi: 10.1182/blood-2010-12-327437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barnes DJ, Melo JV. Primitive, quiescent and difficult to kill: The role of non-proliferating stem cells in chronic myeloid leukemia. Cell Cycle. 2006 Dec;5(24):2862–6. doi: 10.4161/cc.5.24.3573. [DOI] [PubMed] [Google Scholar]
- 52.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011 Jan;121(1):396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002 Aug;2(2):117–25. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 54.Branford S, Rudzki Z, Walsh S, Parkinson I, Grigg A, Szer J, et al. Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (p-loop) are associated with a poor prognosis. Blood. 2003 Jul 1;102(1):276–83. doi: 10.1182/blood-2002-09-2896. [DOI] [PubMed] [Google Scholar]
- 55.Hughes TP, Saglio G, Quintás-Cardama A, Mauro MJ, Kim D-W, Lipton JH, et al. BCR ABL1 mutation development during first-line treatment with dasatinib or imatinib for chronic myeloid leukemia in chronic phase. Leukemia. 2015 Sep;29(9):1832–8. doi: 10.1038/leu.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: Understanding longevity. Nat Rev Mol Cell Biol. 2005 Apr;6(4):298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 57.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009 Mar 1;69(5):1702–5. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 58.Singh SK, Williams CA, Klarmann K, Burkett SS, Keller JR, Oberdoerffer P. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J Exp Med. 2013 May 6;210(5):987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ou X, Chae H-D, Wang R-H, Shelley WC, Cooper S, Taylor T, et al. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood. 2011 Jan 13;117(2):440–50. doi: 10.1182/blood-2010-03-273011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Yuan H, Roth M, Stark JM, Bhatia R, Chen WY. SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene. 2013 Jan 31;32(5):589–98. doi: 10.1038/onc.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yi J, Luo J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochim Biophys Acta. 2010 Aug;1804(8):1684–9. doi: 10.1016/j.bbapap.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009 Feb;9(2):123–8. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent dna-damage responses. Cell. 2005 Nov 4;123(3):437–48. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 64.Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, et al. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of sirt1. Mol Endocrinol. 2009 Nov;23(11):1876–84. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan H, Wang Z, Li L, Zhang H, Modi H, Horne D, et al. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 2012 Feb 23;119(8):1904–14. doi: 10.1182/blood-2011-06-361691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li L, Wang L, Li L, Wang Z, Ho Y, McDonald T, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012 Feb 14;21(2):266–81. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008 May;13(5):454–63. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z, Chen C-C, Chen W. CD150(−) side population defines leukemia stem cells in a BALB/c mouse model of CML and is depleted by genetic loss of SIRT1. Stem Cells. 2015 Oct 15; doi: 10.1002/stem.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015 Sep 16;373(12):1136–52. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 70.Marcucci G, Mrózek K, Bloomfield CD. Molecular heterogeneity and prognostic biomarkers in adults with acute myeloid leukemia and normal cytogenetics. Curr Opin Hematol. 2005 Jan;12(1):68–75. doi: 10.1097/01.moh.0000149608.29685.d1. [DOI] [PubMed] [Google Scholar]
- 71.Gaidzik V, Döhner K. Prognostic implications of gene mutations in acute myeloid leukemia with normal cytogenetics. Semin Oncol. 2008 Aug;35(4):346–55. doi: 10.1053/j.seminoncol.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 72.Scholl S, Fricke H-J, Sayer HG, Höffken K. Clinical implications of molecular genetic aberrations in acute myeloid leukemia. J Cancer Res Clin Oncol. 2009 Apr;135(4):491–505. doi: 10.1007/s00432-008-0524-x. [DOI] [PubMed] [Google Scholar]
- 73.Marcucci G, Haferlach T, Döhner H. Molecular genetics of adult acute myeloid leukemia: Prognostic and therapeutic implications. J Clin Oncol. 2011 Feb 10;29(5):475–86. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 74.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994 Feb 17;367(6464):645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 75.Sanders MA, Valk PJ. The evolving molecular genetic landscape in acute myeloid leukaemia. Curr Opin Hematol. 2013 Mar;20(2):79–85. doi: 10.1097/MOH.0b013e32835d821c. [DOI] [PubMed] [Google Scholar]
- 76.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013 May 30;368(22):2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bodini M, Ronchini C, Giacò L, Russo A, Melloni GEM, Luzi L, et al. The hidden genomic landscape of acute myeloid leukemia: Subclonal structure revealed by undetected mutations. Blood. 2015 Jan 22;125(4):600–5. doi: 10.1182/blood-2014-05-576157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996 Dec;10(12):1911–8. [PubMed] [Google Scholar]
- 79.Sallmyr A, Fan J, Datta K, Kim K-T, Grosu D, Shapiro P, et al. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: Implications for poor prognosis in AML. Blood. 2008 Mar 15;111(6):3173–82. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- 80.Kindler T, Lipka DB, Fischer T. FLT3 as a therapeutic target in AML: Still challenging after all these years. Blood. 2010 Dec 9;116(24):5089–102. doi: 10.1182/blood-2010-04-261867. [DOI] [PubMed] [Google Scholar]
- 81.Thiede C, Steudel C, Mohr B, Schaich M, Schäkel U, Platzbecker U, et al. Analysis of flt3- activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002 Jun 15;99(12):4326–35. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 82.Fröhling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K, et al. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: A study of the AML study group ulm. Blood. 2002 Dec 15;100(13):4372–80. doi: 10.1182/blood-2002-05-1440. [DOI] [PubMed] [Google Scholar]
- 83.Meshinchi S, Woods WG, Stirewalt DL, Sweetser DA, Buckley JD, Tjoa TK, et al. Prevalence and prognostic significance of flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood. 2001 Jan 1;97(1):89–94. doi: 10.1182/blood.v97.1.89. [DOI] [PubMed] [Google Scholar]
- 84.Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: Analysis of 854 patients from the united kingdom medical research council AML 10 and 12 trials. Blood. 2001 Sep 15;98(6):1752–9. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 85.Kiyoi H, Towatari M, Yokota S, Hamaguchi M, Ohno R, Saito H, Naoe T. Internal tandem duplication of the FLT3 gene is a novel modality of elongation mutation which causes constitutive activation of the product. Leukemia. 1998 Sep;12(9):1333–7. doi: 10.1038/sj.leu.2401130. [DOI] [PubMed] [Google Scholar]
- 86.Mizuki M, Fenski R, Halfter H, Matsumura I, Schmidt R, Müller C, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the ras and STAT5 pathways. Blood. 2000 Dec 1;96(12):3907–14. [PubMed] [Google Scholar]
- 87.Fenski R, Flesch K, Serve S, Mizuki M, Oelmann E, Kratz-Albers K, et al. Constitutive activation of FLT3 in acute myeloid leukaemia and its consequences for growth of 32D cells. Br J Haematol. 2000 Feb;108(2):322–30. doi: 10.1046/j.1365-2141.2000.01831.x. [DOI] [PubMed] [Google Scholar]
- 88.Kiyoi H, Ohno R, Ueda R, Saito H, Naoe T. Mechanism of constitutive activation of FLT3 with internal tandem duplication in the juxtamembrane domain. Oncogene. 2002 Apr 11;21(16):2555–63. doi: 10.1038/sj.onc.1205332. [DOI] [PubMed] [Google Scholar]
- 89.Tse KF, Novelli E, Civin CI, Bohmer FD, Small D. Inhibition of flt3-mediated transformation by use of a tyrosine kinase inhibitor. Leukemia. 2001 Jul;15(7):1001–10. doi: 10.1038/sj.leu.2402199. [DOI] [PubMed] [Google Scholar]
- 90.Hayakawa F, Towatari M, Kiyoi H, Tanimoto M, Kitamura T, Saito H, Naoe T. Tandem- duplicated flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in il-3-dependent cell lines. Oncogene. 2000 Feb 3;19(5):624–31. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- 91.Yoshimoto G, Miyamoto T, Jabbarzadeh-Tabrizi S, Iino T, Rocnik JL, Kikushige Y, et al. FLT3-ITD up-regulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via flt3-itd-specific STAT5 activation. Blood. 2009 Dec 3;114(24):5034–43. doi: 10.1182/blood-2008-12-196055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chu SH, Heiser D, Li L, Kaplan I, Collector M, Huso D, et al. FLT3-ITD knockin impairs hematopoietic stem cell quiescence/homeostasis, leading to myeloproliferative neoplasm. Cell Stem Cell. 2012 Sep;11(3):346–58. doi: 10.1016/j.stem.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grundler R, Thiede C, Miething C, Steudel C, Peschel C, Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003 Jul 15;102(2):646–51. doi: 10.1182/blood-2002-11-3441. [DOI] [PubMed] [Google Scholar]
- 94.Knapper S, Burnett AK, Littlewood T, Kell WJ, Agrawal S, Chopra R, et al. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006 Nov 15;108(10):3262–70. doi: 10.1182/blood-2006-04-015560. [DOI] [PubMed] [Google Scholar]
- 95.Weisberg E, Boulton C, Kelly LM, Manley P, Fabbro D, Meyer T, et al. Inhibition of mutant FLT3 receptors in leukemia cells by the small molecule tyrosine kinase inhibitor PKC412. Cancer Cell. 2002 Jun;1(5):433–43. doi: 10.1016/s1535-6108(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 96.Levis M, Ravandi F, Wang ES, Baer MR, Perl A, Coutre S, et al. Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse. Blood. 2011 Mar 24;117(12):3294–301. doi: 10.1182/blood-2010-08-301796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Levis M. Quizartinib for the treatment of FLT3/ITD acute myeloid leukemia. Future Oncol. 2014;10(9):1571–9. doi: 10.2217/fon.14.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Swords R, Freeman C, Giles F. Targeting the fms-like tyrosine kinase 3 in acute myeloid leukemia. Leukemia. 2012 Oct;26(10):2176–85. doi: 10.1038/leu.2012.114. [DOI] [PubMed] [Google Scholar]
- 99.Smith CC, Wang Q, Chin C-S, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012 May 10;485(7397):260–3. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li L, Osdal T, Ho Y, Chun S, McDonald T, Agarwal P, et al. SIRT1 activation by a c-myc oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell. 2014 Oct 2;15(4):431–46. doi: 10.1016/j.stem.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin Z, Yang H, Kong Q, Li J, Lee S-M, Gao B, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell. 2012 May 25;46(4):484–94. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 102.Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PNG, Böhmer FD, et al. Mislocalized activation of oncogenic rtks switches downstream signaling outcomes. Mol Cell. 2009 Oct 23;36(2):326–39. doi: 10.1016/j.molcel.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 103.Kim K-T, Baird K, Ahn J-Y, Meltzer P, Lilly M, Levis M, Small D. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in flt3-mediated cell survival. Blood. 2005 Feb 15;105(4):1759–67. doi: 10.1182/blood-2004-05-2006. [DOI] [PubMed] [Google Scholar]
- 104.Sasca D, Hähnel PS, Szybinski J, Khawaja K, Kriege O, Pante SV, et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood. 2014 Jul 3;124(1):121–33. doi: 10.1182/blood-2013-11-538819. [DOI] [PubMed] [Google Scholar]
- 105.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997 Nov 7;278(5340):1059–64. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 106.Speck NA, Stacy T, Wang Q, North T, Gu TL, Miller J, et al. Core-binding factor: A central player in hematopoiesis and leukemia. Cancer Res. 1999 Apr 1;59(7 Suppl):1789s–93s. [PubMed] [Google Scholar]
- 107.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002 Jul;2(7):502–13. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 108.Liu PP, Wijmenga C, Hajra A, Blake TB, Kelley CA, Adelstein RS, et al. Identification of the chimeric protein product of the CBFB-MYH11 fusion gene in inv(16) leukemia cells. Genes Chromosomes Cancer. 1996 Jun;16(2):77–87. doi: 10.1002/(SICI)1098-2264(199606)16:2<77::AID-GCC1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 109.Liu P, Tarlé SA, Hajra A, Claxton DF, Marlton P, Freedman M, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993 Aug 20;261(5124):1041–4. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 110.Liu PP, Hajra A, Wijmenga C, Collins FS. Molecular pathogenesis of the chromosome 16 inversion in the m4eo subtype of acute myeloid leukemia. Blood. 1995 May 1;85(9):2289–302. [PubMed] [Google Scholar]
- 111.Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996 Nov 15;87(4):687–96. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 112.Castilla LH, Garrett L, Adya N, Orlic D, Dutra A, Anderson S, et al. The fusion gene cbfb myh11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999 Oct;23(2):144–6. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 113.Kuo Y-H, Landrette SF, Heilman SA, Perrat PN, Garrett L, Liu PP, et al. Cbf beta-smmhc induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006 Jan;9(1):57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 114.Kuo Y-H, Gerstein RM, Castilla LH. Cbfbeta-SMMHC impairs differentiation of common lymphoid progenitors and reveals an essential role for RUNX in early b-cell development. Blood. 2008 Feb 1;111(3):1543–51. doi: 10.1182/blood-2007-07-104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao L, Cannons JL, Anderson S, Kirby M, Xu L, Castilla LH, et al. CBFB-MYH11 hinders early t-cell development and induces massive cell death in the thymus. Blood. 2007 Apr 15;109(8):3432–40. doi: 10.1182/blood-2006-10-051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kanno Y, Kanno T, Sakakura C, Bae SC, Ito Y. Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor alpha (cbfalpha) subunit by the leukemia-related PEBP2/cbfbeta-smmhc fusion protein inhibits PEBP2/cbf- mediated transactivation. Mol Cell Biol. 1998 Jul;18(7):4252–61. doi: 10.1128/mcb.18.7.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (cbfbeta)-smooth-muscle myosin heavy chain sequesters cbfalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998 Dec;18(12):7432–43. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lutterbach B, Hou Y, Durst KL, Hiebert SW. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc Natl Acad Sci U S A. 1999 Oct 26;96(22):12822–7. doi: 10.1073/pnas.96.22.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Durst KL, Lutterbach B, Kummalue T, Friedman AD, Hiebert SW. The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol Cell Biol. 2003 Jan;23(2):607–19. doi: 10.1128/MCB.23.2.607-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kuo Y-H, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. Runx2 induces acute myeloid leukemia in cooperation with cbfbeta-smmhc in mice. Blood. 2009 Apr 2;113(14):3323–32. doi: 10.1182/blood-2008-06-162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kamikubo Y, Zhao L, Wunderlich M, Corpora T, Hyde RK, Paul TA, et al. Accelerated leukemogenesis by truncated CBF beta-smmhc defective in high-affinity binding with RUNX1. Cancer Cell. 2010 May 18;17(5):455–68. doi: 10.1016/j.ccr.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Goyama S, Schibler J, Cunningham L, Zhang Y, Rao Y, Nishimoto N, et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J Clin Invest. 2013 Sep 3;123(9):3876–88. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ben-Ami O, Friedman D, Leshkowitz D, Goldenberg D, Orlovsky K, Pencovich N, et al. Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep. 2013 Sep 26;4(6):1131–43. doi: 10.1016/j.celrep.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 124.Hyde RK, Zhao L, Alemu L, Liu PP. Runx1 is required for hematopoietic defects and leukemogenesis in cbfb-myh11 knock-in mice. Leukemia. 2015 Mar 6; doi: 10.1038/leu.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Britos-Bray M, Ramirez M, Cao W, Wang X, Liu PP, Civin CI, Friedman AD. CBFbeta-SMMHC, expressed in m4eo acute myeloid leukemia, reduces p53 induction and slows apoptosis in hematopoietic cells exposed to dna-damaging agents. Blood. 1998 Dec 1;92(11):4344–52. [PubMed] [Google Scholar]
- 126.Qi J, Singh S, Hua W-K, Cai Q, Chao S-W, Li L, et al. HDAC8 inhibition specifically targets inv(16) acute myeloid leukemic stem cells by restoring p53 acetylation. Cell Stem Cell. 2015 Nov;17(5):597–610. doi: 10.1016/j.stem.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Buggy JJ, Sideris ML, Mak P, Lorimer DD, McIntosh B, Clark JM. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem J. 2000 Aug 15;350(Pt 1):199–205. [PMC free article] [PubMed] [Google Scholar]
- 128.Van den Wyngaert I, de Vries W, Kremer A, Neefs J, Verhasselt P, Luyten WH, Kass SU. Cloning and characterization of human histone deacetylase 8. FEBS Lett. 2000 Jul 28;478(1-2):77–83. doi: 10.1016/s0014-5793(00)01813-5. [DOI] [PubMed] [Google Scholar]
- 129.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, et al. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000 May 19;275(20):15254–64. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 130.Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in t-cell lymphomas. Leukemia. 2008 May;22(5):1026–34. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- 131.Huang W-J, Wang Y-C, Chao S-W, Yang C-Y, Chen L-C, Lin M-H, et al. Synthesis and biological evaluation of ortho-aryl n-hydroxycinnamides as potent histone deacetylase (HDAC) 8 isoform-selective inhibitors. ChemMedChem. 2012 Oct;7(10):1815–24. doi: 10.1002/cmdc.201200300. [DOI] [PubMed] [Google Scholar]
- 132.Prébet T, Boissel N, Reutenauer S, Thomas X, Delaunay J, Cahn J-Y, et al. Acute myeloid leukemia with translocation (8;21) or inversion (16) in elderly patients treated with conventional chemotherapy: A collaborative study of the french CBF-AML intergroup. J Clin Oncol. 2009 Oct 1;27(28):4747–53. doi: 10.1200/JCO.2008.21.0674. [DOI] [PubMed] [Google Scholar]
- 133.Ustun C, Marcucci G. Emerging diagnostic and therapeutic approaches in core binding factor acute myeloid leukaemia. Curr Opin Hematol. 2015 Mar;22(2):85–91. doi: 10.1097/MOH.0000000000000124. [DOI] [PubMed] [Google Scholar]