Abstract
Non-specific adsorption (NSA) of proteins on surface is a critical issue in polydimethylsiloxane (PDMS)-based microfluidics, which may either considerably decrease the efficiency of a continuous flow reaction or cause a large background noise in a heterogeneous sensing. This work introduced a new method to restrain NSA of protein by caulking PDMS with Parylene C, i.e., forming a Parylene C-caulked PDMS (pcPDMS) surface. The caulking depth of Parylene C inside PDMS matrix was characterized by laser scanning confocal microscopy based on a detectable autofluorescence intensity difference between Parylene C and PDMS after being annealed at 270 °C for 2 h in nitrogen. NSA of bovine serum albumin (BSA) on the inner surfaces of PDMS and pcPDMS microchannels was experimentally compared. The results indicated that the adsorbed BSA on the pcPDMS surface were 35.2% of that on the pristine PDMS surface after the BSA solution flowing through the microchannels at a flow rate of 2000 nL/min, a typical scenario of the continuous flow reaction. In a case mimicking the heterogeneous sensing, after a 60 min washing of phosphate buffered saline flow on a pre-saturated BSA adsorbed surface, the residual BSA on the pcPDMS surface was only 4.5% of that on the pristine PDMS surface. Adsorption/desorption coefficients of BSA on the PDMS and the pcPDMS surfaces were extracted from the experimental results based on the first-order Langmuir model, which indicated that the pcPDMS has a lower adsorption coefficient (Ka) and a higher desorption coefficient (Kd), compared to those of the pristine PDMS. A preliminary experiment also indicated that Taq polymerase kept 93.0% activity after flowing through a pcPDMS microchannel, while only 28.9% activity was left after passing a pristine PDMS microchannel under the same operation condition.
I. INTRODUCTION
Polydimethylsiloxane (PDMS) is the most widely used material in microfluidics, thanks to its good characteristics as being low-cost, gas permeable, transparent, biocompatible, and easy to fabricate. However, despite the above advantages, there are still some issues when PDMS is applied in many biological studies,1,2 such as the undesired non-specific adsorption (NSA) of protein on the surface. In a continuous flow microfluidic reactor, like microfluidic polymerase chain reaction (PCR) chip, the undesired non-specific adsorption of functional protein (e.g., DNA polymerase in the PCR) on the PDMS surface will significantly decrease the reaction efficiency, even resulting in a complete failure.3–5 Meanwhile, in a heterogeneous sensing such as the DNA microarray6 or the immunoassay7 biosensor, the NSA of labelled molecules (dyes) may also limit the detection effectiveness. A typical biosensor consists of probe molecules on the surface and a detectable signal will be generated upon the binding between the probes and target molecules. NSA of dyes on PDMS surface causes a high background (noise) signal and degrades the sensing performance.6,7 Efforts have been made to passivate the PDMS surface and restrain the NSA on top. The commonly used surface passivation could be classified into two types. One is the dynamic passivation,4,8–15 where the blocking reagent, e.g., BSA, was added into the reaction system to competitively adsorb onto the PDMS surface and restrain the NSA of the target enzyme.4,12 Optimization of the reaction system was thereby required. However, it should be noticed that this optimized reaction system may not be the best for real applications, where usually low NSA materials, like PS or PMMA, will be used for mass production.4 The other one is the static passivation, where surface treatments of PDMS were conducted before the experiment. The anti-NSA properties of various static passivation methods were compared in Table I. As shown in Table I, the static passivation has different surface modification strategies.16,17 The commonly used modifications include: biomolecule coating, such as ionic complementary peptides17,18 and polysaccharide-based coating;19–21 polymer grafting,22–33 such as PEG (Polyethylene glycol) mediated modification,22,23 fluoro-containing coating,25–27 epoxy based polymers,28,29 plasma polymer,30 polyacrylamide,31 hybrid inorganic/organic polymer,32 and multilayer polyelectrolyte;33 In Refs. 34 and 35, the microchannel inner surface was coated with a glass-like layer by a sol-gel technique,34,35 while in Ref. 36, chemical vapor deposition techniques to deposit SiOx36 onto inner surface of a pre-assembled PDMS microchannel were developed. As shown in Table I, the static passivation usually relied on heavy and time-consuming manual operations. The coating layers are easily degraded or removed in the real environments because of temperature variations, mechanical impacts, or solvolytic interactions.17 In some cases, the size or even the shape of microchannel was changed after the treatment.34 An easy, reliable, and processable method is thereby in high demand for surface passivation of the PDMS surface.
TABLE I.
Comparison of anti-NSA performance of various surface passivation methods of PDMS.
| Ref./Year | PDMS surface modification | Analytes | Adsorption and desorption coefficient | Comments |
|---|---|---|---|---|
| 18/2015 | Self-assembled peptides coating | BSAa and LYZb | … | The adsorption of BSA and LYZ was reduced by 96%–98%, and there was no difference between BSA and LYZ. The coating could only be conducted after chip assembly and was realized in the phosphate buffer at a specific pH of 9.5. Practical application: adsorption of platelet from whole blood was detected |
| 30/2012 | Plasma polymer coating (tetraglyme, ppTTg) | FINc | … | 4-step ppTTg coating process could resist protein adsorption (<10 ng/cm2), while the requirement for the surface to be open for the coating process may be a limitation, and all devices sealing were relied on a clamp system and operational pressure |
| 21/2010 | Photocatalyzed Polysaccharides coating: carboxymethyl cellulose (CMC), carboxymethyl −1, 3-dextran (CMD), and alginic acid (AA) | BSA and LYZ | … | Selectively (isoelectric point of protein (PI) dependent) protein repelling property was demonstrated, and the modification could only be conducted after PDMS bonding |
| 23/2010 | Self-assembled alkanethiol monolayers (SAM) | LYZ and FIN | … | The SAM reduced the adsorption by around 75%, and there was no difference between LYZ and FIN. The modification was conducted after PDMS chip assembly and required a complex surface pretreatment. |
| 24/2007 | Coating of 2-methacryloyloxyethyl phosphorylcholine (MPC) polymers | BSA | … | The MPC coating on PDMS inhibit the adsorption by ∼75%. The modification was carried out after PDMS microchannel preparation |
| 29/2006 | Grafting epoxy-modified hydrophilic polymers | BSA and LYZ | … | Surface adsorption of BSA and LYZ was reduced to less than 10% relative to that on the native PDMS surface, but 10%–15% increase in protein adsorption was observed after 6 days. There is no difference between BSA and LYZ. The coating was carried out after PDMS device preparation with plasma bonding |
| 20/2005 | n-Dodecyl-b-d-Maltoside (DDM) | BSA | … | The adsorption was reduced by a factor of 100. The coating process is complex, and could only be conducted after chip assembly when a small percentage of DDM is tolerated in the reaction system. Practical application: adsorption of cytochrome c (an antibody) and β2 adrenergic receptor (a transmembrane protein) were tested |
| 22/2005 | Platinum-catalyzed covalent immobilization of polyethylene oxide (PEO) | BSA and FIN | … | The adsorption was reduced by more than 90% and no difference for BSA and FIN, but molecular weight of PEO may affect protein. The PEO coating process was conducted on the PDMS specimen and bonding for device assembly was not tested |
| 25/2005 | Fluorous surfactant treatment | BSA and FIN | Adsorption rate | The adsorption of BSA and FIN could be prevented and no significances, while the coating was conducted after microfluidic channel assembly |
| 31/2004 | Polyacrylamide coating through atom-transfer radical polymerization | BSA | … | Surface modification reduced both reversible and irreversible adsorption of BSA, while a rigorous operation was required for the coating. Practical application: demonstrated separation of LYZ and cytochrome c in 35 s showed evidence of no adsorption |
| 27/2003 | Polytetrafluoroethylene (PTFE) cover | BSA | … | Adsorption of BSA was dropped on the PTFE covered surface. But the formation of fine structures required 115 °C under pressure of 40 kPa after cover |
| 37/2003 | Chemical vapor deposition (CVD) of poly(p-xylylene carboxylic acid pentafluorophenolester-co-p-xylylene)26 (PPX-PPF) | … | … | Immobilization of biotin and streptavidin demonstrated the decreased protein adsorption on the PPX-PPF modified PDMS surface, and the process is compatible with complex biological application, while bonding of the microchannel required oxidization-facilitated adhesive epoxy bond |
| This work | Parylene C caulking inside PDMS matrix via chemical vapor deposition (CVD) | BSA | Yes | pcPDMS process was able to reduce the NSA of BSA by 64.8% in a continuous flow reaction case (at a flow rate of 2000 nL/min) and 95.5% in a heterogeneous sensing case. The pcPDMS bonding could be realized via easily available oxygen plasma treatment. Practical application: Taq polymerase for PCR |
Bovine serum albumin.
Lysozyme.
Fibrinogen.
Parylene C, a well-known biocompatible and processable polymer, usually functions as a passivation coating to achieve low adhesion and good barrier properties against moisture, and inorganic and organic molecules. Some works have been performed to study the anti-NSA performance of the Parylene C coating in the PDMS-based microfluidic chips.37–39 Shin et al.38 showed that by coating the PDMS-based micro PCR chips with Parylene C, it was possible to successfully amplify DNA in chip without changing the reaction system. However, the penetration capability of parylene deposition inside microstructures is limited. For a straight microchannel, the length that could be coated by parylene was usually less than a hundred times of the hydraulic diameter of the channel.39 This means that it will be difficult to coat the whole microchannel surface just by depositing parylene after finishing the microfluidic device preparation, i.e., bonding the devices. Sealing PDMS surfaces by parylene before the bonding operation has also attracted many attention. Sawano et al. initiated a MEMS-compatible process for low-permeability PDMS by directly depositing Parylene C onto the PDMS, so-named Parylene C-caulked PDMS (pcPDMS).40 Parylene C monomers effectively penetrated into the bulk PDMS matrix and caulked the permeable sites. The unavoidable Parylene C layer on top of the PDMS surface was removed by oxygen plasma etching to restore the elastic features of PDMS, getting the pcPDMS ready for flexible actuator applications. Our previous work further solved the bonding issue of pcPDMS by employing buffered hydro-fluoride acid (BHF) bath to remove the silica-like layer generated during the long oxygen plasma etching.41,42 It has been demonstrated the pcPDMS can successfully suppress the small molecule diffusion into the PDMS matrix, while the anti-NSA performance of the pcPDMS surface is still unknown.
This work used bovine serum albumin (BSA) as a model protein to study the anti-NSA property of the pcPDMS. Both the continuous flow mode and the heterogeneous sensing mode were taken into consideration. Adsorption/desorption coefficients of BSA on the PDMS and the pcPDMS surfaces were extracted from the experimental measurements. A preliminary experiment was also carried out to investigate the NSA of the functional proteins on pcPDMS and PDMS surfaces in real applications. Furthermore, a noticeable difference of the autofluorescences of Parylene C and PDMS after being annealed at 270 °C for 2 h in nitrogen was used to characterize the caulking status of Parylene C inside the PDMS matrix for the first time.
II. METHODS
A. Chip fabrication
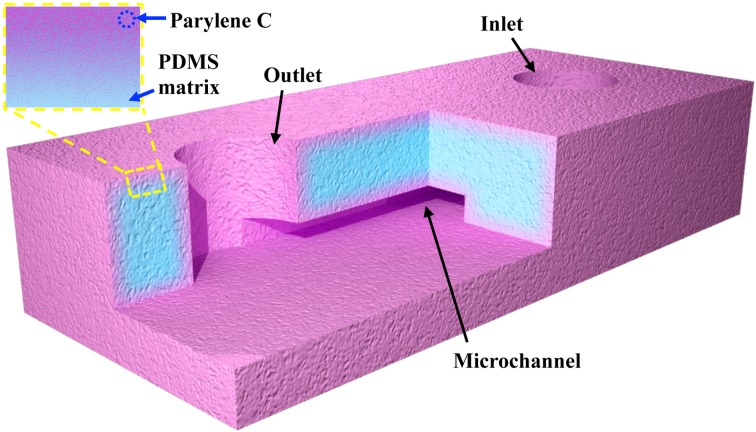
Fabrication of a pcPDMS microfluidic device is described elsewhere.41,42 Briefly, PDMS structures were prepared with a standard soft lithography technique (Sylgard 184, 10:1, 70 °C for 1 h), and then, the prepared PDMS matrix was caulked with our previously reported pcPDMS process.41,42 Finally, the prepared pcPDMS was bonded via a 5 s oxygen plasma treatment to assemble a microfluidic chip. The schematic illustration of a pcPDMS microchannel is shown in Figure 1. A straight microchannel with the dimension of 1000 μm (length) × 170 μm (width) × 20 μm (depth) was used as a microfluidic reactor for the continuous flow test, while a serpentine microchannel with a cross-sectional size of 60 μm (wide) × 50 μm (deep) was adopted to mimic the microfluidic heterogeneous sensing channel. Pristine PDMS devices were also prepared and tested in parallel.
FIG. 1.
Schematic illustration of pcPDMS microfluidic chip with a straight channel, and the inset shows the cross-section view of pcPDMS surface (not to scale).
B. Characterization of the Parylene C caulking status
A laser scanning confocal microscopy was used to characterize the so-prepared pcPDMS and study the caulking status of Parylene C inside the PDMS matrix based on the strong autofluorescence of the annealed Parylene C.43 The pcPDMS samples were annealed in nitrogen at 270 °C for 2 h and then imaged under a laser scanning confocal microscopy (Leica, Germany). The scanning had a step size of 0.5 μm in the z direction (normal to the surface), and the sample was excited at 340–380 nm and detected at 435–485 nm (Gate voltage: 845.8 V, Smart offset: −1.2%, and Pinhole: 60.63 μm). The fluorescent intensities at different z positions were extracted to calculate the distribution of Parylene C caulked inside the PDMS matrix.
C. BSA adsorption test
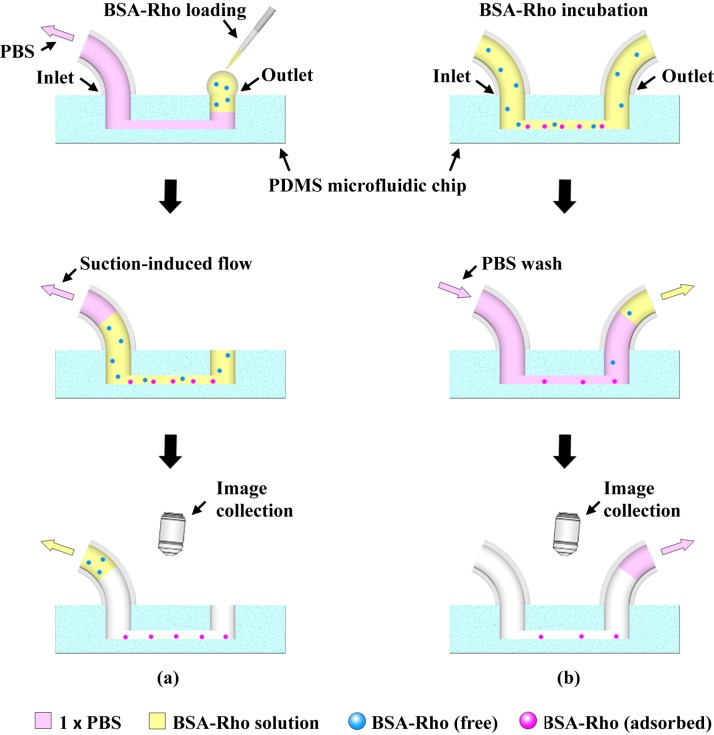
NSA of BSA under the continuous flow mode and the heterogeneous sensing mode was experimentally tested in the pcPDMS and pristine PDMS chips with the aforementioned channel designs. The BSA used here was conjugated with Rhodamine B (Biosynthesis Biotechnology Co., Ltd., Beijing, China) and dissolved in 1× phosphate buffer solution (PBS) for the convenience of observation under a fluorescent microscope (Olympus BX51, Tokyo, Japan).
The experimental processes for the continuous flow mode and the heterogeneous sensing mode were schematically illustrated in Figs. 2(a) and 2(b), respectively. In the experiment under the continuous flow mode, as shown in Fig. 2(a), first, a 5 μL 1× PBS was added to the outlet, and the microchannel was perfused with the PBS solution by applying a negative pressure from the inlet through a syringe pump (Longer, Baoding, Hebei Province, P. R. China) at a flow rate of 500 nL/min. Then, 1 μL 10 μM BSA solution was added into the outlet and flew through the microchannel at different suction rates. After all the BSA solution was sucked out, the bottom surface of the microchannel was imaged under the fluorescent microscopy to record the fluorescence signals generated by the non-specifically adsorbed BSAs. For the test under the heterogeneous sensing mode, as shown in Fig. 2(b), 10 μM BSA was first injected by a syringe pump to fill the microchannels and incubated for 30 min at room temperature, and then 1× PBS was continuously pumped into the channel from the inlet at a flow rate of 1 μL/min to wash the non-specifically adsorbed BSAs away from the surface. The 1× PBS flow rinsing operation was equivalent to the washing in the conventional heterogeneous sensing experiments. Fluorescent images were taken to record the residual BSAs adsorbed on the channel surface every 10 min (60 min in total). The gray scale values were extracted from the fluorescent images to represent the amount of adsorbed BSAs on the surface.
FIG. 2.
Schematic illustrations of experiment processes under the continuous flow mode (a) and the heterogeneous sensing mode (b).
D. Taq polymerase adsorption test
To evaluate the NSA of real functional proteins on the PDMS and pcPDMS surfaces, activity of Taq polymerase was measured before and after flowing through PDMS and pcPDMS microchannels. 10 μL Taq polymerase with the concentration of 50 U/mL (the commonly used concentration in a real PCR reaction system) in the Taq buffer (B33, Thermo Fisher Scientific, including 75 mM Tris-HCl (pH 8.8 at 25 °C), 20 mM (NH4)2SO4, and 0.01% (v/v) Tween 20) was injected into the chip and flew through the channels at a flow rate of 500 nL/min (a typical flow rate in a continuous flow PCR chip44). The fluid collected after flowing through the microchannel was mixed with the detection system, including 60 nM probe (sequence of 5′-FAM-CCCCCC CGCACCTAAAGGGTGCG-3′, Sangon Co., China) and 50 μM dNTPs (TIANGEN, China) to initiate the according nucleic acid polymerization processes. The polymerization was quantitatively detected by a designed photoelectron transfer (PET) nucleic acid probe reported previously45 to evaluate the activity of Taq polymerase inside the reaction system. The activity of polymerase without passing through any microchannel was just deposited in ambient environment at the same time and was also tested as a control case.
III. RESULTS AND DISCUSSIONS
A. pcPDMS microchip
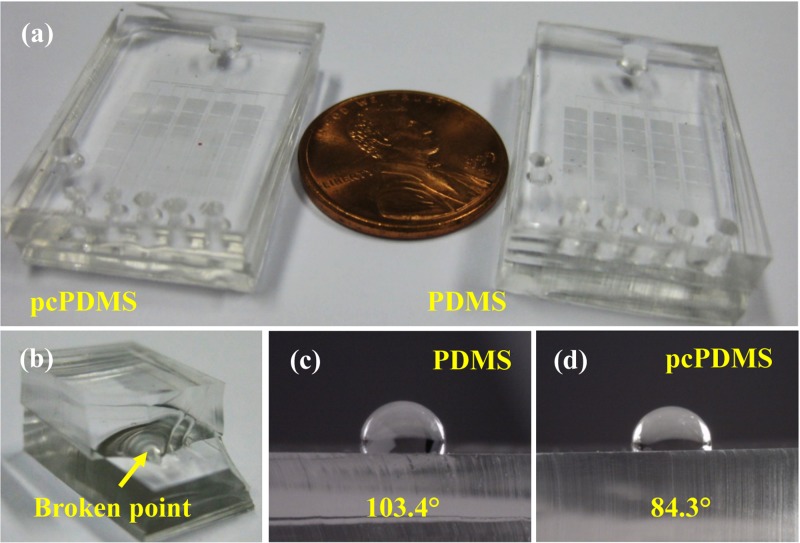
Typical pcPDMS and PDMS chips were shown in Fig. 3(a), indicating that the transparency of pcPDMS did not degrade compared to that of the pristine PDMS. The bonding strength of pcPDMS was also evaluated by a manually peeling test, as exhibited in Fig. 3(b). The break happened in the bulk rather than the bonding interface, which indicated a good irreversible bonding was successfully achieved. Figs. 3(c) and 3(d) showed the contact angles of water on the pristine PDMS and the so-prepared pcPDMS surface were 103.4° and 84.3°, respectively.
FIG. 3.
Characterizations of pcPDMS and PDMS chips in the present work. (a) Typical PDMS and pcPDMS chips, (b) a manually peeling test to evaluate the bonding strength, and (c) and (d) images of contact angles of water on the PDMS and pcPDMS surfaces, respectively.
B. Characterization of the caulking status of Parylene C
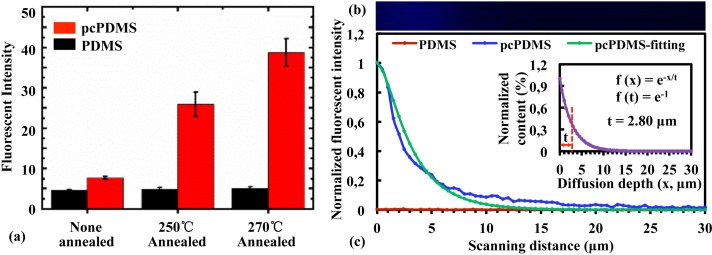
Although it has been reported that Parylene C monomers did penetrate into the PDMS matrix during the deposition,41–43 an effective method to characterize the Parylene C caulking status inside the PDMS matrix is still in lack. It has been reported that Parylene C had a stronger autofluorescence under 340–380 nm excitation compared to that of the PDMS.46 However, the contrast of autofluorescence intensity between the untreated Parylene C and PDMS is not significant. The ratio of the autofluorescence intensity was around 1.69. This work found that annealing at 270 °C for 2 h in nitrogen dramatically increased (5.03 times of that at the room temperature) the autofluorescence intensity of Parylene C while that of PDMS (1.13 times) almost unchanged, and the autofluorescence intensity ratio between annealed Parylene C and PDMS is increased to 7.54, shown in Fig. 4(a). This provided us an alternative way to characterize the caulking status of the Parylene C inside the PDMS. The intensity of the focus spot was assumed to follow a Gaussian distribution, . The pcPDMS (without oxygen plasma etching for the LSCM (Laser scanning confocal microscopy) imaging) had 1 μm thick pure Parylene C on the surface and an exponentially decayed distribution into the PDMS matrix from the surface, . The fluorescent intensities (blue line in Fig. 4(c)) extracted from the LSCM images (Fig. 4(b)) were used to fit the Parylene C distribution (the inset of Fig. 4(c), assuming to be an exponential relationship) based on the convolution operation of the above two distributions. The result indicated that Parylene C monomers penetrated into the PDMS matrix with a characteristic depth (t) of 2.8 μm, as shown in the inset of Fig. 4(c).
FIG. 4.
Characterization of the caulking status of Parylene C inside PDMS. (a) Fluorescent intensities of PDMS and pcPDMS after annealing at different temperatures for 2 h in nitrogen. (b) Typical fluorescent image of pcPDMS (cross-sectional view) generated by the 3D scanning and reconstruction under confocal microscopy. (c) The distribution of fluorescent intensity of pcPDMS along the z direction (blue line) in comparison with that of PDMS (red line). The green line showed the fitted fluorescent intensity of Parylene C inside the pcPDMS. The inset shows the normalized content of Parylene C inside PDMS matrix.
C. Non-specific adsorption of BSA
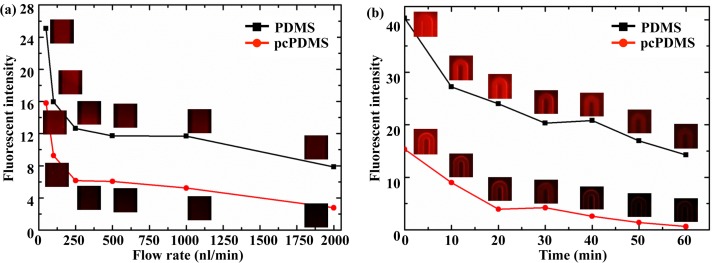
The fluorescent intensities, implying the amount of adsorbed BSA on the surface, at different flow rates in the tests under the continuous flow mode were shown in Fig. 5(a). For the heterogeneous mode test, the variation of fluorescent intensities with the PBS washing time was shown in Fig. 5(b). Because the autofluorescence difference in PDMS and pcPDMS was not obvious without the annealing treatment and the BSA itself is too large to penetrate into the PDMS matrix, the interfering background fluorescences caused by Parylene C or dyed BSA molecules inside the pcPDMS were both neglected. The intensity showed in Figs. 5(a) and 5(b) were both normalized with the background signals accordingly.
FIG. 5.
NSA of BSA on the pcPDMS and PDMS surfaces under the continuous flow mode (a) and the heterogeneous sensing mode (b), respectively. The insets were the typical fluorescent images corresponding to each data point.
In the test under the continuous flow mode, NSA of BSA was considerably restrained on the pcPDMS surface compared to that on the PDMS surface, as shown in Fig. 5(a). The fluorescent intensity of adsorbed BSAs on the surface of pcPDMS was around 51.7% of that in the PDMS chip at the flow rate of 500 nL/min and 35.2% at the flow rate of 2000 nL/min. The fluorescent intensity of adsorbed BSAs was inversely proportional to the flow rate. At a smaller flow rate, the BSA has more time to contact with the surface of microchannel and easier to be adsorbed onside. This indicated that the undesired adsorption of reacting molecules could be decreased by increasing the flow rate. However, a high flow rate requires a high driving power and will reduce the reaction time within the reactor. In the test under the heterogeneous sensing mode, the adsorbed BSA was dramatically washed away by a 60 min PBS rinsing, as shown in Fig. 5(b). The fluorescence intensity decreased to only 4.14% of the saturated value for the pcPDMS sample, while still 35.54% of the start value for the PDMS one after a 60 min washing. Overall, the NSA of BSA in pcPDMS microchannels was only 4.5% of that in PDMS microchannels after a 60 min washing by directly comparing the fluorescence intensities of two channels. This indicated BSA had a low affinity to pcPDMS and can be easily washed away.
To quantitatively compare the NSA of BSA on the PDMS and pcPDMS surfaces, theoretical analysis based on the first-order Langmuir model was conducted. There are three parameters in this adsorption process: (1) concentration of adsorbate, BSA molecules (A), dissolved in the PBS [A]; (2) surface concentration of the adsorption sites (B) on the PDMS/pcPDMS surfaces [B]; and (3) surface concentration of the adsorbed complex AB [AB]. The reversible adsorption and desorption processes can be written as
where Ka and Kd are the adsorption and desorption coefficients, respectively. θ is the percentage of the adsorption sites occupied by BSA at saturation ([AB]eq) to the initial adsorption sites ([B]0), as shown in Fig. 2(b), i.e., [AB]eq/[B]0.
By fitting with the experimental measurement, Ka, Kd, [B]0, and θ for pcPDMS and PDMS are obtained and listed in Table II.
TABLE II.
The fitted Ka, Kd, [B]0, and θ for pcPDMS and PDMS.
| Parameter | Ka (103 M−1 s−1) | Kd (10−4 s−1) | [B]0 (M m−1) | θ (%) |
|---|---|---|---|---|
| pcPDMS | 2.17 | 8.17 | 43.04 | 36 |
| PDMS | 6.20 | 2.50 | 46.39 | 87 |
The values of [B]0 for pcPDMS and PDMS are nearly the same, indicating that Parylene C caulked in the PDMS matrix does not influence the adsorption sites on the surface. This is reasonable as the top surface of pcPDMS was almost the same with PDMS because Parylene C was over-etched during the chip fabrication and only PDMS is exposed on the top surface. However, the adsorption/desorption coefficients and the saturated coverage of BSA on the pcPDMS are 35%/326.8% and 41.4% of those on the PDMS, respectively. This implied that the adsorption/desorption process might be influenced by the caulked Parylene C inside the PDMS matrix perhaps through the varied wettability because of the different surface energies of PDMS and Parylene C. As reported in the previous studies,17,18,22,24,29 the BSA had a low NSA on hydrophilic surfaces. The contact angle measurements shown in Figs. 3(c) and 3(d) indicated that the pcPDMS had a higher surface energy, which means the affinity between BSA and pcPDMS was weaker compared to that between BSA and PDMS.
D. Non-specific adsorption of Taq polymerase
The activity of polymerase after flowing through the pcPDMS microchannel was almost the same (93.0%) as that in the control test. Meanwhile, the activity of Taq polymerase collected from the pristine PDMS microchannels was only 28.9% of that obtained from the pcPDMS chip. This result was consistent with the aforementioned BSA adsorption test. Less Taq polymerases were non-specifically adsorbed on the pcPDMS surface than that on the PDMS one. It should be mentioned that besides the decreased amount of Taq polymerase caused by the NSA, the observed decrement of the polymerization efficiency might also come from the degradation of the intrinsic activity of Taq polymerase molecule after flowing through and contacting with the PDMS surface, i.e., adsorbed and then desorbed from the surface because the adsorption/desorption process could vary the structure and conformation17 of the Taq polymerase, which is critical for the activity maintenance.
IV. CONCLUSION
This work studied the non-specific BSA adsorption on the pcPDMS and PDMS surfaces in two typical microfluidic operation modes. The results showed that the NSA of BSA on the pcPDMS surface was reduced to 35.2% of that on the PDMS surface with the flow rate of 2000 nL/min under the continuous flow mode. While under the heterogeneous sensing mode, after a 60 min rinsing, the non-specific adsorbed BSA in the pcPDMS microchannel was only 4.5% (i.e., reduced by 95.5%) of that in the pristine PDMS channel. The adsorption/desorption coefficients (Ka/Kd) and the saturated coverage (θ) of BSA on the pcPDMS and PDMS surfaces extracted from the experimental measurement also indicated that pcPDMS has a low protein affinity and tends to effectively restrain the NSA. The Taq polymerase activity test results further confirmed the low NSA performance of pcPDMS. A temperature-dependent autofluorescence intensity of Parylene C was used to characterize the Parylene C caulking status inside the PDMS matrix. Preliminary result indicated that the depth of Parylene C penetrated into the PDMS matrix was around 2.8 μm. In short, the pcPDMS is an alternative but an effective surface passivation method for PDMS-based soft lithography to fulfill the devices with a low NSA of proteins.
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 81471750 and 91023045) and the Major State Basic Research Development Program (973 Program) (Grant No. 2015CB352100).
References
- 1. Berthier E., Young E. W. K., and Beebe D., Lab Chip 12, 1224 (2012). 10.1039/c2lc20982a [DOI] [PubMed] [Google Scholar]
- 2. Mukhopadhyay R., Anal. Chem. 79, 3248 (2007). 10.1021/ac071903e [DOI] [PubMed] [Google Scholar]
- 3. Li N., Schwartz M., and Zanetti C. I., J. Biomol. Screening 14, 194 (2009). 10.1177/1087057109346446 [DOI] [PubMed] [Google Scholar]
- 4. Kodziusa R., Xiao K., Wu J., Yi X., Gong X., Foulds I. G., and Wen W., Sens. Actuators, B 161, 349 (2012). 10.1016/j.snb.2011.10.044 [DOI] [Google Scholar]
- 5. Toepke M. W. and Beebe D. J., Lab Chip 6, 1484 (2006). 10.1039/b612140c [DOI] [PubMed] [Google Scholar]
- 6. Cattani-Scholz A., Pedone D., Blobner F., Abstreiter G., Schwartz J., Tornow M., and Andruzzi L., Biomacromolecules 10, 489 (2009). 10.1021/bm801406w [DOI] [PubMed] [Google Scholar]
- 7. Cesaro-Tadic S., Dernick G., Juncker D., Buurman G., Kropshofer H., Michel B., Fattinger C., and Delamarche E., Lab Chip 4, 563 (2004). 10.1039/b408964b [DOI] [PubMed] [Google Scholar]
- 8. Makamba H., Kim J. H., Lim K., Park N., and Hahn J. H., Electrophoresis 24, 3607 (2003). 10.1002/elps.200305627 [DOI] [PubMed] [Google Scholar]
- 9. Zhou J., Khodakov D. A., Ellis A. V., and Voelcker N. H., Electrophoresis 33, 89 (2012). 10.1002/elps.201100482 [DOI] [PubMed] [Google Scholar]
- 10. Wong I. and Ho C. M., Microfluid. Nanofluid. 7, 291 (2009). 10.1007/s10404-009-0443-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim J. A., Lee J. Y., Seong S., Cha S. H., Lee S. H., Kim J. J., and Park T. H., Biochem. Eng. J. 29, 91 (2006). 10.1016/j.bej.2005.02.032 [DOI] [Google Scholar]
- 12. Crabtree H. J., Lauzon J., Morrissey Y. C., Taylor B. J., Liang T., Johnstone R. W., Stickel A. J., Manage D. P., Atrazhev A., Backhouse C. J., and Pilarski L. M., Microfluid. Nanofluid. 13, 383 (2012). 10.1007/s10404-012-0968-9 [DOI] [Google Scholar]
- 13. Xia Y., Hua Z., Srivannavit O., Bilge Ozel A., and Gulari E., J. Chem. Technol. Biotechnol. 82, 33 (2007). 10.1002/jctb.1631 [DOI] [Google Scholar]
- 14. Lou X., Panaro N. J., Wilding P., Fortina P., and Kricka L. J., BioTechniques 36(2), 248 (2004). [DOI] [PubMed] [Google Scholar]
- 15. Shoffner M. A., Cheng J., Hvichia G. E., Kricka L. J., and Wilding P., Nucl. Acids Res. 24, 375 (1996). 10.1093/nar/24.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Priest C., Biomicrofluidics 4, 032206 (2010). 10.1063/1.3493643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang H. and Chiao M., J. Med. Biol. Eng. 35, 143 (2015). 10.1007/s40846-015-0029-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu X., Xiao J., and Dang F., Langmuir 31, 5891 (2015). 10.1021/acs.langmuir.5b01085 [DOI] [PubMed] [Google Scholar]
- 19. McArthur S. L., McLean K. M., Kingshott P., St John H. A. W., Chatelier R. C., and Griessera H. J., Colloids Surf., B 17, 37 (2000). 10.1016/S0927-7765(99)00086-7 [DOI] [Google Scholar]
- 20. Huang B., Wu H., Kim S., and Zare R. N., Lab Chip 5, 1005 (2005). 10.1039/b509251e [DOI] [PubMed] [Google Scholar]
- 21. Yang L., Li L., Tu Q., Ren L., Zhang Y., Wang X., Zhang Z., Liu W., Xin L., and Wang J., Anal Chem. 82, 6430 (2010). 10.1021/ac100544x [DOI] [PubMed] [Google Scholar]
- 22. Chen H., Zhang Z., Chen Y., Brook M. A., and Sheardown H., Biomaterials 26, 2391 (2005). 10.1016/j.biomaterials.2004.07.068 [DOI] [PubMed] [Google Scholar]
- 23. Choi S. and Chae J., J. Micromech. Microeng. 20, 075015 (2010). 10.1088/0960-1317/20/7/075015 [DOI] [Google Scholar]
- 24. Sibarani J., Takai M., and Ishihara K., Colloids Surf., B 54, 88 (2007). 10.1016/j.colsurfb.2006.09.024 [DOI] [PubMed] [Google Scholar]
- 25. Roach L. S., Song H., and Ismagilov R. F., Anal. Chem. 77, 785 (2005). 10.1021/ac049061w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li M., Li S., Wu J., Wen W., Li W., and Alici G., Microfluid. Nanofluid. 12, 751 (2012). 10.1007/s10404-011-0917-z [DOI] [Google Scholar]
- 27. Kanai M., Uchida D., Sugiura S., Shirasaki Y., Go J. S., Nakanishi H., Funatsu T., and Shoji S., in MicroTAS 2003, 5–9 October, Squaw Valley, California, USA, 2003, p. 429. [Google Scholar]
- 28. Wu D., Qin J., and Lin B., Lab Chip 7, 1490 (2007). 10.1039/b708877a [DOI] [PubMed] [Google Scholar]
- 29. Wu D., Zhao B., Dai Z., Qin J., and Lin B., Lab Chip 6, 942 (2006). 10.1039/b600765a [DOI] [PubMed] [Google Scholar]
- 30. Forster S. and McArthur S. L., Biomicrofluidics 6, 036504 (2012). 10.1063/1.4754600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiao D., Le T. V., and Wirth M. J., Anal. Chem. 76, 2055 (2004). 10.1021/ac035254s [DOI] [PubMed] [Google Scholar]
- 32. Kim B. Y., Hong L. Y., Chung Y. M., Kim D. P., and Lee C. S., Adv. Funct. Mater. 19, 3796 (2009). 10.1002/adfm.200901024 [DOI] [Google Scholar]
- 33. Schmolke H., Demming S., Edlich A., Magdanz V., Buttgenbach S., Lara E. F., Krull R., and Klages C. P., Biomicrofluidics 4, 044113 (2010). 10.1063/1.3523059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abate A. R., Lee D., Do T., Holtze C., and Weitz D. A., Lab Chip 8, 516 (2008). 10.1039/b800001h [DOI] [PubMed] [Google Scholar]
- 35. Roman G. T. and Culbertson C. T., Langmuir 22, 4445 (2006). 10.1021/la053085w [DOI] [PubMed] [Google Scholar]
- 36. Riaz A., Gandhiraman R. P., Dimov I. K., Desmonts L. B., Ducree J., Daniels S., Riccoa A. J., and Lee L. P., Lab Chip 12, 4877 (2012). 10.1039/c2lc40296c [DOI] [PubMed] [Google Scholar]
- 37. Lahann J., Balcells M., Lu H., Rodon T., Jensen K. F., and Langer R., Anal. Chem. 75, 2117 (2003). 10.1021/ac020557s [DOI] [PubMed] [Google Scholar]
- 38. Shin Y. S., Cho K., Lim S. H., Chung S., Park S. J., Chung C., Han D. C., and Chang J. K., J. Micromech. Microeng. 13, 768 (2003). 10.1088/0960-1317/13/5/332 [DOI] [Google Scholar]
- 39. Wang W., Kang D., and Tai Y. C., in Transducers 2015, 21–25 June, Alaska, Anchorage, USA, 2015, p. 1307. [Google Scholar]
- 40. Sawano S., Naka1 K., Werber A., Zappe H., and Konishi S., in MEMS 2008, 13–17 January, Tucson, AZ, USA, 2008, p. 419. [Google Scholar]
- 41. Lei Y., Liu Y., Wang W., Wu W., and Li Z., Lab Chip 11, 1385 (2011). 10.1039/c0lc00486c [DOI] [PubMed] [Google Scholar]
- 42. Lei Y., Liu Y., Wang W., Wu W. G., and Li Z. H., in MEMS 2011, 23–27 January, Cancun, Mexico, 2011, p. 1123. [Google Scholar]
- 43. Zhang L., Liu Y., Yang F., Wang W., Zhang D., and Li Z., in MEMS 2015, 18–22 January, Estoril, Portugal, 2015, p. 358. [Google Scholar]
- 44. Kopp M. U., de Mello A. J., and Manz A., Science 280, 1046 (1998). 10.1126/science.280.5366.1046 [DOI] [PubMed] [Google Scholar]
- 45. Song C., Zhang C., and Zhao M., Biosens. Bioelectron. 25, 301 (2009). 10.1016/j.bios.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 46. Lu B., Zheng S., Quacha B. Q., and Tai Y. C., Lab Chip 10, 1826 (2010). 10.1039/b924855b [DOI] [PubMed] [Google Scholar]