Abstract
Purpose
Retinitis pigmentosa (RP) is a clinically and genetically heterogeneous group of inherited retinal degenerations characterized by progressive loss of photoreceptor cells and RPE functions. More than 70 causative genes are known to be responsible for RP. This study aimed to identify the causative gene in a patient from a consanguineous family with childhood-onset severe retinal dystrophy.
Methods
To identify the defective gene, whole exome sequencing was performed. Candidate causative variants were selected and validated using Sanger sequencing. Segregation analysis of the causative gene was performed in additional family members. To verify that the mutation has an effect on protein synthesis, an expression vector containing the first ten amino acids of the mutant protein fused with the DsRed2 fluorescent protein was constructed and transfected into HEK293T cells. Expression of the fusion protein in the transfected cells was measured using fluorescence microscopy.
Results
By filtering against public variant databases, a novel homozygous missense mutation (c.3G>A) localized in the start codon of the MERTK gene was detected as a potentially pathogenic mutation for autosomal recessive RP. The c.3G>A mutation cosegregated with the disease phenotype in the family. No expression of the first ten amino acids of the MerTK mutant fused with the DsRed2 fluorescent protein was detected in HEK293T cells, indicating that the mutation affects the translation initiation site of the gene that may lead to loss of function of the MerTK signaling pathway.
Conclusions
We report a novel missense mutation (c.3G>A, p.0?) in the MERTK gene that causes severe vision impairment in a patient. Taken together with previous reports, our results expand the spectrum of MERTK mutations and extend our understanding of the role of the MerTK protein in the pathogenesis of retinitis pigmentosa.
Introduction
Retinitis pigmentosa (RP) is a clinically and genetically heterogeneous group of inherited retinal degenerations characterized by dysfunction of the rod and cone photoreceptor cells and the RPE leading to night blindness and peripheral visual field loss, followed by variable changes in central vision. Progressive loss of vision can eventually lead to complete blindness in some cases [1]. The age of onset varies among patients with RP from early childhood to the fifth or sixth decade of life [2]. The worldwide prevalence of RP ranges from 1 in 3,500 to 1 in 5,000 [1]. RP can be inherited in autosomal dominant (adRP), autosomal recessive (arRP), or X-linked recessive (xlRP) modes. To date, mutations in 79 genes distributed among all modes of inheritance, i.e., 27 genes in adRP, 55 genes in arRP, and three genes in xlRP, have been reported (RetNet, accessed on January 3, 2016). Among these 79 genes, six have been shown to be involved in both adRP and arRP.
RPE cells have several functions in the vertebrate retina. Daily phagocytosis of shed photoreceptor outer segments (POS) by the RPE is essential for visual function and photoreceptor survival. Abnormal phagocytic function causes an excessive accumulation of POS debris in the subretinal space resulting in less efficient oxygen molecule and nutrient transport to the photoreceptor cells [3]. Mutations in the MER-proto-oncogene, tyrosine kinase (MERTK) gene (Gene ID: 10461; OMIM 604705) are responsible for defects in phagocytosis, resulting in autosomal recessive retinal degeneration. The MERTK gene is composed of 19 exons and encodes the MerTK protein, consisting of 999 amino acids. The MerTK protein is a member of the Tyro3, Axl, and Mertk (TAM) family of receptor tyrosine kinases and is highly expressed in the RPE, macrophages, ovary, prostate, testis, lung, and kidney [4]. MERTK-associated retinal disease was first reported in rodents with loss of MERTK gene function [5] and was subsequently described in three patients with RP who carried different mutations [6]. The MERTK gene has also been shown to be upregulated in canines with retinal degeneration [7].
In humans, mutations in the MERTK gene are relatively rare, accounting for approximately 1% of autosomal recessive retinal dystrophy cases [8]. Patients with MERTK mutations commonly display childhood-onset severe retinal dystrophy with defects in the macula [6,9-11]. Among the missense mutations of the MERTK gene [9,10,12,13], p.Arg844Cys has been shown to cause loss of protein function by making the MerTK signaling pathway malfunction due to an increase in protein degradation in HEK293T cells [9].
In this study, we report a novel homozygous missense mutation (c.3G>A, p.0?) in the MERTK gene detected with whole exome sequencing in a patient with isolated RP. To the best of our knowledge, this is the first report of an initiation codon mutation in the MERTK gene. We also show that the start codon mutation affects the translation start site of the gene, which could lead to the disease.
Methods
Subject
The proband, a 35-year-old male, was evaluated at the Department of Ophthalmology, Siriraj Hospital, in Bangkok, Thailand. The patient underwent a complete ophthalmological examination, including best-corrected visual acuity (BCVA), fundus examination, visual-evoked potential (VEP), full-field flash electroretinography (ERG), optical coherence tomography (OCT), and fundus autofluorescence (FAF). This study was approved by the Institutional Review Board of Siriraj Hospital Mahidol University and adhered to the tenets of the Declaration of Helsinki and the ARVO statement on human subjects. Peripheral blood was collected from the patient, his parents, and unaffected siblings after written informed consent was obtained. Genomic DNA was extracted using the salting-out method [14].
Whole exome sequencing (WES)
Exome capture was performed using solution hybrid selection with the SureSelectXT Human All Exon Automated Target Enrichment Kit (Agilent Technologies, Santa Clara, CA) for Illumina paired-end multiplexed sequencing. Exome capture libraries of 2×100 bp paired-end reads were sequenced on the Illumina HiSeq 2000 at the Department of Medical Genome Sciences, The University of Tokyo.
Exome sequence data analysis
Reads were mapped and aligned to the human reference genome (hg19 build GRCh37), obtained from UCSC, using the Burrows-Wheeler Aligner (BWA) program. SAMtools was used to remove duplicate PCR reads. Single nucleotide polymorphism (SNP) and indel calling were performed according to the Genome Analysis Toolkit (GATK) software package. Due to the extreme heterogeneity, both clinically and genetically, of RP, all genes listed in RetNet associated with non-syndromic retinal disease were examined to determine the causative gene. Sequence variations were classified as deleterious based on filtering strategies similar to those previously described [15]. Variant annotation and filtering were performed using the wANNOVAR web server [16]. This server provides several functional prediction programs, such as PolyPhen-2, Sorting Intolerant From Tolerant (SIFT), MutationTaster, likelihood ratio test (LRT), and PhyloP for non-synonymous variants. Visual inspection of the coverage and base calls of the target variants was performed using the Integrative Genomics Viewer (IGV, Broad Institute, Cambridge, MA). ATGpr, a web-based program, was used to determine the presence of all possible translational initiation sites (TISs) present in the cDNA sequence of the MERTK gene [17].
Data validation
Sanger sequencing was performed to confirm the variants identified in the candidate gene. Sequencing primers were designed using Primer3 to span at least 60 bp upstream and downstream of the putative pathogenic variant in the MERTK gene (RefSeq accession number NM_006343.2). To verify the pathogenicity of the novel variants, denaturing high-performance liquid chromatography (DHPLC) screening of 130 DNA samples from unrelated control subjects was performed on the WAVE DNA Fragment Analysis System (Transgenomic, Inc., San Jose, CA).
Construction of the pcDNA3-IRES-EYFPnuc vector
The internal ribosome entry site–enhanced yellow fluorescent protein with nuclear localization signal IRES-EYFPnuc sequence was amplified from original plasmid DNA (a gift from Dr.Kazuhiro Oka, Baylor College of Medicine) by using the primers IRES-EYFPnuc-F: 5′-AAT TGC GGC CGC GGC CGC AAT TGA TCC GCC-3′ and IRES-EYFPnuc-R: 5′-AAT TAG ATC TAT CCG GTG GAG CCT ACC TT-3′, which contain NotI and XbaI sites, respectively (restriction sites are shown in italics). This fragment contains 1,448 bp of the IRES-EYFPnuc sequence and was ligated into the NotI and XbaI sites of the pcDNA3 vector (Invitrogen, Carlsbad, CA), generating the pcDNA3-IRES-EYFPnuc vector.
Construction of the fusion ten amino acid MerTK (wild-type or mutant) and DsRed2 vectors
The fusion ten amino acid MerTK and red fluorescent protein (DsRed2) sequence was generated by replacement of the first three base pairs, ATG, of the DsRed2 cDNA with 30 bp (encoding ten amino acids, MGPAPLPLLL) upstream of the MERTK gene. The fusion sequence of the wild-type was amplified using the primer EcoRI_10 aa MerTK Wt_DsRed2_F 5′-AAT TGA ATT CGC CAC CAT GGG GCC GGC CCC GCT GCC GCT GCT GCT GGC CTC CTC CGA GGA CGT CAT-3′, which contains an EcoRI site (italics) followed by the Kozak sequence (underlined), 30 bp of the MERTK cDNA (bold), and 20 bp upstream (without the start codon) of the DsRed2 cDNA, respectively. The reverse primer, DsRed2-NotI 5′-AAT TGC GGC CGC CTA CAG GAA CAG GTG GTG G 3′, contains a NotI site (italics) followed by a stop codon (bold) after the last amino acid of the DsRed2 cDNA. The PCR product (724 bp) was cloned into the EcoRI and NotI sites of the pcDNA3-IRES-EYFPnuc vector to generate the pcDNA3–10 aa MerTK Wt-DsRed2-IRES-EYFPnuc vector. For construction of the MERTK mutant (Mut) vector, the fragment was amplified from the pcDNA3–10 aa MerTK Wt-DsRed2-IRES-EYFPnuc vector using the primers EcoRI_10 aa MerTK Mut_DsRed2_F 5′-AAT TGA ATT CGC CAC CAT AGG GCC GGC CCC GCT G-3′, which contains an EcoRI site (italics) followed by the Kozak sequence (underlined), 30 bp of the MERTK cDNA where the third base was changed from G to A (bold), respectively, and the reverse primer DsRed2-NotI. The amplified fragment was ligated into the EcoRI and NotI sites of the pcDNA3-IRES-EYFPnuc vector to generate the pcDNA3–10 aa MerTK Mut-DsRed2-IRES-EYFPnuc vector. All constructs were verified with Sanger sequencing.
Cell culture and transient transfection
Human embryonic kidney cell line HEK293T (originally from ATCC; no. CRL-3216, Manassas, VA; kindly provided by Prof. Dr. Pa-thai Yenchitsomanus), were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen), supplemented with 10% fetal bovine serum (Biochrom, Berlin, Germany). Cells were grown at 37 °C in a humidified incubator containing 5% CO2. Transient transfection was performed using the calcium phosphate precipitation method. Six hours post-transfection, the medium was changed and replaced with fresh medium. Protein expression of the Wt and Mut vectors was measured 48 h after transfection. The expression of fluorescent proteins was visualized under an inverted Olympus IX70 fluorescence microscope (Olympus America Inc., Center Valley, PA) equipped with filters for fluorescein isothiocyanate (FITC) or Red.
Results
Clinical description
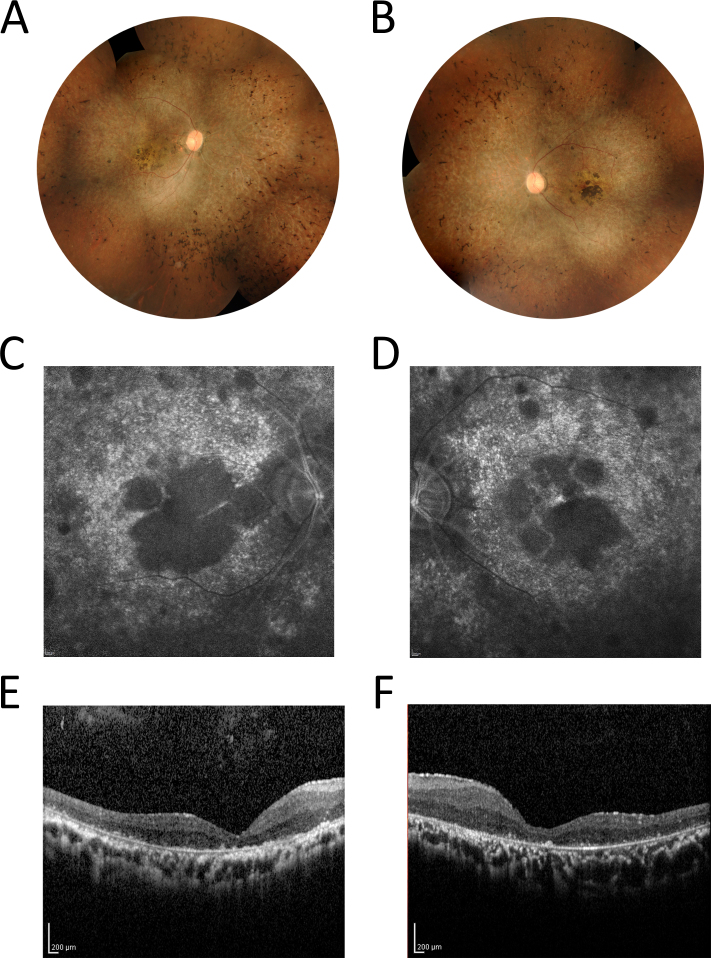
A 35-year-old male patient, with consanguineous parents, was diagnosed with isolated RP at the age of 27. There was no family history of retinal dystrophy. The patient reported difficulty seeing at night since age 7 and blurred distant vision after age 16. At the age of 35, his BCVA was reduced to light perception in the right eye and hand motion in the left eye. Ophthalmological examination results are shown in Figure 1. Fundus examination demonstrated typical RP changes in both eyes with pale optic discs and attenuated retinal vessels, generalized pigmentary granularity, moderate bone-like spicules, and clumps of pigment accumulation in the macula. FAF imaging of the RPE layer showed bilateral decreased autofluorescence corresponding to the areas of pigment accumulation in the center of the macula. No evidence of cystoid macular edema was detected with OCT. ERG response was non-recordable in both eyes. VEP showed markedly decreased amplitudes in both eyes.
Figure 1.
Fundus photographs, FAF, and OCT results of the patient. Color montage fundus photographs of the right eye (A) and the left eye (B) of the patient at age 35 years. Fundus examination demonstrated changes typical of retinitis pigmentosa (RP) in both eyes, including pale optic discs with attenuated retinal vessels, generalized pigmentary granularity, moderate bone-like spicules in four quadrants, and macular atrophy with pigment accumulation. Fundus autofluorescence (FAF) imaging of the right eye (C) and the left eye (D) showed bilateral decreased autofluorescence corresponding to the areas of pigment accumulation in the center of the macula. These areas were encircled by spotty autofluorescence along the temporal vascular arcades. The optical coherence tomography (OCT) scans of the right eye (E) and the left eye (F) revealed decreased macular thickness.
WES analysis revealed a novel start codon mutation in the MERTK gene
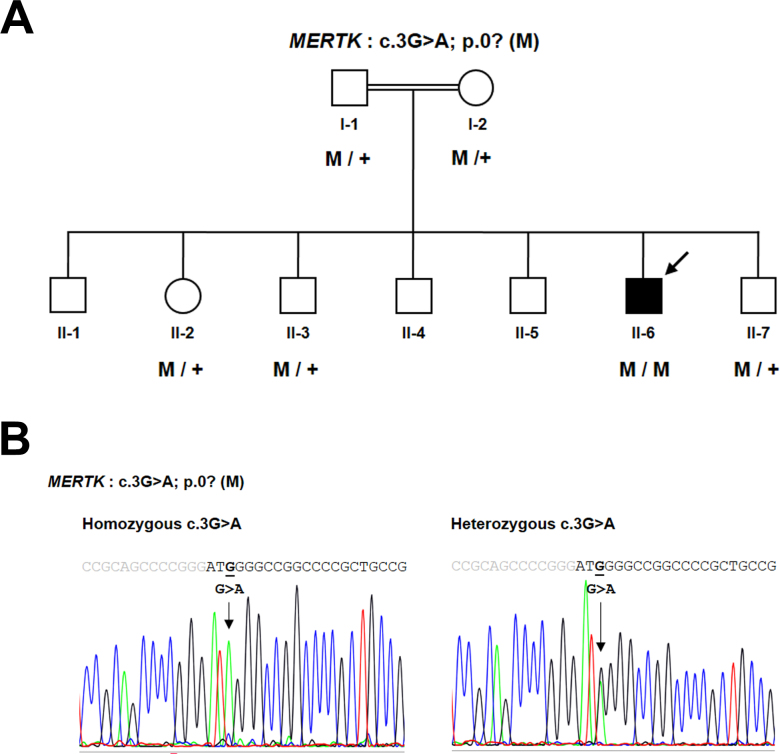
To identify the RP causative gene, WES was performed on genomic DNA extracted from the proband’s peripheral blood. The variant files from WES were subjected to wANNOVAR for annotation [16]. After several filtering steps, a novel homozygous variant, c.3G>A, located in the translation initiation codon ATG of the MERTK gene, was identified (Appendix 1). This variant was predicted to be deleterious and was not observed in the 1000 Genomes Project, dbSNP137, the Exome Variant Server (EVS), the Exome Aggregation Consortium (ExAC), or 260 chromosomes from ethnically matched control samples. Sequence analysis confirmed that the proband (I:1) was homozygous, whereas unaffected individuals (I:1, I:2, II:2, II:3, and II:7) were heterozygous carriers for the initiation codon variant, c.3G>A (Figure 2).
Figure 2.
Pedigree and DNA sequence chromatogram of the family. A: Pedigree of the consanguineous family who participated in this study. Genotype data are presented below the patient and members, where applicable, of his family. The filled symbol with an arrow indicates the index case. Squares: males. Circles: females. Normal alleles are indicated by “+.” The potentially pathogenic mutation, p.0?, is indicated by “M.” B: Sequence analysis of the MERTK gene revealed a missense variant, c.3G>A, located in the start codon of the MerTK protein, homozygous in the patient and heterozygous in other family members. The gray and black letters indicate the 5′ untranslated region (UTR) and exon 1 of the MERTK gene, respectively.
No expression of the mutant fusion protein in HEK293T cells
There is the possibility that protein synthesis occurs using alternative TISs, both upstream and downstream of the canonical initiation codon [18]. A single alternative in-frame TIS at codon 257 downstream of the canonical start codon of MERTK was predicted by ATGpr. The protein sequence encoded by this predicted alternative TIS results in a shorter polypeptide that lacks N-terminal residues. This truncated protein is more likely to lead to loss of MerTK function because the protein lacks two conserved regions, namely, Ig-like C2-type 1 (IgL1) and Ig-like C2-type 2 (IgL2).
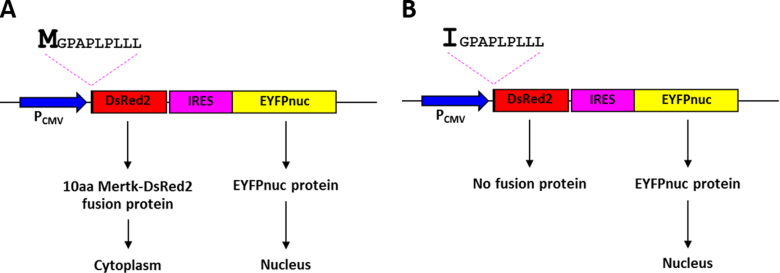
Although we speculate that the start codon mutation may affect protein synthesis, two expression vectors that contain a wild-type or a mutant (c.3G>A) of the first ten amino acids of MerTK fused with the DsRed2 fluorescent protein were designed and examined to support this concept (Figure 3). Both vector constructs contain an IRES sequence and an EYFPnuc sequence. The IRES sequence leads to cap-independent initiation of translation, so the ribosome is able to bind to the IRES sequence directly, resulting in translation initiation of the gene downstream of the IRES sequence [19]. Downstream of the IRES sequence is the EYFPnuc sequence that encodes the nuclear localizing protein. Expression of the EYFPnuc sequence was used as a control for confirming successful transfection. The DsRed2 sequence located in both vector constructs has no start codon; therefore, expression of the fusion protein depends solely on the initiation codon situated at the first amino acid of the MerTK fragment. The DsRed2 sequence encodes a red fluorescent protein that is localized to the cytoplasm (Figure 4). Forty-eight hours after transfection, expression of the wild-type fusion protein was detected in the cytoplasm of the transfected cells. In contrast, cells transfected with the mutant construct showed no expression of the fusion protein (Figure 5).
Figure 3.
Schematic diagram depicting the cloning strategy for construction of the vectors employed in this study. A: pcDNA3-IRES-EYFPnuc. B: pcDNA3–10 aa MerTK Wt_DsRed2-IRES-EYFPnuc. C: pcDNA3–10 aa MerTK Mut_DsRed2-IRES-EYFPnuc. Blue boxes indicate the cytomegalovirus (CMV) promoter (PCMV) sequence, pink boxes indicate the internal ribosome entry site (IRES) sequence, yellow boxes indicate the enhanced yellow fluorescent protein with nuclear localization signal (EYFPnuc) sequence, red boxes indicate the DsRed2 sequence, and the black arrow indicates the 30 bp MERTK sequence that is a different nucleotide at position +3: (B) G in the Wt vector causing codon 1 to encode Met, and (C) A in the Mut vector causing codon 1 to encode Ile.
Figure 4.
Schematic representations of the pcDNA3–10 aa MerTK Wt and Mut-DsRed2-IRES-EYFPnuc vectors and their expected protein expression patterns. The drawings illustrate the pcDNA3–10 aa MerTK Wt-DsRed2-IRES-EYFPnuc vector (A) and the pcDNA3–10 aa MerTK Mut-DsRed2-IRES-EYFPnuc vector (B). Blue arrows indicate the cytomegalovirus (CMV) promoter, which efficiently drives the expression of the bicistronic RNA. The pink dashed lines indicate the first ten amino acid (aa) sequence of the MerTK protein that is a different amino acid at codon 1: Met (M) in 10 aa MerTK Wt (A) and Ile (I) in 10 aa MerTK Mut (B). The red boxes correspond to the DsRed2 sequence, and the pink and yellow boxes indicate the IRES-EYFPnuc sequence. Proteins produced from the wild-type construct vector are predicted to be expressed in the nucleus and the cytoplasm, while protein expression from the mutant construct vector is predicted to be found only in the nucleus.
Figure 5.
Identification of fluorescence proteins in HEK293T cells. Protein expression of the Wt or Mut vectors was examined 48 h after transfection. Images were captured using a 40X objective lens. A–C: Images of HEK293T cells transfected with the Wt construct encoding an enhanced green fluorescent protein that localizes to the nucleus (A), the fusion protein of 10 aa MerTK Wt-DsRed2 is present in the cytoplasm (B). Merged images (A and B) confirm the fusion protein of the Wt construct is expressed only in the cytoplasm (C). D–F: Images of HEK293T cells transfected with the Mut construct encoding an enhanced green fluorescent protein that localizes to the nucleus (D). E: No fluorescence fusion protein from the Mut construct was detectable in the cytoplasm. The merged images (D and E) indicate no fluorescence fusion protein expressed, except the enhanced green fluorescent protein that was observed in the nucleus (F).
Discussion
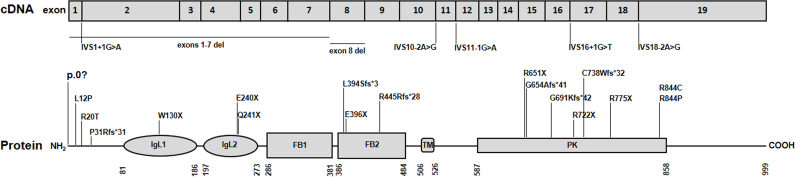
Since the first MERTK mutations related to human retinal degeneration were reported in 2000 [6], only 25 (including the present report) different mutations have been identified scattered along the entire gene (Figure 6), including five missense mutations [9,10,12,13], seven nonsense mutations [6,9,20-24], five mutations resulting in defective splicing [6,11,23,25,26], six small deletions [6,10,12,24,26,27], and two gross deletions [11,28]. This suggests that mutations in the MERTK gene are a rare cause of retinal degeneration.
Figure 6.
Schematic representation of the MERTK gene at the transcript level and functional domains of the MerTK protein. The MERTK gene consists of 19 exons. Mutations in the MERTK gene are distributed along the entire gene. MerTK is a 999 amino acid protein that contains several conserved domains: Ig-like C2-type 1 (IgL1), Ig-like C2-type 2 (IgL2), fibronectin type-III 1 (FB1), fibronectin type-III 2 (FB2), transmembrane (TM), and protein kinase (PK). Numbers under the protein line indicate the boundaries of each domain. Nucleotide 1 is A of the ATG initiation codon according to RefSeq NM_006343.2. The amino acid residues are numbered according to RefSeq NP_006334.2, starting at the initiator methionine residue.
Using WES, we identified a novel homozygous missense mutation, c.3G>A, in the MERTK gene. Clinical observations of the retinal phenotypes in our patient, who developed severe progressive retinal dystrophy, are consistent with previous studies reporting the age at onset from early childhood to the age of 12 and the age at diagnosis from 3 to 45 years [6,9-12,20,25,28].
In this study, we found a missense mutation, c.3G>A, in the initiation codon of the MERTK gene. This mutation was predicted to abolish the TIS of the MERTK gene. Based on a previous report, effective mRNA translation depends on the nucleotide context of the Kozak sequence (gccRccAUGg; where R is a purine, and AUG is the start codon) [18]. To test the concept that a mutation in the start codon may affect translation, two fusion protein models, the first ten amino acids of the MerTK wild-type or mutant, fused with the DsRed2 protein were generated and transfected separately into HEK293T cells. The results confirm that translation of the first ten amino acids of the Mertk-DsRed2 fusion protein can occur only from the initiation codon in the MerTK sequence of the wild-type model. Therefore, our results indicate that the start codon mutation, c.3G>A, affects the TIS of the MERTK gene resulting in loss of protein synthesis.
To date, more than 400 distinct single-nucleotide substitutions, embedded within the TISs of human genes, have been reported to cause inherited human diseases [29]. These substitutions are predicted to abolish the Kozak consensus sequence, leading to loss of protein synthesis. Although alternative start sites located downstream of the canonical start codon have been predicted in some cases, translation initiation by alternative initiation codons are likely to yield N-terminal truncated proteins. Normally, secretory proteins and membrane proteins contain an N-terminal signal sequence (typically 15–30 amino acids in length), which plays an important role in protein targeting to the endoplasmic reticulum membrane [30]. Thus, the production of an N-terminal truncated protein lacking its N-terminal signal peptide is likely to result in the intracellular misrouting of the nonfunctional protein, subsequently leading to the development of disease [31,32].
From our analysis, at least three findings support the possibility that c.3G>A tends to be a pathogenic mutation that affects the function of the MerTK protein. First, the homozygous c.3G>A mutation in the MERTK gene identified in the proband is consistent with an autosomal recessive mode of inheritance and cosegregates in the patient’s family. Second, the c.3G>A mutation changes an amino acid in the start codon that has a deleterious effect on the TIS of the MERTK gene, resulting in an inability to synthesize the mature MerTK protein. Although translation can occur via an alternative TIS located downstream of the original initiation codon, an N-terminally truncated MerTK protein generated from this translation will produce a non-functional protein. Third, this mutation is located within the signal peptide sequence (amino acids 1–20), which is necessary for translocation of the MerTK protein across the plasma membrane. The N-terminally truncated protein is most likely translocated to the cytosol, resulting in elimination via the proteasome [31,32].
In conclusion, as a result of our experimental model, we have demonstrated that the MERTK mutation, p.0?, involved in translation initiation, results in no MerTK synthesis. Taken together, the lack of the MerTK protein may result in impaired phagocytosis functioning, leading to the RP phenotype.
Acknowledgments
The authors are grateful to the patient and the family members who participated in this study. We also thank Fusano Todokoro and all staff at the Department of Medical Genome Sciences, The University of Tokyo, and all staff at the Department of Clinical Pathology, Mahidol University, for their technical assistance. We wish to thank Dr. James Hejna of Kyoto University for his helpful advice about cloning. We also thank Dr. Wichit Suthammarak of Mahidol University for his helpful advice regarding the test for gene expression. This study was supported by the operational expenditure fund of RIKEN (TDT); MEXT KAKENHI Grant 221S0002; the Royal Golden Jubilee PhD Program, Grant PHD/0102/2552 (WJ); the Siriraj Foundation (L-OA); the Chalermphrakiat Grant, Faculty of Medicine Siriraj Hospital, Mahidol University (L-OA, NP, WT, CL, PL, PS).
Appendix 1. Identification of homozygous c.3G>A variant in the MERTK gene by whole exome sequencing, as visualized by the Integrative Genomics Viewer (IGV).
This IGV snapshot shows the position of the homozygous G to A transition at nucleotide 3 (c.3G>A) in exon 1, which corresponds to genomic position 112,656,315 at chromosome 2. Vertical bars indicate read coverage. Horizontal bars represent 100 bp paired-end reads that mapped onto the human reference genome (hg19 build GRCh37). To access the data, click or select the words “Appendix 1.”
References
- 1.Anasagasti A, Irigoyen C, Barandika O, Lopez de Munain A, Ruiz-Ederra J. Current mutation discovery approaches in Retinitis Pigmentosa. Vision Res. 2012;75:117–29. doi: 10.1016/j.visres.2012.09.012. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23022136&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 2.Tsujikawa M, Wada Y, Sukegawa M, Sawa M, Gomi F, Nishida K, Tano Y. Age at onset curves of retinitis pigmentosa. Arch Ophthalmol. 2008;126:337–40. doi: 10.1001/archopht.126.3.337. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18332312&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 3.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–81. doi: 10.1152/physrev.00021.2004. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15987797&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 4.Linger RM, Keating AK, Earp HS, Graham DK. TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv Cancer Res. 2008;100:35–83. doi: 10.1016/S0065-230X(08)00002-X. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18620092&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Cruz PM, Yasumura D, Weir J, Matthes MT, Abderrahim H, LaVail MM, Vollrath D. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Genet. 2000;9:645–51. doi: 10.1093/hmg/9.4.645. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10699188&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 6.Gal A, Li Y, Thompson DA, Weir J, Orth U, Jacobson SG, Apfelstedt-Sylla E, Vollrath D. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet. 2000;26:270–1. doi: 10.1038/81555. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11062461&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 7.Ahonen SJ, Arumilli M, Seppala E, Hakosalo O, Kaukonen MK, Komaromy AM, Lohi H. Increased expression of MERTK is associated with a unique form of canine retinopathy. PLoS One. 2014;9:e114552. doi: 10.1371/journal.pone.0114552. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=25517981&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strick DJ, Vollrath D. Focus on molecules: MERTK. Exp Eye Res. 2010;91:786–7. doi: 10.1016/j.exer.2010.05.006. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20488176&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McHenry CL, Liu Y, Feng W, Nair AR, Feathers KL, Ding X, Gal A, Vollrath D, Sieving PA, Thompson DA. MERTK arginine-844-cysteine in a patient with severe rod-cone dystrophy: loss of mutant protein function in transfected cells. Invest Ophthalmol Vis Sci. 2004;45:1456–63. doi: 10.1167/iovs.03-0909. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=15111602&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 10.Tschernutter M, Jenkins SA, Waseem NH, Saihan Z, Holder GE, Bird AC, Bhattacharya SS, Ali RR, Webster AR. Clinical characterisation of a family with retinal dystrophy caused by mutation in the Mertk gene. Br J Ophthalmol. 2006;90:718–23. doi: 10.1136/bjo.2005.084897. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16714263&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackay DS, Henderson RH, Sergouniotis PI, Li Z, Moradi P, Holder GE, Waseem N, Bhattacharya SS, Aldahmesh MA, Alkuraya FS, Meyer B, Webster AR, Moore AT. Novel mutations in MERTK associated with childhood onset rod-cone dystrophy. Mol Vis. 2010;16:369–77. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20300561&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 12.Collin RW, van den Born LI, Klevering BJ, de Castro-Miro M, Littink KW, Arimadyo K, Azam M, Yazar V, Zonneveld MN, Paun CC, Siemiatkowska AM, Strom TM, Hehir-Kwa JY, Kroes HY, de Faber JT, van Schooneveld MJ, Heckenlively JR, Hoyng CB, den Hollander AI, Cremers FP. High-resolution homozygosity mapping is a powerful tool to detect novel mutations causative of autosomal recessive RP in the Dutch population. Invest Ophthalmol Vis Sci. 2011;52:2227–39. doi: 10.1167/iovs.10-6185. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21217109&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 13.Tada A, Wada Y, Sato H, Itabashi T, Kawamura M, Tamai M, Nishida K. Screening of the MERTK gene for mutations in Japanese patients with autosomal recessive retinitis pigmentosa. Mol Vis. 2006;12:441–4. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=16710167&dopt=Abstract [PubMed] [Google Scholar]
- 14.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=3344216&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinda W, Taylor TD, Suzuki Y, Thongnoppakhun W, Limwongse C, Lertrit P, Suriyaphol P, Trinavarat A, Atchaneeyasakul LO. Whole exome sequencing in Thai patients with retinitis pigmentosa reveals novel mutations in six genes. Invest Ophthalmol Vis Sci. 2014;55:2259–68. doi: 10.1167/iovs.13-13567. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24618324&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 16.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49:433–6. doi: 10.1136/jmedgenet-2012-100918. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22717648&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salamov AA, Nishikawa T, Swindells MB. Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics. 1998;14:384–90. doi: 10.1093/bioinformatics/14.5.384. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=9682051&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12459250&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–40. doi: 10.4161/cc.10.2.14472. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21220943&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ksantini M, Lafont E, Bocquet B, Meunier I, Hamel CP. Homozygous mutation in MERTK causes severe autosomal recessive retinitis pigmentosa. Eur J Ophthalmol. 2012;22:647–53. doi: 10.5301/ejo.5000096. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22180149&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 21.Shahzadi A, Riazuddin SA, Ali S, Li D, Khan SN, Husnain T, Akram J, Sieving PA, Hejtmancik JF, Riazuddin S. Nonsense mutation in MERTK causes autosomal recessive retinitis pigmentosa in a consanguineous Pakistani family. Br J Ophthalmol. 2010;94:1094–9. doi: 10.1136/bjo.2009.171892. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20538656&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srilekha S, Arokiasamy T, Srikrupa NN, Umashankar V, Meenakshi S, Sen P, Kapur S, Soumittra N. Homozygosity Mapping in Leber Congenital Amaurosis and Autosomal Recessive Retinitis Pigmentosa in South Indian Families. PLoS One. 2015;10:e0131679. doi: 10.1371/journal.pone.0131679. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26147992&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Guan L, Shen T, Zhang J, Xiao X, Jiang H, Li S, Yang J, Jia X, Yin Y, Guo X, Wang J, Zhang Q. Mutations of 60 known causative genes in 157 families with retinitis pigmentosa based on exome sequencing. Hum Genet. 2014;133:1255–71. doi: 10.1007/s00439-014-1460-2. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24938718&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 24.Wang F, Wang H, Tuan HF, Nguyen DH, Sun V, Keser V, Bowne SJ, Sullivan LS, Luo H, Zhao L, Wang X, Zaneveld JE, Salvo JS, Siddiqui S, Mao L, Wheaton DK, Birch DG, Branham KE, Heckenlively JR, Wen C, Flagg K, Ferreyra H, Pei J, Khan A, Ren H, Wang K, Lopez I, Qamar R, Zenteno JC, Ayala-Ramirez R, Buentello-Volante B, Fu Q, Simpson DA, Li Y, Sui R, Silvestri G, Daiger SP, Koenekoop RK, Zhang K, Chen R. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2014;133:331–45. doi: 10.1007/s00439-013-1381-5. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24154662&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebermann I, Walger M, Scholl HP, Charbel Issa P, Luke C, Nurnberg G, Lang-Roth R, Becker C, Nurnberg P, Bolz HJ. Truncating mutation of the DFNB59 gene causes cochlear hearing impairment and central vestibular dysfunction. Hum Mutat. 2007;28:571–7. doi: 10.1002/humu.20478. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17301963&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 26.Siemiatkowska AM, Arimadyo K, Moruz LM, Astuti GD, de Castro-Miro M, Zonneveld MN, Strom TM, de Wijs IJ, Hoefsloot LH, Faradz SM, Cremers FP, den Hollander AI, Collin RW. Molecular genetic analysis of retinitis pigmentosa in Indonesia using genome-wide homozygosity mapping. Mol Vis. 2011;17:3013–24. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22128245&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 27.Aldahmesh MA, Safieh LA, Alkuraya H, Al-Rajhi A, Shamseldin H, Hashem M, Alzahrani F, Khan AO, Alqahtani F, Rahbeeni Z, Alowain M, Khalak H, Al-Hazzaa S, Meyer BF, Alkuraya FS. Molecular characterization of retinitis pigmentosa in Saudi Arabia. Mol Vis. 2009;15:2464–9. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19956407&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 28.Ostergaard E, Duno M, Batbayli M, Vilhelmsen K, Rosenberg T. A novel MERTK deletion is a common founder mutation in the Faroe Islands and is responsible for a high proportion of retinitis pigmentosa cases. Mol Vis. 2011;17:1485–92. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21677792&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf A, Caliebe A, Thomas NS, Ball EV, Mort M, Stenson PD, Krawczak M, Cooper DN. Single base-pair substitutions at the translation initiation sites of human genes as a cause of inherited disease. Hum Mutat. 2011;32:1137–43. doi: 10.1002/humu.21547. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21681852&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 30.Johnson AE, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10611978&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 31.Mentrup B, Marschall C, Barvencik F, Amling M, Plendl H, Jakob F, Beck C. Functional characterization of a novel mutation localized in the start codon of the tissue-nonspecific alkaline phosphatase gene. Bone. 2011;48:1401–8. doi: 10.1016/j.bone.2011.03.676. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21419245&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 32.Vijayasarathy C, Sui R, Zeng Y, Yang G, Xu F, Caruso RC, Lewis RA, Ziccardi L, Sieving PA. Molecular mechanisms leading to null-protein product from retinoschisin (RS1) signal-sequence mutants in X-linked retinoschisis (XLRS) disease. Hum Mutat. 2010;31:1251–60. doi: 10.1002/humu.21350. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20809529&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]