Abstract
We report a genome-wide association study (GWAS) of cutaneous squamous cell carcinoma (SCC) conducted among non-Hispanic white (NHW) members of the Kaiser Permanente Northern California (KPNC) health care system. The study includes a genome-wide screen of 61,457 members (6,891 cases and 54,566 controls) genotyped on the Affymetrix Axiom European array and a replication phase involving an independent set of 6,410 additional members (810 cases and 5600 controls). Combined analysis of screening and replication phases identified ten loci containing single-nucleotide polymorphisms (SNPs) with P-values < 5×10-8. Six loci contain genes in the pigmentation pathway; SNPs at these loci appear to modulate SCC risk independently of the pigmentation phenotypes. Another locus contains HLA class II genes studied in relation to elevated SCC risk following immunosuppression. SNPs at the remaining three loci include an intronic SNP in FOXP1 at locus 3p13, an intergenic SNP at 3q28 near TP63, and an intergenic SNP at 9p22 near BNC2. These findings provide insights into the genetic factors accounting for inherited SCC susceptibility.
Introduction
SCC is among the most common and costly malignancies in populations of European ancestry (Housman at al., 2003). Its primary cause is ultraviolet radiation (UVR) exposure, which causes DNA damage in keratinocytes (Dessinioti et al., 2011; Roewert-Huber, 2007), for which melanin provides a protective filter An inherited basis for SCC risk is supported by the increased risk among first-degree relatives of SCC cases (Hemminki at al., 2003; Hussain et al., 2009), but the specific genetic factors that determine susceptibility are not well understood. Although GWAS have identified susceptibility loci for other skin cancers, e.g., cutaneous malignant melanoma (CMM) (Barrett et al., 2011; Bishop et al., 2009), basal cell carcinoma (BCC) (Stacey et al., 2008; Nan et al., 2011) and BCC and SCC combined as nonmelanoma skin cancer (NMSC) (Stacey et al., 2009), we are unaware of previous GWASs focused solely on SCC. While the GWAS of NMSC investigated the SNPs identified in the combined analysis for their individual effects on BCC and SCC, the power to detect SNPs specifically related to SCC was limited by the number of SCC cases. We describe an internally validated GWAS based on data from 67,867 Non-Hispanic White (NHW) individuals (7,701 SCC cases and 60,166 controls) enrolled in the Kaiser Permanente Research Program on Genes, Environment, and Health (RPGEH).
Results
We identified ten loci containing SNPs with combined P-values meeting the genomewide threshold of 5×10-8 (Pe'er et al., 2008) (Figure 1). Table 1 shows per-allele odds-ratios (ORs) and P-values for screening phase, replication phase and combined data for the most significant SNPs at these 10 loci. Six of the ten loci encompass genes that play established roles in the pigmentation pathway (Scherer and Kumar, 2010); SNPs at these loci have been associated with skin cancers and/or pigment-related phenotypes such as eye, hair or skin color, tanning, burning or freckles (Gerstenblith et al., 2010).
Figure 1.
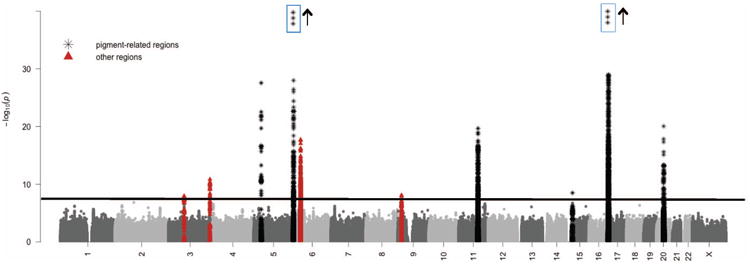
Manhattan plot showing −log10 P-values of squared Cochran-Armitage trend statistics. The horizontal line represents the threshold P-value of 5×10-8. Markers within 50 kb of SNPs with P-values < 5×10-8 are indicated with black asterisks for those in pigment-related regions and in red triangles for those in other regions. The y-axis is truncated at P = 10-30, although three SNPs at the 6p25 locus have P-values between 10-30 and 10-97, and 72 SNPs at the 16q24 locus have P-values between 10-30 and 10-44.
Table 1. Genome-wide association and replication for ten SCC loci *.
| Locus | SNP† | Gene | Minor Allele | MAF‡ | Info\\ | Initial Screen | Replication Phase | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR§ | CI§ | P-value¶ | OR | CI | P-value | OR | CI | P-value | ||||||
| A. Pigment-related Loci | ||||||||||||||
| Chr 5p13 | rs16891982 | SLC45A2 | C | 0.45 | typed | 0.53 | 0.47-0.60 | 1.64 ×10-24 | 0.48 | 0.34-0.68 | 2.88 ×10-5 | 0.52 | 0.47-0.59 | 2.77 ×10-28 |
| Chr 6p25 | rs12203592 | IRF4 | T | 0.17 | 1.00 | 1.54 | 1.48-1.61 | 2.45 ×10-83 | 1.71 | 1.49-1.95 | 5.95 ×10-15 | 1.56 | 1.49-1.62 | 8.29 ×10-97 |
| Chr 11q14 | rs1126809 | TYR | A | 0.28 | 1.00 | 1.19 | 1.16-1.25 | 1.20×10-20 | 1.08 | 0.96-1.22 | 2.09×10-1 | 1.19 | 1.15-1.24 | 2.18×10-20 |
| Chr 15q13 | rs12916300 | HERC2 | C | 0.26 | 1.00 | 0.89 | 0.85-0.93 | 1.84 ×10-7 | 0.82 | 0.72-0.93 | 2.40 ×10-3 | 0.88 | 0.85-0.92 | 3.30 ×10-9 |
| Chr 16q24 | rs4268748 | DEF8 | C | 0.26 | 0.85 | 1.34 | 1.28-1.40 | 3.24 ×10-41 | 1.28 | 1.13-1.45 | 7.85 ×10-5 | 1.33 | 1.28-1.39 | 1.75 ×10-44 |
| Chr 20q11 | rs6059655 | RALY | A | 0.08 | 0.99 | 1.30 | 1.22-1.38 | 5.18 ×10-17 | 1.49 | 1.25-1.78 | 1.14 ×10-5 | 1.32 | 1.24-1.39 | 9.03 ×10-21 |
| B. Other Loci | ||||||||||||||
| Chr 3p13 | rs62246017 | FOXP1 | A | 0.33 | 0.94 | 1.12 | 1.08-1.16 | 1.70×10-8 | 1.07 | 0.95-1.2 | 0.26×10-1 | 1.11 | 1.07-1.16 | 1.16 ×10-8 |
| Chr 3q28 | rs6791479 | TPRG1/TP63** | T | 0.43 | 1.00 | 1.12 | 1.08-1.16 | 2.57 ×10-9 | 1.21 | 1.09-1.35 | 5.03 ×10-4 | 1.13 | 1.09-1.16 | 1.47 ×10-11 |
| Chr 6p21 | rs4455710 | HLA-DQA1 | T | 0.38 | 1.00 | 1.17 | 1.12-1.21 | 1.80 ×10-16 | 1.18 | 1.06-1.32 | 2.28 ×10-3 | 1.17 | 1.13-1.21 | 1.86 ×10-18 |
| Chr 9p22 | rs74664507 | BNC2/CNTLN** | T | 0.44 | 0.95 | 0.90 | 0.87-0.93 | 3.64 ×10-8 | 0.91 | 0.81-1.01 | 8.69 ×10-2 | 0.90 | 0.87-0.93 | 8.24 ×10-9 |
regions containing SNPs with combined P-values < 5×10-8;
SNP with strongest combined P-value in the region;
MAF = minor allele frequency among control subjects;
OR = odds-ratio per minor allele, CI=95% confidence interval;
based on squared Cochran-Armitage trend test;
for imputed data, the info is the IMPUTE-2 information measure for imputation accuracy (Marchini and Howie, 2010);
two flanking genes of an intergenic SNP.
Pigmentation loci
The first three pigment-related SNPs in Table 1A have been associated previously with SCC, BCC and CMM. SNP rs16891982 at locus 5p13, a nonsynonymous SNP (Phe374Leu) in SLC45A2, has been associated with SCC (Stacey et al., 2009), BCC (Stacey et al., 2009), and CMM (Barrett et al., 2011; Stacey et al., 2009; Guedj et al., 2008; Duffy et al., 2010b; Ibarrola-Villava et al., 2012; Kosiniak-Kamysz et al., 2014; Fernandez et al., 2008) as well as eye, hair and skin color (Duffy et al., 2010b; Branicki et al., 2009; Eriksson et al., 2010; Stokowski et al., 2007; Soejima and Koda, 2007; Liu et al., 2015). SLC45A2 encodes a membrane-associated transporter enzyme, and loss of SLC45A2 activity has been found to disrupt post-Golgi-level trafficking of tyrosinase to the melanosomes (Newton et al., 2007) where melanin is synthesized and stored. Mutations in SLC45A2 cause type four oculocutaneous albinism (OCA) syndrome, which is characterized by failure to synthesize melanin (Simeonov et al. 2013). SNP rs12203592 at locus 6p25, an intronic SNP in IRF4, has been associated with SCC, BCC and CMM (Barrett et al., 2011; Han et al., 2011; Zhang et al., 2013; Stefanaki et al., 2013; Duffy et al., 2010a), and with pigmentation phenotypes (Duffy et al., 2010b; Soejima and Koda, 2007; Sulem et al., 2007; Han et al., 2008; Visser et al., 2015; Jacobs et al., 2015; Nan et al., 2009a). The T allele of this SNP reduces expression of IRF4 (Praetorius et al., 2013), which encodes a transcription factor that, in cooperation with MITF, activates expression of TYR and is used by melanocytes to produce & store melanin. SNP rs1126809 at locus 11q14 is a non-synonymous SNP (Arg402Gln) in TYR, which encodes the enzyme tyrosinase that catalyzes multiple steps in the conversion of tyrosine to melanin. SNP rs1126809 has been associated with SCC (Nan et al., 2011), BCC (Nan et al., 2011) and CMM (Bishop 2009; Nan et al., 2011; Duffy et al., 2010b; Ibarrola-Villava et al., 2012; Gudbjartsson et al., 2008; Hu et al., 2011).
The remaining three SNPs in Table 1A lie near genes that play established roles in the pigmentation pathway and that have been associated with pigmentation traits and/or skin cancer risk. However the relationships of these SNPs to SCC risk are uncertain. For example, SNP rs12916300 at 15q13 (Figure S1) lies within intron 68 of HERC2, which encodes a ubiquitin-protein ligase recruited to sites of DNA damage from ionizing radiation, and which has been associated with BCC (Han et al., 2011) and pigmentation phenotypes (Han et al., 2011; Eiberg et al., 2008). This SNP is in high linkage disequilibrium (LD) (R2 = 0.85) with rs12913832, another intronic SNP in HERC2 that has been associated with pigmentation phenotypes (Sturm et al., 2008; Han et al., 2008; Eiberg et al., 2008; Branicki et al., 2009; Nan et al., 2009b) and that modifies expression of OCA2, a nearby gene encoding a melanosomal enzyme needed for melanin synthesis (Visser et al., 2012). However SNP rs12916300 remained associated with SCC risk with genome-wide significance after adjustment for rs12913832, suggesting that its effects are not merely due to its correlation with rs12913832. Currently there is no specific evidence that rs12916300 disrupts any transcription factor binding sites, although it is located two base pairs from the binding motif of the transcription factor FOXI1 (Pique-Regi et al 2011 PMID21106904, and http://regulomedb.org/snp). In data from the GTEx Consortium (2015), rs12916300 was not associated with OCA2 expression in sun-exposed skin (P = 0.056 compared to a p-value of 0.013 for rs12913832) or in non sun-exposed skin (P = 0.44), even without adjustment for LD with rs12913832. None of the established pigment-related SNPs in HERC2/OCA2 met genome-wide significance for association in these data, despite their imputation with high accuracy (Table S1A). However nine additional SNPs (three in HERC2 and six in OCA2) achieved genome-wide significance after adjustment for all ten SNPs in Table 1. (Table S1B) Further research is needed to clarify the roles in SCC risk of these nine SNPs, whose pairwise correlations range from weak to strong (Figure S2).
SNP rs4268748 at 16q24 is intronic in DEF8, which encodes an activator of intracellular signal transduction. This SNP is associated with expression levels of CDK10 (a gene critical for cell cycle progression) in sun-exposed skin (P = 2.3×10-11) and in non-exposed (supra-pubic) skin (P = 1.4×10-8) (GTEx Consortium 2015). The role of rs4268748 in SCC is complicated by its proximity (39 kb) to MC1R, a gene that encodes a receptor for melanocyte stimulating hormone and that contains several nonsynonymous SNPs associated with skin pigmentation (Liu et al., 2015; Valverde et al., 1995) and skin cancer (Barrett et al., 2011; Bishop 2009; Duffy et al., 2010b; Ibarrola-Villava et al., 2012; Sulem et al., 2007; Palmer et al., 2000; Box et al., 2001) (Table S2). In particular, SNPs in MC1R meeting genome-wide significance in this study include rs258322, which has been associated with CMM (Barrett et al., 2011; Bishop 2009; Stefanaki et al., 2013) and pigment-related traits (Valverde et al., 1995), and the nonsynonymous SNPs rs1805007 (Arg151Cys) and rs1805008 (Arg160Trp), which have been associated with CMM (Duffy et al., 2010b; Ibarrola-Villava et al., 2012; Sulem et al., 2007; Palmer et al., 2000), NMSC (Box et al., 2001) and, among renal transplant patients, SCC (Andresen et al., 2013). However, none of these three SNPs maintained genome-wide significance after adjusting for genotypes at rs4268748, which has been associated with MC1R expression with borderline statistical significance (P = 0.012) in sun-exposed skin (GTEx consortium 2015). Furthermore, two additional SNPs at this locus (rs35063026 in an untranslated region of SPATA33, and rs78703231 in an intron in SPIRE2) were independently associated with SCC risk after adjustment for rs4268748 (Figure 2 and Supplementary Materials). These three SNPs (rs4268748, rs35063026 and rs78703231) lie at most 249 kb downstream of MC1R, and their SCC associations could reflect correlation with one or more of the nonsynonymous MC1R SNPs previously associated with pigmentation and/or skin cancer risk. Indeed, after adjustment for the subset of these SNPs with MAF greater than 1% in the present study (Supplementary Materials and Table S3), rs78703231 lost statistical significance, with pre- and post-adjustment risk ratios of 1.21, P = 1.77×10-17 and 1.05, P = 8.21×10-2. However strong associations persisted for both rs4268748 (post-adjustment risk ratio = 1.22, P = 1.96×10-16) and rs35063026 (post-adjustment risk ratio = 1.33, P = 5.38×10-11). These findings suggest the existence of multiple independent SCC susceptibility loci in the 16q24 region whose precise roles in SCC pathogenesis remain to be elucidated.
Figure 2.
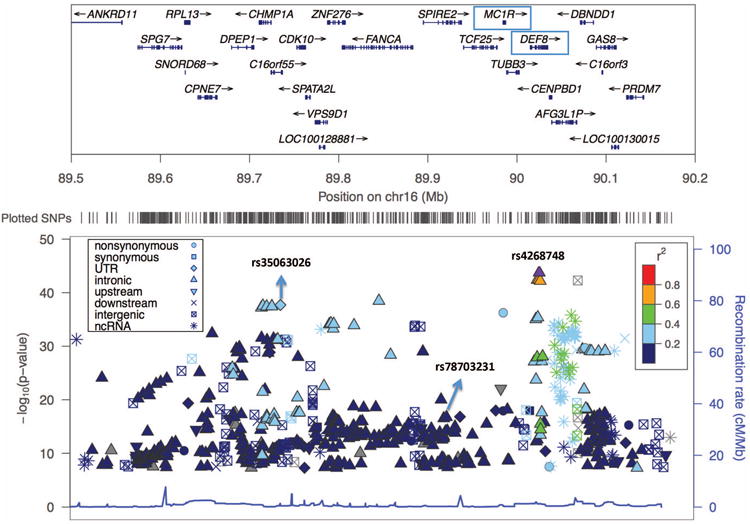
Manhattan plot enlargement showing 0.7 Mb region at the 16q24 locus.
The upper panel shows the name and location of genes in the region, with an arrow indicating the transcribed strand of a gene and ticks indicating exons. The genes DEF8 (containing the top SNP rs4268748) and MC1R (associated with skin pigmentation and skin cancer) are shown in boxes. The lower panel shows the significance levels of the SNPs in this region, with labels for the three SNPs that are independently associated with SCC risk.
Finally, SNP rs6059655 at 20q11, which is intronic in RALY, lies 182 kb upstream of ASIP, a gene that encompasses SNPs associated with pigmentation traits (Liu et al., 2015; Jacobs et al., 2015), CMM (Barrett et al., 2011; Duffy et al., 2010b; Brown et al., 2008; Amos et al., 2011; Maccioni et al., 2013) and NMSC (Nan et al, 2010; Binstock et al., 2014; Nan et al., 2009a, Lin et al 2011) (Figure S3). Only three of these SNPs (rs4911442, rs910873 & rs1885120) met the genome-wide significance threshold for association with SCC in the present data, and none maintained genome-wide significance after adjustment for rs6059655. A haplotype in ASIP containing the T allele of rs4911414 and the G allele of rs1015362 has been associated with pigment-related traits and with CMM and BCC (Sulem et al., 2007; Gudbjartsson et al., 2008). However adjustment for this haplotype did not eliminate the observed association between SCC and rs6059655 (OR: 1.40, P = 1.31×10-10 for rs6059655 and 1.30, P = 2.0×10-16 for the haplotype). This finding, coupled with the observed association between rs6059655 and ASIP expression levels in sun-exposed skin (P = 5.3×10-9) and non sun-exposed skin (P = 4.4×10-4) (GTEx Consortium, 2015), lends support to the possibility that rs6059655 (or a nearby functional SNP) affects ASIP activity independently of the ASIP haplotype.
In summary, the observed associations between SCC risk and SNPs at 15q13, 16q24 and 20q11 could not be explained by their proximity to previously studied SNPs in the pigmentation and skin cancer genes HERC2/OCA2, MC1R and ASIP, respectively. Further work on the functions of these SNPs is needed to elucidate their potential causal roles and mechanisms in SCC.
We checked whether subjects' total counts of risk alleles of the six pigment-related SNPs in Table 1A (called their risk indices) influence their SCC risk independently of their pigment-related phenotypes (skin color, tanning and sun sensitivity). Since we lacked self-reported or clinical data on these pigment-related phenotypes, we estimated them using subjects' total allele counts for SNPs associated with fair skin, poor tanning ability and tendency to sunburn (see Supplementary Materials and Table S4 for details on how we assembled these pigmentation scores). Table S5 shows the joint distribution of subjects by tertiles of risk index and of skin phenotype score, with tertiles determined by the distributions in controls. The table also shows joint and marginal odds-ratios relating SCC risk to tertiles of risk index and pigmentation score. Because some SNPs were used to calculate both the risk indices and the pigmentation scores, the odds-ratios may be attenuated toward unity. Nevertheless, consistently with the findings of others (Andresen et al., 2013, Bastiaens et al., 2001; Kennedy et al., 2001; Gerstenblith et al., 2010, Nan et al., 2009a, Palmer et al., 2000), we observed increased SCC risk with increasing risk index, independently of the estimated pigmentation phenotypes. Specifically, Table S5 shows strong trends of increased SCC risk with increasing risk index within each tertile of the skin pigmentation scores, as well as overall, after adjustment for pigmentation score. Moreover, the data suggest trends of increasing risk index odds-ratios with increasingly sun-sensitive pigmentation phenotypes, with significance levels of P = 0.85, P = 1.15×10-7 and P = 0.002 for skin color, sun sensitivity and tanning ability, respectively). We also found similar trends of higher SCC risk with increasing pigmentation score within each tertile of risk index, and overall, after adjustment for risk index, suggesting independent contributions of genetic risk index and pigmentation phenotypes in determining SCC susceptibility.
Other loci
Four additional SCC loci met the genome-wide significance threshold (Table 1B), that to our knowledge, have limited or no relation to the pigmentation pathway. Among the four, the HLA locus at 6p21 was most strongly associated with SCC risk, with a per-allele risk ratio of 1.17, P =1.86×10-8) for the most significant SNP, rs4455710 (Figure 1). This SNP is intronic in the class II HLA gene HLADQA1 (Figure S4), which binds peptides derived from antigens that access the endocytic route of antigen-presenting cells and presents them on the cell surface for recognition by CD4 T-cells. No other SNP in the region maintained genome-wide significance after adjusting for rs4455710. This SNP is also strongly associated with expression levels of nearby genes, including HLA-DQA1 (P = 3.3×10-7 and P = 4.2×10-6 in sun-exposed and non-exposed skin respectively), HLA-DQB1 (P = 1.1×10-6 in sun-exposed skin), and HLA-DRB5 (P = 4.3×10-12 and P = 2.6×10-9 in sun-exposed and non-exposed skin respectively) (GTEx Consortium, 2015). A role for HLA antigens and immune response in SCC is supported by findings of elevated SCC risk in immunocompromised individuals (Bouwes Bavinck et al., 1991), by data relating NMSCs to specific HLA alleles (Bouwes Bavinck and Claas, 1994), and by elevated SCC risk in cigarette smokers (Leonardi-Bee et al, 2012), since smoking is associated with changes in circulating markers for immune response (Shiels et al., 2014). However we did not find strong evidence that the risk ratio per-allele of rs4455710 varies by history of smoking or immunosuppression (Table S6).
The remaining three SNPs in Table 1B open avenues for further study. SNP rs62246017 at 3p13 is intronic in FOXP1, which encodes a transcription repressor with important roles in immune response, organ development and the pathogenesis of epithelial malignancies. Alterations in FOXP1 expression have been implicated in the pathogenesis of squamous cell cancers of the lung and of the head and neck (Yang et al., 2012; Feng et al., 2012).
The intergenic SNP rs6791479 at 3q28 lies 144 kb upstream of TP63, which encodes a member of the p53 family of transcription factors (Poligone et al., 2015). One of its two main isoforms (ΔNp63) is involved in multiple functions during skin development and in adult stem/progenitor cell regulation (Crum and McKeon, 2010; Leonard et al., 2011). TP63 immuno-staining has utility for differentiating subtypes of head and neck SCCs. In other tissues, TP63 is helpful in distinguishing poorly differentiated SCC (strong staining) from small cell carcinoma or adenocarcinoma (no staining). SNP rs6791479 is associated with expression of TP63 in non sun-exposed skin (P = 0.0036) (GTEx Consortium, 2015).
Finally, the inter-genic SNP rs74664507 at 9p22 lies 43 kb upstream of BNC2, which encodes a zinc-finger protein thought to be involved in mRNA processing (Vanhoutteghem and Djian, 2006) and to function as a transcription factor (Romano et al., 2004). The role of BNC2 in human pigmentation is unclear, despite experimental evidence suggesting that the gene influences pigmentation in mice (Smyth et al., 2006) and zebrafish (Patterson and Parichy, 2013), and despite its elevated expression levels in human epidermal skin samples and melanocytic cell lines from dark-skinned donors (Visser et al., 2014). Functional data indicate that BNC2 expression is influenced by the inter-genic SNP rs12350739 located in a BNC2 enhancer (Visser et al., 2014). This finding, coupled with the high LD at this locus, has suggested that rs12350739 is responsible for the observed associations of intronic SNP rs10756819 with skin color (Jacobs et al., 2013) and of intronic SNP rs215327 with freckling (Eriksson et al., 2010). Interestingly, SNPs rs74664507 and rs12350739 are also in high LD (R2= 0.84). Thus not surprisingly, univariate regression of SCC risk against genotypes of rs12350739 showed borderline genome-wide significance (P = 7.62×10-08), while regression of SCC risk against both SNPs showed attenuated association (P = 0.04) for rs74664507 and no association (P = 0.92) for rs12350739. Additional replication and functional work are needed to elucidate the role of BNC2 in SCC risk.
Allelic effects of SNPs in Table 1
For each of the SNPs at the 10 susceptibility loci in Table 1, we used a likelihood ratio statistic (LRS) to evaluate how well allelic effects on SCC risk were captured by a one-degree-of-freedom (DF) log-additive logistic model relative to a co-dominant (two DF) model. The LRS has a chi-square distribution with one DF under the null hypothesis that the log-additive allelic model fits well. We found significant improvement in fit for the co-dominant model relative to the log-additive model only for SNP rs1126809 in TYR at 11q24 (P = 3.6×10-3). SCC odds-ratios for heterozygote and homozygote carriers of the A-allele of this SNP were 1.13 (95% confidence interval: 1.07-1.19) and 1.51 (1.39-1.64), respectively.
Genetic and nongenetic interactions
We found no evidence that odds-ratios for the SNPs in Table 1 varied by gender, but some varied by age at RPGEH enrollment, as classified into three categories based on the control tertiles and coded ordinally. Specifically, we found significant interaction P-values for SNPs rs12203592 in IRF4 (P = 3.52×10-6), rs1126809 in TYR (P = 0.04), and SNP rs4268748 in DEF8 (P = 0.01). In all three analyses the odds-ratio estimates decreased with increasing age. We also evaluated potential epistatic effects on SCC risk among the 45 pairs of the 10 SNPs in Table 1. We observed evidence for superadditivity (on the log odds scale) of effects at pairs of loci 20q11 and 11q14 (P = 0.017) and for sub-additivity for five other pairs: 5p13 and 6p25 (P = 0.013), 6p21 and 15q13 (P = 0.023), 20q11 and 5p13 (P = 0.032), and 20q11 and 16q24 (P = 0.041) (Table S7). In addition, we evaluated the effects of SNP rs4455710 at 6p21 by history of smoking and immunosuppression. Specifically, because immunosuppressed recipients of major organ transplants have elevated SCC risk (Andresen 2013), we examined whether the log odds of SCC varied between subjects with and without a history of immunosuppression, as indicated by a history of organ transplantation, chronic lymphocytic (CLL) or HIV infection. Similarly, because cigarette smokers have altered immune status compared to nonsmokers (Shiels et al 2014), we compared SNP odds-ratios in ever- vs. never-smokers. Table S6 shows no evidence of effect-modification at this SNP by either smoking status or history of immunosuppression.
Discussion
This large SCC GWAS has confirmed existing associations between SCC risk and three SNPs in genes known to be related to the pigmentation pathway. These include nonsynonymous SNPs in SLC45A2 on chromosome 5p13 and in TYR on chromosome 11q14, and a functional intronic SNP in IRF4 on chromosome 6p25. It also has identified previously unreported SCC-associated SNPs at three other loci (15q13, 16q24 and 20q11) containing pigmentation genes associated with skin cancer risk; and we were unable to attribute these associations to correlation with putative causal variants in these genes. Finally, we have identified four additional loci previously unrelated to SCC risk.
Strengths of this study include its large sample size and its use of a demographically diverse NHW cohort characterized by comprehensive clinical data. The findings need replication in a NHW population having demographic characteristics and ancestries that differ from those of the Northern California population studied here. If the associations are replicated, functional research will be needed to better understand the roles of the associated SNPs in the pigment-related regions 15q13, 16q24 and 20q11, and in the four nonpigment-related regions. In summary, the present findings can facilitate the development of SCC prediction models that identify subgroups in need of more intensive screening. They also can motivate future investigations of genetic factors that modify UVR exposures and other lifestyle characteristics in conferring SCC risk.
Materials and Methods
Study population
Potentially eligible study subjects were RPGEH members who at cohort entry reported NHW race/ethnicity, were at least 18 years of age, and had no diagnostic codes for rare genetic disorders associated with increased SCC risk (e.g., xeroderma pigmentosum, oculocutaneous albinism, dystrophic epidermolysis bullosa, epidermodysplasia verruciformis, Fanconi anemia, dyskeratoses congenital, Rothmund-Thomson syndrome, Bloom syndrome, Werner Syndrome, and Ferguson Smith syndrome).
Potentially eligible cases were subjects whose pathology records were consistent with incident SCC (invasive or in-situ, excluding ano-genital and mucosal SCCs) during the period from survey completion until either December 31, 2012 or departure from the KPNC healthcare system, whichever occurred first. (See Supplementary Materials for details on SCC case verification by pathology review). Potentially eligible controls (n = 64,218) were subjects with no RPGEH-survey-reported history of skin cancer and no pathology records consistent with skin cancer in the KPNC database. After QC was applied to all potentially eligible subjects (as described in Supplementary Materials and Table S8), a total of 67,867 subjects were eligible for analysis.
Genotyping
Saliva samples from eligible subjects were genotyped on four custom Affymetrix Axiom arrays optimized for individuals of European (EUR), East Asian (EAS), Latino (LAT) and African-American (AFR) race/ethnicity (Hoffmann et al., 2011a, 2011b for the other arrays [http://www.ncbi.nlm.nih.gov/pubmed/21903159]). Genotyping and SNP quality control (QC) have been described previously (dbGaP phs000674.v1.p1) (Kvale et al., 2015; Banda et al., 2015; Hoffmann et al., 2011a and 2011b). SNP imputation to the 1000 Genomes Project is described in the Supplementary Materials.
Internal validation
For the screening phase we used the genotypes of the 61,457 subjects (6891 cases and 54,566 controls) who had been typed on the EUR array, and for the replication phase we used the remaining 6,410 NHW subjects (810 cases and 5600 controls) who had been typed erroneously on a nonEUR array. Table S9 shows classification of all subjects by case-control status, gender and study phase (screening or replication). Subjects' genotypes for untyped SNPS were imputed to the 1000 Genomes Project.
eQTL evaluation
We also examined the public GTEx database for evidence that SNPs of interest regulate the expression of nearby genes in skin tissue. (Supplementary Materials).
Statistical Analysis
To adjust for population stratification, we determined ancestry principal components (PCs) for the combined data from screening and replication phases using the smartpca program in the EIGENSOFT4.2 software package (Hemminki and Czene, 2003), as has been described elsewhere (dbGaP phs000674.v1.p1). For each of the screening and replication phases, we then evaluated SCC-association with each SNP's allelic count using the Armitage-Cochran trend statistic, adjusted for gender, the first 10 principal components of ancestry, and for the genotyping array and reagent kit. We combined regression coefficients and P-values obtained from screening and replication phases using Cochran's method (Cochran 1954; Zhou et al., 2011), and identified as significant those SNPs with combined P-values < 5×10-8 (Pe'er et al., 2008). We checked for residual population stratification by examining the inflation factor λGC (Devlin and Roeder 1999), and Q-Q plots of percentiles of the observed distribution of test statistics versus those expected under the global null hypothesis (see Supplementary Materials). Further adjustment for age and immunosuppressive status resulted in negligible changes in the odds-ratios for the top 10 SNPs in Table 1.
Data accessibility
Genotypes used in this GWAS have been registered with dbGAP; (Study Accession: phs000674.v1.p1).
All study procedures were approved by the Institutional Review Board of the Kaiser Foundation Research Institute.
Supplementary Material
Acknowledgments
This research was supported by NIH grant R01CA166672, by the GO grant RC2 AG036607, and by the Wayne and Gladys Valley Foundation, the Ellison Medical Foundation, the Robert Wood Johnson Foundation, and Kaiser Permanente Community Benefit Program. We thank Neil Risch, University of California at San Francisco, and Cathy Schaefer, KPNC Division of Research, for establishing the RPGEH cohort and processing and providing the genetic data. We also thank Julia L Kay and Dilrini K. Ranatunga, KPNC Division of Research, for their help in documenting and transferring the genetic and nongenetic data and providing the background information needed for the analysis.
Footnotes
Conflict of Interest: The authors state no conflicts of interest.
Supplementary Materials: The Supplementary Materials section is linked to the online version of the paper at http://www.nature.com/jid
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amos CI, Wang LE, Lee JE, Gershenwald JE, Chen WV, Fang S, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet. 2011;20(24):5012–23. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen PA, Nymoen DA, Kjaerheim K, Leivestad T, Helsing P. Biomark Cancer. Vol. 5. New Zealand: 2013. Susceptibility to Cutaneous Squamous Cell Carcinoma in Renal Transplant Recipients Associates with Genes Regulating Melanogenesis Independent of their Role in Pigmentation; pp. 41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banda Y, Kvale MN, Hoffmann TJ, et al. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1285–95. doi: 10.1534/genetics.115.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011;43(11):1108–13. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaens MT, ter Huurne JA, Kielich C, Gruis NA, Westendorp RG, Vermeer BJ, et al. Melanocortin-1 receptor gene variants determine the risk of nonmelanoma skin cancer independently of fair skin and red hair. Am J Hum Genet. 2001;68(4):884–94. doi: 10.1086/319500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binstock M, Hafeez F, Metchnikoff C, Arron ST. Single-nucleotide polymorphisms in pigment genes and nonmelanoma skin cancer predisposition: a systematic review. Br J Dermatol. 2014;171(4):713–21. doi: 10.1111/bjd.13283. [DOI] [PubMed] [Google Scholar]
- Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, et al. Nat Genet. Vol. 41. United States: 2009. Genome-wide association study identifies three loci associated with melanoma risk; pp. 920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwes Bavinck JN, Claas FH. The role of HLA molecules in the development of skin cancer. Hum Immunol. 1994;41(3):173–9. doi: 10.1016/0198-8859(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Bouwes Bavinck JN, Vermeer BJ, van der Woude FJ, Vandenbroucke JP, Schreuder GM, Thorogood J, et al. Relation between skin cancer and HLA antigens in renal-transplant recipients. N Engl J Med. 1991;325(12):843–8. doi: 10.1056/NEJM199109193251203. [DOI] [PubMed] [Google Scholar]
- Box NF, Duffy DL, Irving RE, Russell A, Chen W, Griffyths LR, et al. Melanocortin-1 receptor genotype is a risk factor for basal and squamous cell carcinoma. J Invest Dermatol. 2001;116(2):224–9. doi: 10.1046/j.1523-1747.2001.01224.x. [DOI] [PubMed] [Google Scholar]
- Branicki W, Brudnik U, Wojas-Pelc A. Ann Hum Genet. Vol. 73. England: 2009. Interactions between HERC2, OCA2 and MC1R may influence human pigmentation phenotype; pp. 160–70. [DOI] [PubMed] [Google Scholar]
- Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40(7):838–40. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–29. [Google Scholar]
- Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol. 2010;5:349–71. doi: 10.1146/annurev-pathol-121808-102117. [DOI] [PubMed] [Google Scholar]
- Dessinioti C, Tzannis K, Sypsa V, Nikolaou V, Kypreou K, Antoniou C, et al. Epidemiologic risk factors of basal cell carcinoma development and age at onset in a Southern European population from Greece. Exp Dermatol. 2011;20(8):622–6. doi: 10.1111/j.1600-0625.2011.01275.x. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Duffy DL, Iles MM, Glass D, Zhu G, Barrett JH, Hoiom V, et al. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am J Hum Genet. 2010;87(1):6–16. doi: 10.1016/j.ajhg.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DL, Zhao ZZ, Sturm RA, Hayward NK, Martin NG, Montgomery GW. Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J Invest Dermatol. 2010;130(2):520–8. doi: 10.1038/jid.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiberg H, Troelsen J, Nielsen M, Mikkelsen A, Mengel-From J, Kjaer KW, et al. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet. 2008;123(2):177–87. doi: 10.1007/s00439-007-0460-x. [DOI] [PubMed] [Google Scholar]
- Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, Saxonov S, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6(6):e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhang X, Zhu H, Wang X, Ni S, Huang J. High expression of FoxP1 is associated with improved survival in patients with non-small cell lung cancer. Am J Clin Pathol. 2012;138(2):230–5. doi: 10.1309/AJCPDHQFNYJZ01YG. [DOI] [PubMed] [Google Scholar]
- Fernandez LP, Milne RL, Pita G, Aviles JA, Lazaro P, Benitez J, et al. SLC45A2: a novel malignant melanoma-associated gene. Hum Mutat. 2008;29(9):1161–7. doi: 10.1002/humu.20804. [DOI] [PubMed] [Google Scholar]
- Gerstenblith MR, Shi J, Landi MT. Genome-wide association studies of pigmentation and skin cancer: a review and meta-analysis. Pigment Cell Melanoma Res. 2010;23(5):587–606. doi: 10.1111/j.1755-148X.2010.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Sulem P, Stacey SN, Goldstein AM, Rafnar T, Sigurgeirsson B, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40(7):886–91. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- Guedj M, Bourillon A, Combadieres C, Rodero M, Dieude P, Descamps V, et al. Variants of the MATP/SLC45A2 gene are protective for melanoma in the French population. Hum Mutat. 2008;29(9):1154–60. doi: 10.1002/humu.20823. [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4(5):e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Qureshi AA, Nan H, Zhang J, Song Y, Guo Q, et al. Cancer Res. Vol. 71. United States: 2011. A germline variant in the interferon regulatory factor 4 gene as a novel skin cancer risk locus; pp. 1533–9. 2011 Aacr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki K, Zhang H, Czene K. Familial invasive and in situ squamous cell carcinoma of the skin. Br J Cancer. 2003;88(9):1375–80. doi: 10.1038/sj.bjc.6600909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, Kvale MN, Hesselson SE, Zhan Y, Aquino C, Cao Y, et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98(2):79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, Zhan Y, Kvale MN, Hesselson SE, Gollub J, Iribarren C, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98(6):422–30. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housman TS, Feldman SR, Williford PM, Fleischer AB, Jr, Goldman ND, Acostamadiedo JM, et al. Skin cancer is among the most costly of all cancers to treat for the Medicare population. J Am Acad Dermatol. 2003;48(3):425–9. doi: 10.1067/mjd.2003.186. [DOI] [PubMed] [Google Scholar]
- Hu HH, Guedj M, Descamps V, Jouary T, Bourillon A, Ezzedine K, et al. Assessment of tyrosinase variants and skin cancer risk in a large cohort of French subjects. J Dermatol Sci. 2011;64(2):127–33. doi: 10.1016/j.jdermsci.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Hussain SK, Sundquist J, Hemminki K. The effect of having an affected parent or sibling on invasive and in situ skin cancer risk in Sweden. J Invest Dermatol. 2009;129(9):2142–7. doi: 10.1038/jid.2009.31. [DOI] [PubMed] [Google Scholar]
- Ibarrola-Villava M, Hu HH, Guedj M, Fernandez LP, Descamps V, Basset-Seguin N, et al. MC1R, SLC45A2 and TYR genetic variants involved in melanoma susceptibility in southern European populations: results from a meta-analysis. Eur J Cancer. 2012;48(14):2183–91. doi: 10.1016/j.ejca.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Jacobs LC, Hamer MA, Gunn DA, Deelen J, Lall JS, van Heemst D, et al. A Genome-Wide Association Study Identifies the Skin Color Genes IRF4, MC1R, ASIP, and BNC2 Influencing Facial Pigmented Spots. J Invest Dermatol. 2015;135(7):1735–42. doi: 10.1038/jid.2015.62. [DOI] [PubMed] [Google Scholar]
- Jacobs LC, Wollstein A, Lao O, Hofman A, Klaver CC, Uitterlinden AG, et al. Comprehensive candidate gene study highlights UGT1A and BNC2 as new genes determining continuous skin color variation in Europeans. Hum Genet. 2013;132(2):147–58. doi: 10.1007/s00439-012-1232-9. [DOI] [PubMed] [Google Scholar]
- Kennedy C, ter Huurne J, Berkhout M, Gruis N, Bastiaens M, Bergman W, et al. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol. 2001;117(2):294–300. doi: 10.1046/j.0022-202x.2001.01421.x. [DOI] [PubMed] [Google Scholar]
- Kosiniak-Kamysz A, Marczakiewicz-Lustig A, Marcinska M, Skowron M, Wojas-Pelc A, Pospiech E, et al. Increased risk of developing cutaneous malignant melanoma is associated with variation in pigmentation genes and VDR, and may involve epistatic effects. Melanoma Res. 2014;24(4):388–96. doi: 10.1097/CMR.0000000000000095. [DOI] [PubMed] [Google Scholar]
- Kvale MN, Hesselson S, Hoffmann TJ, Cao Y, Chan D, Connell S, et al. Genotyping informatics and quality control for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1051–60. doi: 10.1534/genetics.115.178905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard MK, Kommagani R, Payal V, Mayo LD, Shamma HN, Kadakia MP. DeltaNp63alpha regulates keratinocyte proliferation by controlling PTEN expression and localization. Cell Death Differ. 2011;18(12):1924–33. doi: 10.1038/cdd.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi-Bee J, Ellison T, Bath-Hextall F. Smoking and the risk of nonmelanoma skin cancer: systematic review and meta-analysis. Arch Dermatol. 2012;148(8):939–46. doi: 10.1001/archdermatol.2012.1374. [DOI] [PubMed] [Google Scholar]
- Lin W, Qureshi AA, Kraft P, Nan H, Guo Q, Hu FB, et al. ASIP genetic variants and the number of non-melanoma skin cancers. Cancer Causes Control. 2011;22(3):495–501. doi: 10.1007/s10552-010-9724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Visser M, Duffy DL, Hysi PG, Jacobs LC, Lao O, et al. Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Hum Genet. 2015;134(8):823–35. doi: 10.1007/s00439-015-1559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccioni L, Rachakonda PS, Scherer D, Bermejo JL, Planelles D, Requena C, et al. Variants at chromosome 20 (ASIP locus) and melanoma risk. Int J Cancer. 2013;132(1):42–54. doi: 10.1002/ijc.27648. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Nan H, Kraft P, Hunter DJ, Han J. Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. Int J Cancer. 2009;125(4):909–17. doi: 10.1002/ijc.24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H, Kraft P, Qureshi AA, Guo Q, Chen C, Hankinson SE, et al. Genome-wide association study of tanning phenotype in a population of European ancestry. J Invest Dermatol. 2009;129(9):2250–7. doi: 10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H, Qureshi AA, Han J. Br J Dermatol. Vol. 162. England: 2010. Melanoma susceptibility variants on chromosome 20q11.22 are associated with pigmentary traits and the risk of nonmelanoma skin cancer; pp. 461–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H, Xu M, Kraft P, Qureshi AA, Chen C, Guo Q, et al. Genome-wide association study identifies novel alleles associated with risk of cutaneous basal cell carcinoma and squamous cell carcinoma. Hum Mol Genet. 2011;20(18):3718–24. doi: 10.1093/hmg/ddr287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RA, Cook AL, Roberts DW, Leonard JH, Sturm RA. Post-transcriptional regulation of melanin biosynthetic enzymes by cAMP and resveratrol in human melanocytes. J Invest Dermatol. 2007;127(9):2216–27. doi: 10.1038/sj.jid.5700840. [DOI] [PubMed] [Google Scholar]
- Palmer JS, Duffy DL, Box NF, Aitken JF, O'Gorman LE, Green AC, et al. Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet. 2000;66(1):176–86. doi: 10.1086/302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LB, Parichy DM. Interactions with iridophores and the tissue environment required for patterning melanophores and xanthophores during zebrafish adult pigment stripe formation. PLoS Genet. 2013;9(5):e1003561. doi: 10.1371/journal.pgen.1003561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, et al. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–55. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poligone B, Gilmore ES, Alexander CV, Oleksyn D, Gillespie K, Zhao J, et al. PKK suppresses tumor growth and is decreased in squamous cell carcinoma of the skin. J Invest Dermatol. 2015;135(3):869–76. doi: 10.1038/jid.2014.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, Robinson KC, et al. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell. 2013;155(5):1022–33. doi: 10.1016/j.cell.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roewert-Huber J, Lange-Asschenfeldt B, Stockfleth E, Kerl H. Epidemiology and aetiology of basal cell carcinoma. Br J Dermatol. 2007;157(Suppl 2):47–51. doi: 10.1111/j.1365-2133.2007.08273.x. [DOI] [PubMed] [Google Scholar]
- Romano RA, Li H, Tummala R, Maul R, Sinha S. Identification of Basonuclin2, a DNA-binding zinc-finger protein expressed in germ tissues and skin keratinocytes. Genomics. 2004;83(5):821–33. doi: 10.1016/j.ygeno.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Scherer D, Kumar R. Genetics of pigmentation in skin cancer--a review. Mutat Res. 2010;705(2):141–53. doi: 10.1016/j.mrrev.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Shiels MS, Katki HA, Freedman ND, Purdue MP, Wentzensen N, Trabert B, et al. Cigarette smoking and variations in systemic immune and inflammation markers. J Natl Cancer Inst. 2014;106:dju294. doi: 10.1093/jnci/dju294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonov DR, Wang X, Wang C, Sergeev Y, Dolinska M, Bower M, et al. DNA variations in oculocutaneous albinism: an updated mutation list and current outstanding issues in molecular diagnostics. Hum Mutat. 2013;34(6):827–35. doi: 10.1002/humu.22315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth IM, Wilming L, Lee AW, Taylor MS, Gautier P, Barlow K, et al. Genomic anatomy of the Tyrp1 (brown) deletion complex. Proc Natl Acad Sci U S A. 2006;103(10):3704–9. doi: 10.1073/pnas.0600199103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soejima M, Koda Y. Population differences of two coding SNPs in pigmentation-related genes SLC24A5 and SLC45A2. Int J Legal Med. 2007;121(1):36–9. doi: 10.1007/s00414-006-0112-z. [DOI] [PubMed] [Google Scholar]
- Stacey SN, Gudbjartsson DF, Sulem P, Bergthorsson JT, Kumar R, Thorleifsson G, et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet. 2008;40(11):1313–8. doi: 10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41(8):909–14. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanaki I, Panagiotou OA, Kodela E, Gogas H, Kypreou KP, Chatzinasiou F, et al. Replication and predictive value of SNPs associated with melanoma and pigmentation traits in a Southern European case-control study. PLoS One. 2013;8(2):e55712. doi: 10.1371/journal.pone.0055712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokowski RP, Pant PV, Dadd T, Fereday A, Hinds DA, Jarman C, et al. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 2007;81(6):1119–32. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL, Zhao ZZ, Leite FP, Stark MS, Hayward NK, et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet. 2008;82(2):424–31. doi: 10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39(12):1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11(3):328–30. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- Vanhoutteghem A, Djian P. Basonuclins 1 and 2, whose genes share a common origin, are proteins with widely different properties and functions. Proc Natl Acad Sci U S A. 2006;103(33):12423–8. doi: 10.1073/pnas.0605086103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22(3):446–55. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M, Palstra RJ, Kayser M. Human skin color is influenced by an intergenic DNA polymorphism regulating transcription of the nearby BNC2 pigmentation gene. Hum Mol Genet. 2014;23(21):5750–62. doi: 10.1093/hmg/ddu289. [DOI] [PubMed] [Google Scholar]
- Visser M, Palstra RJ, Kayser M. Allele-specific transcriptional regulation of IRF4 in melanocytes is mediated by chromatin looping of the intronic rs12203592 enhancer to the IRF4 promoter. Hum Mol Genet. 2015;24(9):2649–61. doi: 10.1093/hmg/ddv029. [DOI] [PubMed] [Google Scholar]
- Yang MH, Lin BR, Chang CH, Chen ST, Lin SK, Kuo MY, et al. Connective tissue growth factor modulates oral squamous cell carcinoma invasion by activating a miR-504/FOXP1 signalling. Oncogene. 2012;31(19):2401–11. doi: 10.1038/onc.2011.423. [DOI] [PubMed] [Google Scholar]
- Zhang M, Song F, Liang L, Nan H, Zhang J, Liu H, et al. Genome-wide association studies identify several new loci associated with pigmentation traits and skin cancer risk in European Americans. Hum Mol Genet. 2013;22(14):2948–59. doi: 10.1093/hmg/ddt142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Shi J, Whittemore AS. Optimal methods for meta-analysis of genome-wide association studies. Genet Epidemiol. 2011;35(7):581–91. doi: 10.1002/gepi.20603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotypes used in this GWAS have been registered with dbGAP; (Study Accession: phs000674.v1.p1).
All study procedures were approved by the Institutional Review Board of the Kaiser Foundation Research Institute.


