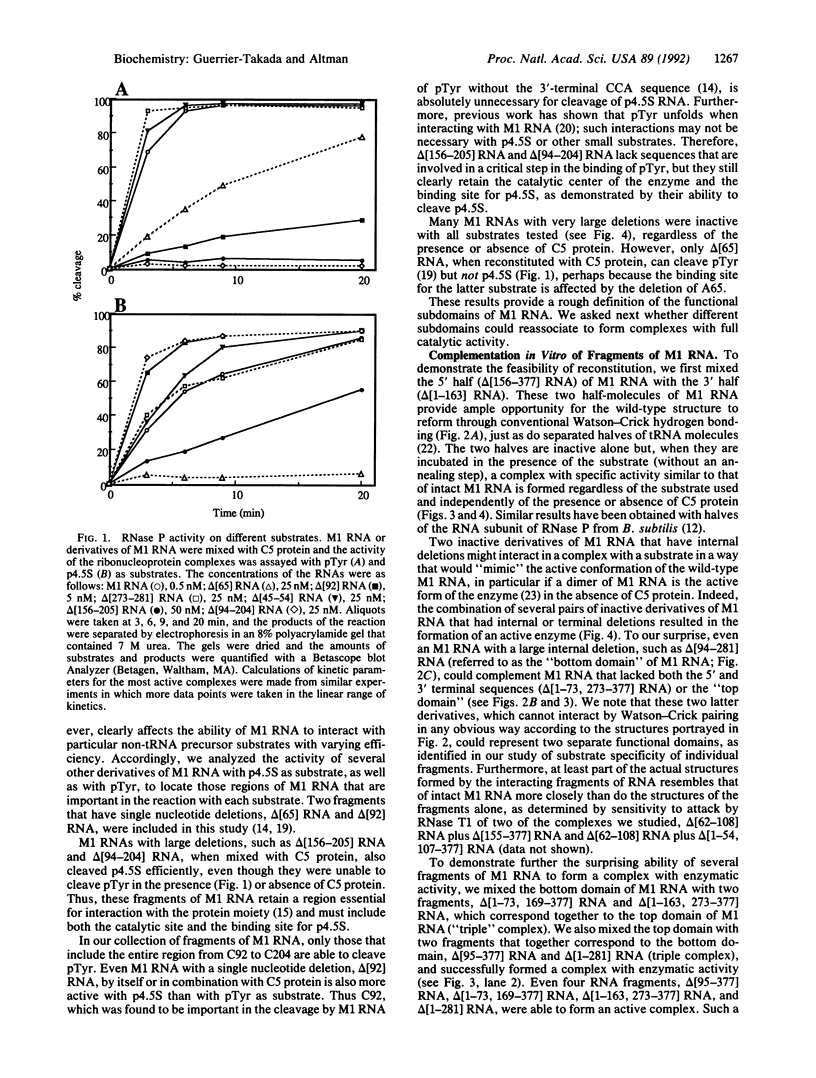
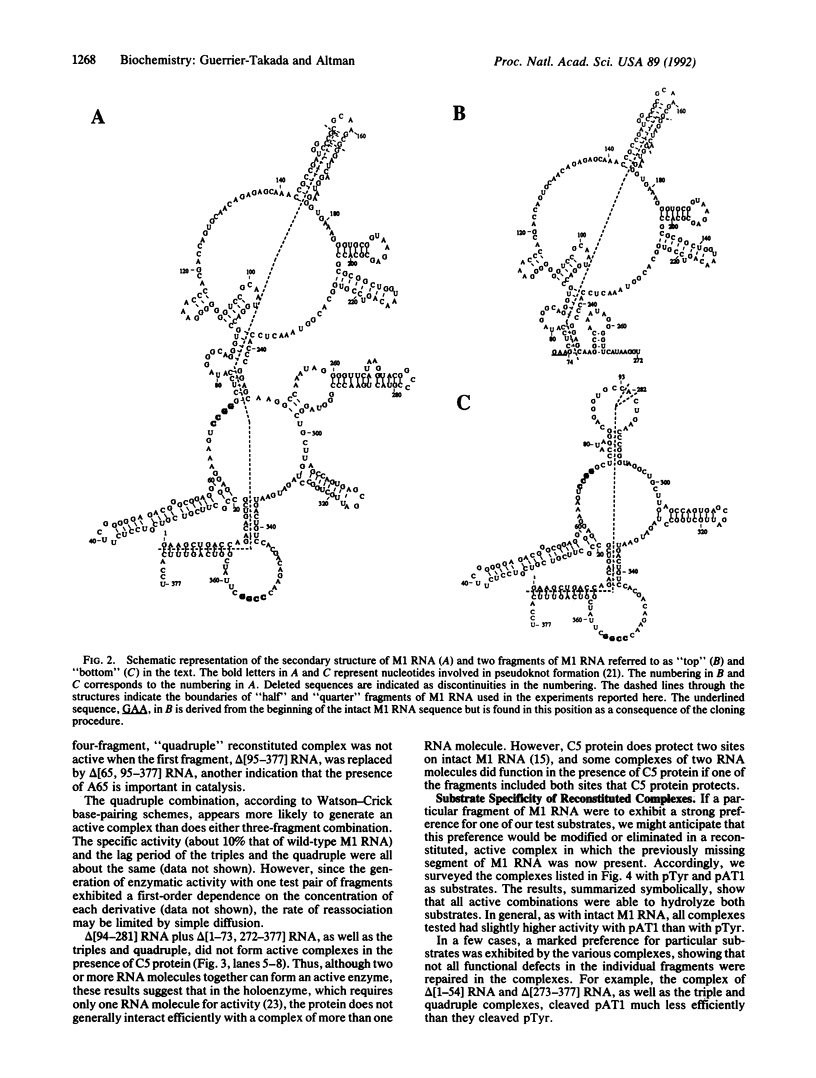
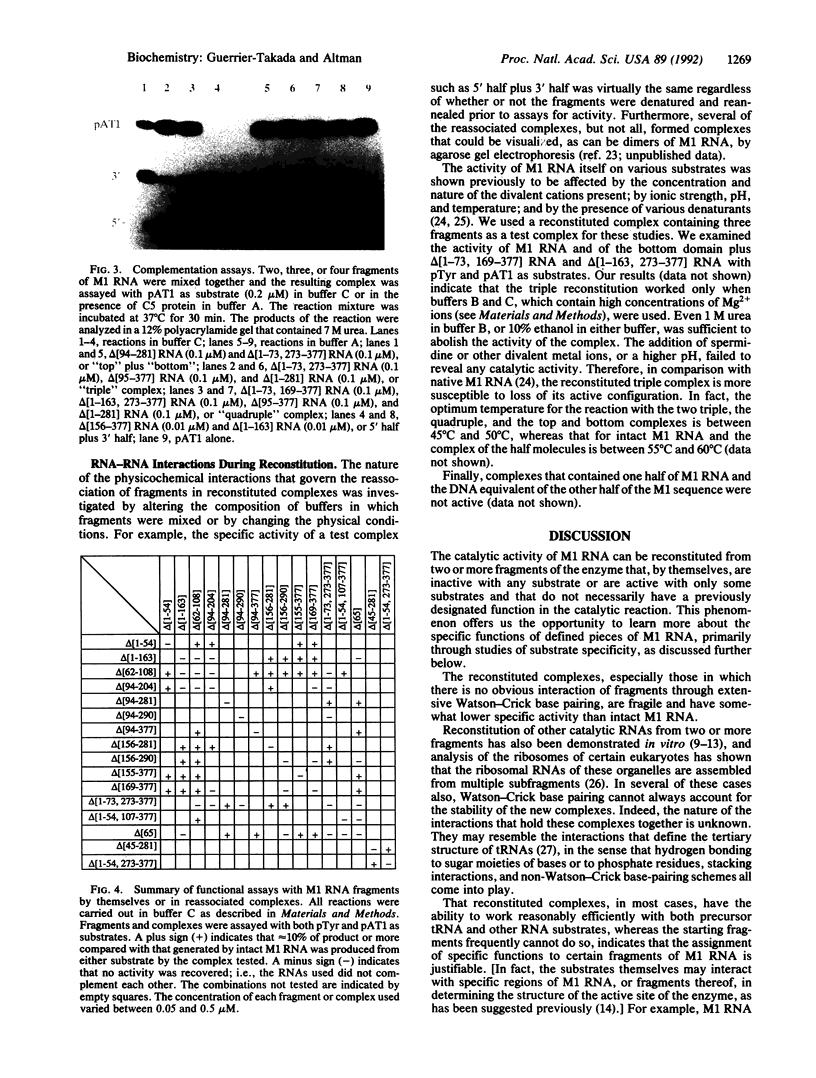
Abstract
Certain fragments of M1 RNA, the catalytic subunit of RNase P from Escherichia coli, either have no enzymatic activity at all or have altered substrate specificity compared with that of the intact catalytic RNA. After simple mixing in vitro, many of these fragments of M1 RNA can reassociate with other fragments to form complexes that have enzymatic activity typical of wild-type M1 RNA. Furthermore, inactive M1 RNA molecules with internal deletions can be complemented in vitro by other inactive derivatives of M1 RNA that have nonoverlapping deletions. Thus, two inactive molecules of M1 RNA can interact to form an active RNA enzyme. Functional attributes can be assigned to various regions of M1 RNA when the reconstitution process is combined with assays for activity with different substrates.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S. Ribonuclease P. Postscript. J Biol Chem. 1990 Nov 25;265(33):20053–20056. [PubMed] [Google Scholar]
- Altman S. Ribonuclease P: an enzyme with a catalytic RNA subunit. Adv Enzymol Relat Areas Mol Biol. 1989;62:1–36. doi: 10.1002/9780470123089.ch1. [DOI] [PubMed] [Google Scholar]
- Doudna J. A., Couture S., Szostak J. W. A multisubunit ribozyme that is a catalyst of and template for complementary strand RNA synthesis. Science. 1991 Mar 29;251(5001):1605–1608. doi: 10.1126/science.1707185. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990 Aug 10;62(3):407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. M1 RNA with large terminal deletions retains its catalytic activity. Cell. 1986 Apr 25;45(2):177–183. doi: 10.1016/0092-8674(86)90381-8. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Haydock K., Allen L., Altman S. Metal ion requirements and other aspects of the reaction catalyzed by M1 RNA, the RNA subunit of ribonuclease P from Escherichia coli. Biochemistry. 1986 Apr 8;25(7):1509–1515. doi: 10.1021/bi00355a006. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Lumelsky N., Altman S. Specific interactions in RNA enzyme-substrate complexes. Science. 1989 Dec 22;246(4937):1578–1584. doi: 10.1126/science.2480641. [DOI] [PubMed] [Google Scholar]
- Haas E. S., Morse D. P., Brown J. W., Schmidt F. J., Pace N. R. Long-range structure in ribonuclease P RNA. Science. 1991 Nov 8;254(5033):853–856. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- Hansen F. G., Hansen E. B., Atlung T. Physical mapping and nucleotide sequence of the rnpA gene that encodes the protein component of ribonuclease P in Escherichia coli. Gene. 1985;38(1-3):85–93. doi: 10.1016/0378-1119(85)90206-9. [DOI] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Liu J. S., Pace N. R. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell. 1988 Jan 15;52(1):19–26. doi: 10.1016/0092-8674(88)90527-2. [DOI] [PubMed] [Google Scholar]
- Jarrell K. A., Dietrich R. C., Perlman P. S. Group II intron domain 5 facilitates a trans-splicing reaction. Mol Cell Biol. 1988 Jun;8(6):2361–2366. doi: 10.1128/mcb.8.6.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakov S., Altman S. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9193–9197. doi: 10.1073/pnas.88.20.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knap A. K., Wesolowski D., Altman S. Protection from chemical modification of nucleotides in complexes of M1 RNA, the catalytic subunit of RNase P from E coli, and tRNA precursors. Biochimie. 1990 Nov;72(11):779–790. doi: 10.1016/0300-9084(90)90187-l. [DOI] [PubMed] [Google Scholar]
- Kole R., Baer M. F., Stark B. C., Altman S. E. coli RNAase P has a required RNA component. Cell. 1980 Apr;19(4):881–887. doi: 10.1016/0092-8674(80)90079-3. [DOI] [PubMed] [Google Scholar]
- Lumelsky N., Altman S. Selection and characterization of randomly produced mutants in the gene coding for M1 RNA. J Mol Biol. 1988 Aug 5;202(3):443–454. doi: 10.1016/0022-2836(88)90277-x. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Lastity D., Levina E. S., Bayev A. A. Localization of two recognition sites in yeast valine tRNA I. Nat New Biol. 1971 Jan 6;229(1):21–22. doi: 10.1038/newbio229021a0. [DOI] [PubMed] [Google Scholar]
- Odai O., Hiroaki H., Sakata T., Tanaka T., Uesugi S. The role of a conserved guanosine residue in the hammerhead-type RNA enzyme. FEBS Lett. 1990 Jul 2;267(1):150–152. doi: 10.1016/0014-5793(90)80311-6. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Reich C., James B. D., Olsen G. J., Pace B., Waugh D. S. Structure and catalytic function in ribonuclease P. Cold Spring Harb Symp Quant Biol. 1987;52:239–248. doi: 10.1101/sqb.1987.052.01.029. [DOI] [PubMed] [Google Scholar]
- Peck-Miller K. A., Altman S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J Mol Biol. 1991 Sep 5;221(1):1–5. doi: 10.1016/0022-2836(91)80194-y. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Sakano H., Yamada S., Ikemura T., Shimura Y., Ozeki H. Temperature sensitive mutants of Escherichia coli for tRNA synthesis. Nucleic Acids Res. 1974 Mar;1(3):355–371. doi: 10.1093/nar/1.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl P., Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare M. N., Gray M. W. Sixteen discrete RNA components in the cytoplasmic ribosome of Euglena gracilis. J Mol Biol. 1990 Sep 5;215(1):73–83. doi: 10.1016/S0022-2836(05)80096-8. [DOI] [PubMed] [Google Scholar]
- Vioque A., Arnez J., Altman S. Protein-RNA interactions in the RNase P holoenzyme from Escherichia coli. J Mol Biol. 1988 Aug 20;202(4):835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- van der Horst G., Christian A., Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]