Abstract
To precisely diagnose metastasis state is important for tailoring treatments for gastric cancer patients. However, the routinely employed radiological and pathologic tests for tumour metastasis have considerable high false negative rates, which may retard the identification of reproducible metastasis-related molecular biomarkers for gastric cancer. In this research, using three datasets, we firstly shwed that differentially expressed genes (DEGs) between metastatic tissue samples and non-metastatic tissue samples could hardly be reproducibly detected with a proper statistical control when the metastatic and non-metastatic samples were defined by TNM stage alone. Then, assuming that undetectable micrometastases are the prime cause for recurrence of early stage patients with curative resection, we reclassified all the “non-metastatic” samples as metastatic samples whenever the patients experienced tumour recurrence during follow-up after tumour resection. In this way, we were able to find distinct and reproducible DEGs between the reclassified metastatic and non-metastatic tissue samples and concordantly significant DNA methylation alterations distinguishing metastatic tissues and non-metastatic tissues of gastric cancer. Our analyses suggested that the follow-up recurrence information for patients should be employed in the research of tumour metastasis in order to decrease the confounding effects of false non-metastatic samples with undetected micrometastases.
Tumour metastasis is the primary cause of recurrence and mortality of early stage gastric cancer patients after curative surgery1,2,3,4. Therefore, accurate diagnosis of distant and lymph node metastasis is essential for predicting prognosis and tailoring treatment strategies for gastric cancer patients5,6. However, current preoperative imaging techniques such as computed tomography (CT) and endoscopic ultrasound (EUS) are lack of accuracy7,8 and especially tend to produce a high rate of false negative clinical reports due to the poor identification of tiny lesions or micrometastases4,7,8. The lymph node metastasis is routinely detected by hematoxylin-eosin (H&E) staining of one section containing the largest dimension of the lymph node6, which also tends to produce a high rate of false negative clinical reports because of the random distribution of tumour cells throughout the lymph node4,6,9,10. More lymph node sections may decrease the false negative rate of H&E staining but the workload of surgeons and pathologists will be increased greatly4,6. The same problem exists when immunohistochemistry is used for detecting lymph node metastasis4,6. Consequently biomarkers for predicting the metastasis state for individual patients are in urgent need to avoid over- or inadequate-treatment owing to the misdiagnosis.
Because gene expression profiling has the advantage of exploring the tumour progression systematically based on the multiple gene disorders, many researches have exploited the high throughout data to study the transcriptional characteristics of metastasis and identify transcriptional biomarkers for metastasis11,12,13. Epigenomics data has also been taken into consideration by researchers and some methylation loci related to gastric metastasis have been reported14,15. However, the results of different studies showed inconsistency and lacked independent validation16. The same irreproducibility problem may exist for the basic task of extracting differentially expressed genes (DEGs) between the metastasis and non-metastasis samples17, which might make it unreliable to investigate the metastasis based on DEGs.
In this research, using three datasets of gene expression profiles for gastric cancer, we firstly showed that DEGs between metastatic tissue samples and non-metastatic tissue samples could hardly be reproducibly detected with a proper statistical control when the metastasis and non-metastasis samples were defined by TNM stage alone. Because micrometastases not found by the routine pathology diagnosis could be the major cause for recurrence after curative surgery18,19,20, we could hypothesize that the patients diagnosed as non-metastasis cases but subsequently suffered the recurrence should have developed micrometastases before the surgery. According to this hypothesis, we reclassified all the “non-metastatic” samples of patients, defined according to TNM stage, as metastatic samples whenever the patients experienced tumour recurrence during follow-up after tumour resection. By this strategy, we were able to find distinct and reproducible DEGs and concordant DNA methylation alterations between the reclassified metastatic and non-metastatic tissue samples with a proper statistical control false discovery rate (FDR) of less than 20%.
Results
Detecting reproducible metastasis-associated DEGs with the recurrence information
The TNM stage of the samples in the three datasets analysed in this study were diagnosed according to the 6th edition (GSE15459 and GSE62254) or 7th edition (TCGA batch 220) of the AJCC Cancer Staging Manual21, where the two editions have the same definition for metastasis and non-metastasis. According to TNM stage, the non-metastasis group consisted of samples without lymph node metastasis (N0) nor distant metastasis (M0), while the metastasis group included samples with lymph node metastasis (N+) and /or distant metastasis (M+) (Table 1). Using Wilcoxon rank-sum test with FDR < 10%, 126 and 1687 DEGs were detected between the metastasis group and non-metastasis group for the GSE15459 and GSE62254 datasets, respectively. The two lists of DEGs shared only 7 genes and the concordance score (see Materials and Methods) was 57.1% (p = 0.5). With FDR < 10%, 69 DEGs were found in TCGA batch 220 by the edgeR package (see Materials and Methods), of which only 1 and 3 DEGs were shared by GSE15459 and GSE62254, respectively (Supplementary Table 1). With FDR < 20%, 660 and 3371 DEGs were detected in GSE15459 and GSE62254 respectively. The two lists of DEGs had only 94 overlapped genes, and the concordance scores was as low as 54.3% (p = 0.24). With FDR < 20%, 124 DEGs were detected in the TCGA batch 220, of which only 3 and 15 DEGs were also detected as DEGs in GSE15459 and GSE62254 and the concordance scores were as low as 0% and 33.3% (p = 0.94), respectively. The low concordance scores and small overlaps between DEGs identified from independent datasets indicated that differential gene expression signals were weak and poorly reproducible in the three datasets when the samples were grouped by the TNM stage only (Supplementary Table 2), possibly due to confounding factors such as false negatives and/or false positive samples.
Table 1. Samples classified as metastasis and non-metastasis groups according to different criteria.
| Dataset | Metastasis group | Non-metastasis group |
|---|---|---|
| Gene expression profiles (grouped by TNM stage) | ||
| GSE15459 | 139 | 35 |
| GSE62254 | 263 | 37 |
| TCGA batch 220 | 33 | 19 |
| Gene expression profiles (regrouped by TNM stage and recurrence information) | ||
| GSE15459 | 94 | 27 |
| GSE62254 | 132 | 27 |
| TCGA batch 220 | 18 | 11 |
| Methylation profile (regrouped by TNM stage and recurrence information) | ||
| TCGA batch 220 | 21 | 14 |
Considering that micrometastases undetectable by the routine pathology diagnosis could be the major cause for recurrence after curative surgery, we reclassified the samples by taking into account the recurrence information. GSE15459 provided the adjuvant treatment information for individual patients. Some patients experiencing no recurrence after curative surgery might benefit from the adjuvant treatment. Accordingly, only patients who were diagnosed as non-metastasis (N0M0) and did not recur under the condition of without adjuvant treatment were defined as the non-metastasis group. Because the information on adjuvant treatment were not explicitly provided in both GSE62254 and TCGA batch 220, the non-metastasis samples were defined as the ones who were diagnosed as non-metastasis (N0M0) and did not recur. For all these three datasets, the metastasis group consisted of the patients who were diagnosed as distant metastasis and the ones without distant metastasis but suffered from recurrence. In order to exclude the potential non-distant metastasis samples (false positive samples), we ignored the samples who were diagnosed as lymph node metastasis (N + M0) but did not recur after curative resection. After this reclassification, we obtained 94 metastasis samples and 27 non-metastasis samples in GSE15459, 132 metastasis samples and 27 non-metastasis samples in GSE62254, 18 metastasis samples and 11 non-metastasis samples in TCGA batch 220 respectively (Table 1). With FDR < 20%, the DEGs between the regrouped metastasis and non-metastasis samples were separately detected by the Wilcoxon rank-sum test for the GSE15459 and GSE62254 datasets and by the edgeR package for the data of the TCGA batch 220. After the sample reclassification, both the overlaps and concordance scores between every two lists of DEGs identified from the three independent datasets increased greatly (Supplementary Table 3). For GSE62254 and TCGA batch 220, the concordance score increased to 92.9% (p < 2.20 × 10−16) and the consistent DEGs increased to 2625. The concordance score between GSE15459 and GSE62254 increased to 90.5% (p < 1.11 × 10−4) and the score between the GSE15459 and TCGA batch 220 increased to 92.3% (p < 1.71 × 10−3), respectively, although they still had small numbers of overlapped DEGs. The high and statistically significant concordance scores verified that the reclassification of recurred samples was a powerful practice to extract reliable DEGs related to gastric cancer metastasis (Table 2). Part of the misjudged samples had been regrouped in accord with their actual metastasis status by this practice.
Table 2. Concordance scores between DEGs detected from different datasets (FDR < 20%).
| Datasets | TNM stage1 | TNM stage and recurrence2 | ||
|---|---|---|---|---|
| overlap(CS3) | p value | overlap(CS) | pvalue | |
| GSE15459 vs. GSE62254 | 94 (54.3%) | 0.24 | 21 (90.5%) | 1.11 × 10−4 |
| GSE62254 vs. TCGA batch 220 | 15 (33.3%) | 0.94 | 2827 (92.9%) | <2.2 × 10−16 |
| GSE15459 vs. TCGA batch 220 | 3 (0%) | >0.99 | 13 (92.3%) | 1.71 × 10−3 |
Note: 1results for sample classified by the TNM stage alone.
2Results for sample classified by TNM stage and recurrence information.
3CS denotes for concordant score.
Functional enrichment analysis further supported that the DEGs consistently detected in GSE62254 and TCGA batch 220 were correlated with metastasis. With FDR < 20%, the DEGs up-regulated in the metastasis samples compared with the non-metastasis samples were significantly enriched in some typical tumour metastasis-associated signalling pathways, such as ECM-receptor interaction22, focal adhesion23,24,25 and cGMP-PKG signalling pathways26,27 (Supplementary Table 4). In contrast, the DEGs down-regulated in the metastasis samples were significantly enriched in pathways involved in cell metabolism, such as biosynthesis of amino acids, carbon metabolism, pyrimidine metabolism and many other pathways, such as homologous recombination, DNA replication and mismatch repair (Supplementary Table 4).
Distinct epigenomic characteristics of metastasis
After reclassifying the samples with methylation data of TCGA batch 220 by the same rule used for the gene expression profiles, we compared the methylation profiles between the metastasis and the non-metastasis samples. Using the Wilcoxon rank-sum test with FDR < 20%, 447 and 233 genes were found to be hypermethylated and hypomethylated in the metastasis samples compared with the non-metastasis samples, respectively. Among the 447 hypermethylated genes, 62 genes were also identified as DEGs between the two groups, among which 90.3% were concordantly down-regulated in the metastasis samples compared with the non-metastasis samples, which was unlikely to be observed by chance (p < 1.49 × 10−11). These results suggested that hypermethylation of gene promoters may play a major role in inducing gene down-regulations in the metastasis tissues, and thus could be a major driver for the gastric cancer metastasis. Some of the concordant genes play important roles in the process of tumour cell migration. For example, IFNG in the regulation of autophagy pathway, which was both hypermethylated and down-regulated in metastasis tissues, might reduce cell epithelial apoptosis and decrease cell proliferation via autophagy28.
Similarly, 207 out of 233 hypomethylated genes were identified as DEGs between the two groups, among which 57.5% were concordantly up-regulated in the metastasis samples compared with the non-metastasis samples, which was also unlikely to be observed by chance (p < 0.02). Although the correlation between hypomethylation of gene promoters and gene overexpression was weak, DNA hypomethylation might also play a role in the metastasis. For example, we found that the COL4A3 annotated in the ECM-receptor interaction pathway was both hypomethylated and up-regulated in metastasis tissues, which might play a role in tumour metastasis29.
Discussion
Our analyses demonstrated that the metastasis-associated differential gene expression signals were very weak and thus poorly reproducible in independent data when the samples were classified simply according to the TNM stage. The high recurrence rates of non-metastasis samples used in this study indicated high false negative rates, which might blur the difference between the metastasis and non-metastasis samples. This problem might exist for many studies on cancer metastasis mechanisms or predictive signatures, including both the high- and low-throughput researches. In order to reduce the interference of the false negative samples, we suggest making use of follow-up information of samples when researches on gastric cancer metastasis are conducted. Our results showed that distinct metastasis-associated DEGs could be reproducibly detected in independent data when the samples were regrouped based on both the TNM stage and recurrence information. With the help of recurrence information, classical metastasis-associated pathways significantly enriched with metastasis-associated DEGs could be readily detected, including focal adhesion, ECM-receptor interaction and metabolism pathways. The functional analysis results also provided extra evidence for the authenticity of the metastasis-associated DEGs identified between the reclassified metastasis and non-metastasis groups.
Gene expression alterations are usually caused by epigenomic and/or genomic lesions30,31. Metastasis-associated DNA methylation alterations which were significantly concordant with differential gene expressions were indeed observed between the reclassified metastasis and non-metastasis groups. Our results showed that hypermethylation of CpG loci in genes’ promoter regions could contribute to genes’ down-regulations in metastasis samples, indicating that DNA methylation alternation might be an important factor promoting cancer metastasis. However, we were unable to detect copy number alternations and gene mutations with significantly different frequencies between the metastasis and non-metastasis samples by the Fisher’s exact test with FDR control (FDR < 20%). The failure in finding genomic events characterizing the metastasis samples might indicate the existence of a certain percentage of misjudged samples in the datasets analysed in this study even after reclassifying some potential false negative samples. The undetected misjudged samples could be possibly due to the confounded effect of adjuvant treatment and short-term follow-up.
In summary, the false negative problem lays a major barrier for detecting reproducible metastasis-associated DEGs, let alone the identification of signatures for predicting metastasis. The same problem should exist in studies for other cancers and thus we suggest that the follow-up information should be taken into consideration for studying cancer metastasis.
Materials and Methods
Data acquisition and pre-processing
Gastric cancer gene expression profiles of the GSE15459 and GSE62254 datasets were downloaded from the GEO. The raw data (.CEL files) were normalized using the robust multi-array average method (RMA) in the Bioconductor package32,33,34. If multiple probes were mapped to the same gene, the expression value for the gene was summarized as the arithmetic mean of the values of the multiple probes (on the log2 scale). After data preprocess, 20283 genes were remained for analysis for both GSE15459 and GSE62254.
The multi-omic data for gastric cancer were derived from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/). In order to avoid the batch effect, we restricted our analysis to samples of batch 220 which had comprehensive clinical information. The count data of RNA-seq were downloaded from the TCGA Web Portal. After excluding the unknown transcripts, we kept the data of 22509 genes for the following analysis. The methylation beta-values of samples measured by the Infinium HumanMethylation450 platform were downloaded from the TCGA Web Portal. Because the correlation between gene body methylation and gene expression is not clearly understood until now35,36, we focused on analysing the 27,578 CpG loci within the promoters for 14,495 protein-coding genes, which were defined in the Infinium HumanMethylation27 platform37. It has been widely recognized that there is a negative correlation between the promoter methylation and transcription activity, especially the hypermethylation of CpG loci in a gene promoter could lead to silence in gene transcription38,39. After excluding the loci with missing values, 22,432 CpG loci within the promoters for 14,495 protein-coding genes were analysed. CNVs data of level 4 of the TCGA samples analysed by GISTIC 2.0 were downloaded from Firehose (https://confluence.broadinstitute.org/display/GDAC/Download). A total of 36 significant amplification peaks and 53 deletion peaks were obtained.
Identification of DEGs and differentially methylated genes
The two-tailed Wilcoxon rank-sum test was used to select DEGs and differentially methylated (DM) genes between metastasis and non-metastasis samples for microarray data and methylation data40. The R package of edgeR41 for RNA-seq data was conducted to exact DEGs between two kinds of samples. All the p values in this paper were adjusted by the Benjamini-Hochberg FDR procedure42.
Analysis of epigenetic data
Only the CpG loci within the gene promoters for 14,495 protein-coding genes, as defined in the Infinium HumanMethylation27 platform37, were analysed. If a gene had both hypermethylated and hypomethylated CpG loci, this gene was excluded from subsequent analysis43. A gene with at least one DM locus in its promoter was termed a DM gene. By comparing the mean beta values of DM CpG loci between metastasis and non-metastasis samples, we classified the DM genes as hypermethylated genes or hypomethylated genes.
Concordance scores
Suppose a couple of DEGs lists extracted separately from two datasets shared k genes, of which s genes showed the same deregulation directions (up- or down-regulation). In this case the concordance score was calculated as s/k × 100%. This score was used to evaluate the consistence of DEGs extracted from independent datasets.
If k genes are both significantly altered down-regulated (or up-regulation) in gene expression and methylated in the metastasis samples, of which s genes were hypermethylated (or hypomethylated) and correspondingly down-regulated (or up-regulation), then the concordance score was calculated as s/k × 100%. This score was used to evaluate the concordance of hypermethylation (or hypomethylation) with down-regulation (or up-regulation).
The probability of observing a concordance score of s/k by chance was evaluated by the cumulative binomial distribution model as follows:
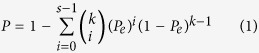 |
where Pe is the probability of one gene having the concordant relationship between the two lists of genes by chance (here, Pe = 0.5).
Functional enrichment analysis
The functional enrichment analysis was conducted based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database44. The biological pathways in this database are described in KEGG Markup Language (KGML) files including nodes (genes and compounds) and edges (functional links). The KGML data files were obtained manually from the KEGG website in July, 2014. After removing the pathways without functional links between genes, we obtained 217 pathways. Functional KEGG enrichment analyses were performed separately for up- and down-regulated genes for the reason that it was more powerful than analysing all the DEGs together45. The biological pathways that were significantly enriched with genes of interest were determined by the hypergeometric distribution model. If k genes were identified as interesting genes (such as DEGs) from n genes in a dataset and x of them were annotated in a pathway with m genes, then the probability of observing at least x genes in this pathway by chance can be appropriately modelled by the cumulative hypergeometric distribution model as follows:
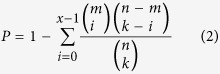 |
Additional Information
How to cite this article: Li, M. et al. Identifying Reproducible Molecular Biomarkers for Gastric Cancer Metastasis with the Aid of Recurrence Information. Sci. Rep. 6, 24869; doi: 10.1038/srep24869 (2016).
Supplementary Material
Acknowledgments
This work was supported by the Natural Science Foundation of China [grant numbers: 81372213, 81572935, 81501215 and 81501829]. The authors thanked Dr. Ju-Seog Lee of Department of Pathology at The University of Texas MD Anderson Cancer Center for providing clinical data of GSE15459.
Footnotes
Author Contributions All authors meet the authorship requirements. Z.G., M.L. and H.L. designed the study. G.H. downloaded the KGML data files. J.C. collected the gene expression datasets and X.L. collected the CNVs data. M.L. collected the RNA-seq and the methylation data and performed all the data analyses. H.C., Q.G. and M.T. interpreted the function annotations. M.L. drafted the manuscript. Z.G., H.L., M.L., G.H. and J.L. revised the manuscript critically for important intellectual content. Z.G., H.L. and M.L. were agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
References
- Yamamichi K., Uehara Y., Kitamura N., Nakane Y. & Hioki K. Increased expression of CD44v6 mRNA significantly correlates with distant metastasis and poor prognosis in gastric cancer. International journal of cancer. Journal international du cancer 79, 256–262 (1998). [DOI] [PubMed] [Google Scholar]
- Saka M., Katai H., Fukagawa T., Nijjar R. & Sano T. Recurrence in early gastric cancer with lymph node metastasis. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association 11, 214–218, 10.1007/s10120-008-0485-4 (2008). [DOI] [PubMed] [Google Scholar]
- Sano T., Sasako M., Kinoshita T. & Maruyama K. Recurrence of early gastric cancer. Follow-up of 1475 patients and review of the Japanese literature. Cancer 72, 3174–3178 (1993). [DOI] [PubMed] [Google Scholar]
- Arigami T. et al. Clinical significance of lymph node micrometastasis in gastric cancer. Annals of surgical oncology 20, 515–521, 10.1245/s10434-012-2355-x (2013). [DOI] [PubMed] [Google Scholar]
- Hirakawa S. et al. VEGF-C-induced lymphangiogenesis in sentinel lymph nodes promotes tumor metastasis to distant sites. Blood 109, 1010–1017, 10.1182/blood-2006-05-021758 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai K. et al. Multicenter study evaluating the clinical performance of the OSNA assay for the molecular detection of lymph node metastases in gastric cancer patients. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association 17, 273–280, 10.1007/s10120-013-0271-9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cidon E. U. & Cuenca I. J. Gastric Adenocarcinoma: Is Computed Tomography (CT) Useful in Preoperative Staging? Clinical medicine. Oncology 3, 91–97 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q. et al. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncology reports 31, 1863–1870, 10.3892/or.2014.3004 (2014). [DOI] [PubMed] [Google Scholar]
- Yonemura Y. et al. Evaluation of lymphatic invasion in primary gastric cancer by a new monoclonal antibody, D2–40. Human pathology 37, 1193–1199, 10.1016/j.humpath.2006.04.014 (2006). [DOI] [PubMed] [Google Scholar]
- Kahn H. J. & Marks A. A new monoclonal antibody, D2–40, for detection of lymphatic invasion in primary tumors. Laboratory investigation; a journal of technical methods and pathology 82, 1255–1257 (2002). [DOI] [PubMed] [Google Scholar]
- Weiss M. M. et al. Genomic profiling of gastric cancer predicts lymph node status and survival. Oncogene 22, 1872–1879, 10.1038/sj.onc.1206350 (2003). [DOI] [PubMed] [Google Scholar]
- Cristescu R. et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nature medicine 21, 449–456, 10.1038/nm.3850 (2015). [DOI] [PubMed] [Google Scholar]
- Marchet A. et al. Gene expression profile of primary gastric cancer: towards the prediction of lymph node status. Annals of surgical oncology 14, 1058–1064, 10.1245/s10434-006-9090-0 (2007). [DOI] [PubMed] [Google Scholar]
- Shigematsu Y. et al. Identification of a DNA methylation marker that detects the presence of lymph node metastases of gastric cancers. Oncology letters 4, 268–274, 10.3892/ol.2012.708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J., Tanaka T., Kitajima Y., Noshiro H. & Miyazaki K. Methylation-mediated gene silencing as biomarkers of gastric cancer: a review. World journal of gastroenterology 20, 11991–12006, 10.3748/wjg.v20.i34.11991 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H. et al. CD44v6, MMP-7 and nuclear Cdx2 are significant biomarkers for prediction of lymph node metastasis in primary gastric cancer. Oncology reports 22, 745–755 (2009). [DOI] [PubMed] [Google Scholar]
- Zou J. et al. Revealing weak differential gene expressions and their reproducible functions associated with breast cancer metastasis. Computational biology and chemistry 39, 1–5, 10.1016/j.compbiolchem.2012.04.002 (2012). [DOI] [PubMed] [Google Scholar]
- Maehara Y. et al. Clinical significance of occult micrometastasis lymph nodes from patients with early gastric cancer who died of recurrence. Surgery 119, 397–402 (1996). [DOI] [PubMed] [Google Scholar]
- Lee C. M. et al. Should lymph node micrometastasis be considered in node staging for gastric cancer?: the significance of lymph node micrometastasis in gastric cancer. Annals of surgical oncology 22, 765–771, 10.1245/s10434-014-4073-z (2015). [DOI] [PubMed] [Google Scholar]
- Doekhie F. S. et al. Clinical relevance of occult tumor cells in lymph nodes from gastric cancer patients. The American journal of surgical pathology 29, 1135–1144 (2005). [DOI] [PubMed] [Google Scholar]
- Washington K. 7th edition of the AJCC cancer staging manual: stomach. Annals of surgical oncology 17, 3077–3079, 10.1245/s10434-010-1362-z (2010). [DOI] [PubMed] [Google Scholar]
- Olson M. F. & Sahai E. The actin cytoskeleton in cancer cell motility. Clinical & experimental metastasis 26, 273–287, 10.1007/s10585-008-9174-2 (2009). [DOI] [PubMed] [Google Scholar]
- Kurayoshi M. et al. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer research 66, 10439–10448, 10.1158/0008-5472.CAN-06-2359 (2006). [DOI] [PubMed] [Google Scholar]
- Thiery J. P. Epithelial-mesenchymal transitions in development and pathologies. Current opinion in cell biology 15, 740–746 (2003). [DOI] [PubMed] [Google Scholar]
- Chang W. et al. Identification of novel hub genes associated with liver metastasis of gastric cancer. International journal of cancer. Journal international du cancer 125, 2844–2853, 10.1002/ijc.24699 (2009). [DOI] [PubMed] [Google Scholar]
- Browning D. D. Protein kinase G as a therapeutic target for the treatment of metastatic colorectal cancer. Expert opinion on therapeutic targets 12, 367–376, 10.1517/14728222.12.3.367 (2008). [DOI] [PubMed] [Google Scholar]
- Babykutty S. et al. Insidious role of nitric oxide in migration/invasion of colon cancer cells by upregulating MMP-2/9 via activation of cGMP-PKG-ERK signaling pathways. Clinical & experimental metastasis 29, 471–492, 10.1007/s10585-012-9464-6 (2012). [DOI] [PubMed] [Google Scholar]
- Tu S. P. et al. IFN-gamma inhibits gastric carcinogenesis by inducing epithelial cell autophagy and T-cell apoptosis. Cancer research 71, 4247–4259, 10.1158/0008-5472.CAN-10-4009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X. C. et al. COL4A3 expression correlates with pathogenesis, pathologic behaviors, and prognosis of gastric carcinomas. Human pathology 44, 77–86, 10.1016/j.humpath.2011.10.028 (2013). [DOI] [PubMed] [Google Scholar]
- Jaenisch R. & Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics 33 Suppl, 245–254, 10.1038/ng1089 (2003). [DOI] [PubMed] [Google Scholar]
- Stranger B. E. et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315, 848–853, 10.1126/science.1136678 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264, 10.1093/biostatistics/4.2.249 (2003). [DOI] [PubMed] [Google Scholar]
- Irizarry R. A. et al. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research 31, e15 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M. & Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003). [DOI] [PubMed] [Google Scholar]
- Yang X. et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer cell 26, 577–590, 10.1016/j.ccr.2014.07.028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jjingo D., Conley A. B., Yi S. V., Lunyak V. V. & Jordan I. K. On the presence and role of human gene-body DNA methylation. Oncotarget 3, 462–474, 10.18632/oncotarget.497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird P. W. Principles and challenges of genomewide DNA methylation analysis. Nature reviews. Genetics 11, 191–203, 10.1038/nrg2732 (2010). [DOI] [PubMed] [Google Scholar]
- Tate P. H. & Bird A. P. Effects of DNA methylation on DNA-binding proteins and gene expression. Current opinion in genetics & development 3, 226–231 (1993). [DOI] [PubMed] [Google Scholar]
- Baylin S. B. DNA methylation and gene silencing in cancer. Nature clinical practice. Oncology 2 Suppl 1, S4–11, 10.1038/ncponc0354 (2005). [DOI] [PubMed] [Google Scholar]
- Adjaye J. et al. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem cells 23, 1514–1525, 10.1634/stemcells.2005-0113 (2005). [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. H. Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc B 57, 289–300 (1995). [Google Scholar]
- Kim E. H., Park A. K., Dong S. M., Ahn J. H. & Park W. Y. Global analysis of CpG methylation reveals epigenetic control of the radiosensitivity in lung cancer cell lines. Oncogene 29, 4725–4731, 10.1038/onc.2010.223 (2010). [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M. & Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids research 40, D109–114, 10.1093/nar/gkr988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G., Zhang W., Li H., Shen X. & Guo Z. Separate enrichment analysis of pathways for up- and downregulated genes. Journal of the Royal Society, Interface/the Royal Society 11, 20130950, 10.1098/rsif.2013.0950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


