Significance
Eubacterial protein synthesis entails formation of an unlocked preinitiation complex consisting of the 30S ribosomal subunit, initiation factors, mRNA, and initiator tRNA. A conformational change in the subunit accompanies mRNA–tRNA codon–anticodon base-pairing generating a locked 30S complex. If correctly formed, this complex associates with the 50S ribosomal subunit forming a 70S complex, and the initiation factors are ejected. We show that the translational inhibitor GE81112 targets this essential step, hampering formation of a canonical codon–anticodon interaction and stalling the 30S in an unlocked state. Moreover, in the presence of GE81112 three rRNA helices, h44/h45/h24a, are stabilized in a disengaged conformation, suggesting that their conformation is associated with tRNA/mRNA decoding and transition of the 30S from unlocked to locked state.
Keywords: GE81112, ribosome, X-ray crystallography, protein synthesis, antibiotics
Abstract
In prokaryotic systems, the initiation phase of protein synthesis is governed by the presence of initiation factors that guide the transition of the small ribosomal subunit (30S) from an unlocked preinitiation complex (30S preIC) to a locked initiation complex (30SIC) upon the formation of a correct codon–anticodon interaction in the peptidyl (P) site. Biochemical and structural characterization of GE81112, a translational inhibitor specific for the initiation phase, indicates that the main mechanism of action of this antibiotic is to prevent P-site decoding by stabilizing the anticodon stem loop of the initiator tRNA in a distorted conformation. This distortion stalls initiation in the unlocked 30S preIC state characterized by tighter IF3 binding and a reduced association rate for the 50S subunit. At the structural level we observe that in the presence of GE81112 the h44/h45/h24a interface, which is part of the IF3 binding site and forms ribosomal intersubunit bridges, preferentially adopts a disengaged conformation. Accordingly, the findings reveal that the dynamic equilibrium between the disengaged and engaged conformations of the h44/h45/h24a interface regulates the progression of protein synthesis, acting as a molecular switch that senses and couples the 30S P-site decoding step of translation initiation to the transition from an unlocked preIC to a locked 30SIC state.
The majority of known antibiotics target the translational apparatus (1–4), but very few (e.g., kasugamycin, edeine, pactamycin, thiostrepton) are inhibitors of the initiation phase of protein synthesis, and even fewer display specificity for either the target or the kingdom of life in which they are effective (2–8). Two notable exceptions are Furvina (9) and GE81112 (6, 10), which inhibit bacterial protein synthesis by interfering with initiator fMet–tRNA binding to the 30S ribosomal subunit, thereby preventing the formation of the 30S initiation complex (30SIC).
Bacterial initiation progresses through a series of kinetic checkpoints that determine the overall efficiency and fidelity of translation initiation of a given mRNA (11–13). The initial step involves the recruitment of initiation factors (IFs) IF1, IF2⋅GTP, and IF3, mRNA, and fMet–tRNA to the 30S subunit that leads to the formation of the 30S preinitiation complex (30S preIC) in which initiator tRNA and mRNA are both ribosome-bound but not mutually interacting (12). Subsequently, a first-order isomerization of the 30S preIC leads to peptidyl (P)-site codon–anticodon pairing and yields a “locked” 30SIC. This 30S preIC→30SIC transition represents the first kinetic checkpoint of translational fidelity, because this process is under the kinetic control of the IFs, and during this step IF3, assisted by IF1, “inspects” the correctness of the 30S ligands, in particular the tRNA anticodon stem loop (ASL) and the mRNA start codon; any complex containing noncanonical components or having a geometry unfavorable for docking to the 50S ribosomal subunit is preferentially dissociated by IF3. The subsequent subunit-association step and the concomitant ejection of the IFs occur very rapidly only if the 30SIC contains canonical ligands and is geometrically fit for successful docking by the 50S. The subunit association step itself and the subsequent initial events that mark the 30SIC→70SIC transition until IF3 and IF1 are dissociated and the 70S complex stabilized represent the second and third kinetic checkpoint, respectively. Together they allow only a correctly formed 30SIC to proceed into the elongation phase of translation (11–14).
GE81112 (6, 10) is a highly hydrophilic chlorine-containing, noncyclic, nonribosomal tetrapeptide constituted by four nonproteinogenic l-amino acids. It is found in nature in three structural variants (A, B, and B1 with molecular masses of 643–658 Da). The most active of these variants proved to be variant B (658 Da), which was used throughout this study and whose structure is shown in Fig. S1. Fourteen biosynthetic genes (getA–N) involved in the synthesis of GE81112 have been identified within a larger biosynthetic gene cluster, which has been cloned from Streptomyces sp. l-49973, sequenced, and partially characterized (15). Because it belongs to a structurally unique class of antibiotics and inhibits an underexploited target within the translational apparatus, GE821112 seems to be a promising pharmacophore from which one could derive a new class of anti-infective agents for which no resistance has yet developed in nature.
Fig. S1.
Structure of GE81112. Structure of variant B of GE81112 (658 Da) as determined by NMR spectroscopy. The molecule consists of four amino acids: 3-hydroxypipecolic acid, 2-amino-5-[(aminocarbonyl)oxy]-4-hydroxypentanoic acid, 5-amino-histidine, and 5-chloro-2-imidazolylserine (10).
The aim of this work is to characterize the ribosomal binding site of GE81112 and gain a deeper understanding of the mechanism by which P-site binding of the initiator tRNA is inhibited by this molecule. By combining biochemical and structural methods, we aim to understand the action of GE81112 within the context of the translational initiation pathway.
Results
X-Ray Diffraction Studies Show That GE81112 Stabilizes the P Site ASL in a Distorted Conformation and Prevents the Formation of a Codon–Anticodon Interaction.
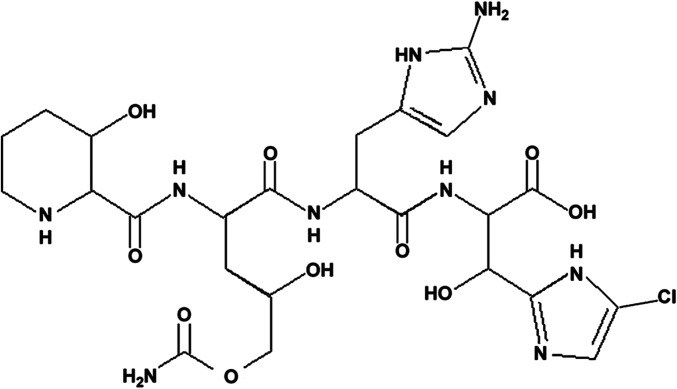
After ascertaining that GE81112 can inhibit fMet–tRNA binding to Thermus thermophilus 30S ribosomal subunits, X-ray crystallography of the T. thermophilus 30S-GE81112 complex was used to obtain high-resolution structural information on the binding site of the antibiotic (Fig. 1) and to understand the molecular basis of its mechanism of action. The initial Fo-Fc difference map showed a region of positive density in a position where the tip of the spur (h6) of a symmetry-related 30S subunit packs into the P site, mimicking the ASL of P-site–bound tRNA (Fig. 1 A and B and Fig. S2A) (16–19). To facilitate interpretation and further improve the quality of the difference maps, we used a bulk solvent modeling protection approach (20, 21) that prevents uninterpreted regions of interest from being incorporated into the solvent mask (Materials and Methods).
Fig. 1.
GE81112 binds in the ribosomal P site and distorts the ASL mimic. (A) An overview showing the binding site of GE81112 on the 30S subunit with the relevant 16S rRNA helices labeled. (Inset) Two symmetry-related 30S subunits are packed in the crystal such that h6 (blue) of one 30S subunit (red) inserts into the P site of a second subunit (gray) to mimic a P-tRNA. h6 in both subunits is circled. (B) Two orthogonal views of the ASL mimic showing a shift of the backbone between the 30S apo and 30S-GE81112 conformations (red and blue, respectively). Note that U81–U84 of the ASL mimic correspond to U33–U36 in the anticodon loop of the initiator tRNA. The initial unbiased positive Fo-Fc map is contoured at 4 σ (green mesh). (C) The C-terminal tail (K121–R125) of T. thermophilus S13, generally disordered and not visible in 30S crystal structures, becomes structured in the presence of GE81112. The Fo-Fc map is shown. (D) GE81112 is modeled in a binding pocket between S13 and the distorted ASL. (E) Fo-Fc map in the ASL and GE81112 region (Inset) with a zoom-in on the GE81112 electron densities where the ASL density is masked out for clarity. Maps in C and E are rendered at 3 σ and used the bulk solvent modeling protection approach (21).
Fig. S2.
Comparison and alignment of P-site tRNAs and ASL. (A) In 30S crystals h6 of one subunit is inserted into the P-site of a symmetry-related subunit mimicking the ASL of the P-site–bound tRNA. The figure in the panel presents the structure alignment of initiator fMet–tRNA as seen in the 70S structure (PDB ID code 2J00) and the ribosomal spur (h6) as seen in the 30S crystals in the absence of GE81112 (PDB ID code 2ZM6). The structures were aligned using 16S rRNA residues surrounding the P-tRNA. (B) The alignment between the tip of the ASL mimic distorted in the presence of GE81112 with the initiator P-site–bound tRNA as seen in the crystal structure of the 70S ribosome (PDB ID code 2J00) illustrates how the conformational change precludes codon–anticodon interaction. The residues involved in the canonical codon–anticodon interaction are labeled in green (A16–G18) and gray (C34–U36). The distorted ASL mimic (h6) as seen in the presence GE81112 is colored red. The structural alignment was preformed using 16S rRNA elements surrounding the P-tRNA.
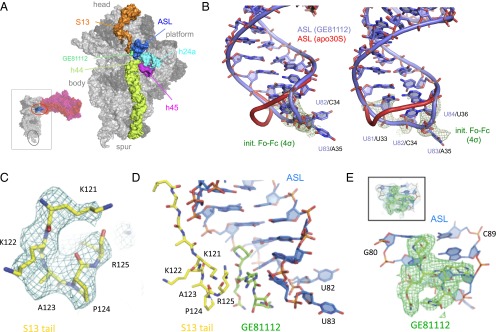
Analysis of the resulting maps indicates that GE81112 binds in a pocket formed by the C-terminal tail of S13, the distorted tip of the ASL mimic, the tip of h44 (C1400), and the loop of h31 (G960) (Figs. 1D and 2A). Accordingly, in the T. thermophilus 30S subunit, the accommodation of the drug in this pocket involves (i) the ordering of the flexible C-terminal tail of S13, which is generally disordered in the structures of the T. thermophilus 30S subunits but sometimes is seen between the A- and P-tRNAs (22, 23), and (ii) the stabilization of a conformational change in the ASL mimic. With regard to the S13 tail, residues K120–R125 become ordered so that they can be putatively modeled within the electron density (Fig. 1C). However, this ordering in the presence of GE81112 may play a role in only a subset of bacterial species, because in T. thermophilus the C-terminal tail of S13 (126 aa) is extended relative to that in Escherichia coli (118 aa). As seen in Fig. 1 C and E, although the ligand GE81112 and S13 can be putatively modeled into the electron density, it is not possible at the resolution of the map to distinguish the last C-terminal residues of S13 unambiguously from the four nonproteinogenic amino acid moieties of GE81112, and therefore alternative arrangements are possible. The limited possibilities for the drug to interact with the 16S rRNA in its pocket explain why the RNA-probing experiments to determine the GE81112 localization on the 30S subunit have been inconclusive (see Fig. S7 and ref. 6).
Fig. 2.
Overview of conformational changes observed in the 30S subunit in the presence of GE81112. (A) In the GE81112 ribosomal complex, several 16S rRNA residues assume alternative conformations. At the interface of h24a (light blue) and h45 (orange) residues U793, A1519, A1518, G1517, and G1516 are colored red and are drawn in the alternative conformation unique to the GE81112 structure. Similarly residues U81–A85 of the ASL are colored red and drawn as seen only in the presence of GE81112. Note that residues G1491–A1493, although not in an alternative conformation, show significant disorder. The crystallographic refinement indicates that the residues of the ASL, h24a, and h45 that are colored red have occupancies of about 60% and 40%, corresponding to the conformations seen in the presence and absence (PDB ID code 2ZM6) of GE81112. S13 was omitted, because it is not likely to be involved in GE81112 binding in all bacterial species. (B and C) Key residues at the interface of h44/h45/h24a are illustrated in the engaged (B) and disengaged (C) conformation. Although both conformations are observed in the presence of GE81112, the disengaged conformation is predominant in the presence of GE81112, and the engaged conformation is predominant in the apo30S subunits (PDB ID codes 2ZM6 and 1J5E).
Fig. S7.
Effect of GE81112 on the in situ accessibility of 16S rRNA to hydroxyl radical cleavage. Primer extension analysis of the 16S rRNA cleavage sites generated by hydroxyl radicals within the 30S subunit (9) in the presence of increasing concentrations of GE81112 (10–100 μM; increasing concentrations indicated with a triangle). (A and B) Analyses of the 16S rRNA residues 650–710 and 1,340–1,380, respectively. The G and A lanes contain the sequencing reactions. Bases whose accessibility to the cleavage is affected by GE81112 are indicated on the right side of the gels. (C) The positions of 16S rRNA bases becoming more (red) or less (blue) exposed to cleavage in the presence of GE81112 are indicated within the in the 3D 30S structure.
Taken together, these results indicate that upon binding to the 30S subunit GE81112 occupies a position corresponding to the P-site tRNA and stabilizes a conformational change of the ASL. In fact, GE81112 would overlap with the ASL, as observed in the apo30S, and the residues forming the ASL tip (U81–A85) are displaced relative to the position that they occupy in the native 30S crystals. In particular, the phosphate backbone of U82 and U83, which correspond to and hereafter are referred to as “C34” and “A35” of the initiator tRNA anticodon (Fig. S2A), are shifted by ∼9 Å (Fig. 1B). C34 and A35 decode the third and second positions of the AUG start codon, and their displacement, as seen in the GE81112 ribosomal complex, would preclude them from forming a correct codon–anticodon interaction with the mRNA (Fig. S2B). This significant conformational change of the ASL tip is allowed because the ribosome does not monitor or interact directly with the P-site tRNA anticodon aside from a stacking interaction between the third position of the codon–anticodon duplex and C1400/G966 of 16S rRNA (17). The conformation of the remaining residues of the ASL mimic that contact the 30S subunit near h24a (A790) and h42 (1338–1339) (Fig. 2A) is changed only slightly in the presence of GE81112.
The Presence of GE81112 and Absence of P-Site Codon–Anticodon Interaction Stabilizes the h44/h45/h24a Interface in the Disengaged Conformation.
The electron density map of the 30S⋅GE81112 complex reveals that, in addition to the effects on the structure of the ASL mimic and the consequent impairment in the formation of a codon–anticodon interaction in the P site, the interface of h44, h45, and h24a also undergoes a substantial conformational switch (Fig. 2). Here the highly conserved GGAA tetraloop of h45 (G1516–A1519) is observed in two alternative conformations: a weaker engaged conformation (Fig. 2B) characterized by a potential hydrogen bond network between h45 (G1517–A1519) and h44 (C1496–1497) and a predominant disengaged conformation (Fig. 2C) in which the h45 loop withdraws from h44, disrupting the hydrogen bond network (compare Fig. 2 B and C). The withdrawal of the h45 loop is brought about by a conformational change that shifts G1517 and A1518 by 4.5 and 4.7 Å, respectively, disrupting the internal hydrogen bonds that close the GGAA tetraloop. The shift of A1518 brings the dimethylated N6 of this base into the proximity of U793 (h24a) so that the latter base is shifted (Fig. 2 B and C), in turn causing a slight perturbation of the positions of the neighboring bases A792 and A794. These conformational changes are consistent with DMS probing experiments (6) that showed that the reactivity of bases in h24a (A792, C795, and C796) is altered by GE81112 binding.
GE81112 Inhibits the Formation of a Locked 30SIC.
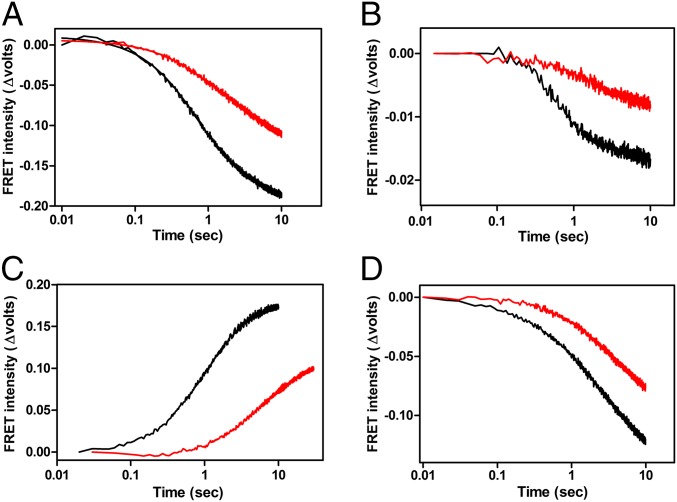
To understand how the structural changes described above play a role in the translational initiation pathway, we used fluorescence stopped-flow analysis and rapid nitrocellulose filtration to quantify the effect of GE81112 on the kinetics of fMet–tRNA binding to 30S ribosomal subunits. In the first approach the increase in the FRET signal generated by the proximity of fMet–tRNA8-fluo (acting as donor) and IF3166-ALEXA555 (acting as acceptor) is recorded as a function of the time elapsed after mixing the fluorescent initiator tRNA with mRNA-programmed 30S subunits bearing IF1, IF2⋅GTP, and fluorescent IF3. No effect of the antibiotic on the fMet–tRNA binding kinetics was detected in these experiments, even at a very high antibiotic concentration (100 µM), because superimposable tracings were obtained in the presence or absence of GE81112 (Fig. 3A). Similar results were obtained when the antibiotic was preincubated with the 30S subunits or offered to the subunits together with fMet–tRNA. Furthermore, no GE81112 inhibition of the ribosomal binding of fMet–tRNA could be seen when the binding kinetics was monitored using IF14-ALEXA555 as the FRET acceptor (Fig. S3A) or by the quenching of the IF2757-ALEXA488 fluorescence by the approaching fMet–tRNA8-QSY35 (Fig. S3B). Because these experiments monitor the initial binding of fMet–tRNA to mRNA-programmed 30S ribosomal subunits to form a 30S preIC, it can be concluded that the antibiotic has no effect on this early step of the translation initiation pathway. However, when the binding kinetics were analyzed by rapid filtration, a method that detects the formation of a “locked” 30SIC produced by a first-order isomerization of the 30S preIC (12, 13), a clear inhibition of fMet–tRNA binding by even low (1.5 μM) concentrations of GE81112 was observed (Fig. 3B). These results indicate that GE81112 interferes with the isomerization of the 30S complex that leads to a stable interaction between mRNA and initiator tRNA and marks the transition from the 30S preIC to the 30SIC (Fig. 3C).
Fig. 3.
Inhibition of 30SIC and 70SIC formation by GE81112. (A) Kinetics of 30S preIC formation in the presence (red tracing) and absence (black tracing) of 100 µM GE81112. The ribosomal binding kinetics of fMet–tRNA were followed with a fluorescence stop-flow apparatus making observable the FRET signal generated by the proximity of fMet–tRNA8-fluo, acting as a donor to IF3166-ALEXA555. (B) Kinetics of locked 30SIC formation in the absence (red trace) or in the presence (black trace) of 1.5 µM GE81112. Binding of f[35S]Met-tRNA to 022 mRNA-programmed 30S subunits to form a 30SIC were measured by rapid nitrocellulose filter binding as described (29). (C) Schematic representation of the 30S preIC complex→30SIC transition. In the 30S preIC complex both mRNA and initiator tRNA are ribosome-bound, but no proper codon–anticodon interaction takes place. A subsequent locking step (inhibited by GE81112) entails P-site codon–anticodon base-pairing that stabilizes the mRNA–initiator tRNA interaction yielding a 30SIC. (D) Stopped-flow kinetics of a 50S subunit docking to 30SIC to produce a 70SIC in the absence (black tracing) or presence (red tracing) of 100 µM GE81112. The association of the 50S subunits with a 30SIC was monitored using the variation in the light-scattering signal as an observable. a.u., arbitrary units.
Fig. S3.
GE81112 does not affect the kinetics of 30S preIC formation. Binding kinetics of fMet–tRNA to the 30S ribosomal subunit in the absence (black tracing) or presence (red tracing) of 100 µM GE81112 measured by a fluorescence stop-flow apparatus making use of two types of observables, namely the FRET signals generated by the proximity of fMet–tRNA8fluo acting as a donor and IF1Alexa555 as acceptor (A) and the quenching of IF2757Alexa488 fluorescence by the approaching fMet–tRNA8QSY35 (B).
This locking step is under the kinetic control of IF3 and IF1 and depends upon the nature of the 30S ligands (canonical vs. noncanonical) (12, 24). Therefore, experiments were carried out to investigate if the inhibition by GE81112 might be accompanied by an altered interaction of the IFs with the ribosome and to determine if the inhibition of 30SIC formation might depend upon the nature of the 30S ligands, as is the case for the other powerful P-site inhibitor Furvina, whose inhibition displays initiation codon bias (9).
The results of these experiments demonstrated that GE81112 inhibits 30SIC formation (Fig. S4A) (25, 26) and mRNA translation (Fig. S4B) to the same extent, regardless of the canonical or noncanonical nature of the start codon (AUG and AUU, respectively) and the presence or absence of a 5′ UTR (leadered and leaderless mRNA, respectively). Furthermore, the formation of a “pseudoinitiation complex” in which the fMet–tRNA analog NAc–Phe–tRNA is bound to poly(U)-programmed 30S ribosomal subunits (27) was found to be inhibited by GE81112, albeit at an antibiotic concentration ∼60-fold higher than that necessary to inhibit fMet–tRNA binding to the same extent (Fig. S4C). Thus, the inhibition by GE81112 is neither codon nor amino acid–tRNA specific. Taken together, these results indicate that GE81112 and Furvina inhibit P-site binding of initiator tRNA by different mechanisms and that the target of GE81112 inhibition is not a particular ribosomal ligand but instead is the molecular mechanism that underlies the locking step allowing the 30S preIC→30SIC transition.
Fig. S4.
Inhibition of translation initiation by GE81112 does not depend on the nature of the initiation codon or the nature of the aminoacyl-tRNA and template. (A) GE81112 concentration-dependent inhibition of [35S]fMet–tRNA binding to 30S ribosomal subunits programmed with 022AUGmRNA (■) or 022AUU mRNA(●) measured by filtration through nitrocellulose filters as described previously (25). (B) In vitro translation of an E. coli cell-free system programmed with 022AUGmRNA (■) or 022AUUmRNA (●). (C) Binding of NAc[3H]–Phe–tRNA to poly(U)-programmed 30S ribosomal subunits to form a pseudoinitiation complex. The reaction was carried out at 5 mM (●) or 15 mM (■) of magnesium acetate (26).
GE81112 Stabilizes an Altered Ribosomal Positioning of IFs, fMet–tRNA, and mRNA.
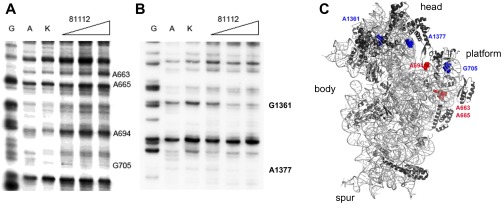
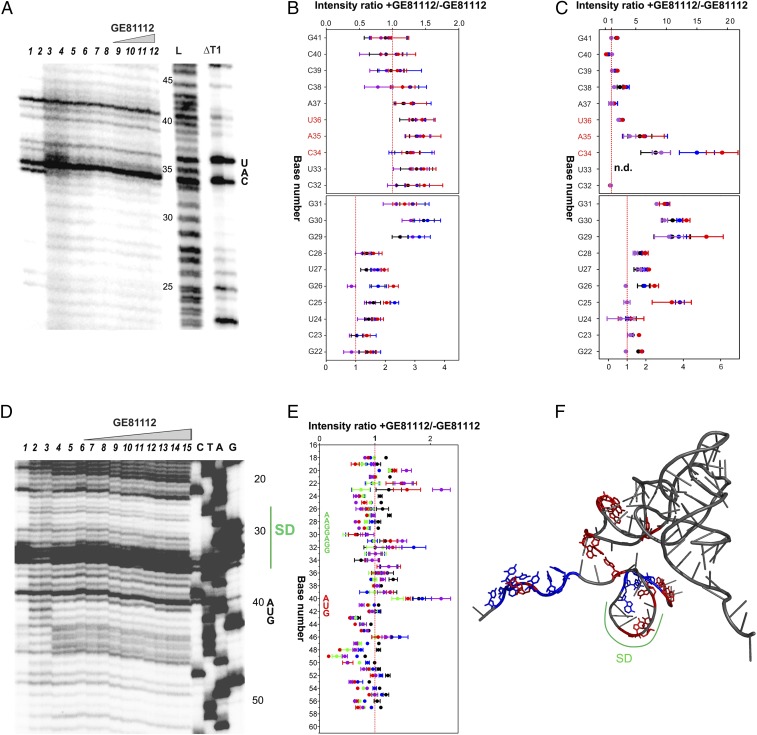
Because the structural results indicated that GE81112 stabilizes the ASL of the P-site substrate in an altered conformation, we tested the in situ accessibility of fMet–tRNA and mRNA to hydroxyl radical cleavage within the 30SIC in the presence and absence of the antibiotic. In the absence of GE81112 the cleavage pattern of 30SIC-bound fMet–tRNA displays typical features of ribosome-bound tRNA. In particular, bases G29, G30, and G31 are strongly protected from cleavage (Fig. 4A, lanes 7 and 8, and Fig. 4 B and C). These protections presumably are caused by the extensive interactions occurring between the consecutive GC pairs of the ASL and the universally conserved nucleotides G1338 and A1339 of 16S rRNA that line one side of the P site (17, 28). Additional interactions that may contribute to the observed protection are those involving the sugar–phosphate backbone of nucleotides A1229–C1230 of 16S rRNA and possibly the C-terminal tail of S13 (17, 28). However, these protections are partially lost in the complex formed in the absence of mRNA (Fig. 4A, lanes 5 and 6) and in the presence of increasing amounts of GE81112 (Fig. 4A, lanes 9–12), as evidenced by the ratios of intensities of the electrophoretically resolved bands (Fig. 4B). In comparison with these effects, the changes in accessibility induced by the antibiotic in the anticodon region are more difficult to detect because of the intrinsic susceptibility of this portion of the tRNA molecule to spontaneous cleavages. Thus, although electrophoretically purified, intact tRNA was used in these experiments, the anticodon loop and other single-stranded regions of the tRNA underwent spontaneous hydrolysis, and fragments derived from cleaved tRNA were again produced (Fig. 4A, lanes 1 and 2). The repeated, harsh phenol-chloroform-SDS extractions necessary to remove the ribosomal proteins that could interfere with the subsequent electrophoretic separation also worsen the situation. Nevertheless, although bands generated by these spontaneous cleavages contribute to increasing the background of the bands produced by hydroxyl radical cleavage and partly mask the effects of the antibiotic, a GE81112-induced increased exposure of the anticodon region is visible upon inspection of the gel (Fig. 4A, compare lanes 7 and 8 with lanes 9–12). The effects caused by GE81112 are even more clear when the ratios of the band intensities observed in the presence and absence of the antibiotic are determined. Fig. 4 B and C displays plots of the ratios of the intensities of the individual bands before (Fig. 4B) and after (Fig. 4C) subtraction of the band intensities of lanes 1 and 2 that are assumed to represent a background insofar as they indicate the approximate extent of the spontaneous cleavages occurring in the tRNA used to form the complexes. In conclusion, these experiments give a clear indication that the bases of the anticodon loop, C34 and A35 in particular, become more exposed in the presence of GE81112, in full agreement with the aforementioned effects observed by crystallography (Fig. 2A).
Fig. 4.
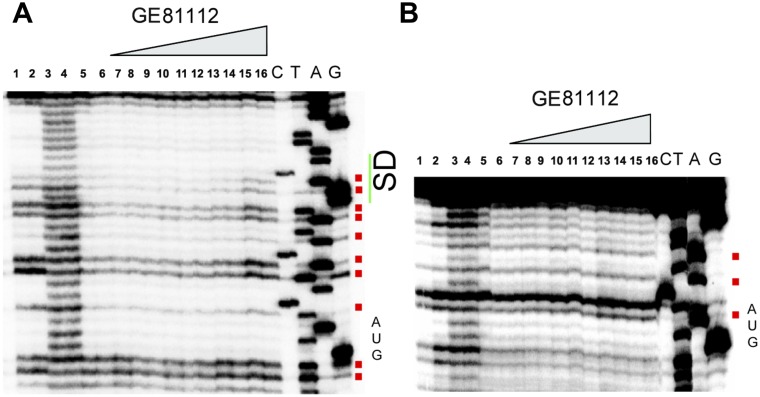
Accessibility to hydroxyl radical cleavage of fMet–tRNA in the 30S complexes formed with and without GE81112. (A) Electrophoretic migration of uncleaved (lanes 1 and 2) or hydroxyl radical-cleaved (lanes 3–12) [32P]fMet–tRNA in the unbound form (lanes 3 and 4), within a 30S complex lacking mRNA (lanes 5 and 6), within a complete 30SIC without GE81112 (lanes 7 and 8), or within a complete 30SIC with increasing amounts (i.e., 1.5, 12, 25, and 50 µM) of GE81112 (lanes 9–12). The L and ΔT1 lanes correspond to the ladders obtained upon alkaline hydrolysis and RNaseT1 digestion of tRNA, respectively. The intensity of the bands obtained upon cleavage in the absence (lanes 7 and 8) or in the presence of 1.5 (black), 12 (blue), 25 (red), and 50 µM (purple) of GE81112 (lanes 9–12) were quantified by densitometry. (B and C) The ratios of the intensities obtained with and without GE81112 are plotted; the anticodon bases are indicated in red. The ratios were calculated using the absolute intensity values (B) or the intensities after subtraction of the averaged intensities of the bands of lanes 1 and 2 considered to represent the spontaneously cleaved background (C). The intensities ratio of U33 could not be calculated. Because of the different extents of the recorded effects, the scales of the ordinate for bases G41–C32 and for G31–G22 are different in both plots. (D) Primer extension analysis of the 002 mRNA sites cleaved by hydroxyl radicals in the presence and absence of GE81112. Uncleaved mRNA (lane 1), mRNA cleaved in the unbound form (lanes 2 and 3) and with a complete 30SIC assembled in the absence (lanes 4 and 5) or in the presence of 0.5 (lanes 6 and 7), 1.5 (lanes 8 and 9), 12 (lanes 10 and 11), 25 (lanes 12 and 13), and 50 µM (lanes 14 and 15) GE81112. Lanes C, T, A, and G are the sequencing ladders. (E) Plot of the ratio of the densitometrically quantified, averaged intensities of bands 6,7/4,5 (black); 8,9/4,5 (blue); 10,11/4,5 (green); 12,13/4,5 (red), and 14,15/4,5 (purple). The SD sequence (green) and the initiation codon AUG (red) are indicated. (F) 3D structure of fMet–tRNA–mRNA interaction (PDB ID code 1YL4) highlighting the tRNA and mRNA regions that are more (red) or less (blue) exposed to cleavage in the presence of GE81112. Further details are given in Materials and Methods.
30S-mRNA Interaction.
GE81112 was shown to have only a marginal effect on the binding kinetics of either 5′ leadered (Fig. S5C) or leaderless (Fig. S5D) mRNA to the 30S subunit. However, the antibiotic had profound effects on the accessibility of 30S-bound mRNA to hydroxyl radical cleavage. Both protection and exposure in the presence of GE81112 were observed with 002 mRNA (Fig. 4 D and E), 003 mRNA (Fig. S6A), and 022 mRNA (Fig. S6B), three model mRNAs that differ in the length of the SD sequence and spacer (29). The cleavage pattern indicates that the bases of the SD region are more exposed when GE81112 is present (Fig. 4 D and E and Fig. S6A). Furthermore, GE81112 makes the first base of the start codon of 002 mRNA (Fig. 4 D and E) and 003 mRNA (Fig. S6B) more accessible. The effect on the start codon of 022 mRNA can be hardly detected, because this mRNA region is strongly protected by the 30S subunit under all conditions (Fig. S6A). In addition to the aforementioned increased exposures, some regions of the mRNAs, both upstream and downstream of the initiation triplet, become more protected in the presence of GE81112 (Fig. 4 D and E). Understandably, the cleavage pattern in the 5′ region upstream of the start codon is different in the three different mRNAs and reflects the different natures and strengths of their interaction with the 30S subunit, possibly resulting from the different lengths of their SD sequences (29, 30).
Fig. S5.
Effect of GE81112 on the rate of exchange between 30S-bound and free IF1 and IF2 and on the kinetics of mRNA binding to the 30S subunit. (A and B) Two types of complexes were prepared in the presence (red tracings) and absence (black tracings) of GE81112. One complex contained fluorescently labeled IF1 4-ALEXA555 and fluorescein-labeled fMet–tRNA; IF1 dissociation is monitored by the reduction of the FRET signal between the two fluorescent ligands upon the addition of a 10-fold excess of nonfluorescent IF1 (A). The other complex contained fluorescently labeled IF2 757-Alexa488 and fMet–RNA8-QSY35 (B). Upon the addition of a 10-fold excess of nonfluorescent factor, the dissociation of IF1 (A) causes a decrease in fluorescence intensity (because of the loss of the FRET signal), whereas IF2 dissociation (B) causes an increase in fluorescence because of diminished quenching. (C and D) Fluorescence stopped-flow binding kinetics of 3′ fluorescein semithiocarbazide-labeled (24) leadered (C) and leaderless (D) mRNA to 30S subunits (black and red tracings) or to 30S subunits containing all components of the initiation complex except for the mRNA (green and blue tracings) in the absence (black and green tracings) or in the presence (red and blue tracings) of 100 µM GE81112.
Fig. S6.
GE81112 alters the mRNA position on the 30S subunit. Primer extension analysis of the 022 mRNA (A) and 003 mRNA (B) sites cleaved by hydroxyl radicals. Uncleaved mRNA (lanes 1 and 2), mRNA cleaved in the unbound form (lanes 3 and 4) and within a complete 30SIC assembled in the absence (lanes 5 and 6) or in the presence of 0.5 µM (lanes 7 and 8), 1.5 µM (lanes 9 and 10), 12 µM (lanes 11 and 12), 25 µM (lanes 13 and14), and 50 µM (lanes 15 and 16) of GE81112. Lanes C, T, A, and G are the sequencing ladders. Bases becoming more exposed in the presence of the antibiotic are indicated by red squares. The positions of the initiation triplet AUG and of the SD sequence (in the case of 022 mRNA) are indicated on the right side of the gels.
Overall, these findings indicate that although fMet–tRNA and mRNA are also ribosome-bound in the presence of GE81112, they occupy noncanonical positions on the 30S subunit so that the interactions between the fMet–tRNA and P site, between the mRNA and 30S subunit, and between the fMet–tRNA and mRNA start codon are either lost or weakened in the presence of the antibiotic (Fig. 4F). This conclusion is confirmed by the finding that IF3 is more tightly bound to the 30S complex containing GE81112, because it is exchanged more slowly with the unbound factor (Fig. 5A). Indeed, although IF3 does not dissociate from the 30S subunit until the docking of the 50S is complete, formation of a canonical 30SIC weakens its affinity for the 30S subunit, but the affinity remains high when the complex formed is not canonical (24, 31). In contrast to IF3, neither IF1 nor IF2 is more tightly bound to the 30S subunit in the presence of GE81112 (Fig. S5 A and B). Additional evidence for the noncanonical structure of the 30S complex formed in the presence of GE81112 is its slower docking with the 50S ribosomal subunits (Fig. 3D) and the slower dissociation of the three IFs during the 30SIC→70SIC transition; the rates of IF1 (Fig. 5B) and IF3 (Fig. 5D) dissociation are reduced approximately two-fold, whereas the rate of IF2 dissociation is reduced approximately six-fold (Fig. 5C).
Fig. 5.
Ribosomal affinity and dissociation of the IFs in the presence of GE81112. (A) Rate of exchange between fluorescent 30S-bound and nonfluorescent free IF3 in the presence (red tracing) and absence (black tracing) of GE81112. The dissociation of IF3 was determined from the rate of exchange measured by a fluorescence stopped-flow apparatus monitoring the decrease of fluorescence intensity of IF3166-ALEXA555 upon the addition of a 10-fold excess of nonfluorescent factor. (B–D) Dissociation of the IFs upon association of 30SIC with 50S subunits during the 30SIC→70SIC transition in the presence (red tracing) and absence (black tracing) of GE81112. The dissociation of IF1 (B) and IF3 (D) from 30SIC complexes containing an Alexa 555-labeled factor, and fluorescein-labeled fMet–tRNA was monitored by the decrease in the FRET signal between the two fluorophores. (C) The dissociation of IF2 was monitored by an increase in the fluorescence resulting from the reduced quenching caused by the loss of proximity between IF2757-ALEXA488 and fMet–RNA8-QSY35.
The in situ hydroxyl radical cleavages of 16S rRNA are consistent with the premise that GE81112 affects the conformation of the 30S subunit. The results of these experiments indicate that GE81112 has generalized effects on the head and platform of the 30S subunit (Fig. S7) that indicate extensive GE81112-induced conformational changes in the subunit.
Discussion
Together with Furvina, GE81112 probably is the most effective and specific inhibitor of the bacterial initiation phase of protein synthesis known thus far (6, 9). Because previous studies have shown that GE81112 interferes with the P-site function of the 30S ribosomal subunit, in this study we examined in more depth the mechanism by which this inhibition takes place using a biochemical and structural approach. The data presented here demonstrate that GE81112 does not inhibit the initial binding of fMet–tRNA and mRNA to the 30S ribosomal subunit that yields a 30S preIC; namely, it allows the formation of a complex in which both ligands are ribosome-bound but do not interact stably with each other (12). However, the antibiotic interferes with the subsequent step, namely the first-order isomerization of the 30S complex that marks the transition of 30S preIC to 30SIC (12). Thus, it seems clear that the mechanism of action of GE81112 is different from that of Furvina, which inhibits the initial binding of fMet–tRNA that yields a 30S preIC complex (9). Furthermore, unlike Furvina (9), inhibition by GE81112 is not biased by the nature of the initiation codon, tRNA or mRNA.
The present data indicate that a 30S complex formed in the presence of GE81112 is characterized by a “non–best-fit” structure, as shown by IF3 remaining more tightly bound and the complex being docked more slowly by the 50S subunit to form a 70SIC with a corresponding decrease in the rate at which all three IFs are dissociated during this process. At the molecular level, it was observed that GE81112 binds into a pocket within the P site near the tip of the ASL mimic, stabilizing residues corresponding to C34 and A35 of initiator tRNA in an altered conformation, away from the canonical codon position. Thus, it is possible to conclude that the primary mechanism by which GE81112 inhibits 30SIC formation is through the stabilization of a distorted conformation of the anticodon loop of the initiator tRNA so that it loses, at least partially, the possibility to form a canonical codon–anticodon interaction with the AUG start codon (Fig. 1B and Fig. S2). The latter premise is supported by the results of in situ probing the accessibility of fMet–tRNA and mRNA to hydroxyl radical cleavage (Fig. 4). Aside from this effect on codon–anticodon pairing and on the accessibility of one side of the anticodon stem (Fig. 4 A–C), the remaining interactions of the tRNA with the ribosomal P site are mostly unaffected by the drug (Fig. 2A and Fig. S8A), explaining why, in the presence of GE81112, fMet–tRNA can bind to the 30S subunit and form a 30S preIC (Fig. 3A) and why the tRNA cleavage pattern displays the typical features of ribosome-bound tRNA (Fig. 4A).
Fig. S8.
Comparison of h44/h45/h24a in the engaged and disengaged conformation. The overall structure of the P-site region encompassing h44 (green), h45 (orange), h24a (cyan), the ASL (blue), and the mRNA (yellow) acquires a different conformation in the presence (A) or absence (B and C) of GE81112. The drug stabilizes the ASL tip in an altered conformation so that the mRNA cannot be positioned properly, and P-site codon–anticodon interaction is hampered. The architecture of the adjacent helices h44, h45, and h24a is altered, reflecting the formation of a different hydrogen bonding network that in the absence of GE8112 involves the mRNA and two additional Mg2+ ions (yellow spheres in B and C). In the two complexes, the h44/h45/h24a conformation is either disengaged (A) or engaged (B and C) (also see also Fig. 2 B and C). (C) A close-up and rotated view of B illustrating the potential hydrogen bond network formed by the tip of h44, the mRNA codon, and the ASL. This network could contribute to the stability of the h44 residues that are part of the h44/h45/h24a interface. The mRNA sequence is 5′-AGAAAGGAGGGUUUGGAAUGAACGAGC-3′. The residues most affected by the presence of GE81112 are colored in red (rms value higher than 2 Å) or pink (1 ≤ rmsd < 2 Å).
In addition to the distortion of the ASL tip, the GE81112 complex shows conformational changes of the ribosomal subunit (Fig. S7), the most relevant of which involves the highly conserved GGAA tetraloop of h45 (G1516–A1519). In this region of the 16S rRNA GE81112 favors the disengaged over the engaged configuration of the h44/h45/h24a interface (Fig. 2). Previous studies have shown that in the apo30S subunit the h44/h45/h24a interface is flexible and can exist in two alternative conformations, i.e., engaged and disengaged (32). Switching between these two conformations alters the hydrogen bonding network between h24a, h45, and h44 (Fig. 2 B and C), and minimal alterations in the elements present in this region of the ribosome can stabilize one form or the other. Indeed, transitions between the engaged and disengaged conformations of h44/h45/h24a have been observed in the crystal structure of the 30S-lacking dimethyl groups on A1518 and A1519 (i.e., a ksgA− mutant) and, more recently, in the presence of aminoglycoside antibiotics, near-cognate A-site mimics and rRNA mutations in fully methylated 30S subunits (18, 32, 33). On the other hand, in the 70S ribosome h44/h45/h24a elements take part in the formation of the intersubunit bridges B2a, B2b, and B3 and, unlike in the 30S subunit, only the engaged conformation has been observed on the 70S (Fig. S9) (34–39).
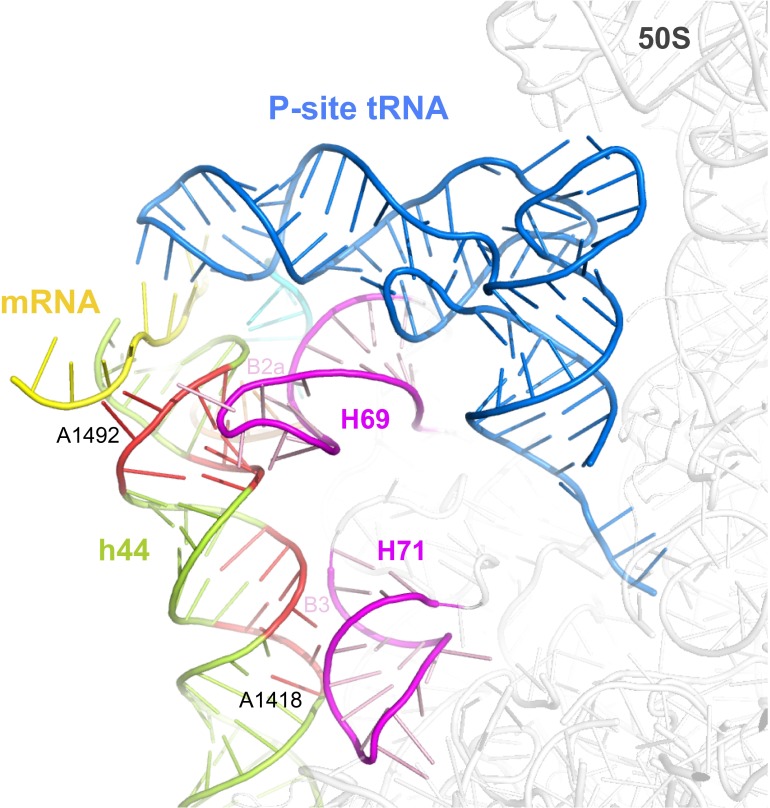
Fig. S9.
The h44 nucleotides affected by GE81112 take part in the formation of bridges B2a and B3. Representative structure of the subunit interface of the 70S ribosome [PDB ID codes 2J00 (30S, including tRNAs and mRNA) and 2J01 (50S)] is shown in cartoon representation; the mRNA is colored in yellow, the P-site tRNA in blue, H69 and H79 of the 50S subunit in purple, h44 of the 30S subunit in green, and its nucleotides (1406–1409, 1491–1495, and 1417–1419) affected by GE81112 and involved in the switch to the disengaged conformation are in red. These nucleotides are either close to or directly involved in the formation of two intersubunit bridges (B2a and B3) that control and stabilize the interaction between the 30S and 50S subunits. In particular, the conformation of the h44 nucleotides (1409–1410 and 1495–1496) involved in the formation of B2a may have a significant role in the formation of a stable 70SIC, because the tip of H69 on the 50S, which participates in the formation of this bridge, is completely disordered in the free 50S subunit (e.g., PDB ID codes 3CC2 and 2ZJR) (34, 35), but it acquires a stable conformation by induced fit upon binding to the 30S subunit and interaction with h44. In the disengaged conformation, h44 in this region has the highest conformational distortion (see also Fig. 2 and Fig. S8), suggesting that this conformation in the 30SIC would hamper the formation of the bridge B2a, but the formation of this bridge would be favored by the engaged conformation, which is the only one observed in the 70S ribosome, and on the 30S in the presence of a proper accommodation of the P-site ligands. H69 also interacts with the P-site tRNA (blue), further suggesting that in the presence of an incorrect positioning of the mRNA and nonproper codon–anticodon interaction in the P site, the stability of the P-site tRNA would be compromised and possibly explaining why, under these conditions, the fMet tRNA dissociates from the complex. This hypothesis finds support from fMet–tRNA binding experiments in the presence of streptomycin, which induces the disengaged conformation of h44/h45/h24a and causes the release of fMet–tRNA from the 70S ribosome (36–39). Similarly, in the absence of methylation of 1518 and 1519, which also causes the stabilization of the disengaged conformation, the association between 30S and 50S is weakened (39).
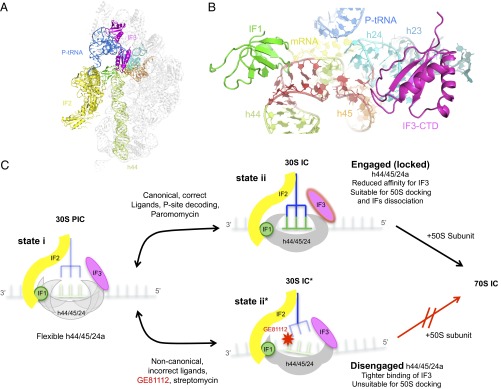
Comparison of the h44/h45/h24a interface conformation in the presence and absence of GE81112 suggests that although the correct P-site positioning of mRNA and initiator tRNA favors the formation of the engaged conformation, the noncanonical positioning of these ligands, or their absence, stabilizes the alternative disengaged conformation of h44/h45/h24a (Fig. 2 and Fig. S8). This finding and the fact that the primary target of GE81112 inhibition is the 30S preIC→30SIC transition suggest that the first-order isomerization that underlies this transition entails a shift of the h44/h45/h24a interface from the disengaged to the engaged conformation. The present data also suggest that the conformational transition of the h44/h45/h24a interface represents a molecular device for monitoring the P-site decoding function whereby the correct accommodation of fMet–tRNA and mRNA in the P site is sensed by the acquisition of the engaged conformation in the h44/h45/h24a interface and is transmitted to the subsequent steps of the initiation phase (Figs. 2 and 6 and Fig. S8).
Fig. 6.
Structural model on the initial phase of translation initiation and the effect of GE81112 during the transition of a preIC to a 30SIC. (A) Overview on the interaction of the three IFs on the 30S subunit as observed in the cryo-EM structure of a 30S in complex with IF1 (green), IF2 (yellow), IF3 (magenta), P-tRNA (blue), and mRNA (60). (B) Zoom-in on the binding site of IF1 and IF3, near helices h44 (light green), h23 (pale blue), h24a (cyan), and h45 (orange) of 16S rRNA; the residues that acquire an alternative conformation in the presence of GE81112 are in red. (C) Scheme illustrating the molecular events suggested by the findings of the present study. (Left) A 30S preIC containing the three IFs and a not yet base-paired initiator tRNA and mRNA bears a flexible h44/h45/h24a interface. (Upper Center) If tRNA and mRNA are canonical, P-site decoding occurs to yield a bona fide 30SIC in which the h44/h45/h24a interface is engaged (locked). (Right) The affinity of this complex for IF3 is reduced, and the complex is amenable for docking by the 50S subunit and for undergoing 30SIC→70SIC transition with concomitant dissociation of the three IFs. (Lower Center) If the ligands in the 30S preIC are noncanonical or if GE81112 (or streptomycin) is present, a structurally faulty complex 30SIC* is formed in which proper codon–anticodon interaction does not occur. This complex contains a disengaged h44/h45/h24a structure, binds IF3 more tightly, and is unsuitable for docking with 50S subunits. Further details can be found in the text.
Furthermore, the available data suggest that the conformational switch of h44/h45/h24a also could be monitored by IF3 in its fidelity function. In particular, we suggest that the switch in IF3 affinity for the 30S subunit that marks the 30S preIC→30SIC transition may depend upon the h44/h45/h24a conformation. Indeed, compared with the canonical 30SIC, IF3 displayed a tighter binding to the faulty 30S complex formed in the presence of GE81112 in which the h44/h45/h24a interface is predominantly in the disengaged conformation; its affinity for the 30S subunit is somewhat lower in the canonical 30SIC (24, 31). In this context it should be recalled that previous structural and biochemical studies indicate that there is a physical proximity between the IF3 binding site and the h44/h45/h24a interface and that the IF affects the structure of this region. In fact, binding of IF3 disrupts a crosslink between U793 (h24a) and G1517 (h45) (40), two bases that, together with A1518, are among the residues with highest conformational variability during the disengaged→engaged transition (Fig. 2 and Fig. S8). Furthermore, 16S rRNA probing using hydroxyl radicals (41), 1-cyclohexyl-(2-morpholinoethyl) carbodiimide metho-p-toluenesulfonate (CMCT) (42), and kethoxal (43) showed that IF3, in addition to protecting G700 and G703 (in h23b) completely, also protects G791 and U793 in h24a. Likewise, binding of IF3C, the domain of the factor capable of performing all the activities of intact IF3 (44), protects bases in h23b and exposes other h24a bases (e.g., 783, 784, 790, and 794) flanking h45 (45). Finally, the point mutations A790G, U789C, and G791A in h24a were found to alter both the ribosomal affinity and fidelity functions of IF3 (46, 47).
In conclusion, in light of the present findings and of a large body of published data, we propose the mechanism schematically presented in Fig. 6C. Namely in the 30S preIC the disengaged and engaged conformations of the h44/h45/h24a interface are in a dynamic equilibrium, so that in the absence of P-site codon–anticodon interaction, as observed in the 30S–GE81112 complex, the disengaged form prevails. In contrast, upon proper accommodation of mRNA and P-site tRNA, h44/h45/h24a instead adopts the engaged conformation. IF3 and IF1 favor the formation of the disengaged conformation, unless the h44/h45/h24a conformation is switched to the engaged conformation by the presence of canonical 30S ligands and proper P-site codon–anticodon interaction. In turn, this switch results in the weakening of the 30S–IF3 interaction and in a best-fit docking of the 50S subunit.
Materials and Methods
Preparation of mRNAs and Translational Tests.
The 022, 003, and 002 mRNAs (48) were prepared by in vitro transcription with bacteriophage T7 RNA polymerase of a DNA template derived from the amplification of the corresponding E. coli coding genes cloned in pTZ18R using primers that anneal to nucleotides 143–160 (forward primer 5′-GCTTCCGGCTCGTATGTTGTGTG-3′) and 297–319 (reverse primer 5′-GTAAAACGACGGCCAGT-3′) of the vector (25, 29). Translational tests were carried out as described (25, 29) using E. coli ribosomes, ribosomal subunits, initiator tRNA, and translational factors prepared as described (49).
Fluorescence-Labeling of IF1, IF2, IF3, and tRNAfMet.
E. coli IF1, IF2, and IF3 genetically modified to introduce single cysteines were labeled essentially as previously described (49). Reducing agents (i.e., 2-mercaptoethanol or DTT) were removed by extensive dialysis at 4 °C against 50 mM Tris⋅HCl (pH 7.1) buffer containing 100 mM NH4Cl and 0.1 mM EDTA. Disulfide bond formation was prevented by 10-min treatment at 37 °C with a 10-fold molar excess of Tris (2-carboxyethyl) phosphine hydrochloride (Sigma), a reducing agent that does not compete with thiol modifications. Labeling was carried out with a 20-fold molar excess of Alexa 488 or Alexa 555 (Invitrogen) as appropriate. E. coli initiator tRNAfMet [65 A260 units/mL of 12 mM Hepes-KOH (pH 8.2), containing 80% DMSO] was labeled at the thio-uridine in position 8 by 2-h incubation at 50 °C in the dark with 3.5 mM 50-IAF (iodoacetamide derivatives of fluorescein). Labeling efficiency approached 100%. The reaction was stopped by the addition of 0.3 M K-acetate (pH 5.0) and the labeled tRNAfMet was precipitated with 2.5 volumes of cold ethanol. For other experiments tRNAfMet was labeled with the fluorescence quencher QSY-35 as described (49).
Kinetics of IF Dissociation from 30SIC.
All experiments were carried out at 20 °C in a KinTek SF-2004 stopped-flow apparatus in 10 mM Tris⋅HCl (pH 7.7), 60 mM NH4Cl, 7 mM Mg acetate. The 30SIC complex was allowed to assemble at 37 °C for 10 min in the presence or absence of GE81112. The 30SIC in syringe A contained 0.2 μM 30S subunits, 0.4 μM 022 mRNA, 0.5 mM GTP, 0.2 μM each of IF1, IF2, and IF3, one of which was fluorescently labeled (IF1ALEXA555, IF3ALEXA555, or IF2ALEXA488), as specified in each case, and 0.4 μM fMet–tRNAfluo. The kinetics of IF dissociation from 30S subunits was monitored by promoting an exchange between fluorescent ribosome-bound and free nonfluorescent factors. For these experiments syringe A was loaded with the preformed 30SIC, and syringe B was filled with a single unlabeled IF (as specified) in a 10-fold stoichiometric excess with respect to the corresponding 30S-bound fluorescent IF. The kinetics of IF dissociation during formation of the 70SIC was monitored upon mixing aliquots of preformed 30SIC in syringe A with equal volumes of 0.6 μM 50S in syringe B. The reactions were followed for the times indicated in each case.
Light Scattering.
Equal volumes (20 μL) of 30SIC (0.2 μM) prepared as described above, with or without 100 µM of GE81112 (syringe A) or 0.6 μM 50S (syringe B) were allowed to mix at 20 °C in a stopped-flow apparatus. Changes in light scattering were recorded by exciting at 430 nm and measuring the scattered light at 90° with respect to the incident beam without a filter. In a single experiment, 1,000 data points were acquired in logarithmic sampling mode.
tRNAfMet 5′-End Dephosphorylation and Labeling with 32P γ-ATP.
The 5′ phosphate of tRNA was removed essentially as described (50) by mixing tRNA (∼50 µg) with 0.6 U/µL calf intestinal alkaline phosphatase (CIAP) in 1× CIAP buffer (Amersham). After 1-h incubation at 37 °C the dephosphorylated tRNA was subjected to 10% PAGE in 8 M urea; the electrophoretically resolved band corresponding to the tRNA identified by UV254 nm shadowing was excised, and the tRNA was eluted by shaking the gel slice overnight in 350 µL of elution buffer and 70 µL of phenol. The dephosphorylated tRNA then was precipitated with three volumes of ethanol, resuspended in H2O, quantified from its A260 nm, and stored at −80 °C. The dephosphorylated tRNAfMet and tRNAfMet-fluo (2 µg) were 5′-end–labeled in 1× polynucleotide kinase (PNK) buffer (Fermentas) with 15 units of PNK enzyme and 5 μL 32P γ-ATP 3000 Ci/mmol for 75 min at 37 °C. The reaction was stopped by the addition of 1/10 volume of 3 M Na acetate (pH 5.2) and three volumes of cold absolute ethanol.
32P tRNAfMet and tRNAfMet-fluo Charging and Formylation.
The labeled tRNAs were charged with methionine in 150 µL of 10 mM Mg acetate, 30 mM imidazole (pH 7.5), 6 mM 2-mercaptoethanol, and 100 mM KCl in the presence of 5 mM ATP, 5 mM phosphoenolpyruvate, 0.005 µg/μL methionine, 0.5 mM citrovorum factor, 0.06 µg/µL methionine transferase, and 0.03 µg/µL formyl methionine transferase. At the end of the incubation, the labeled fMet–tRNAs were subjected to phenol-chloroform extraction and precipitated for 1 h at −20 °C with 1/10 volume 3 M Na acetate and three volumes absolute ethanol. After centrifugation the pellets were resuspended in 50 µL of H2O.
Probing 32P-End–Labeled fMet–tRNA and 022 mRNA by Hydroxyl Radical Cleavage.
These experiments were carried out as described (51). 30SIC was allowed to assemble by incubation in 40 µL of 20 mM Na cacodylate (pH 7.7), 50 mM KCl, 10 mM MgCl2, 15 pmol 30S subunit, 30 pmol mRNA, 15 pmol each of IF1, IF2, and IF3, ∼25,000 cpm 32P-end–labeled fMet–tRNA, and the indicated amounts of GE81112. After 10 min at 37 °C, H2O2 was added (0.15% final concentration), and cleavage was started by adding Fe(II)-EDTA (3 mM final concentration). Cleavage was allowed to proceed for 45 s at 37 °C before the addition of 126 µL quenching solution containing 0.3 M Na acetate (pH 5.2) in absolute ethanol. The precipitated samples were resuspended in H2O, extracted with phenol-chloroform, and reprecipitated with cold 0.3 M Na acetate (pH 5.2) in absolute ethanol. The cleavage products then were resuspended in 5 µL 8 M urea containing loading dye and were analyzed on 10% PAGE in 8 M urea in parallel with an RNase T1-digested sample and an alkaline-digested ladder (51). The gel was dried, and the radioactivity therein was detected and quantified by a Bio-Rad model GS-250 molecular imager. Hydroxyl radical cleavage of 002 mRNA, 022 mRNA, and 003 mRNA was performed on 30SIC prepared by incubation in 50 µL of 20 mM Na cacodylate (pH 7.7) buffer, 50 mM KCl, 10 mM MgCl2, 40 pmol 30S subunit, 5 pmol mRNA, 40 pmol each of IF1, IF2, and IF3, 40 pmol fMet–tRNA, and the indicated amounts of GE81112. After 10 min incubation at 37 °C, H2O2 (0.075% final concentration) was added, and cleavage was started by adding Fe(II)-EDTA (3 mM final concentration). After 45 s at 37 °C, 155 µL of quenching solution containing 0.3 M Na acetate (pH 5.2) in absolute ethanol was added, and the precipitated samples were resuspended in H2O, extracted with phenol, phenol-chloroform, and chloroform, and finally precipitated with cold 0.3 M Na acetate (pH 5.2) in absolute ethanol. The precipitated reaction products, resuspended in 3 µL of sterile H2O, then were subjected to primer extension analysis at 42 °C for 45 min in 15 µL of reaction mixtures containing 0.33 mM dNTP, 0.13 units of avian myeloblastosis virus reverse transcriptase (Roche), and 0.1 pmol of 32P-labeled oligonucleotide no. 022: 5′-AGCTTGCATGCCTGC-3′, which anneals 38 nucleotides downstream of the initiation triplet of the mRNAs. The reactions products then were precipitated by the addition of 0.3 M Na acetate (pH 5.2) in absolute ethanol, resuspended in 8 M urea loading dye, and analyzed on 8% PAGE in 8 M urea in parallel with a dideoxy chain termination sequencing reaction (11). The gels were dried, and the radioactivity therein was detected and quantified by a Bio-Rad model GS-250 molecular imager.
Structure Determination by X-Ray Crystallography.
The 30S ribosomal subunit was purified from T. thermophilus HB8 (52) and crystallized (19, 52) in the presence of an RNA oligonucleotide (Dharmacon) with the sequence 5′-GAAAGGAGG-3′, which is complementary to the anti–Shine–Dalgarno sequence present at the 3′ end of the T. thermophilus 16S rRNA, as described previously (7, 53). Crystals were soaked overnight with 45 µM GE81112 and were flash-frozen in liquid nitrogen. Diffraction data were collected at the X06SA beamline of the Swiss Light Source and were processed with the XDS (54) and CCP4 (55) program packages. The native structure of the 30S subunit [Protein Data Bank (PDB) ID code: 2ZM6] was refined against the structure factor amplitudes of the 30S–GE81112 complex using the PHENIX program package (56). Conformational changes in the ribosomal binding pocket of GE81112 were detected by the difference Fourier technique where, specifically, we used a bulk solvent modeling protection approach as previously described (20, 21). Subsequently, GE81112 and the described conformational changes were modeled into the residual electron density using Coot (57), and the resulting 30S–GE81112 structure was further refined to convergence. For the calculation of the free R-factor, 5% of the data were omitted throughout refinement. Crystallographic data of the fully refined structure are shown in Table 1. Figures containing structural formula and 3D structures were illustrated with MarvinSketch (ChemAxon) and PyMOL (58), respectively. rRNA residues were numbered according to the E. coli scheme, and helices are indicated using standard nomenclature (59) throughout the article.
Table 1.
Data collection and refinement statistics
| Value | |
| Data collection | |
| Space group | P41212 |
| Cell dimensions | |
| a, b, c, Å | 413.14, 413.14, 173.63 |
| α, β, γ, ° | 90, 90, 90 |
| Resolution, Å | 160–3.50 (3.69–3.50) |
| Completeness, % | 99.9 (99.6) |
| Rsym, % | 16.5 (237.9) |
| I/σI | 9.79 (1.46) |
| Multiplicity | 11.3 (11.6) |
| Refinement | |
| Resolution | 3.5 |
| No. of reflections | 187167 |
| Rwork/Rfree | 16.9/21.7 |
| Total no. of atoms | 52,121 |
| Ribosome | 51,887 |
| Ligand | 46 |
| Ions | 188 |
| Rmsd | |
| Bond lengths, Å | 0.019 |
| Bond angles, ° | 0.92 |
The dataset was obtained from three crystals. Values in parentheses refer to the highest-resolution bin.
Acknowledgments
The GE81112 molecule used in this study was obtained from Biosearch Italia S.p.A. (a biotech company that no longer exists), where it was discovered, prepared, and purified. We thank our colleagues at RIKEN and the Center for Cooperative Research in Biosciences for their support of this study, in particular Chieko Naoe and Masahito Kawazoe of RIKEN for their participation in the preparation of ribosome crystals and Neha Dhimole and Shu Zhou for diffraction data collection. These studies could not have been performed without the expert assistance of the staff at the X06SA, BL-5A, ID29, and XALOC beamlines (Swiss Light Source, Photon Factory, European Synchrotron Radiation Facility, and ALBA respectively). Initial funding for this work was provided by European Commission Contract QLRT-2001-00892 ‘‘Ribosome inhibitors’’ (to C.O.G. and L.B.) and the Deutsche Forschungsgemeinschaft Grant FU579 1-3 (to P.F.). Subsequent work in Camerino was made possible by generous gifts of chemicals and fine biochemical precursors and other support from colleagues and friends around the world. Additional support was provided by Bizkaia:Talent and the European Union’s Seventh Framework Program (Marie Curie Actions; COFUND) (to S.R.C., A.S., and T.K.), Marie Curie Action Career Integration Grant PCIG14-GA-2013-632072 (to P.F.), and Ministerio de Economía Y Competitividad Grant CTQ2014-55907-R (to P.F. and S.R.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this article have been deposited in the Protein Data Bank (PDB) (ID code 5IWA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521156113/-/DCSupplemental.
References
- 1.Gale EF. The Molecular basis of Antibiotic Action. Wiley; London: 1981. [Google Scholar]
- 2.Brandi L, Fabbretti A, Pon CL, Dahlberg AE, Gualerzi CO. Initiation of protein synthesis: A target for antimicrobials. Expert Opin Ther Targets. 2008;12(5):519–534. doi: 10.1517/14728222.12.5.519. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44(6):393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 4.Fabbretti A, Gualerzi CO, Brandi L. How to cope with the quest for new antibiotics. FEBS Lett. 2011;585(11):1673–1681. doi: 10.1016/j.febslet.2011.04.029. [DOI] [PubMed] [Google Scholar]
- 5.Brandi L, et al. The translation initiation functions of IF2: Targets for thiostrepton inhibition. J Mol Biol. 2004;335(4):881–894. doi: 10.1016/j.jmb.2003.10.067. [DOI] [PubMed] [Google Scholar]
- 6.Brandi L, et al. Specific, efficient, and selective inhibition of prokaryotic translation initiation by a novel peptide antibiotic. Proc Natl Acad Sci USA. 2006;103(1):39–44. doi: 10.1073/pnas.0507740102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schluenzen F, et al. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol. 2006;13(10):871–878. doi: 10.1038/nsmb1145. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12(1):35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 9.Fabbretti A, et al. The antibiotic Furvina® targets the P-site of 30S ribosomal subunits and inhibits translation initiation displaying start codon bias. Nucleic Acids Res. 2012;40(20):10366–10374. doi: 10.1093/nar/gks822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandi L, et al. Novel tetrapeptide inhibitors of bacterial protein synthesis produced by a Streptomyces sp. Biochemistry. 2006;45(11):3692–3702. doi: 10.1021/bi052540k. [DOI] [PubMed] [Google Scholar]
- 11.Milón P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. Real-time assembly landscape of bacterial 30S translation initiation complex. Nat Struct Mol Biol. 2012;19(6):609–615. doi: 10.1038/nsmb.2285. [DOI] [PubMed] [Google Scholar]
- 12.Gualerzi C, Risuleo G, Pon CL. Initial rate kinetic analysis of the mechanism of initiation complex formation and the role of initiation factor IF-3. Biochemistry. 1977;16(8):1684–1689. doi: 10.1021/bi00627a025. [DOI] [PubMed] [Google Scholar]
- 13.Milon P, et al. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 2010;11(4):312–316. doi: 10.1038/embor.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigoriadou C, Marzi S, Pan D, Gualerzi CO, Cooperman BS. The translational fidelity function of IF3 during transition from the 30 S initiation complex to the 70 S initiation complex. J Mol Biol. 2007;373(3):551–561. doi: 10.1016/j.jmb.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binz TM, Maffioli SI, Sosio M, Donadio S, Müller R. Insights into an unusual nonribosomal peptide synthetase biosynthesis: Identification and characterization of the GE81112 biosynthetic gene cluster. J Biol Chem. 2010;285(43):32710–32719. doi: 10.1074/jbc.M110.146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407(6802):327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 17.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 18.Demirci H, et al. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA. 2010;16(12):2319–2324. doi: 10.1261/rna.2357210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schluenzen F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102(5):615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 20.Urzhumtsev AG. Local improvement of electron-density maps. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 5):540–543. doi: 10.1107/S0907444997004885. [DOI] [PubMed] [Google Scholar]
- 21.Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol. 2014;21(9):787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Fernandez D, et al. 4′-O-substitutions determine selectivity of aminoglycoside antibiotics. Nat Commun. 2014;5:3112. doi: 10.1038/ncomms4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantara WA, Murphy FV, IV, Demirci H, Agris PF. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc Natl Acad Sci USA. 2013;110(27):10964–10969. doi: 10.1073/pnas.1222641110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milon P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Mol Cell. 2008;30(6):712–720. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 25.La Teana A, Pon CL, Gualerzi CO. Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc Natl Acad Sci USA. 1993;90(9):4161–4165. doi: 10.1073/pnas.90.9.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gualerzi C, Pon CL, Kaji A. Initiation factor dependent release of aminoacyl-tRNAs from complexes of 30S ribosomal subunits, synthetic polynucleotide and aminoacyl tRNA. Biochem Biophys Res Commun. 1971;45(5):1312–1319. doi: 10.1016/0006-291x(71)90162-8. [DOI] [PubMed] [Google Scholar]
- 27.Gualerzi CO, et al. Initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harb Symp Quant Biol. 2001;66:363–376. doi: 10.1101/sqb.2001.66.363. [DOI] [PubMed] [Google Scholar]
- 28.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci USA. 2006;103(43):15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandi L, et al. Methods for identifying compounds that specifically target translation. Methods Enzymol. 2007;431:229–267. doi: 10.1016/S0076-6879(07)31012-4. [DOI] [PubMed] [Google Scholar]
- 30.Calogero RA, Pon CL, Canonaco MA, Gualerzi CO. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc Natl Acad Sci USA. 1988;85(17):6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi S, et al. Kinetic analysis of ribosome binding process onto mRNA using a quartz-crystal microbalance. Nucleic Acids Symp Ser (Oxf) 2006;50:49–50. doi: 10.1093/nass/nrl025. [DOI] [PubMed] [Google Scholar]
- 32.Demirci H, et al. A structural basis for streptomycin-induced misreading of the genetic code. Nat Commun. 2013;4:1355. doi: 10.1038/ncomms2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demirci H, et al. The central role of protein S12 in organizing the structure of the decoding site of the ribosome. RNA. 2013;19(12):1791–1801. doi: 10.1261/rna.040030.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harms JM, et al. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30(1):26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Blaha G, Gürel G, Schroeder SJ, Moore PB, Steitz TA. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J Mol Biol. 2008;379(3):505–519. doi: 10.1016/j.jmb.2008.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modolell J, Cabrer B, Parmeggiani A, Vazquez D. Inhibition by siomycin and thiostrepton of both aminoacyl-tRNA and factor G binding to ribosomes. Proc Natl Acad Sci USA. 1971;68(8):1796–1800. doi: 10.1073/pnas.68.8.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modolell J, Vazquez D, Monro RE. Ribosomes, G-factor and siomycin. Nat New Biol. 1971;230(12):109–112. doi: 10.1038/newbio230109a0. [DOI] [PubMed] [Google Scholar]
- 38.Lelong JC, Gros D, Cousin MA, Grunberg-Manago M, Gros F. Streptomycin induced release of fMet-tRNA from the ribosomal initiation complex. Biochem Biophys Res Commun. 1971;42(3):530–537. doi: 10.1016/0006-291x(71)90403-7. [DOI] [PubMed] [Google Scholar]
- 39.Poldermans B, Bakker H, Van Knippenberg PH. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16S ribosomal RNA of Escherichia coli. IV. The effect of the methylgroups on ribosomal subunit interaction. Nucleic Acids Res. 1980;8(1):143–151. doi: 10.1093/nar/8.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapkina TG, Dolan MA, Babin P, Wollenzien P. Initiation factor 3-induced structural changes in the 30 S ribosomal subunit and in complexes containing tRNA(f)(Met) and mRNA. J Mol Biol. 2000;299(3):615–628. doi: 10.1006/jmbi.2000.3774. [DOI] [PubMed] [Google Scholar]
- 41.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell. 2001;8(4):855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 42.Muralikrishna P, Wickstrom E. Escherichia coli initiation factor 3 protein binding to 30S ribosomal subunits alters the accessibility of nucleotides within the conserved central region of 16S rRNA. Biochemistry. 1989;28(19):7505–7510. doi: 10.1021/bi00445a002. [DOI] [PubMed] [Google Scholar]
- 43.Moazed D, Samaha RR, Gualerzi C, Noller HF. Specific protection of 16 S rRNA by translational initiation factors. J Mol Biol. 1995;248(2):207–210. doi: 10.1016/s0022-2836(95)80042-5. [DOI] [PubMed] [Google Scholar]
- 44.Petrelli D, et al. Translation initiation factor IF3: Two domains, five functions, one mechanism? EMBO J. 2001;20(16):4560–4569. doi: 10.1093/emboj/20.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fabbretti A, et al. The real-time path of translation factor IF3 onto and off the ribosome. Mol Cell. 2007;25(2):285–296. doi: 10.1016/j.molcel.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 46.Tapprich WE, Goss DJ, Dahlberg AE. Mutation at position 791 in Escherichia coli 16S ribosomal RNA affects processes involved in the initiation of protein synthesis. Proc Natl Acad Sci USA. 1989;86(13):4927–4931. doi: 10.1073/pnas.86.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin D, Abdi NM, Fredrick K. Characterization of 16S rRNA mutations that decrease the fidelity of translation initiation. RNA. 2007;13(12):2348–2355. doi: 10.1261/rna.715307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawlik RT, Littlechild J, Pon C, Gualerzi C. Purification and Properties of Escherichia coli Translational Initiation Factors. Biochem Int. 1981;2(4):421–428. [Google Scholar]
- 49.Brandi L, Dresios J, Gualerzi CO. Assays for the identification of inhibitors targeting specific translational steps. Methods Mol Med. 2008;142:87–105. doi: 10.1007/978-1-59745-246-5_8. [DOI] [PubMed] [Google Scholar]
- 50.Milon P, et al. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 2007;430:1–30. doi: 10.1016/S0076-6879(07)30001-3. [DOI] [PubMed] [Google Scholar]
- 51.Donis-Keller H, Maxam AM, Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma MR, et al. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol Cell. 2005;18(3):319–329. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 53.Kaminishi T, et al. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure. 2007;15(3):289–297. doi: 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 54.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Schrodinger LLC. 2010. The PyMOL Molecular Graphics System, Version 1.6. Available at www.pymol.org.
- 59.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316(3):725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 60.Julián P, et al. The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol. 2011;9(7):e1001095. doi: 10.1371/journal.pbio.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]