Abstract
BACKGROUND
Untargeted multiomics datasets are obtained for samples in systems, synthetic, and chemical biology by integrating chromatographic separations with ion mobility-mass spectrometry (IM-MS) analysis. The datasets are interrogated using bioinformatics strategies to organize the data for identification prioritization.
CONTENT
The use of big data approaches for data mining of massive datasets in systems-wide analyses is presented. Untargeted biological data across multiomics dimensions are obtained using a variety of chromatography strategies with structural mass spectrometry. Separation timescales for different techniques and the resulting data deluge when combined with IM-MS is presented. Data mining self-organizing map (SOM) approaches are used to rapidly filter the data highlighting those features describing uniqueness to the query. Examples are provided in longitudinal analyses in synthetic biology, human liver exposure to acetaminophen, and in chemical biology, natural product discovery from bacterial biomes.
CONCLUSIONS
Matching separation timescales of different forms of chromatography with IM-MS provides sufficient multiomics selectivity to perform untargeted systems-wide analyses. New data mining strategies provide a means for rapidly interrogating these data sets for feature prioritization and discovery in a range of applications in systems, synthetic, and chemical biology.
Keywords: Systems analysis, ion mobility, mass spectrometry, ion mobility-mass spectrometry, omics, bioinformatics, self-organizing maps, systems biology, synthetic biology, chemical biology
In parallel with big data endeavors in information technology, the past ten years have driven allied pursuits in genomics and genome medicine. Of particular note is the use of broad scale genome-wide association studies (GWAS) to correlate genetic alterations with phenotype. Beyond the genome, these foundational concepts are increasingly utilized in concert with advances in molecular characterization approaches for metabolome-wide association studies (MWAS) to correlate the dynamic molecular complement in tissues or bodily fluids with phenotype (1,2). Systems-wide MWAS strategies are now performed with the primary aim of characterizing, quantifying, and cataloging the biomolecular inventory at specific dimensions of space (e.g., cellular, tissue, or organism levels) and time (e.g., longitudinal exposure, point in the life cycle, healthy vs. diseased state). These studies are broadly facilitated by the emerging capabilities of mass spectrometry (MS), which can provide both targeted quantitative information and/or broad scale untargeted data. The later approaches produce largely hypothesis-independent data, which are then integrated with bioinformatics strategies to derive desired information pertaining to the question at hand. While spatial information is typically obtained using a combination of different microscopy modalities and imaging MS (3,4), temporal information with high time resolution for long durations have proven more challenging, limited by the timescales for liquid chromatography-mass spectrometry (LC-MS) and sample preparation to isolate specific classes of molecules (e.g. proteins for proteomics studies). Importantly new data acquisition strategies coupled with informatics approaches from big data techniques have overcome many of these challenges and are highlighted here. It is anticipated that these emerging strategies will become increasingly utilized in the clinical setting.
Systems-wide analyses necessitate the acquisition of multi-dimensional datasets with separations distinguishing different physical characteristics in each dimension for high molecular selectivity. While quantitation of gene transcription is dominated by array technology, many omics endeavors, such as metabolomics, proteomics, lipidomics, and glycomics are most commonly performed using MS or LC-MS (5). In large part, this is attributed to the necessity of requiring massive numbers of experiments to understand metabolic and molecular networks under different conditions and the information rich data afforded by contemporary MS instrumentation (6,7). Nevertheless, in many contemporary shotgun approaches or LC-MS or GC-MS omics studies, typically the class of molecule of interest (e.g. metabolites, proteins, lipids, etc.) is purified prior to analysis to reduce chemical interference from all others, which restricts molecular breadth in untargeted approaches. While limiting the scope of the analysis to one or several molecular classes is potentially warranted for high quantitation accuracy, large-scale systems-wide experiments motivate the development of measurement strategies that incorporate higher throughput, higher selectivity, and require minimal sample manipulation. A key technology that has shown considerable promise that incorporates aspects of these features is the integration of gas-phase electrophoresis, or ion mobility spectrometry (IMS) with mass spectrometry (IM-MS).
Scaling separations for high-dimensional data acquisition
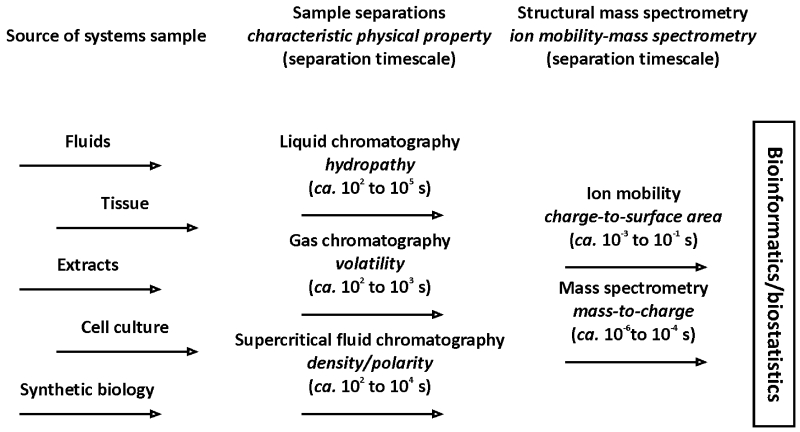
One of the primary challenges to broad scale systems-wide analyses is the menagerie of chemical/physical properties represented in the biomolecular inventory. Clearly this breadth of chemical diversity provides for assorted biological function, but analytically it requires the integration of separation strategies that each provide selectivity based on a different physical characteristic. Figure 1 illustrates several common separation strategies used in the characterization of biological samples with MS and IM-MS detection. While the source of the sample for systems-wide interrogation may arise from bodily fluids, tissues, extracts of organisms, and increasingly from synthetic biological constructs, typically one or more different sample separations steps are used to tease apart the complexity of the sample. Typically LC is used to perform separations on the basis of hydropathy (hydrophilicity versus hydrophobicity) and GC is often used to separate on the basis of volatility. Note that a distinction is made between separations based on chromatography, or a partitioning between phases, and ion mobility and mass spectrometry, for which interphase partitioning does not occur and consequently a partition coefficient cannot be written. This lack of phase partitioning coupled with gas-phase separations distinguishes the IM and MS separation methods by the extremely fast timescales that are used for analysis (8). When phase partitioning is used, the separation timescales are typically on the order of minutes to hours, while in IMS and MS, the separation timescales are usually on the order of microseconds to milliseconds. Thus, integrating the separations of IMS with MS (i.e. IM-MS) does not limit the throughput of the experiment which is dominated by the chromatographic separation. It should be noted that because the enhancement in throughput is attained by performing the separations following ionization, a tradeoff to minimal sample preparation in IM-MS is the challenge of ion suppression effects and potentially concentration-dependent response. Thus, for complex samples IM is oftentimes integrated with pre-ionization chromatographies, most commonly LC-IM-MS, to provide additional molecular selectivity while mitigating ionization effects (9).
Figure 1.
A depiction of the sources of samples used for systems-wide analyses (left), typical chromatographic separations strategies utilized for selectivity and associated separation timescales (middle), and structural mass spectrometry and corresponding timescales for separation and detection (right). The untargeted datasets are then processed using emerging bioinformatics/biostatistical strategies as described.
Although there exist a multitude of arrangements for performing IM-MS, untargeted analyses are commonly accepted to correspond to time-dispersive IM coupled with time-of-flight MS (8,9). The distinction between these methods arises in how the electric fields are applied for IM separation, namely electrodynamic IM fields (10), and electrostatic IM fields (11), respectively. One of the primary reasons time-dispersive IM-MS has been widely adopted is because the drift time across the ion mobility cell, analogous to LC retention time, can predictably be correlated to an observed ion-neutral collision cross section (Ω, Å2), which is a rotationally averaged apparent surface area of the ion. This is achieved through ion-neutral collisions with an inert background buffer gas as ions traverse a drift region under the influence of the defined electric field. Larger surface area molecules experience a larger number of collisions relative to a smaller surface area molecule of the same mass, which results in a longer drift time.
Importantly for untargeted analyses, different classes of molecules distribute into unique regions of IM-MS separations space, or conformational space, depending on the type of molecule it is and the typical density for that class of molecule. As a result, predictable correlations emerge in the dataset (mobility versus mass) related to the types of molecules analyzed and the prevailing intramolecular folding forces for each biomolecular class (12,13). This provides a rapid means for integrating omics data acquisition without the need for extensive sample preparation. Recent studies have also focused on utilizing these correlations within a specific molecular class (e.g. lipids (14-16), carbohydrates (17,18), peptides/proteins (19,21), etc.) for predictive purposes (11,22). Such approaches have been demonstrated in a wide array of emerging applications ranging from systems diagnosis of wound healing (23), to cancer (24,25), to drug discovery efforts (26-28). It is important to note that interlaboratory studies indicate that these mobility-mass correlations exhibit very high reproducibility, making them well suited for integration in systems-wide protocols (29).
Bioinformatics of high dimensional data for rapid target prioritization
One of the primary challenges facing IM-MS in systems-wide analyses is the interrogation of massive high dimensional datasets. To illustrate the data volume in a typical LC-IM-MS experiment, an LC run of less than an hour easily results in the generation of >104 IM separations with >106 corresponding MS spectra. To compound matters, typically MS/MS spectra are acquired continuously across these separation dimensions resulting in ca. 107 to 108 fragmentation spectra for a single LC run. This multidimensional data places particular demands on the bioinformatics and biostatistics that are used to infer desired information from the systems-wide data (9). In the first stage of data processing, it is complicated to extract peak features correlated across high dimensional data. Recently, automated strategies have been developed for feature extraction from such datasets (30,31). Once features have been extracted, philosophically two avenues can be followed for projecting the multidimensional data in a visually instructive manner to guide the biological interpretation and subsequent analyses. For single cell analyses with a means for reducing the number of features per entity, such as those in labeled mass cytometry studies, advanced means have been developed relating projection distance to cell phenotype (32,33). For systems-wide label free characterization, there exist a large proportion of features/molecules within the dataset that do not describe the biological process, disease, or phenotype under investigation, but rather correspond to biological housekeeping and superimposed unconnected biological response to other stimuli or stresses beyond that being investigated. The motivation then is to rapidly unravel those features revealing the molecular consequences specific to the question at hand. For systems-wide feature prioritization, self-organizing map (SOM) strategies have demonstrated great utility in performing this function (34,35). In a generalized framework, a data processing workflow for alignment and feature prioritization to discern molecular response using SOM termed molecular expression dynamics inspector (MEDI) has been described (34).
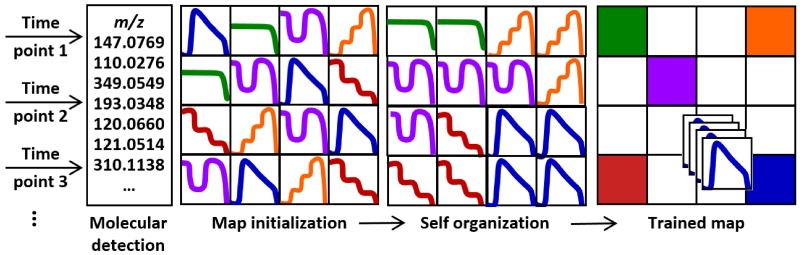
Conceptually, the SOM and MEDI approach is analogous to strategies used in a wide array of big data applications from internet commerce to discerning population behavior in civil engineering or ecology. Similar to these applications, correlations are highlighted across multiple massive datasets. A conceptual depiction of the SOM approach is illustrated in Figure 2. Once data sets are obtained, for example representing different response to different exposures/stimuli or time points of longitudinal response, the features across the datasets are aligned and extracted. Each extracted feature forms a pattern, represented by a tile in Figure 2, most often the signal intensity or relative abundance of the feature as a function of the ordering of the datasets, for example increasing time in longitudinal exposure. There is a separate tile constructed for each feature or molecule. The tiles are then sequentially compared and shifted in a recursive strategy until the tiles form neighborhoods of most similar correlated pattern, i.e. self-organization. These neighborhoods then project the high dimensional data in a straightforward way to highlight groups of molecules that correspond to similar responses. When the initial patterns are constructed to highlight specific responses, e.g. increased/decreased expression level, then the corresponding neighborhood prioritizes those features for subsequent identification from the sea of feature data. It is important to note that the patterns used for SOM are data agnostic and merging disparate data streams can be accomplished, for example combining IM-MS and meta or other forms of omics data such as that derived from sources such as transcriptomics experiments.
Figure 2.
A conceptual workflow for the self-organization of high dimensional data using MEDI. A series of untargeted experiments are performed for which a separate tile is constructed for each molecular feature as a function of abundance extracted for each experimental condition, such as different time points in longitudinal exposure. After molecular detection, the map initialization step randomly generates a map with intensity profiles indicative of the data. During the self organization training process, intensity profiles (or tiles) are grouped based on similarities in an iterative process until they are matched to their closest matching profile. Once the training phase is complete and a grid location determined, heat maps are generated for each sample based upon the intensity of the seeded features within that sample. These self-organized heat maps can then be averaged and/or differentially analyzed to distinguish regions of interest that differ among samples.
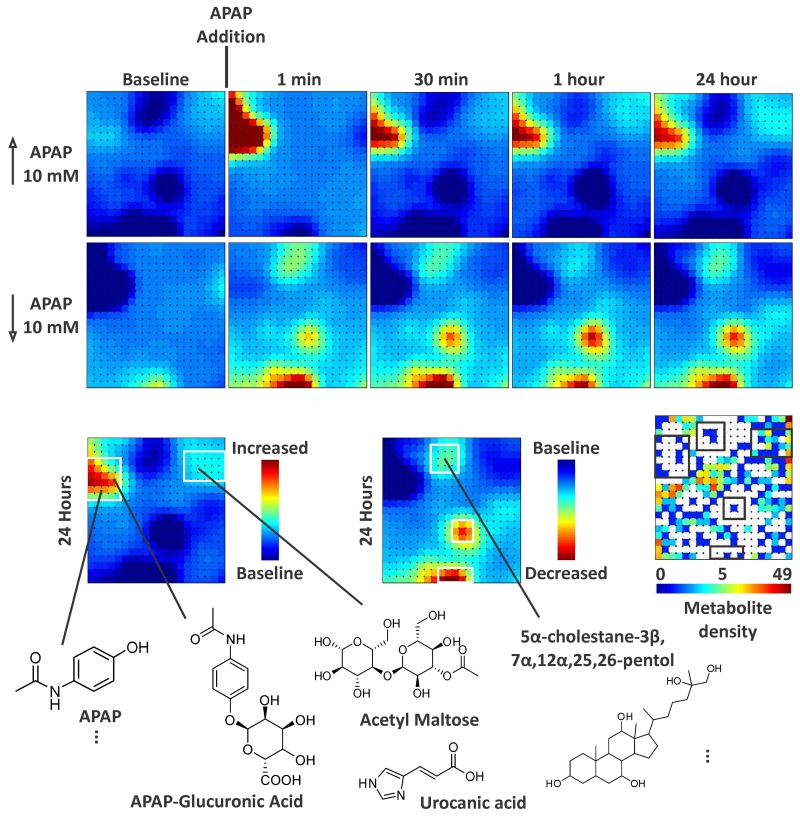
One of the emerging areas in synthetic biology and medicine is the recent development of 3D organotypic chip platforms to emulate human organs with the ultimate aim of constructing the so-called human-on-a-chip (36-38). These efforts are motivated, in part, to perform broad scale toxicology experiments on human organs for system exposure to drugs as a bridge between in vitro and clinical experiments. Figure 3 illustrates MEDI maps for longitudinal molecular mapping from a human liver bioreactor exposed to acetaminophen (APAP). The LBR is seeded for culture from human cadaver liver and are perfused with media and gas distribution via hollow fibers around which the cells form the organ. The waste stream from the organ is interrogated by IM-MS with MEDI data processing over the course of 24 hours following APAP exposure. In the upper portion of the figure, selected time points are illustrated mapping the molecules that are expressed in higher and lower abundance following APAP exposure. The neighborhoods forming the regions of difference direct identification of the species distinguishing APAP from baseline exposure. Representative molecules found in these regions are shown in the bottom of Figure 3 to demonstrate the breadth of molecular characterization. In the seconds following APAP exposure, dysregulation of bile acid production is noted, which is a hallmark of liver stress, and other small molecule indicators of liver health, xenobiotic conjugates, and different forms of liver dysregulation are observed.
Figure 3.
Liver bioreactors were sampled at intervals before and after treatment with acetaminophen (APAP) by extracting perfusate for UPLC-IM-MS analysis. Differential metabolic profiles were generated by MEDI to depict molecules with elevated and decreased abundances from LBRs exposed to 10 mM APAP. The molecules in the regions of interest were subsequently identified using accurate mass measurement and high energy (fragmentation spectra) to search databases for candidate molecules. These representative identifications illustrate the utility of self-organizing map analysis, grouping together metabolic pathways (APAP and APAP conjugates, bile acids, etc) that were generated or perturbed upon treatment. The metabolite density plot indicates the number of molecules residing in the corresponding grid location.
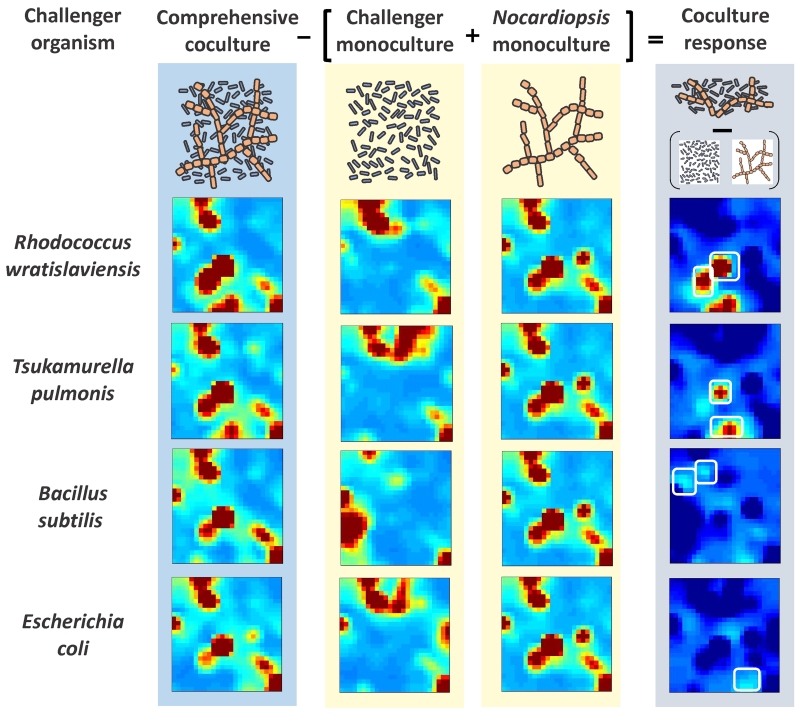
Rapid target prioritization is also critical in chemical biology and drug discovery. Self-organizing data techniques and MEDI approaches have been used to rapidly characterize molecular indicators of drug addiction phenotype (34) and to prioritize candidate molecules in drug discovery efforts from natural products (39,40). The latter demonstrates a different feature of MEDI maps, namely the ability to perform mathematical operations on the untargeted data to drive further interpretive power. For example, Figure 4 illustrates a strategy colloquially termed “bacteria fight club.” In this approach, a system for which the genome is well characterized and known to be a reservoir of gene clusters corresponding secondary metabolites and potentially new drug-like compounds is grown as a monoculture, such as Nocadiopsis in this example. The challenge is to find conditions that promote the expression of these secondary metabolites. While, changes to the environment and cell culture media can be exhaustively examined, coculturing the bacteria with a challenger bacteria that competes for resources can provide an effector for expression of secondary metabolites that adversely influence the challenger. To tease apart the 10s of thousands of features identified in the MEDI maps of the monoculture of challenger and Nocariopsis, the trained maps can provide the difference of the coculturing conditions, where secondary metabolite expression is abundant to “fight” between the two organisms (40). The result is a MEDI map illustrating only neighborhoods corresponding to new compounds expressed under the coculture condition from those of the monoculture conditions. Using this approach a few 100s of potentially new chemical entities are prioritized for subsequent identification from 10s of thousands of features observed in the bacterial cultures.
Figure 4.
Target prioritization of secondary metabolites from bacteria using MEDI. Self-organizing maps of features four cocultures and five monocultures (Nocariopsis and four challenger organisms) were constructed. The difference SOM of the coculture from the monocultures results in a map coculture response map highlighting only those features that are distinct form the monoculture conditions to identify unique and upregulated features. (Adapted from Reference 40).
Conclusions
Systems-wide analysis are facilitated by obtaining high dimensional untargeted data through a combination of different separations strategies with ultrafast separations of IM-MS. This approach allows the integration of omics analyses without sample pretreatment to isolate classes of molecules of interest. Importantly, these datasets can be coupled with emerging bioinformatics strategies to self-organize the data to prioritize which features contain the desired information from massive datasets to prioritize which features warrant identification to answer the query at hand. Such approaches are opening new avenues of inquiry in biology, medicine, and clinical diagnostics using systems-wide approaches.
Acknowledgements
The authors gratefully acknowledge our collaborators in the untargeted studies of the liver bioreactor data illustrated Figure 3 - Rashi Iyer (Los Alamos National Laboratory), Katrin Zeilinger (Charité - Universitätsmedizin Berlin, (CUB)), Marc Luebberstedt (CUB), Georg Damm (CUB), and Cody R. Goodwin (Vanderbilt University) – and our collaborators in natural product discovery illustrated in Figure 4 with Brian O. Bachmann (Vanderbilt University). Financial support for aspects of this work was provided by the National Institutes of Health/National Center for Advancing Translational Sciences (4UH3TR000491), the National Institutes of Health/National Institute for General Medical Sciences (5R01GM092218), the National Science Foundation Major Research Instrumentation program (NSF/MRI CHE-1229341), the Defense Threat Reduction Agency (CBMXCEL-XLI-2-0001), the Vanderbilt Institute of Chemical Biology; the Vanderbilt Institute for Integrative Biosystems Research and Education; and Vanderbilt University. This is an un-copyedited authored manuscript copyrighted by the American Association for Clinical Chemistry (AACC). This may not be duplicated or reproduced, other than for personal use or within the rule of ‘Fair Use of Copyrighted Materials’ (section 107, Title 17, U.S. Code) without permission of the copyright owner, AACC. The AACC disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties. The final publisher-authenticated version of the article will be made available at http://www.clinchem.org 12 months after its publication in Clinical Chemistry.
Abbreviations
- GC
Gas chromatography
- LC
Liquid chromatography
- MS
Mass spectrometry
- IMS
Ion mobility spectrometry
- IM-MS
Ion mobility-mass spectrometry
- LC-MS
Liquid chromatography-mass spectrometry
- GC-MS
Gas chromatography-mass spectrometry
- LC-IM-MS
Liquid chromatography-ion mobility-mass spectrometry
- SOM
Self-organizing map
- MEDI
Molecular expression dynamics inspector
- LBR
Liver bioreactor
- APAP
Acetaminophen
References
- 1.Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature. 2012;491:384–92. doi: 10.1038/nature11708. [DOI] [PubMed] [Google Scholar]
- 2.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang J, Dorrestein PC. Emerging mass spectrometry techniques for the direct analysis of microbial colonies. Curr Opin Microbiol. 2015;19:120–9. doi: 10.1016/j.mib.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva LP, Northern TR. Exometabolomics and MSI: deconstructing how cells interact to transform their small molecule environment. Curr Opin Biotechnol. 2015;34:209–16. doi: 10.1016/j.copbio.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–3. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 6.Fuhrer T, Zamboni N. High-throughput discovery metabolomics. Curr Opin Biotech. 2015;31:73–8. doi: 10.1016/j.copbio.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–9. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.May JC, McLean JA. Ion mobility-mass spectrometry: time dispersive instrumentation. Anal Chem. 2015;87:1422–36. doi: 10.1021/ac504720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.May JC, Goodwin CR, McLean JA. Ion mobility-mass spectrometry strategies for untargeted systems, synthetic, and chemical biology. Curr Opin Biotechnol. 2015;31:117–21. doi: 10.1016/j.copbio.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun Mass Spectrom. 2004;18:2401–14. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 11.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, et al. Conformational ordering of biomolecules in the gas-phase: nitrogen collision cross-sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal Chem. 2014;86:2107–16. doi: 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenn LS, McLean JA. Biomolecular structural separations by ion mobility-mass spectrometry. Anal Bioanal Chem. 2008;391:905–9. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- 13.Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA. Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples. Anal Bioanal Chem. 2009;394:235–44. doi: 10.1007/s00216-009-2666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kliman M, May JC, McLean JA. Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim Biophys Acta. 2011;1811:935–45. doi: 10.1016/j.bbalip.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castro-Perez J, Roddy TP, Nibbering NM, Shah V, McLaren DG, Previs S, et al. Localization of fatty acyl and double bond positions in phosphatidylcholines using a dual stage CID fragmentation coupled with ion mobility mass spectrometry. J Am Soc Mass Spectrom. 2011;22:1552–67. doi: 10.1007/s13361-011-0172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wenk MR. Lipidomics: new tools and applications. Cell. 2010;143:888–95. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 17.Both P, Green AP, Gray CJ, Sardzík R, Voglmeir J, Fontana C, et al. Discrimination of epimeric glycans and glycopeptides using IM-MS and its potential for carbohydrate sequencing. Nat Chem. 2014;6:65–74. doi: 10.1038/nchem.1817. [DOI] [PubMed] [Google Scholar]
- 18.Harvey DJ, Sobott F, Crispin M, Wrobel A, Bonomelli C, Vasiljevic S, et al. Ion mobility mass spectrometry for extracting spectra of N-glycans directly from incubation mixtures following glycan release: application to glycans from engineered glycoforms of intact, folded HIV gp120. J Am Soc Mass Spectrom. 2010;22:568–81. doi: 10.1007/s13361-010-0053-0. [DOI] [PubMed] [Google Scholar]
- 19.Bush MF, Campuzano IDG, Robinson CV. Ion mobility mass spectrometry of peptide ions: effects of drift gas and calibration strategies. Anal Chem. 2012;84:7124–30. doi: 10.1021/ac3014498. [DOI] [PubMed] [Google Scholar]
- 20.Shliaha PV, Bond NJ, Gatto L, Lilley KS. Effects of traveling wave ion mobility separation on data Independent acquisition in proteomics studies. J Proteome Res. 2013;12:2323–39. doi: 10.1021/pr300775k. [DOI] [PubMed] [Google Scholar]
- 21.Jia C, Lietz CB, Yu Q, Li L. Site-specific characterization of (D)-amino acid containing peptide epimers by ion mobility spectrometry. Anal Chem. 2014;86:2972–2981. doi: 10.1021/ac4033824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean JA. The mass-mobility correlation redux: the conformational landscape of anhydrous biomolecules. J Am Soc Mass Spectrom. 2009;20:1775–81. doi: 10.1016/j.jasms.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Hines KM, Ashfaq S, Davidson JM, Opalenik SR, Wikswo JP, McLean JA. Biomolecular signatures of diabetic wound healing by structural mass spectrometry. Anal Chem. 2013;85:3651–59. doi: 10.1021/ac303594m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker ES, Burnum-Johnson KE, Jacobs JM, Diamond DL, Brown RN, Ibrahim YM, et al. Advancing the high throughput identification of liver fibrosis protein signatures using multiplexed ion mobility spectrometry. Mol Cell Proteomics. 2014;13:1119–27. doi: 10.1074/mcp.M113.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hines KM, Ballard BR, Marshall DR, McLean JA. Structural mass spectrometry of tissue extracts to distinguish cancerous and non-cancerous breast diseases. Mol Biosyst. 2014;10:2827–37. doi: 10.1039/c4mb00250d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodwin CR, Fenn LS, Derewacz DK, Bachmann BO, McLean JA. Structural mass spectrometry: rapid methods for separation and analysis of peptide natural products. J Nat Prod. 2012;75:48–53. doi: 10.1021/np200457r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esquenazi E, Daly M, Bahrainwala T, Gerwick WH, Dorrestein PC. Ion mobility mass spectrometry enables the efficient detection and identification of halogenated natural products from cyanobacteria with minimal sample preparation. Bioorg Med Chem. 2011;19:6639–44. doi: 10.1016/j.bmc.2011.06.081. [DOI] [PubMed] [Google Scholar]
- 28.Derewacz DK, Goodwin CR, McNees CR, McLean JA, Bachmann BO. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proc Natl Acad Sci USA. 2013;110:2336–41. doi: 10.1073/pnas.1218524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paglia G, Williams JP, Menikarachchi L, Thompson JW, Tyldesley-Worster R, Halldórsson S, et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal Chem. 2014;86:3985–93. doi: 10.1021/ac500405x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crowell KL, Slysz GW, Baker ES, LaMarche BL, Monroe ME, Ibrahim YM, et al. LC-IMS-MS feature finder: detecting multidimensional liquid chromatography, ion mobility and mass spectrometry features in complex datasets. Bioinformatics. 2013;29(1):2804–5. doi: 10.1093/bioinformatics/btt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivalingam GN, Yan J, Sahota H, Thalassinos K. Amphitrite: A program for processing travelling wave ion mobility mass spectrometry data. Int J Mass Spectrom. 2013;345-347:54–62. doi: 10.1016/j.ijms.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nat Biotech. 2012;30:639–47. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 33.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr., Bruggner RV, Linderman MD, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotech. 2011;29:886–91. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodwin CR, Sherrod SD, Marasco CC, Bachmann BO, Schramm-Sapyta N, Wikswo JP, McLean JA. Phenotypic mapping of metabolic profiles using self-organizing maps of high-dimensional mass spectrometry data. Anal Chem. 2014;86:6563–71. doi: 10.1021/ac5010794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patterson AD, Li H, Eichler GS, Krausz KW, Weinstein JN, Fornace AJ, Jr, et al. UPLC-ESI-TOFMS-based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Anal Chem. 2008;80:665–74. doi: 10.1021/ac701807v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wikswo JP, Block FE, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, et al. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans Biomed Eng. 2013;60:682–90. doi: 10.1109/TBME.2013.2244891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcendor DJ, Block FE, Cliffel DE, Daniels JS, Ellacott KLJ, Goodwin CR, et al. Neurovascular unit on a chip: implications for translational applications. Stem Cell Res Ther. 2013;4(S18):1–5. doi: 10.1186/scrt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi M, Majumdar D, Gao Y, Brewer BM, Goodwin CR, McLean JA, et al. Glia co-culture with neurons in microfluidic platforms promotes the formation and stabilization of synaptic contacts. Lab on a Chip. 2013;13:3008–21. doi: 10.1039/c3lc50249j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodwin CR, Covington BC, Derewacz DK, McNees CR, Wikswo JP, McLean JA, et al. Structuring microbial metabolic responses to multiplexed stimuli via self-organizing metabolomics maps. Chemistry and Biology. 2015;22:661–70. doi: 10.1016/j.chembiol.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derewacz DK, Covington BC, McLean JA, Bachmann BO. Mining microbe biomes for induced natural product discovery. ACS Chem Biol. 2015;10 doi: 10.1021/acschembio.5b00001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]