Abstract
Background & Aims
Exosomes are small membrane vesicles involved in intercellular communication. Hepatocytes are known to release exosomes, but little is known about their biological function. We sought to determine if exosomes derived from hepatocytes contribute to liver repair and regeneration after injury.
Methods
Exosomes derived from primary murine hepatocytes were isolated and characterized biochemically and biophysically. Using cultures of primary hepatocytes, we tested whether hepatocyte exosomes induced proliferation of hepatocytes in vitro. Using models of ischemia/reperfusion injury and partial hepatectomy, we evaluated whether hepatocyte exosomes promote hepatocyte proliferation and liver regeneration in vivo.
Results
Hepatocyte exosomes, but not exosomes from other liver cell types, induce dose-dependent hepatocyte proliferation in vitro and in vivo. Mechanistically, hepatocyte exosomes directly fuse with target hepatocytes and transfer neutral ceramidase and sphingosine kinase 2 (SK2) causing increased synthesis of sphingosine-1-phosphate (S1P) within target hepatocytes. Ablation of exosomal SK prevents the proliferative effect of exosomes. After ischemia/reperfusion injury, the number of circulating exosomes with proliferative effects increases.
Conclusions
Our data shows that hepatocyte-derived exosomes deliver the synthetic machinery to form S1P in target hepatocytes resulting in cell proliferation and liver regeneration after ischemia/reperfusion injury or partial hepatectomy. These findings represent a potentially novel new contributing mechanism of liver regeneration and have important implications for new therapeutic approaches to acute and chronic liver disease.
Keywords: Liver injury, Sphingolipids, Sphingosine kinase, Ischemia/reperfusion, Transplantation
Introduction
Liver regeneration is a compensatory process that replaces functional liver mass lost as a result of injury or disease. The mechanisms governing this process are highly complex and have been the focus of investigation for decades. The specific functions of numerous hormones, growth factors and cytokines have been identified over the temporal course of the regenerative process [1]. However, whether other forms of intercellular communication, such as exosomes, may participate in this process has not been investigated.
Exosomes are membrane nanovesicles (30–100 nm) released by cells into the extracellular environment upon fusion of multivesicular bodies with the plasma membrane [2–4]. Exosomes contain membrane components but also contain proteins, micro-RNAs and mRNAs [4–7]. A variety of cell types, including hepatocytes, secrete exosomes into body fluids such as blood and urine [5,8–10]. Previous studies have shown that exosome released from cells depends on the activity of neutral sphingomyelinase, releasing ceramide from sphingomyelin and thereby controlling the process of exosome formation and release [11,12]. An emerging interest in exosomes has focused on their potential roles as a form of intercellular communication via delivery of exosomal contents and modulation of cellular activities in recipient cells [5,6,13]. For example, B-lymphocytes and dendritic cells have been shown to release exosomes that can stimulate T cell proliferation and contribute to a robust immune response [14]. In contrast, tumor cells have been shown to release exosomes that promote tumor growth and metastasis [15], and hepatocytes infected with hepatitis C virus (HCV) have been shown to release exosomes that contain and transmit HCV to other hepatocytes [16–18]. As such, it appears that exosomes serve as a fundamental mechanism of cell communication for basic homeostasis that can be hijacked by malignant and virus-infected cells.
Here, we investigated the role of exosomes as a mode of intercellular communication in the processes of liver repair and regeneration. We demonstrate that exosomes produced by hepatocytes, but not other liver cell types, promote hepatocyte proliferation in vitro and induce liver regeneration in vivo. The mechanism of this effect was found to be due to sphingosine kinase 2 (SK2), which was contained within hepatocytes exosomes, but was absent in exosomes from other liver cell types. Delivery of SK2 by exosomes induced proliferation in target hepatocytes via synthesis of intracellular sphingosine-1-phosphate (S1P). This represents a novel mechanism by which hepatocytes signal via exosomes in a paracrine fashion to promote liver repair and regeneration after injury.
Materials and methods
Mice
Male C57Bl/6 mice and SK2-knockout mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice used for experiments were 6–8 weeks of age. All animal use and procedures described were reviewed and approved by the University of Cincinnati Animal Care and Use Committee and was in compliance with the National Institutes of Health guidelines. All animals and resulting samples were assigned a number that did not reveal the group allocation so that analyses were performed by blinded investigators.
Liver cells and exosome isolation and characterization
Hepatocytes and Kupffer cells were isolated from mice as previously described [19], and 2 × 106 cells/5 ml Williams media supplemented with 5% exosome-free FBS (System Biosciences) were cultured in 50 mm plates. Cells were cultured overnight and then media was changed. Forty-eight hours later, media was harvested for exosome isolation. Primary liver sinusoidal endothelial cells from C57Bl/6 mice were purchased from Cell Biologics (Chicago, IL). 1 × 107 cells/10 ml endothelial cell medium (Cell Biologics) supplemented with 5% exosome-free FBS were cultured in T75 flasks. Media was changed when cells reached confluence and 48 h later media was harvested for exosome isolation. Exosomes were isolated using differential centrifugation and sucrose density-gradient methods as previously described [20,21].
Serum exosomes were isolated with Exoquick according to the manufacturer’s protocol (System Biosciences). Briefly, blood was obtained by cardiac puncture and 125 μl of serum was collected and mixed with Exoquick. Samples were centrifuged at 1500 g for 30 min, followed by incubation overnight at 4 °C. The supernatant was decanted and the exosome pellet was resuspended in phosphate-buffered saline (PBS).
Exosome size was determined using a Zetasizer Nano (Malvern Instruments, Malvern, UK) and the number of exosomes was assessed by the CD81-antigen ELISA kit (System Biosciences). Exosome purity was assessed by electron microscopy and by Western blot of markers of early endosome (EEA-1), endoplasmic reticulum (Grp78), and exosome (Tsg101, CD81, and CD63) compartments, respectively.
Electron microscopy
Exosomes in PBS were fixed in 1.5 M sodium cacodylate buffer (pH 7.4) and were absorbed onto copper-mesh formvar grids (Electron microscopy Sciences, Hatfield, PA) and negatively stained by 2% uranyl acetate. Samples were observed using a H7650 transmission electron microscope (Hitachi, Tokyo, Japan) operated at an accelerating voltage of 80 kV. Images were taken with an AMT digital camera for data acquisition.
Hepatocyte proliferation
Hepatocyte proliferation in vitro was determined by DNA incorporation of 5-bromo-2-deoxyuridine (BrdU). Hepatocytes were treated with hepatocyte-derived exosomes for 24 h prior to assessing BrdU incorporation. Data were normalized by the amount of viable cells and expressed as a ratio compared with medium alone. A commercial BrdU cell proliferation ELISA system (Abcam, Cambridge, UK) was used for this assay.
For histologic analysis of hepatocyte proliferation, tissue samples were fixed in 10% neutral-buffered formalin and embedded in paraffin prior to immunohistochemical staining for proliferating cell nuclear antigen (PCNA) or phosphorylated histone H3 (H3-P). Staining of PCNA was performed as previously described [22], and staining of H3-P was performed according to the manufacturer’s instructions (Ser10; dilution of 1:200, Cell Signaling Technology, Danvers, MA). Sections were counterstained with hematoxylin and quantitation was performed based on the percentage of positive nuclei of 400–600 hepatocytes from 4–6 positive fields at high power (400×).
Hepatic ischemia/reperfusion (I/R) injury and partial hepatectomy
Mice were randomly assigned to undergo either sham surgery, I/R, or partial hepatectomy as previously described [23,24]. For I/R injury, sham mice underwent the same procedure without vascular occlusion. Mice were injected intravenously with exosomes or saline (vehicle control) 24 and 48 h after reperfusion. For partial hepatectomy, mice were injected intravenously with exosomes or saline (vehicle control) immediately after and 24 h after hepatectomy.
Exosome-hepatocyte fusion
Exosomes were labeled with 2 μM PKH67 (Sigma-Aldrich) for 5 min, washed and incubated for 24 h with cultured hepatocytes. The samples were washed and counterstained with DAPI, and analyzed by fluorescence microscopy.
Measurement of sphingolipid substrates and enzymes
Ceramide was quantified by kinase assays exactly as previously described [25].
Quantification of S1P in hepatocytes was determined by ELISA and mass spectrometry. An S1P ELISA (Echelon Biosciences) was performed according to the manufacturer’s instructions. For mass spectrometry, S1P was extracted by a modified two-step lipid extraction. Briefly, cells were transferred into a glass tube and resuspended in 1 ml of medium. Then, 100 pmol C17-S1P as internal standard, 100 μl of a 3N NaOH solution, 1 ml of chloroform and 1 ml of methanol/HCl (99.8:0.2 v/v) were added. After separation, the aqueous phase was acidified with 100 μl concentrated HCl and extracted with 1.5 ml chloroform. The organic phase was evaporated and the dried lipids were resolved in 200 μl methanol. Sample analysis was performed by rapid resolution liquid chromatography/tandem mass spectrometry using a quadrupole time of flight 6530 mass spectrometer (Agilent Technologies, Waldbronn, Germany) operating in the positive electrospray ionization mode. Chromatographic separations were performed by an X-Bridge column (C18, 4.6 × 150 mm, 3.5 μm particle size, 138 Å pore size, Waters GmbH, Eschborn, Germany). Elution was performed using a gradient consisting of eluent A (water/formic acid 100:0.1 v/v) and eluent B (acetonitril/tetrahydrofuran/for mic acid 50:50:0.1 v/v/v). The precursor ions of S1P (m/z 380.2560) and C17-S1P (m/z 366.2404) were cleaved into the fragment ions of m/z 264.2700 and m/z 250.2529 respectively. Quantification was performed with Mass Hunter Software.
Neutral sphingomyelinase activity was measured by incubation of samples with 0.05 μCi per sample [14C]sphingomyelin in 100 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2.5 mM DTT, 0.2% Triton, 10 μg/ml each of aprotinin and leupeptin for 60 min at 37 °C. The [14C]sphingomyelin was dried prior to analysis, resuspended in the assay buffer, sonicated for 10 min and an aliquot was added to the samples. The reactions were analyzed as above.
Neutral ceramidase activity was measured by incubating samples in 100 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2.5 mM DTT, 0.2% Triton, 10 μg/ml each of aprotinin and leupeptin and 0.1 μCi micellar [14C16]-ceramide (ARC0831, 55 mCi/mmol). The substrate was dried prior to use, resuspended in the assay buffer and bath-sonicated for 10 min. Samples were extracted after 60 min in 200 μl H2O and CHCl3:CH3OH:HCl (100:100:1, v/v/v). The lower phase was dried, samples were resuspended in CHCl3:CH3OH (1:1, v/v) and separated by thin-layer chromatography (TLC) using CHCl3:CH3OH:ammoniumhydroxide (90:20:0.5, v/v/v) as the developing solvent. The plates were analyzed using a Fuji-Imager and ceramidase activity was determined by conversion of radioactive ceramide into sphingosine and radioactive fatty acid.
To determine SK activity, samples were incubated with 500 pmol sphingosine in the presence of 50 mM HEPES (pH 7.4), 250 mM NaCl, 30 mM MgCl2, 1 mM ATP and 10 μCi [32P]γATP for 60 min at 30 °C. Samples were extracted by addition of 20 μl 1N HCl, 800 μl CHCl3/CH3OH/1N HCl (100:200:1, v/v/v), 240 μl CHCl3 and 2 M KCl. Phases were separated, the lower phase was collected, dried, dissolved in 20 μl of CHCl3:CH3OH (1:1, v/v) and separated on Silica G60 TLC plates using CHCl3/CH3OH/acetic acid/H2O (90:90:15:5, v/v/v/v). The TLC plates were analyzed employing a phosphoimager. Sphingosine was quantified by comparison with a standard curve of C18-sphingosine and SK activity was calculated from the conversion of the standards to S1[32P].
Western blot studies
For Western blots of Tsg101, CD81, CD63, EEA-1, Grp78, α-actin, neutral ceramidase and SK in exosomes as well as SK in hepatocytes, the samples were resuspended in 5x SDS sample buffer, separated by 4–20% SDS-PAGE and developed using anti-TSG101 (ab83, Abcam Inc.), anti-CD81 (Eat2, Abd Serotec, Oxford, UK), anti-CD63 (AD1, BD Biosciences, Mountain View, CA), anti-EEA-1 (EEA-1, BD Biosciences Inc.), anti-Grp78 (BiP, BD Biosciences Inc.), α-actin (ab8227, Abcam Inc.), anti-neutral ceramidase (H300, Santa Cruz Inc.), anti-SK1 (M209, Santa Cruz Inc.), and anti-SK2 (ab37977, Abcam Inc.) antibodies followed by ECL.
siRNA-knockdown and inhibition of SK2 in exosomes
SK2 siRNA sequences was 5′-GGCUGCUCAUAUUGGUCAATT-3′ and non-targeting siRNA was the control siRNA (both from Qiagen). The siRNA duplexes were introduced into primary hepatocytes according to the manufacturer’s instructions. Briefly, hepatocytes were distributed onto 6-well plates at a concentration of 2 × 105 cells/well and incubated overnight to allow cell adherence. Transfection of siRNA was performed in the presence of Lipofectamine RNAiMAX (Invitrogen). Aliquots of siRNAs (25 pmol) and 5 μl Lipofectamine RNAiMAX per well were applied in Opti-MEM® I Reduced Serum Medium (Gibco). After 48 h, cells were then lysed and prepared for Western blot analysis. For exosome study, the medium was renewed with Williams media supplemented with 5% exosome-depleted FBS (System Biosciences) and incubated for 48 h.
To irreversibly block SK in exosomes, 20 μg isolated exosomes were treated with 1 mM 5-p-fluorosulfonylbenzoyladenosine (FSBA) for 60 min at 22 °C and for a further 16 h at 4 °C consisting of 50 mM HEPES (pH 7.4), 150 mM NaCl, 5 mM MgCl2 and 10% glycerol. The reaction was stopped by the addition of 1 mM dithiothreitol, exosomes were washed by ultracentrifugation, resuspended in Williams media supplemented with 5% exosome-removed FBS and primary hepatocytes were treated for 24 h to determine cell proliferation via BrdU incorporation.
Statistical analysis
Sample size was calculated with the two-sided Wilcoxon-Mann-Whitney test (software: G*Power Version 3.1.7). Data are expressed as the mean ± standard deviation (SD). Data were analyzed with a one-way analysis of variance, with a subsequent Student t test. Differences were considered significant when p <0.05.
Results
Hepatocyte exosomes, but not exosomes from Kupffer cells or sinusoidal endothelial cells, promote hepatocyte proliferation in vitro and liver regeneration in vivo
To characterize exosomes released by hepatocytes, primary murine hepatocytes were cultured in exosome-free media. The exosomes released into the culture media were isolated and subjected to biochemical and biophysical analyses. Analysis of exosomes by electron microscopy revealed the expected cup-shaped morphology (Fig. 1A). Quantitation of exosome size was determined using a Zetasizer Nano and revealed a mean vesicle diameter of 79 nm (Fig. 1B). While there were particle sizes identified above 100 nm, none of the exosomes observed via electron microscopy were greater than 100 nm, yet we did observe some aggregates of exosomes which may have resulted in the larger particles identified by the Zetasizer Nano. Biochemical analysis of exosomes demonstrated expression of the exosomal proteins Tsg101, CD81, and CD63, and lack of expression of proteins associated with early endosomes (EEA-1), or endoplasmic reticulum (Grp78) (Fig. 1C). All of these analyses are fully consistent with previous characterizations of hepatocyte exosomes [5].
Fig. 1. Characterization of exosomes secreted by hepatocytes.
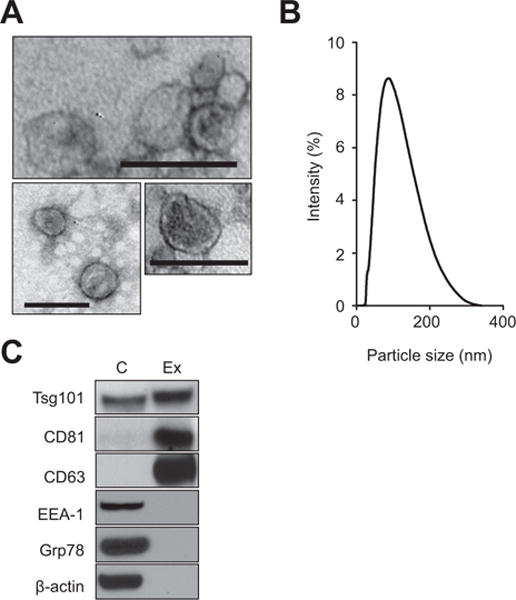
(A) Analysis of exosomes by electron microscopy (bars 100 nm). (B) Size of exosomes isolated from primary hepatocytes (mean: 79 nm). (C) Western blot analysis of cell extracts prepared from primary hepatocytes (C) and exosomes isolated from primary hepatocytes (Ex) shows expression of Tsg101, CD81, CD63, early endosomes (EEA-1), or endoplasmic reticulum (Grp78) and α-actin. Shown are Western blot studies representative for three independent experiments.
To determine if hepatocyte exosomes had any modulatory effect on hepatocyte proliferation, hepatocyte exosomes were added to primary hepatocytes in culture. The addition of hepatocyte exosomes resulted in a dose-dependent increase in hepatocyte proliferation (Fig. 2A). This was not an effect of exosomes in general, as treatment of primary hepatocytes with exosomes derived from Kupffer cells (Fig. 2B) or liver sinusoidal endothelial cells (Fig. 2C) did not induce proliferation. Interestingly, Kupffer cell exosomes actually decreased hepatocyte proliferation (Fig. 2B). Exosomes from Kupffer cells and sinusoidal endothelial cells were also analyzed biochemically and biophysically to confirm their exosome characteristics (Supplementary Fig. 1).
Fig. 2. Hepatocyte-derived exosomes induce hepatocyte proliferation in vitro.
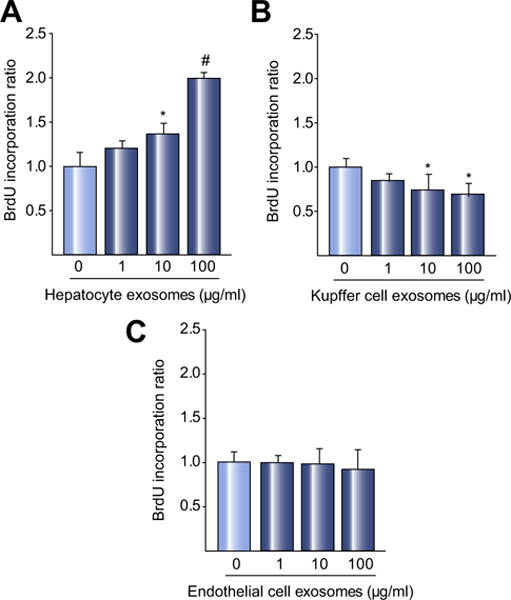
Exosomes isolated from primary hepatocytes (A), Kupffer cells (B), or sinusoidal endothelial cells (C), were added to primary hepatocytes and proliferation measured 24 h later by BrdU incorporation. Exosomes from primary hepatocytes dose-dependently increased hepatocyte proliferation. Kupffer cell exosomes reduced hepatocyte proliferation and exosomes from sinusoidal endothelial cells had no effect on proliferation. For all panels, data are mean ± SD with n = 8 (A, B) or n = 6–8 (C) per group. *p <0.05 compared to media control (0 exosomes), #p <0.05 compared to all other groups.
Because we found that hepatocyte exosomes induced hepatocyte proliferation in vitro, we next sought to determine whether hepatocyte exosomes could induce hepatocyte proliferation in vivo. I/R injury to the liver occurs within the first 24 h after injury, after which the liver begins a reparative process characterized by the clearance of dead hepatocytes and increased hepatocyte proliferation [22]. Therefore, mice were injected intravenously with hepatocyte-derived exosomes 24 and 48 h after reperfusion and hepatocyte proliferation was assessed 72 h after reperfusion. Administration of hepatocyte exosomes dose-dependently increased hepatocyte proliferation, assessed by both PCNA and H3-P staining (Fig. 3A), and had no effect on serum ALT (control: 92.3 ± 23.9, n = 6; exosomes (200 μg): 89.1 ± 49.0, n = 3).
Fig. 3. Hepatocyte-derived exosomes induce hepatocyte proliferation and liver regeneration in vivo.
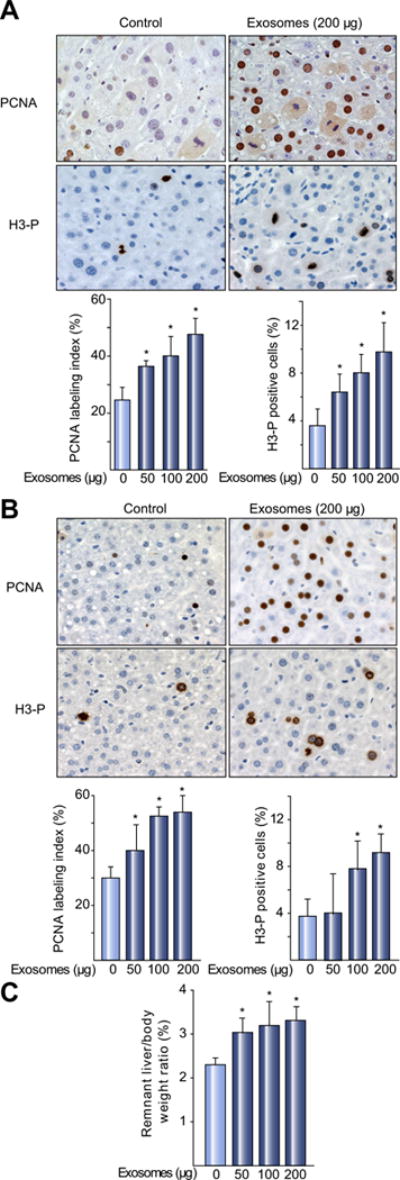
(A) Mice were injected intravenously with saline (control) or hepatocyte exosomes 24 h and 48 h after I/R. Ischemic lobes were taken for immunohistochemical and quantitative analysis of PCNA and H3-P staining 72 h after reperfusion. Original magnification was 400×. (B) Mice were injected intravenously with saline (control) or hepatocyte exosomes immediately after and 24 h after hepatectomy. Liver samples were taken for immunohistochemical and quantitative analysis of PCNA and H3-P staining 48 h after hepatectomy. Original magnification was 400×. (C) Liver regeneration was determined 48 h after hepatectomy by liver to body weight ratio. For all panels, data are the mean ± SD with n = 3–4 (A) or n = 4 (B, C) *p <0.05 compared to control (0 exosomes) mice. (This figure appears in colour on the web.)
To determine if these in vivo effects contributed to liver regeneration, mice undergoing 70% hepatectomy were injected with hepatocyte exosomes immediately after hepatectomy and again 24 h later. Administration of hepatocyte exosomes dose-dependently increased hepatocyte proliferation (Fig. 3B) and liver mass (Fig. 3C) 48 h after hepatectomy.
Hepatocyte exosomes contain specific sphingolipids and sphingolipid enzymes that are delivered to target hepatocytes
To investigate the mechanism by which exosomes induce hepatocyte proliferation, we first examined if treatment of hepatocytes with exosomes resulted in direct transfer of exosome components. Primary hepatocytes were treated with exosomes that had been labeled with PKH67, a cell membrane marker. Fig. 4 shows that exosome membranes are directly incorporated into the hepatocyte plasma membrane as well as membranes of intracellular organelles – most likely endosomes.
Fig. 4. Exosomes directly fuse with and are internalized by target hepatocytes.
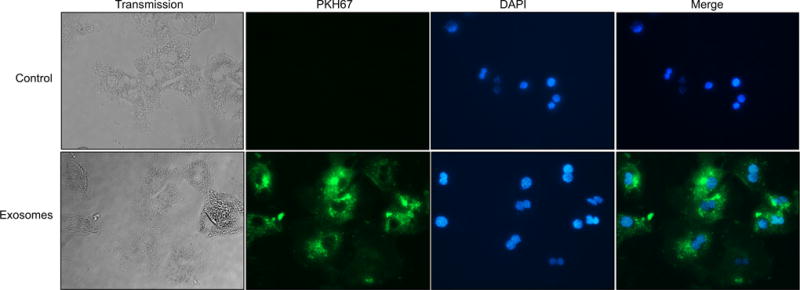
Hepatocyte-derived exosomes were fluorescently labeled with the cell membrane marker, PKH67, prior to incubation with primary hepatocytes for 24 h. After incubation, hepatocytes were washed and counterstained with DAPI to stain the nuclei. Control hepatocytes were treated with PBS vehicle. Shown is a representative result from four independent experiments. (This figure appears in colour on the web.)
The sphingomyelin-ceramide pathway is critical for the generation of exosomes [11]. Furthermore, one of the end products of this pathway, S1P has been linked to proliferation in cell types other than hepatocytes [26–29]. Hepatocyte exosomes were analyzed for their content of sphingolipids and sphingolipid enzymes. We found that exosomes contained abundant amounts of ceramide, but no detectable S1P (Table 1). There was no detectable neutral sphingomyelinase activity, but neutral ceramidase and SK activity was abundant (Table 1). The presence of neutral ceramidase and SK2 were confirmed by Western blot (Fig. 5A), but SK1 was undetectable (data not shown). Exosomes from Kupffer cells or sinusoidal endothelial cells, which lacked proliferative effects, contained no detectable SK activity or SK2 protein (data not shown).
Table 1.
Expression of S1P and ceramide and activity of neutral sphingomyelinase, sphingosine kinase, and neutral ceramidase in hepatocyte exosomes.
| Exosomes | |
|---|---|
| Ceramide [pmol/μg] | 2.81 ± 0.97 |
| S1P [pmol/μg] | n.d. |
| Neutral sphingomyelinase activity [pmol/hr/μg] | n.d. |
| Neutral ceramidase activity [pmol/hr/μg] | 0.30 ± 0.08 |
| Sphingosine kinase activity [fmol/hr/μg] | 125.8 ± 43.0 |
n.d., not detectable. Data are mean ± SD, n = 4.
Fig. 5. Hepatocyte-derived exosomes deliver SK2 to target hepatocytes and induce hepatocyte proliferation via generation of S1P.
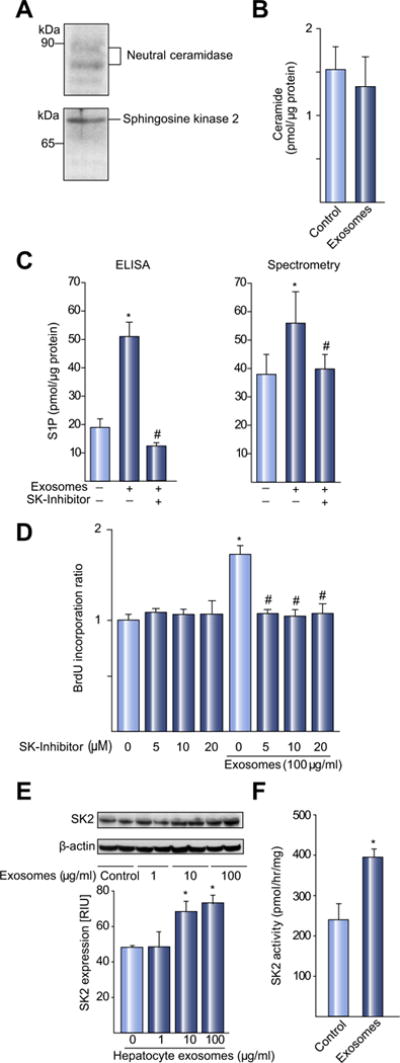
(A) Western blot analysis of hepatocyte-derived exosomes shows expression of neutral ceramidase and SK2 proteins. Shown are Western blot studies representative for four independent experiments. (B) Primary hepatocytes were treated with media (control) or 100 μg/ml hepatocyte exosomes and cellular levels of S1P and ceramide were determined. Ceramide levels in hepatocytes were not changed by treatment with exosomes. (C) Exosomes induced S1P as determined by ELISA and mass spectrometry. The induction of S1P in hepatocytes by exosomes was prevented by pre-incubation with the SK inhibitor SPHK I2 (20 μM). For A–C, data are mean ± SD, n = 4; *p <0.05 compared to control group; #p <0.05 compared to exosome only group. (D) Exosome-induced proliferation of hepatocytes is abrogated by the SK inhibitor, SPHK I2. Cell proliferation was determined via BrdU corporation. Data are mean ± SD, n = 3. *p <0.05 compared to control group (0 exosomes, no SK inhibitor); #p <0.05 compared to exosomes only group. (E) Hepatocyte-derived exosomes deliver SK2 to target hepatocytes. Western blot analysis of SK2 in hepatocytes treated with media (control) or increasing amounts of hepatocyte-derived exosomes. SK2 expression was measured 24 h after treatment and normalized to α-actin and quantitative analysis performed. Data are mean ± SD with n = 4 per group. *p <0.05 compared to control (0 exosomes) hepatocytes. (F) SK activity in target hepatocytes was measured by kinase assays 24 h after treatment with media (control) or 100 μg/ml hepatocyte exosomes. Data are mean ± SD with n = 4 per group. *p <0.05 compared to control hepatocytes.
Because we had evidence of direct transfer of exosome contents into target hepatocytes (Fig. 4), we next measured the content of sphingolipids in hepatocytes after treatment with hepatocyte exosomes. Ceramide levels were not changed by exosome treatment (Fig. 5B). However, intracellular levels of S1P were significantly increased after treatment with exosomes (Fig. 5C). The increase in S1P levels was completely abrogated by co-treatment of hepatocytes with the SK inhibitor, SPHK I2 (Fig. 5C). Furthermore, treatment of hepatocytes with the SK inhibitor also completely inhibited the proliferation of hepatocytes induced by hepatocyte exosomes (Fig. 5D). These results suggest that the delivery of SK2 by exosomes results in the generation of S1P within target hepatocytes resulting in cell proliferation.
In order to test this hypothesis, we examined whether treatment with exosomes resulted in increased expression and activity of SK2 in target hepatocytes. Treatment of hepatocytes with exosomes increased SK2 expression by Western blot (Fig. 5E) and nearly doubled the level of SK2 activity within the hepatocytes (Fig. 5F).
Hepatocyte exosomes transfer S1P synthetic machinery to promote hepatocyte proliferation
To directly test the hypothesis that delivery of SK2 by exosomes is the mechanism associated with the increased S1P levels and subsequent proliferative effects, we conducted three complementary experiments. First, we transfected hepatocytes with scrambled control siRNA or siRNA targeting SK2, the latter which effectively knocked down the expression of SK2 (Fig. 6A). We then isolated exosomes from these cells and measured their ability to induce hepatocyte proliferation. Treatment of hepatocytes with SK2-deficient exosomes resulted in no cellular proliferation (Fig. 6B). Second, we isolated exosomes from hepatocytes from SK2-knockout mice and measured their ability to induce hepatocyte proliferation. Treatment of hepatocytes with exosomes from SK2-knockout hepatocytes resulted in no cellular proliferation (Fig. 6B). The third approach directly blocked the activity of SK2 in exosomes by treating normal hepatocyte exosomes with FSBA, an irreversible inhibitor that binds covalently to the ATP binding site of sphingosine kinases [30]. Biochemical inhibition of exosome SK2 with FSBA completely abrogated the proliferative effect of exosomes on hepatocytes (Fig. 6C). These data prove that exosomes deliver SK2 to the target hepatocytes resulting in intracellular generation of S1P which promotes cell proliferation.
Fig. 6. SK2 mediates the proliferative effects of hepatocyte exosomes.
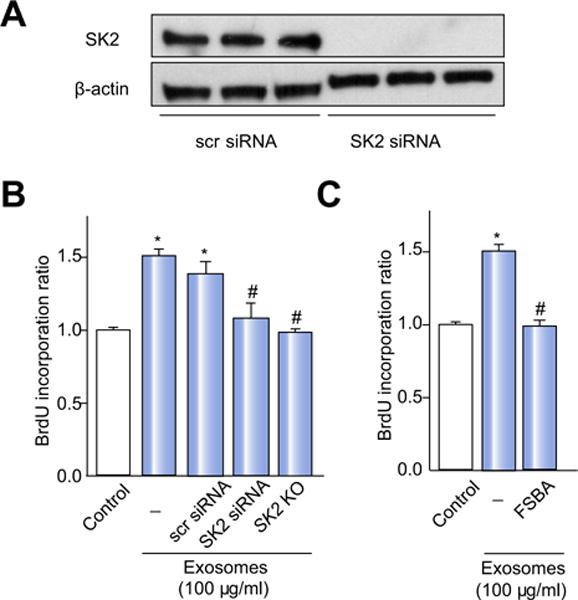
(A) Transfection of hepatocytes with siRNA targeting SK2 downregulates SK2 expression, while a control scrambled (scr) siRNA has no effect. Shown are Western blot studies representative of three independent experiments. (B) Exosomes derived from hepatocytes in which SK2 had been knocked down or SK2-knockout (KO) hepatocytes have no proliferative effect on target hepatocytes in vitro. (C) Biochemical inhibition of SK with FSBA prevents the proliferative effects of exosomes on hepatocytes. For all panels, data are the mean ± SD with n = 4; *p <0.05 compared to control group; #p <0.05 compared to scr siRNA group.
Next, to demonstrate that SK2 contained within exosomes was responsible for hepatocyte proliferation in vivo, we isolated exosomes from hepatocytes in which SK2 had been knocked down with siRNA as well as exosomes from SK2-knockout hepatocytes. Similar to previous experiments, mice were injected with exosomes 24 and 48 h after I/R. As expected, treatment of mice with exosomes from hepatocytes treated with control, scrambled siRNA (Fig. 7A), or with exosomes from wild-type hepatocytes (Fig. 7B), resulted in significant hepatocyte proliferation. In contrast, treatment with exosomes from hepatocytes in which SK2 had been knocked down with siRNA (Fig. 7A) or from SK2-knockout hepatocytes (Fig. 7B) failed to induce hepatocyte proliferation.
Fig. 7. SK2 mediates the proliferative effects of hepatocyte exosomes in vivo.
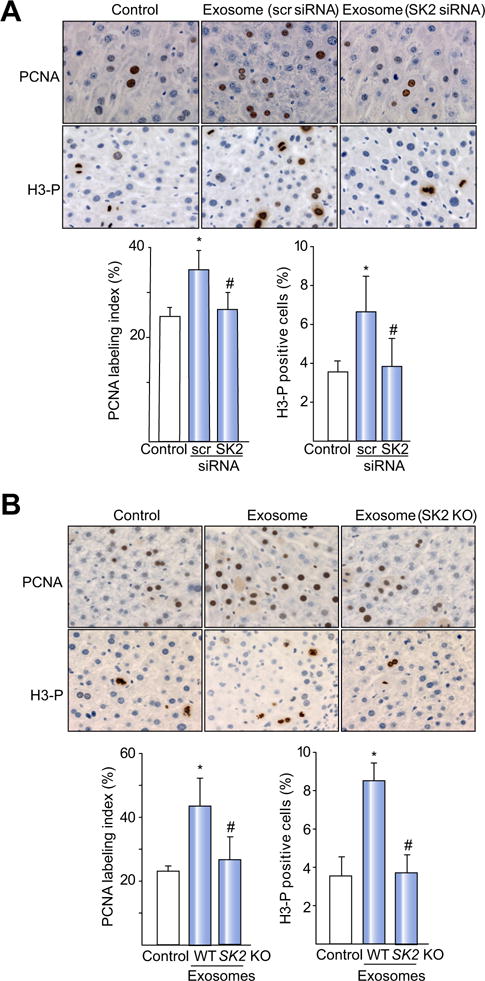
Mice were injected intravenously with saline (control) or hepatocyte-derived exosomes 24 h and 48 h after I/R. (A) Treatment with exosomes isolated from hepatocytes that had been transfected with a scrambled, control siRNA (scr siRNA) or siRNA targeting SK2 (SK2 siRNA). (B) Treatment with exosomes isolated from wild-type (WT) or SK2-knockout (SK2 KO) hepatocytes. Hepatocyte proliferation was determined by immunohistochemical and quantitative analysis of PCNA and H3-P staining 72 h after reperfusion. Original magnification was 400×. For all panels, data are the mean ± SD with n = 4 (A), 4–5 (B); *p <0.05 compared to control group; #p <0.05 compared to scr siRNA group (A). #p <0.05 compared to hepatocyte-derived exosomes group (B). (This figure appears in colour on the web.)
Exosomes released after liver injury have hepatocyte proliferative effects
To test whether liver injury results in a release of exosomes in vivo, we measured the number of exosomes in serum before and after hepatic I/R injury. Exosomes were present in the serum of untreated mice and increased after I/R with maximum levels 24 h after reperfusion and returning to baseline levels 96 h after reperfusion (Fig. 8A). Exosomes isolated from serum 24 h after I/R injury were characterized biophysically and biochemically. Electron microscopy showed the cup-shaped morphology of exosomes (Fig. 8B). Western blot analyses demonstrated that the isolated exosomes expressed the exosome proteins, Tsg101, CD81, and CD63, but lacked the early endosome protein, EAA-1, and the endoplasmic reticulum protein, Grp78 (Fig. 8C). Similar to hepatocyte-derived exosomes, the presence of SK2 was confirmed by Western blot, but SK1 was undetectable (Fig. 8C). Zetasizing of serum exosomes showed a mean diameter of 74 nm (Fig. 8D), which was consistent with our electron micrographs (Fig. 8B). We next determined if serum exosomes induced by I/R injury could stimulate hepatocyte proliferation. Primary hepatocytes treated with exosomes isolated 24 h after I/R injury displayed a dose-dependent increase in cell proliferation (Fig. 8E).
Fig. 8. Hepatic I/R injury induces release of exosomes into the serum.
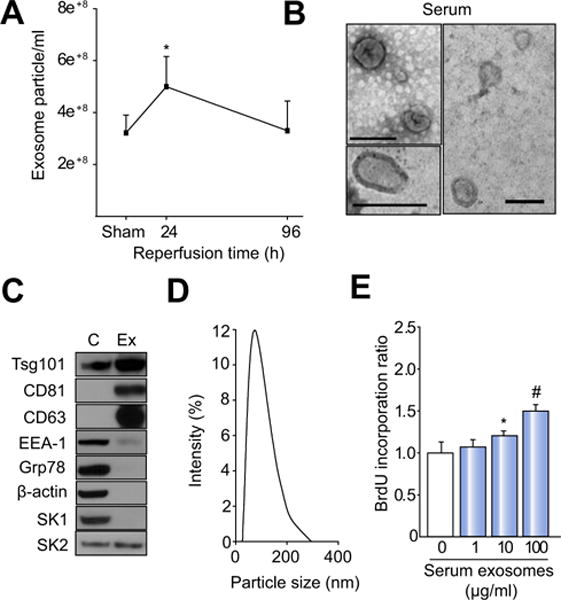
(A) Serum exosome levels over a 96 h time course of reperfusion. Data are mean ± SD with n = 7 per group (sham group) and n = 7 per group (I/R groups). *p <0.05 compared to sham control. (B) Characterization of serum exosomes isolated 24 h after I/R injury by electron microscopy (bars, 100 nm). (C) Western blot analysis of control hepatocyte lysates (C) and exosomes isolated from serum 24 h after I/R injury (Ex) for expression of Tsg101, CD81, CD63, early endosomes (EEA-1), endoplasmic reticulum (Grp78), α-actin, SK1, and SK2. (D) Size of exosomes isolated from serum (mean: 74 nm). (E) Exosomes isolated from serum 24 h after I/R injury were added to primary hepatocytes and proliferation measured 24 h later by BrdU incorporation. Data are the mean ± SD with n = 8; *p <0.05 compared to control group (0 exosomes); #p <0.05 compared to all other groups.
Discussion
Here we describe a novel and highly significant function of hepatocyte exosomes. Our studies show that exosomes released by hepatocytes fuse with, and promote the proliferation of target hepatocytes, both in vitro and in vivo. We found that hepatocyte exosomes transfer the synthetic machinery to produce S1P and that intracellular generation of S1P is required for exosome-induced proliferation. Our findings greatly extend our current understanding of the function of hepatocyte-derived exosomes and suggest they may function to restore homeostasis after liver injury, dysfunction or disease. Furthermore, our data suggest that hepatocyte exosomes may represent a novel therapeutic modality.
A previous study has suggested that hepatocyte-derived exosomes function as potential biomarkers of liver disease [10]. Our data support that concept as we found increased circulating levels of exosomes after hepatic I/R injury. However, our current data reveal that hepatocyte exosomes do much more than simply signal an injury, but may represent a novel mechanism to facilitate tissue repair and regeneration. By packaging specific cargo, hepatocyte exosomes appear to function as an important form of paracrine intercellular communication. The fact that exosomes from other liver cell types lacked the key cargo, SK2, and were unable to induce hepatocyte proliferation demonstrates a specific characteristic of hepatocyte exosomes.
Another report has shown that exosomes isolated from mesenchymal stem cells have proliferative effects on hepatocytes after liver injury induced by carbon tetrachloride [31]. While that study is similar to ours in that exogenously-applied exosomes triggered liver repair, our data suggests that hepatocytes produce exosomes in response to injury as a physiological feedback mechanism to trigger homeostatic liver regeneration. The dose-dependent proliferative effects of hepatocyte exosomes suggests that the increased number of hepatocyte exosomes released after liver injury contribute to liver repair and regeneration. Further, we have identified the mechanism and components by which exosomes induce hepatocyte proliferation allowing the rational design of lipid particles that do not contain other potentially antigenic molecules and could be repeatedly applied to patients with acute and/or chronic liver diseases.
Our studies demonstrate that the proliferative effects of hepatocyte exosomes are mediated by intracellular generation of S1P. It is important to note that our results suggest it is intracellular S1P and not extracellular S1P that mediates these proliferative effects. We show that exosomes fuse with hepatocytes and deliver their contents intracellularly. Furthermore, treatment of hepatocytes with extracellular S1P results in cell death (data not shown). This effect has been confirmed in hepatocytes in vitro and on liver regeneration in vivo by other investigators [32,33]. In contrast, intracellular S1P has been shown to promote cell proliferation in a variety of cell types [26–29]. By delivering both a substrate (ceramide) and modifying enzymes (neutral ceramidase and SK2), exosomes deliver the synthetic machinery required to produce S1P in target hepatocytes resulting in their proliferation.
In conclusion, the current study is the first to describe the manner in which hepatocyte-derived exosomes induce proliferation in hepatocytes in vitro and in vivo. We have delineated the mechanism of this proliferative effect by discovering that exosomes contain components of the sphingolipid pathway that are delivered intracellularly to target hepatocytes leading to intracellular generation of S1P. Knockout, knockdown, or inhibition of the terminal enzyme for S1P synthesis, SK2, in exosomes abrogates their proliferative effect. Collectively, these findings suggest a novel contributory mechanism of liver regeneration, as well as a potential therapeutic application for a variety of liver diseases and transplantation.
Supplementary Material
Acknowledgments
Financial support
This work was supported in part by National Institutes of Health grants DK56029 and AG025881 to A.B.L.
Abbreviations
- SK
sphingosine kinase
- S1P
sphingosine-1-phosphate
- BrdU
5-bromo-2-deoxyuridine
- I/R
ischemia/reperfusion
- ELISA
enzyme-linked immunosorbant assay
- PCNA
proliferating cell nuclear antigen
- H3-P
phosphorylated histone H3
- FSBA
fluorosulfonylbenzoyladenosine
Footnotes
Conflict of interest
The authors who have taken part in this study declared no conflict of interest with respect to this manuscript.
Author’s contributions
Study concept and design (M.J.E., E.G., A.B.L.); acquisition of data (H.N., C.M.F., R.M.S., L.J., B.K.); analysis and interpretation of data (H.N., M.J.E., E.G., A.B.L.); drafting of manuscript (H.N., C.M.F., A.B.L.); critical revision of manuscript (H.N., E.G., A.B.L.).
Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jhep.2015.07.030.
References
- 1.Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Compr Physiol. 2013;3:485–513. doi: 10.1002/cphy.c120014. [DOI] [PubMed] [Google Scholar]
- 2.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 3.Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113:3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- 4.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, Valle M, et al. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J Proteome Res. 2008;7:5157–5166. doi: 10.1021/pr8004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 7.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 9.van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, et al. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121:337–349. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- 10.Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56:1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 12.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- 16.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A. 2013;110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 2014;10:e1004424. doi: 10.1371/journal.ppat.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamai K, Shiina M, Tanaka N, Nakano T, Yamamoto A, Kondo Y, et al. Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology. 2012;422:377–385. doi: 10.1016/j.virol.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Sakai N, Van Sweringen HL, Quillin RC, Schuster R, Blanchard J, Burns JM, et al. Interleukin-33 is hepatoprotective during liver ischemia/reperfusion in mice. Hepatology. 2012;56:1468–1478. doi: 10.1002/hep.25768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012:e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology/editorial board, Juan S Bonifacino [et al] 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3: Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 22.Kuboki S, Shin T, Huber N, Eismann T, Galloway E, Schuster R, et al. Hepatocyte signaling through CXC chemokine receptor-2 is detrimental to liver recovery after ischemia/reperfusion in mice. Hepatology. 2008;48:1213–1223. doi: 10.1002/hep.22471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lentsch AB, Yoshidome H, Cheadle WG, Miller FN, Edwards MJ. Chemokine involvement in hepatic ischemia/reperfusion injury in mice: roles for macrophage inflammatory protein-2 and KC. Hepatology. 1998;27:1172–1177. doi: 10.1002/hep.510270440. [DOI] [PubMed] [Google Scholar]
- 24.Wilson GC, Kuboki S, Freeman CM, Nojima H, Schuster RM, Edwards MJ, et al. CXC chemokines function as a rheostat for hepatocyte proliferation and liver regeneration. PLoS One. 2015;10:e0120092. doi: 10.1371/journal.pone.0120092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pewzner-Jung Y, Tavakoli Tabazavareh S, Grassme H, Becker KA, Japtok L, Steinmann J, et al. Sphingoid long chain bases prevent lung infection by Pseudomonas aeruginosa. EMBO Mol Med. 2014;6:1205–1214. doi: 10.15252/emmm.201404075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olivera A, Kohama T, Edsall L, Nava V, Cuvillier O, Poulton S, et al. Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. J Cell Biol. 1999;147:545–558. doi: 10.1083/jcb.147.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limaye V, Vadas MA, Pitson SM, Gamble JR. The effects of markedly raised intracellular sphingosine kinase-1 activity in endothelial cells. Cell Mol Biol Lett. 2009;14:411–423. doi: 10.2478/s11658-009-0008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebrahimian T, Arfa O, Simeone S, Lemarie CA, Lehoux S, Wassmann S. Inhibition of four-and-a-half LIM domain protein-2 increases survival, migratory capacity, and paracrine function of human early outgrowth cells through activation of the sphingosine kinase-1 pathway: implications for endothelial regeneration. Circ Res. 2014;114:114–123. doi: 10.1161/CIRCRESAHA.113.301954. [DOI] [PubMed] [Google Scholar]
- 29.Van Brocklyn JR, Lee MJ, Menzeleev R, Olivera A, Edsall L, Cuvillier O, et al. Dual actions of sphingosine-1-phosphate: extracellular through the Gi-coupled receptor Edg-1 and intracellular to regulate proliferation and survival. J Cell Biol. 1998;142:229–240. doi: 10.1083/jcb.142.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitson SM, Moretti PA, Zebol JR, Zareie R, Derian CK, Darrow AL, et al. The nucleotide-binding site of human sphingosine kinase 1. J Biol Chem. 2002;277:49545–49553. doi: 10.1074/jbc.M206687200. [DOI] [PubMed] [Google Scholar]
- 31.Li T, Yan Y, Wang B, Qian H, Zhang X, Shen L, et al. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate liver fibrosis. Stem Cells Dev. 2013;22:845–854. doi: 10.1089/scd.2012.0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda H, Satoh H, Yanase M, Inoue Y, Tomiya T, Arai M, et al. Antiproliferative property of sphingosine 1-phosphate in rat hepatocytes involves activation of Rho via Edg-5. Gastroenterology. 2003;124:459–469. doi: 10.1053/gast.2003.50049. [DOI] [PubMed] [Google Scholar]
- 33.Ikeda H, Watanabe N, Ishii I, Shimosawa T, Kume Y, Tomiya T, et al. Sphingosine 1-phosphate regulates regeneration and fibrosis after liver injury via sphingosine 1-phosphate receptor 2. J Lipid Res. 2009;50:556–564. doi: 10.1194/jlr.M800496-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


