Abstract
To better understand the relative contributions of mesenchymal and endothelial progenitor cells to rhBMP-2 induced bone formation, we examined the distribution of lineage-labelled cells in Tie2-Cre:Ai9 and αSMA-creERT2:Col2.3-GFP:Ai9 reporter mice. Established orthopedic models of ectopic bone formation in the hind limb and spine fusion were employed. Tie2-lineage cells were found extensively in the ectopic bone and spine fusion masses, but co-staining was only seen with tartrate-resistant acid phosphatase (TRAP) activity (osteoclasts) and CD31 immunohistochemistry (vascular endothelial cells), and not alkaline phosphatase (AP) activity (osteoblasts). To further confirm the lack of a functional contribution of Tie2-lineage cells to BMP-induced bone, we developed conditional knockout mice where Tie2-lineage cells are rendered null for key bone transcription factor osterix (Tie2-cre:Osxfx/fx mice). Conditional knockout mice showed no difference in BMP-induced bone formation compared to littermate controls. Pulse labelling of mesenchymal cells with Tamoxifen in mice undergoing spine fusion revealed that αSMA-lineage cells contributed to the osteoblastic lineage (Col2.3-GFP), but not to endothelial cells or osteoclast populations. These data indicate that the αSMA+ and Tie2+ progenitor lineages make distinct cellular contributions to bone formation, angiogenesis, and resorption/remodeling.
Keywords: Bone morphogenetic proteins, BMPs, lineage tracking, spine fusion, endothelial cells, Tie2, transgenic reporter mice
Introduction
Bone is a tissue that developmentally arises from the mesoderm. This is regulated by a variety of signals, however expression of the bone morphogenetic proteins (BMPs) are key factors that modulate this process (1). Recombinant human BMPs (rhBMPs) are utilized in orthopedic medicine as potent inducers of new bone formation and are approved for the treatment of open fractures and non-unions (2). When used clinically, cells are exposed to super-physiological concentrations of these factors, which has the potential to transform cells beyond those of the mesenchymal lineages.
In 2009 a study by Lounev et al. examined the contribution of different cell lineages to bone in a model of rhBMP-2 induced ectopic bone formation and in the Nse-BMP4 transgenic mouse (3). A variety of transgenic mouse strains employing the Rosa26R (R26R) reporter were used to assess the relative input of MyoD-lineage, Tie2-lineage, and SMMHC-lineage cells. These were intended to reflect the contributions of myogenic, endothelial and mesenchymal progenitors respectively. Ectopic bone formed readily in muscle, a tissue that is often adjacent to bone and has been speculated to contain numerous types of cellular progenitors that could contribute to bone formation and repair (4,5).
Lounev et al. reported that MyoD-lineage cells made a negligible contribution to rhBMP-2 induced ectopic bone formation (3). This was later contrasted by studies showing greater MyoD-lineage contribution in a variant of the ectopic bone formation model featuring rhBMP-7 as well as increased tissue trauma (6). Intriguingly, Lounev et al. found that up to 50% of cells in the ectopic bone were of the Tie2-lineage. In a subsequent study, the group described a mechanism by which Tie2-lineage cells could undergo an endothelial-mesenchymal transition (EMT) to form osteoblasts (7). While this study focused on describing the genetic disease fibrodysplasia ossificans progressiva, it was unclear whether EMT could also occur when endothelial cells were exposed to high concentrations of rhBMPs.
The potential of endothelial Tie2-lineage cells to contribute to bone repair has been further implied by a range of cell transplantation studies. Purified endothelial progenitor cells (EPCs) have been repeatedly demonstrated to enhance orthopedic repair (8,9,10,11,12), although the precise mechanism for this is unclear. EPCs have been shown to enhance angiogenesis, potentially by secretion of pro-angiogenic growth factors (13), or by direct contribution to new vessels. Transplanted EPCs have also been shown to enhance tissue mineralization (9), raising the possibility of a direct contribution of these cells to bone repair.
In 2012 Wosczyna et al. published a report indicating that the Tie2-lineage cells included multiple sub-populations of cells, some capable and some incapable of contributing to rhBMP-2 induced cartilage and bone formation (14). The populations that formed cartilage and bone were not of endothelial origin, suggesting that EMT was not the underlying mechanism in this model. Both the Lounev and Wosczyna studies utilized a Tie2-cre line that demarks endothelial and hematopoietic, progenitors as well as a subset of mesenchymal-like progenitors (15).
In this study we report the contribution of endothelial cells to rhBMP-2 induced ectopic bone formation and spine fusion using an alternative Tie2-cre mouse, which shows greater specificity for the endothelial lineages (16,17). These cell tracking studies employ a Cre-dependent fluorescent tdTomato reporter that allows for co-labeling with chondrocyte, osteoblastic, osteoclastic, and vascular markers (18). Conditional knockout mouse models were also employed where Tie2-lineage cells selectively inactivated the key osteogenic transcription factor osterix. This approach allowed for the functional assessment of the contribution of cells of the Tie2-lineage to new bone formation. To complement these experiments we examined the contribution of mesenchymal progenitors using mice with an inducible αSMA-creERT2 transgene. Prior studies have shown that this cell lineage directly contributes to bone formation and repair (19,20,21,22).
Materials & Methods
Mouse lines and Genotyping
The Ai9 reporter line (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J) was purchased from Jackson Laboratories (18). The Tie2-cre line (B6.Cg-Tg(Tek-cre)12Flv/J) was sourced from the Garvan Institute for Medical Research with permission from the original laboratory (23). The Osxfx/fx line was sourced from Benoit de Crombrugghe (MD Anderson Cancer Center, Houston, TX, USA) (15). The Tie2-cre × Ai9 line was generated in-house by cross breeding and showed no adverse phenotype; the line specifically labels Tie2-lineage with a fluorescent tdTomato reporter. The Tie2-cre × Osxfx/fx line was generated by crossing for two generations to create homozygous conditional double knockout mice; in this line there is targeted disruption of the key osteoblastic gene osterix (Osx) in all cells of the Tie2-lineage. Mice were genotyped from ear biopsies using real time PCR with specific probes designed for each gene (Transnetyx, Cordova, TN, USA). Experiments using the Tie2-cre cross strains were approved by the CHW/CMRI Animal Ethics Committee (Protocol K248, K303). All strains were on the C57BL6/J background, and hemizygous strains were maintained on wild type mice purchased from the Animal Resources Centre (ARC, Perth, Australia).
αSMA-creERT2 (smooth muscle α-actin promoter) × Ai9 mice, which enable labeling of mesenchymal progenitors (19,20) that also incorporated a GFP transgene under the control of the upstream 2.3kb region of the Col1a1 promoter (Col2.3-GFP transgene) to label osteoblasts (24) were sourced from in-house colonies at the University of Connecticut (Farmington, CT, USA). Experiments using these mice were approved by the UConn animal ethics committees.
Surgical models
Implants were manufactured by adding 20 μl sterile saline containing 5μg recombinant human BMP-2 (Medronic Australasia, North Ryde, Australia) onto collagen-hydroxyapatite sponge prior to surgery. For the ectopic bone formation model, uniform discs manufactured using a surgical tissue punch were surgically implanted into the hind limbs of mice using published methods (25). For the spine fusion model, twin porous collagen sponges (Medronic Australasia) were inserted parallel to the vertebrae as previously described (26). All mice were aged 8–12 weeks. Anesthesia was induced with inhaled isofluorane (hind limb model) or using Ketamine/Xylazine (35mg/kg; 5mg/kg) (spine fusion model). Buprenorphine (0.05–0.1 mg/kg) was administered subcutaneously (s.c.) preoperatively and every 12 hours as required for pain management. Post-surgery saline was given s.c. to prevent dehydration. Mice of the αSMA-creERT2 × Ai9 cross received tamoxifen at a dose of 75 mg/kg on the day of surgery to induce lineage labelling. Mice were euthanized at 7, 14, or 21 days (ectopic bone) or 2, 10, or 17 days (spine fusion) for radiological and/or histological analyses.
Radiological imaging
The formation of rhBMP-2 induced bone was confirmed and visualized by digital X-ray (25 kV, 2× magnification; Faxitron X-ray Corp, Illinois, USA). For studies where ectopic bone was quantified, samples were scanned by micro-computed tomography (microCT) using a SkyScan 1174 compact microCT scanner (Kontich, Belgium) at a pixel resolution of 14.8 μm. All samples were scanned in 70% ethanol, using a 0.5 mm aluminum filter, 50 kV X-ray tube voltage, and 800 μA tube electric current. A global threshold to define bone tissue in pellets was set at a mineral density of 0.3 g/cm3. Images were reconstructed using NRecon, version 1.6.1.7 (SkyScan), and analyzed using CTAnalyser software, version 1.11.8.0 (SkyScan).
Immunofluorescence and Microscopy
The fluorescent signal from the tdTomato reporter strain was captured using either an epi-fluorescent microscope or a Leica TCS SP5 confocal laser scanning microscope. The sections were cover-slipped and the intrinsic fluorescent signal imaged directly. ELF97 Phosphatase Substrate (Molecular Probes) was used to detect alkaline phosphatase (AP) and tartrate-resistant acidic phosphatase (TRAP). AP assay was done according to the manufactures instructions. For TRAP: The classical TRAP buffer (110 mM acetate buffer, pH 5.2, 1.1 mM sodium nitrite, 7.4 mM tartrate) was used followed by 200μM concentration of ELF97 incubation for 5-min. Slides were washed with an EDTA containing wash buffer. The fluorescence was visualized with a DAPI/Hoechst longpass filter set.
For examining co-labelling between tdTomato signal and immunofluorescent staining for lineage markers, the following methods were used. Slides were rehydrated in PBS for 20 min and treated with 0.5% Triton X-100 for 20 min. The sections were blocked in 10% goat serum PBS for 1 h at room temperature prior to incubation with a primary antibody (SOX9, Millipore AB5535; CD31, BD Pharmigen MEC13.3) in blocking buffer overnight at 4°C. After washing the sections were incubated with Alexa-Fluor-647 conjugated secondary antibody (Molecular Probes diluted in PBS. Tissue was counter stained with 200 ng/ml 4',6-diamidino-2-pheylindole (DAPI, molecular probes) for 1 min and washed in PBS and mounted in Aqueous Mounting Medium (DAKO). Images were captured using an epi-fluorescent microscope with appropriate filters or using the Leica TCS SP5 confocal laser scanning microscope
Statistical analyses
Statistical analysis of microCT data was conducted with non-parametric testing as amount of bone formed by these assays has not been determined to follow a normal distribution. Significance differences in bone volume were determined using a Mann Whitney U test with a cutoff of α<0.05 (Graphpad Prism, La Jolla, CA, USA).
Results
Tie2-lineage cells localize to rhBMP-2 induced bone
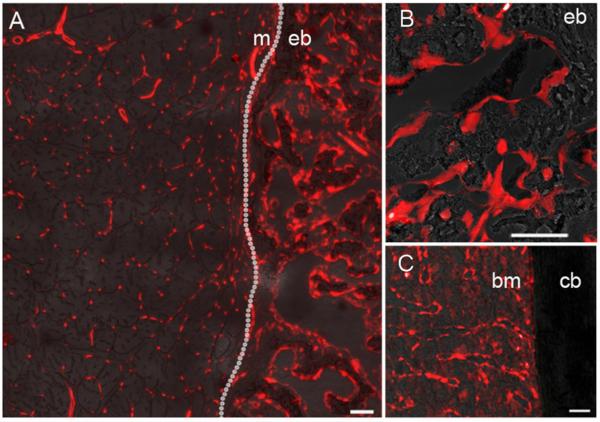
To assess the contribution of Tie2-lineage cells to newly formed bone we performed a model of rhBMP-2 induced ectopic bone formation in Tie2-cre:Ai9 reporter mice. Ectopic bone was present at 2 weeks by X-ray and specimens were analyzed by fluorescent imaging. The muscle adjacent to the ectopic bone had labelling with the appearance of microvessels, and labelling was absent from the muscle fibers (Figure 1A). The ectopic bone showed abundant staining suggestive of a significant involvement of Tie2-lineage cells in the bone formation process. Closer examination revealed an abundance of Tie2-lineage cells on the bone surfaces (Figure 1B).
Figure 1.
Confocal fluorescent images of Tie2-lineage cells (tdTomato, red) in rhBMP-2 induced muscle, native bone, bone marrow, and rhBMP-2 induced ectopic bone. Images are shown at the muscle/ectopic bone interface (A) within an ectopic bone nodule (B), and within the native bone of the femur (C). Text labels, m = muscle, eb = ectopic bone, bm = bone marrow, cb = cortical bone. Scale bar = 50 μm.
Examination of native long bones revealed negligible staining in the cortical bone, although again vessel-like staining was seen in the bone marrow space (Figure 1C).
Tie2-lineage cells contribute to the vasculature and to resorbing osteoclasts in native and rhBMP-2 induced bone
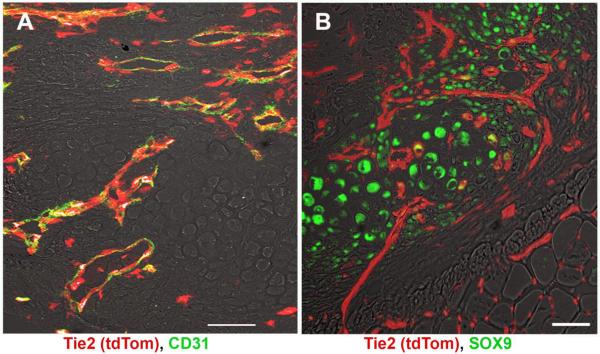
Specimens from d7 were analyzed to look at the contribution of Tie2-lineage cells to early progenitors. Co-staining for endothelial cell marker CD31 revealed that all new vessels were derived from Tie2-lineage cells (Figure 2A). Some cells that were of the Tie2-lineage that did not co-express CD31 were present, often near or adjacent to vessels. Co-staining for cartilage marker SOX9 showed negligible overlap (1–2%) between or co-staining between chondrocytes and Tie2-lineage cells (Figure 2B).
Figure 2.
Fluorescent images of ectopic bone showing Tie2-lineage cells (tdTomato) overlaid with vascular marker CD31 (A) or chondrocyte marker SOX9 (B). Tie2-lineage cells were found to demark the new blood vessels in the ectopic bone but not cartilage islands. Scale bar = 50 μm
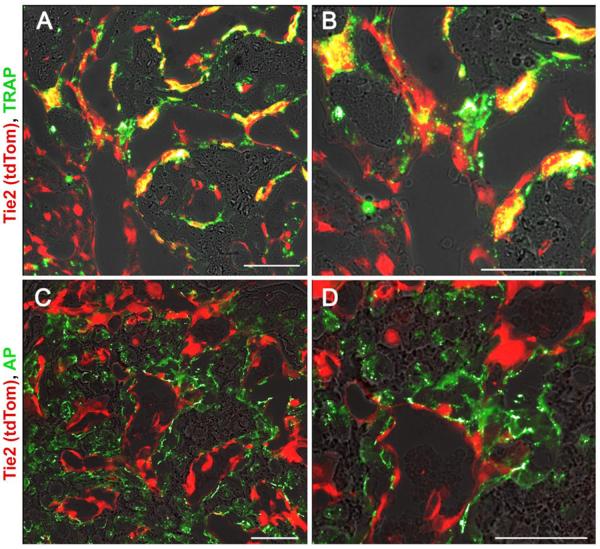
Fluorescent histochemical staining was performed on d14 sections for the osteoclast marker TRAP and the osteoblast marker AP. TRAP+ cells co-labelled with tdTomato, indicating they originated from the Tie2-lineage (Figure 3A–B). In contrast, there was no overlap between tdTomato and AP (Figure 3C–D).
Figure 3.
Fluorescent images of ectopic bone showing Tie2-lineage cells (tdTomato) overlaid with tartrate-resistant acid phosphatase/TRAP activity stain (ELF97, green) to label osteoclasts (A, zoom in B). No overlap was seen between Tie2-lineage cells with an alkaline phosphatase/AP activity stain (ELF97) to label osteoblasts (C, zoom in D). Scale bar = 50 μm.
Targeted deletion of osterix in Tie2-lineage cells does not affect rhBMP-2 induced bone formation
To further examine the functional importance of Tie2-lineage cells in this model, experiments were performed in Tie2-cre:Osxfx/fx conditional knockout mice. Osx has been previously shown to be essential for the osteogenic differentiation of osteochondral progenitors (27) and thus this line could be used to assess whether Tie2-lineage cells make a significant contribution to osteoblastic bone formation. Tie2-cre:Osxfx/fx mice underwent 12 weeks of assessment for health and welfare and no adverse phenotype was found. This included no defects in skeletogenesis or skeletal health (data not shown), implying that the contribution Tie2-lineage cells to osteoblasts is not developmentally important.
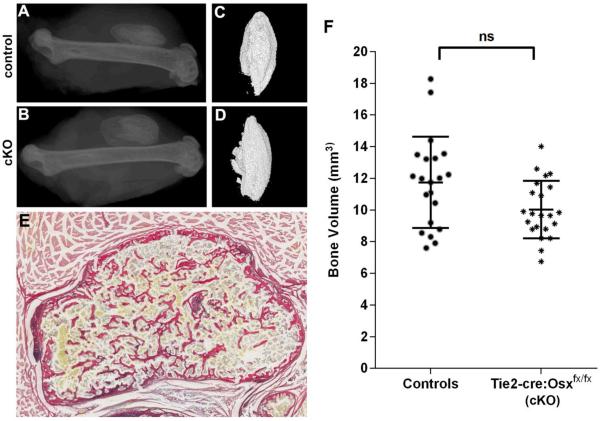
Next, rhBMP-2 induced bone nodules at 21d were imaged by X-ray (Figure 4A–B) and microCT reconstruction (Figure 4C–D) from Tie2-cre:Osxfx/fx mice and control littermates lacking the Tie2-cre transgene (Tie2-cre−/−; Osxfx/fx, Osxfx/+). Ectopic nodules had a standard appearance by tissue histology, with a cortical-like shell and an interior marrow-like space with trabecular-like bony elements (Figure 4E). Quantification by microCT confirmed that there was no significant difference in bone formation between the genotypes (Figure 4F).
Figure 4.
rhBMP-2 induced ectopic bone formation in Tie2-cre-Osxfx/fx mice and littermate controls. Representative XRs (A, B) and microCT reconstructions (C, D) are shown for the specimens corresponding to the median bone volume for each group. Littermate controls (A, C) were compared to Tie2-cre-Osxfx/fx conditional knockout (cKO) mice (B, D). The bone nodules showed a cortical shell with some trabecular-like elements visualized using Picro Sirius Red/Alcian Blue staining (E). MicroCT quantification revealed no significant difference in bone volume of ectopic rhBMP-2 induced bone with deletion of the Osx gene in Tie2-lineage cells (F).
Contribution of Tie2-lineage cells and αSMA-lineage cells in an rhBMP-2 induced spine fusion model
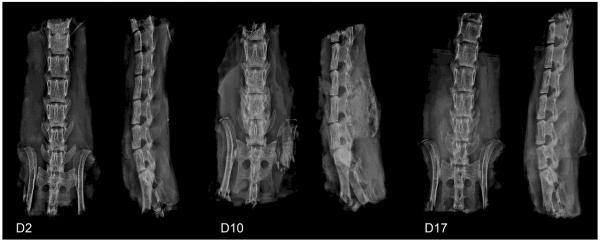
As an alternative model of rhBMP-2 induced bone formation, fusion was induced in the mouse spine as a location that would have a capacity for contribution by soft tissue progenitors and endogenous osteoprogenitors. Fusion occurred rapidly after the surgical procedure, with new bone bridging the vertebral processes apparent within 10 days (Figure 5).
Figure 5.
X-ray images showing the progression of rhBMP-2 induced spine fusion. After implantation of collagen sponges loaded with rhBMP-2, no mineralized bone was seen at D2, but a fusion mass was apparent in all specimens by D10 and D17. Anterior-posterior and lateral views are shown for all time points.
In the Tie2-cre:Ai9 reporter mice, comparable results were seen to the hind limb rhBMP-2 implantation model. Samples obtained from mice culled at day 2 post-operatively had not yet formed any new bone overlying the vertebrae and very few tdTomato+ cells were noted in these areas (Figure 5). However, specimens from this time point showed a pattern of co-localization of tdTomato+ and TRAP+ cells in the original cortical bone of the lumbar spinous process (Supp Figure 1). Abundant tdTomato+ cells were noted in the vertebral fusion masses of Tie2-cre:Ai9 mice by day 10. The majority of cells that expressed tdTomato were co-labelled with TRAP, suggesting that the majority of Tie-2-lineage cells in the fusion mass contribute to the cellular milieu by way of osteoclastogenesis (Supp Figure 1). CD31 staining was also found to significantly overlap with tdTomato expression at d10 (Supp Figure 2), but less so at d17 where the tdTomato+ cells on the cell surface primarily corresponded to osteoclasts.
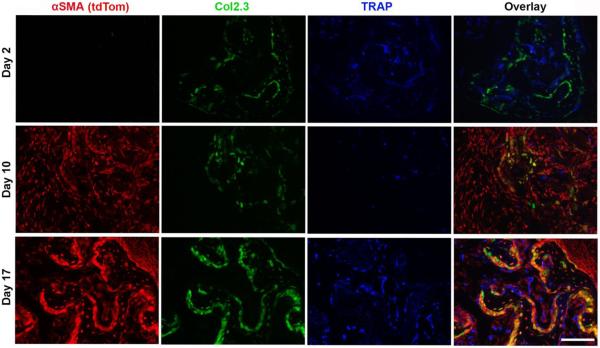
The spine fusion model was also performed in the αSMA-creERT2:Col2.3-GFP:Ai9 mouse line to allowed for the tracking of mesenchymal progenitors after tamoxifen induction (tdTomato) while co-labeling osteoblasts (GFP). Tamoxifen injection was administered at the time of surgery and the contribution of αSMA-lineage progenitors was assessed at day 2, 10 and 17 (Figure 6). At d2, no tdTomato+ cells were observed within implanted sponge as part of the initial inflammatory cellular infiltrate. However, some tdTomato+ cells were observed within the muscle adjacent to the sponge, indicating that Cre-mediated recombination was present at this time point (Supp Fig 3). This is consistent with previous reports (19). By day 10 and day 17, tdTomato+ cells were present throughout the BMP-2 induced fusion mass. All osteoblastic cells expressing Col2.3-GFP were found to be tdTomato+ and thus originate from the αSMA-lineage. A greater proportion of the tdTomato+ expressed the Col2.3-GFP marker over time, and by day 17 a significant proportion of the αSMA cells labelled at surgery expressed the osteoblastic marker (Figure 6). In contrast, αSMA-lineage cells were found to not co-localize with the TRAP activity stain.
Figure 6.
Fluorescent images of spine fusion bone masses with co-labeling for αSMA-lineage cells expressing tdTomato (red), Col2.3-GFP (green), and a TRAP activity stain (blue). At D2, no incorporation of αSMA-lineage progenitors was seen within the native spinous processes (green) or the implanted collagen sponge. At D10 and D17, Col2.3-GFP+ cells in the fusion mass were shown to originate from αSMA-lineage progenitors. No tdTomato+ signal was seen with TRAP+ cells at any time point. Scale bar = 50 μm.
Control animals not receiving tamoxifen showed no leakiness of the tdTomato reporter at any time point (example at day 17, Supp Fig 4).
Discussion
In orthopedic medicine, the cell populations that give rise to osteoblasts that generate new bone remain poorly defined. It is generally accepted that cells from the periosteum can play a major role in fracture repair, particularly in closed fractures where the periosteum is largely intact (28). Conversely, in animal models and clinical practice periosteal stripping is associated with impairment of bone repair (29). Mesenchymal cells from the marrow compartment have been shown to readily differentiate into an osteogenic lineage in vitro, but the importance of these cells in bone repair is less well defined (28,30,31). However, even in fractures with intramedullary nailing, poor marrow availability and significant periosteal stripping, healing can still eventuate. This indirectly evidences a potential contribution by progenitors from non-osseous tissues to new bone formation.
In this study we have aimed to address the potential transdifferentiation of vascular endothelial progenitors under conditions featuring high concentrations of exogenously supplied rhBMP-2. Endothelial to mesenchymal transitions have emerged as a occurring in range of tissue processes, particularly in development and in tissue fibrosis (32). It has also been linked to genetic disease, specifically the abnormal ossification seen in fibrodysplasia ossificans progressiva and mutations in the BMP-receptor ACVR1 (7). This led us to speculate that even if Tie2-lineage cells did not make a significant contribution to normal bone formation or repair (33), excessive BMP signaling could lead to transition of endothelial progenitors into osteoblasts.
Nevertheless, these data comprehensively demonstrate that Tie2-lineage cells contribute to the developing vascular network and not osteoblasts in BMP-induced bone. Prior studies showing extensive Tie2-lineage reporter staining in the bone are likely to be confounded by two issues. Firstly, Tie2 is expressed by hematopoietic lineages as well as endothelial cells, which give rise to osteoclasts. The co-staining of the osteoclastic marker TRAP with tdTomato+ cells reinforces the concept that the cells seen within the bone in prior studies may have been osteoclasts. Prior studies using histochemical markers such as LacZ (3) did not allow for co-staining with other lineage markers and also have intrinsic problems with specificity in post-natal bone (34).
Secondly, other studies demarking cells of the Tie2-lineage in bone utilized an alternative transgenic mouse line that showed a wider pattern of distribution including mesenchymal cells (15). In contrast, our Tie2-cre:Ai9 mouse line demonstrates a more restrictive pattern of expression (16,17), which may explain the lack of contribution to mesenchymal elements. Notably, the study by Wosczyna (14) also used a more endothelial specific VE-Cadherin cre reporter system, and this showed no co-staining with bone or cartilage. This highlights a challenge associated with lineage tracking studies, where the non-specificity and/or leakiness of Cre mouse models can lead to confounding results (35). Thus caution must be made with the interpretation of all tracking studies.
The findings of a lack of contribution of Tie2-lineage cells to osteoblasts was found in both the hind limb rhBMP-2 induced ectopic bone formation model and in the rhBMP-2 induced spine fusion model. In the latter model, all osteoblasts (marked with the Col2.3-GFP transgene) were observed to originate from αSMA-lineage lineage progenitors. This mouse, which enables the inducible labelling of progenitors just prior to intervention, has proven a valuable tool for identifying physiological processes where the primary cellular contribution is mesenchymal (19,20,22).
Finally, confirmation of the lack of a functional importance for Tie2-lineage cells in the direct formation of bone was seen using the novel Tie2-cre:Osxfx/fx mouse line. Loss of the osterix gene in cells of the chondrogenic lineage has been previously reported to disrupt global bone formation (13). In contrast the Tie2-cre:Osxfx/fx were viable with no adverse phenotype, suggesting no significant contribution of Tie2-lineage cells to bone formation. Moreover, in parallel with the findings of the Tie2-cre:Ai9 mouse line showing no reporter expression in osteoblasts, the Tie2-cre:Osxfx/fx line showed no deficit in rhBMP-2 induced ectopic bone formation. Of note, in this mouse line, the staged differentiation of Tie2-lineage cells from chondrocytes to osteoblasts would also be arrested, thus resulting in persistent cartilaginous nodules if this lineage was making a major contribution to BMP-induced cartilage.
In summary, this study uses rigorous methodology to demonstrate that Tie2-lineage and αSMA-lineage cells make distinct contributions to rhBMP-2 induced bone formation. Moreover, despite evidence that BMPs and in particular high doses of rhBMPs may be able to induce endothelial to mesenchymal transitions, we have found no evidence for this in our in vivo bone formation models. We conclude that while this process may be problematic in certain disease states, it does not feature under normal physiological conditions. These data further suggest that the advantages of endothelial progenitor transplantation in orthopedics and bone tissue engineering are likely due to increases in angiogenesis rather than by these cells contributing directly to the local osteoblastic populations.
Supplementary Material
Fluorescent images of Tie2-lineage cells (tdTomato) with TRAP activity (ELF97) in spine fusion masses showing recapitulates the findings seen in ectopic bone nodules (Figure 3). Osteoclasts were observed in the fusion mass from D10 onwards. Scale bar = 50 μm
Fluorescent images of Tie2-lineage cells (tdTomato) with CD31 immunohistochemistry in spine fusion masses showing recapitulates the findings seen in ectopic bone nodules (Figure 2A) at D10, but shows abundant CD31- tdTomato+ cells (osteoclasts) on the bone surface at D17. Scale bar = 50 μm
Images of the spine and implanted collagen sponges at D2 in αSMA-CreERT2Col2.3-GFP:Ai9 mice. The presence of tdTomato+ cells are seen in the muscle adjacent to the fusion mass.
Control fluorescent images of rhBMP-2 induced spine fusion masses in Tamoxifen untreated αSMA-CreERT2Col2.3-GFP:Ai9 mice at D17. (A) DAPI staining (blue) of nuclei, (B) tdTomato (red) is not seen without Tamoxifen treatment, (C) Col2.3-GFP+ expression (green) is seen in the new bone, and (D) an overlay with light microscopy to visualize the bone. Scale bar = 50 μm
Research Highlights.
Tie2-lineage cells were tracked in BMP-2 induced bone using fluorescent report mice
Tie2-lineage cells contributed to vessels and osteoclasts but not osteoblasts
αSMA-lineage (mesenchymal) cells contributed to osteoblasts
Conditional deletion of Osx in Tie2-lineage cells did not affect BMP-2 induced bone
Acknowledgments
This study was directly supported by NHMRC project grant APP1003480 and A Schindeler received salary support from NHMRC project grant APP1003478. I Kalajzic is supported by NIH/NIAMS grant AR055607. The Leica SP5 in the CLEM Suite at KRI was supported by the following grants: Cancer Institute New South Wales Research Equipment [10/REG/1-23], Australian National Health and Medical Research Council [2009-02759], the Ian Potter Foundation [20100508], the Perpetual Foundation [730], Ramaciotti Foundation [3037/2010], and the Sydney Medical School Research Infrastructure Major Equipment Scheme.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wan M, Cao X. BMP signaling in skeletal development. Biochem Biophys Res Commun. 2005;328(3):651–7. doi: 10.1016/j.bbrc.2004.11.067. [DOI] [PubMed] [Google Scholar]
- 2.Axelrad TW, Einhorn TA. Bone morphogenetic proteins in orthopaedic surgery. Cytokine Growth Factor Rev. 2009;20(5–6):481–8. doi: 10.1016/j.cytogfr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Lounev VY, Ramachandran R, Wosczyna MN, Yamamoto M, Maidment AD, Shore EM, Glaser DL, Goldhamer DJ, Kaplan FS. Identification of progenitor cells that contribute to heterotopic skeletogenesis. J Bone Joint Surg Am. 2009;91(3):652–63. doi: 10.2106/JBJS.H.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schindeler A, Liu R, Little DG. The contribution of different cell lineages to bone repair: exploring a role for muscle stem cells. Differentiation. 2009;77(1):12–8. doi: 10.1016/j.diff.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Liu R, Schindeler A, Little DG. The potential role of muscle in bone repair. J Musculoskelet Neuronal Interact. 2010;10(1):71–6. [PubMed] [Google Scholar]
- 6.Liu R, Birke O, Morse A, Peacock L, Mikulec K, Little DG, Schindeler A. Myogenic progenitors contribute to open but not closed fracture repair. BMC Musculoskelet Disord. 2011;12:288. doi: 10.1186/1471-2474-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medici D, Shore EM, Lounev VY, Kaplan FS, Kalluri R, Olsen BR. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med. 2010;16(12):1400–6. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee DY, Cho TJ, Kim JA, Lee HR, Yoo WJ, Chung CY, Choi IH. Mobilization of endothelial progenitor cells in fracture healing and distraction osteogenesis. Bone. 2008;42(5):932–41. doi: 10.1016/j.bone.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Preininger B, Duda G, Gerigk H, Bruckner J, Ellinghaus A, Sass FA, Perka C, Schmidt-Bleek K, Dienelt A. CD133: enhancement of bone healing by local transplantation of peripheral blood cells in a biologically delayed rat osteotomy model. PLoS One. 2013;8(2):e52650. doi: 10.1371/journal.pone.0052650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, Dziak R, Yuan X, Mao K, Genco R, Swihart M, Sarkar D, Li C, Wang C, Lu L, Andreadis S, Yang S. BMP2 genetically engineered MSCs and EPCs promote vascularized bone regeneration in rat critical-sized calvarial bone defects. PLoS One. 2013;8(4):e60473. doi: 10.1371/journal.pone.0060473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atesok K, Li R, Stewart DJ, Schemitsch EH. Endothelial progenitor cells promote fracture healing in a segmental bone defect model. J Orthop Res. 2010;28(8):1007–14. doi: 10.1002/jor.21083. [DOI] [PubMed] [Google Scholar]
- 12.Li R, Atesok K, Nauth A, Wright D, Qamirani E, Whyne CM, Schemitsch EH. Endothelial progenitor cells for fracture healing: a microcomputed tomography and biomechanical analysis. J Orthop Trauma. 2011;25(8):467–71. doi: 10.1097/BOT.0b013e31821ad4ec. [DOI] [PubMed] [Google Scholar]
- 13.Li R, Nauth A, Li C, Qamirani E, Atesok K, Schemitsch EH. Expression of VEGF gene isoforms in a rat segmental bone defect model treated with EPCs. J Orthop Trauma. 2012;26(12):689–92. doi: 10.1097/BOT.0b013e318266eb7e. [DOI] [PubMed] [Google Scholar]
- 14.Wosczyna MN, Biswas AA, Cogswell CA, Goldhamer DJ. Multipotent progenitors resident in the skeletal muscle interstitium exhibit robust BMP-dependent osteogenic activity and mediate heterotopic ossification. J Bone Miner Res. 2012;27(5):1004–17. doi: 10.1002/jbmr.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–42. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 16.Koni PA, Joshi SK, Temann UA, Olson D, Burkly L, Flavell RA. Conditional Vascular Cell Adhesion Molecule 1 Deletion in Mice. Impaired lymphocyte migration to bone marrow. J Exp Med. 2001;193(6):741–54. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlaeger TM, Bartunkova S, Lawitts JA, Teichmann G, Risau W, Deutsch U, Sato TN. Uniform vascular-endothelial-cell-specific gene expression in both embryonic and adult transgenic mice. Proc Natl Acad Sci U S A. 1997;94(7):3058–63. doi: 10.1073/pnas.94.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grcevic D, Pejda S, Matthews BG, Repic D, Wang L, Li H, Kronenberg MS, Jiang X, Maye P, Adams DJ, Rowe DW, Aguila HL, Kalajzic I. In vivo fate mapping identifies mesenchymal progenitor cells. Stem Cells. 2012;30(2):187–96. doi: 10.1002/stem.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roguljic H, Matthews BG, Yang W, Cvija H, Mina M, Kalajzic I. In vivo identification of periodontal progenitor cells. J Dent Res. 2013;92(8):709–15. doi: 10.1177/0022034513493434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews BG, Grcevic D, Wang L, Hagiwara Y, Roguljic H, Joshi P, Shin DG, Adams DJ, Kalajzic I. Analysis of αSMA-labeled progenitor cell commitment identifies notch signaling as an important pathway in fracture healing. J Bone Miner Res. 2014;29(5):1283–94. doi: 10.1002/jbmr.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyment NA, Hagiwara Y, Matthews BG, Li Y, Kalajzic I, Rowe DW. Lineage tracing of resident tendon progenitor cells during growth and natural healing. PLoS One. 2014;9(4):e96113. doi: 10.1371/journal.pone.0096113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T, de Crombrugghe B. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A. 2005;102(41):14665–70. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17(1):15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 25.Schindeler A, Ramachandran M, Godfrey C, Morse A, McDonald M, Mikulec K, Little DG. Modeling bone morphogenetic protein and bisphosphonate combination therapy in wild-type and Nf1 haploinsufficient mice. J Orthop Res. 2008;26(1):65–74. doi: 10.1002/jor.20481. [DOI] [PubMed] [Google Scholar]
- 26.Bobyn J, Rasch A, Little DG, Schindeler A. Posterolateral inter-transverse lumbar fusion in a mouse model. J Orthop Surg Res. 2013;8:2. doi: 10.1186/1749-799X-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, Aburatani H, Nishimura R, Yoneda T. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 2008;283(43):29119–25. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colnot C. Skeletal cell fate decisions within periosteum and bone marrow during bone regeneration. Journal of Bone & Mineral Research. 2009;24:274–82. doi: 10.1359/jbmr.081003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oni OO, Stafford H, Gregg PJ. A study of diaphyseal fracture repair using tissue isolation techniques. Injury. 1992;23(7):467–70. doi: 10.1016/0020-1383(92)90065-z. [DOI] [PubMed] [Google Scholar]
- 30.Colnot C, Huang S, Helms J. Analyzing the cellular contribution of bone marrow to fracture healing using bone marrow transplantation in mice. Biochemical & Biophysical Research Communications. 2006;350:557–61. doi: 10.1016/j.bbrc.2006.09.079. [DOI] [PubMed] [Google Scholar]
- 31.Lu C, Xing Z, Yu YY, Colnot C, Miclau T, Marcucio RS. Recombinant human bone morphogenetic protein-7 enhances fracture healing in an ischemic environment. J Orthop Res. 2010;28(5):687–96. doi: 10.1002/jor.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindeler A, Kolind M, Little DG. Cellular transitions and tissue engineering. Cell Reprogram. 2013;15(2):101–6. doi: 10.1089/cell.2012.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C, Marcucio R, Miclau T. Assessing angiogenesis during fracture healing. Iowa Orthop J. 2006;26:17–26. [PMC free article] [PubMed] [Google Scholar]
- 34.Odgren PR, MacKay CA, Mason-Savas A, Yang M, Mailhot G, Birnbaum MJ. False-positive beta-galactosidase staining in osteoclasts by endogenous enzyme: studies in neonatal and month-old wild-type mice. Connect Tissue Res. 2006;47(4):229–234. doi: 10.1080/03008200600860086. [DOI] [PubMed] [Google Scholar]
- 35.Seime T, Kolind M, Mikulec K, Summers MA, Cantrill L, Little DG, Schindeler A. Inducible cell labelling and lineage tracking during fracture repair. Develop. Growth Differ. 2015;57(1):10–23. doi: 10.1111/dgd.12184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluorescent images of Tie2-lineage cells (tdTomato) with TRAP activity (ELF97) in spine fusion masses showing recapitulates the findings seen in ectopic bone nodules (Figure 3). Osteoclasts were observed in the fusion mass from D10 onwards. Scale bar = 50 μm
Fluorescent images of Tie2-lineage cells (tdTomato) with CD31 immunohistochemistry in spine fusion masses showing recapitulates the findings seen in ectopic bone nodules (Figure 2A) at D10, but shows abundant CD31- tdTomato+ cells (osteoclasts) on the bone surface at D17. Scale bar = 50 μm
Images of the spine and implanted collagen sponges at D2 in αSMA-CreERT2Col2.3-GFP:Ai9 mice. The presence of tdTomato+ cells are seen in the muscle adjacent to the fusion mass.
Control fluorescent images of rhBMP-2 induced spine fusion masses in Tamoxifen untreated αSMA-CreERT2Col2.3-GFP:Ai9 mice at D17. (A) DAPI staining (blue) of nuclei, (B) tdTomato (red) is not seen without Tamoxifen treatment, (C) Col2.3-GFP+ expression (green) is seen in the new bone, and (D) an overlay with light microscopy to visualize the bone. Scale bar = 50 μm