Abstract
Unraveling the first migrations of anatomically modern humans out of Africa has invoked great interest among researchers from a wide range of disciplines. Available fossil, archeological, and climatic data offer many hypotheses, and as such genetics, with the advent of genome-wide genotyping and sequencing techniques and an increase in the availability of ancient samples, offers another important tool for testing theories relating to our own history. In this review, we report the ongoing debates regarding how and when our ancestors left Africa, how many waves of dispersal there were and what geographical routes were taken. We explore the validity of each, using current genetic literature coupled with some of the key archeological findings.
Keywords: Anatomically modern humans, out of Africa model, waves of migration, admixture
Introduction
The origin of humans in Africa was famously proposed in the 19th century by Charles Darwin.1 Based on the presence of chimpanzees and gorillas in Africa and on Huxley’s comparative anatomy studies that showed that modern humans and apes shared a common ancestor,2 Darwin argued that the ancestors of modern humans arose on African soil. Now, over 150 years later, genetics, with the advent of genome-wide genotyping and sequencing techniques coupled with archeological evidence, substantially confirms the African origin of the first modern humans, while highlighting many further complexities. In particular, with the continuing advancement of these genetic methods and through a symbiotic coupling with novel bioinformatic approaches, researchers are attempting to use genetic information to answer questions of our origins in finer resolution than previously obtainable. How and when did our ancestors first leave Africa? Were there multiple dispersal waves or a single one? What geographical routes were taken?
In this review, we present key literature tackling these questions and explore the strength of the conclusions drawn in the context of known genetics (summarized in Table 1) and important archeological findings, as well as highlight the limits of inferences based solely on presently available genetic data.
Table 1.
Classification of the most relevant studies related to the out of Africa event is addressed in this review according to the type of data used along with some examples of the methodology performed.
| EXAMPLES OF INFERENCES | ARCHEOLOGICAL RECORD/FOSSILS | mtDNA AND NRY (ChrY) (SOME RELEVANT REFERENCES) | STR/AUTOSOMAL DNA (SOME RELEVANT REFERENCES) | WHOLE-GENOME GENOTYPING/NEXT-GENERATION SEQUENCING (SOME RELEVANT REFERENCES) |
|---|---|---|---|---|
| Human origin in Africa/serial founder model | Omo I44,45 Herto fossils46 Jebel Irhoud remains53 | 11,16–18,21–26,53–55 | 27–30 | 47,48,50 |
| Possible routes of human dispersal |
Northern route: Skhul and Qafzeh hominins56,57 Southern route: Abdur Reef Limestone75,76 |
Northern route63 Southern route13,66–71 |
Northern route31,32 | Northern route64 |
| Timing of the Out of Africa event |
100–130 Kya: Mount Toba84 80–120 Kya: Daoxian88 70–120 Kya: Zhirendong89 Luna Cave90 Liujiang91 Callao man92 |
40–60 Kya67,80 | 50–60 Kya82,93 | 50–60 Kya94–96 80–100 Kya assuming gene flow up to 20 Kya97 |
| Intermixing between archaics and modern humans | Feldhofer, Vindija and Mezmaiskaya Caves (Neanderthal remains)109,117 Denisova Cave (Neanderthal and Denisova remains)110,111 |
2% DNA from Neanderthals118,119,123,154 Modern human introgression into the ancestors of East Asian Neanderthals 100 Kya124 3–5% DNA from Denisovans111,125 |
||
| Back into Africa migrations | Mota (4,500 years old, no Eurasian admixture)171 | 71,158,159,166–169 | 1–50% ancestry from West Eurasia contributed 4.5 Kya159,160,162,164,165,171 | |
| Some of the main methods used | –Stratigraphy –Radiometric dating –Morphology and comparative anatomy studies |
–Phylogenetic trees –Haplogroup diversity |
–Genetic diversity –Phylogenetic trees –Gene networks |
–Recent admixture detection and dating –Past population sizes estimates –Allele frequency spectrum (AFS) –Identity-by-state (IBS) SNP analysis –Genetic diversity (LD, heterozygosity) –Coalescent-based modeling–Clustering |
Candelabra Versus Replacement Hypothesis
The debate dominating much of the anthropological discourse throughout the second half of the 20th century focused on where and when archaic hominins evolved into modern Homo sapiens, which we refer to as anatomically modern humans (AMH) throughout. Two main hypotheses dominated the discourse: the multiregional and the replacement hypotheses. The original multiregional model was proposed by the anthropologist Weidenreich in 1946 and advocated significant gene flow among subpopulations of Homo erectus living in different parts of the globe throughout the Pleistocene, so that modern humans trace their ancestry to multiple hominin groups living in multiple regions.3 Confusingly, the term “multiregional” model often has been used synonymously with the so-called candelabra model,4 originally proposed by Coon in 1962.5 The candelabra model hypothesizes that our early hominin ancestors, after leaving Africa 1 million years ago and migrating to other continents, independently evolved anatomically modern features. Under this model, the modern human form arose autonomously at multiple times and locations worldwide within the last 1 million years, so that modern non-African populations each primarily descended from separate evolutions of these Homo species.5 This is in contrast to the traditionally proposed multiregional model, which importantly does not propose independent parallel evolution of AMH features (for a detailed discussion of these models, see Ref. 4).
The main fossil evidence in support of the multiregional and candelabra hypotheses was the discovery of the Dali Man in China.6 For multiregionalists or candelabra supporters, the mixture of archaic and modern features was evidence of a midway stage between early and modern hominins. That said, these fossils are poorly preserved, and some authors have suggested that these anatomical characteristics are in fact shared by other Homo worldwide, and thus were not unique to Asia.7,8 Some genetic studies also offered support to a multiregional and candelabra models, inferring the origin of a few genetic loci outside of the African continent. Examples include the oldest haplotype in the human dystrophin gene, which was found to be absent in Africans,9 although this was later explained as resulting from adaptive introgression from Neanderthals rather than providing support for the candelabra model.10
Opposition to the candelabra hypothesis has come from both paleontological and genetic studies. The replacement, or out of Africa (OoA), model proposes a single and relatively recent transition from archaic hominins to AMH in Africa, followed by a later migration to the rest of the world, replacing other extant hominin populations.7,11–15 Under this model, these hominins were driven to extinction, so that most of the genetic diversity in contemporary populations descends from a single or multiple groups of AMH who spread out of Africa sometime in the last 55–200 kya, although debate remains on the precise timings (note that, in light of admixture from extinct hominin groups, the OoA model is consistent with the original multiregional model but not the candelabra model).
The first genetic evidence consistent with the OoA model was provided by the study of mitochondrial DNA (mtDNA) phylogenetic trees, which identified Africa as the source of human mtDNA gene pool.11,16–18 It was shown that all mtDNA haplogroups outside of Africa can be attributed to either the M or N haplogroups, which arose around 60–65 kya in South Asia19 and are thought to descend from the L3 haplogroup postulated to have arisen in East Africa around 80 kya.20 This was supported by further studies of mtDNA,21–23 Y chromosome,24–26 and autosomal regions27–30 that suggested the existence of a common African ancestor. More recently, multilocus studies of genome-wide data have demonstrated that genetic diversity decreases as a function of geographic distance from East or South Africa, for example, as shown by an approximately linear decrease in heterozygosity and increase in linkage disequilibrium (LD),31,32 a finding consistent with the OoA model.
Several further replacement models exist, which differ in their emphasis. Harding and McVean, for example,33 proposed a more complex meta-population system for the origins of the first AMH. The authors highlight evidence suggesting that the ancestral African population from which modern humans arose was genetically structured,34–37 so that extant populations at the time contributed unequally to the gene pool of individuals migrating out of the continent.33,38 The effects of ancient population structure on patterns of modern day genetic variation have also been suggested in several more recent papers,39–41 which assess the impact of ancient hybridization on modern day genetic diversity through modeling and data-driven analyses. In addition to genetic studies, structure in the ancestral African population has also been supported through archeological and palaeoenvironmental models.42,43
Eastern, Southern, or Northern African origin of the first H. sapiens
Although the African origin of AMH is now largely accepted, debate has continued over whether the anatomically modern form first arose in East, South, or North Africa.
Southern or East African origin
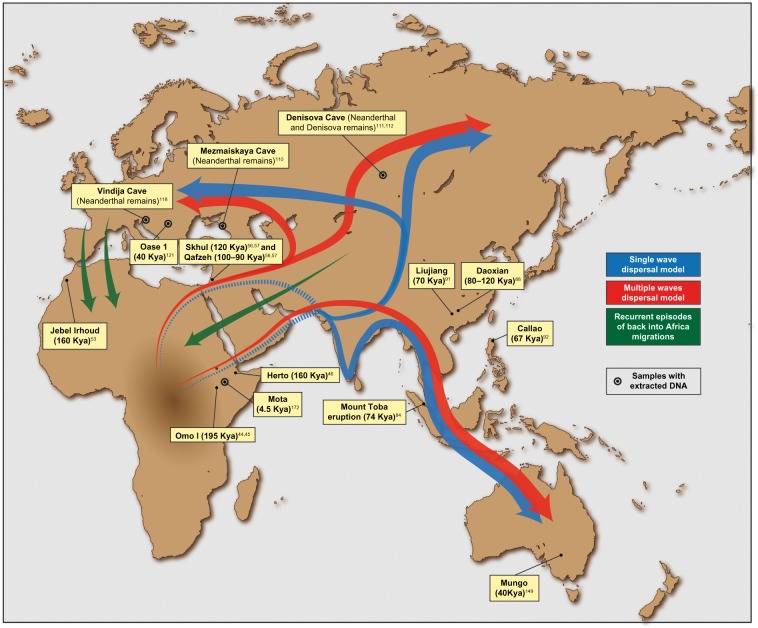
Support for an East African origin is provided by the discovery of the oldest unequivocally modern human fossils to date in Ethiopia: the Omo I from Kibish first discovered by Richard Leakey in 1967 dated to 190 and 200 kya44,45 and the Herto fossils dated to between 160 and 154 kya (Fig. 1).46
Figure 1.
Putative migration waves out of Africa and location of some of the most relevant ancient human remains and archeological sites. The placement of arrows is indicative.
Evidence for a Southern origin has largely been provided by genetic rather than archeological studies. Henn et al analyzed genomes from extant hunter-gatherers from sub-Saharan Africa: pygmies of central Africa, click-speaking populations of Tanzania in East Africa (Hazda and Sandawe), and the Khomani Bushmen of Southern Africa.47 Their analyses of LD and heterozygosity patterns suggested Southern hunter-gatherers were among the most genetically diverse of all human populations, lending support to a Southern African origin of AMH. Schlebusch et al also considered patterns of LD in South Africa, observing the same high levels of genetic diversity, low levels of LD, and shorter runs of heterozygosity as mentioned in the previous studies. However, by incorporating additional samples throughout the rest of Africa, they showed that LD-based statistics fail to pinpoint a specific origin point, as LD levels were similarly low in other parts of the continent besides South Africa, indicating that different groups of individuals within different regions are important. This suggests that the population history within sub-Saharan Africa is likely too complex to localize the origins of H. sapiens using these approaches and available data.48 Similarly, Pickrell et al showed an ancient link between the Southern African Khoe-San and the Hadza and Sandawe of East Africa, suggesting that both Eastern and Southern Africa are equally consistent candidates as an origin locality of modern humans.49
A more recent study jointly analyzed paleo-climatic records and estimates of effective population size from the whole-genome sequences of five Khoe-Sans and one Bantu individual along with 420,000 Single Nucleotide Polymorphisms (SNPs) from worldwide groups.50 Although unable to resolve a Southern or Eastern origin, using coalescent-based modeling, they suggest that Southern African populations have had high effective population sizes throughout their history, which might result in lower levels of LD relative to other parts of Africa, irrespective of where AMH arose. This corresponds well with climatic records, which find that Western and central Africa were affected by a dry climate with increasing aridity, around 100 kya, leading to a decline in the effective population size of West African populations (ancestors of the Bantu-speaking and non-African populations) but not vastly altering the effective size of South Africans (ancestors of the Khoe-San).51 The incorporation of climatic data has also been important in mtDNA analyses aiming to resolve a clear location of origin. In a paper focusing on the distribution of mtDNA haplogroups, Rito et al propose that the most recent common ancestor (MRCA) of modern mtDNA likely arose in Central Africa before splitting into Southern African groups such as the Khoe-San (L0) and central/East African groups (L1’6).52 This is in good agreement with the aforementioned autosomal studies, which postulated an early split between the Khoe-San and more Northern African populations,48,49 although they did not advocate a MRCA in Central Africa.
Northern African origin
Up until recently, little focus has been given to North Africa as a potential origin point of the AMH form, despite the discovery of early AMH, Jebel Irhoud remains in Morocco dated to 160 kya (Fig. 1).53 Focus on the region changed with the publication of a revised Y chromosomal phylogenetic tree based on resequencing of male-specific regions of the Y chromosome from four relevant clades.54 The revised phylogeny found that the deepest clades were rooted in central and Northwest Africa, suggesting this region was more important than previously thought. Fadhlaoui-Zid et al went on to analyze both Y chromosome and genome-wide data from North Africa, the Middle East, and Europe. They found some evidence for a recent origin of human populations in North Africa but stressed how high levels of migration, admixture, and drift in North Africans make interpretation extremely challenging.55
It is important to note that many genetic-based approaches such as these are limited in their ability to answer questions of this nature given that extant human populations are likely poor representatives of populations residing in these regions during pre-AMH time periods. For instance, populations can be quite mobile, and thus the geographic location of modern humans groups may differ from that of their ancestors in the past. Archeological and paleontological approaches, as well as DNA from ancient human remains, can go some way to guiding inference but are also limited given the scarcity of sites. For example, an origin of AMH in Western or central Africa cannot be ruled out but to our knowledge, there is currently no archeological data available to address this as a candidate region.
Potential Routes out of Africa
One of the most intriguing questions regarding the exit of modern humans out of Africa is which geographical route was taken. The consensus view is that if modern humans did exit Africa via a single dispersal, there were two possible routes (not mutually exclusive) at the time: a Northern route, through Egypt and Sinai, and a Southern route, through Ethiopia, the Bab el Mandeb strait, and the Arabian Peninsula (Fig. 1 and Table 1). So far, neither archeological nor genetic evidence has been able to resolve this question with confidence.
Northern route
Some of the earliest remains of AMH anywhere outside of Africa, the Skhul and Qafzeh hominins, were found in the Levant (present-day Israel) and dated to 120 and 100–90 kya, respectively (Fig. 1).56,57 It has been suggested that these fossils represent an early exit of modern humans approximately 120 kya, traveling across the Sinai Peninsula to the Levant.58 The next human remains found in the region include the Manot1 cranium, which was dated to around 55 kya,59 demonstrating a considerable gap in the fossil record of AMH occupation in the Levant. This, in conjunction with climatic records, indicating a global glacial period 90 kya,60 has led some authors to suggest that if the first humans did exit early via the Levant they did not survive, and that the Skhul and Qafzeh hominins are the remnants of this failed exodus.58 Other authors emphasize the possibility that this group could have already left the Levant before the glacial period 90 kya.61 That said, the recent presentation of archeological material, primarily stone tools and assemblages dated to 100–80 kya, from an empty corner of the Arabian Peninsula suggests early settlements may have been widely distributed and that even if Skhul and Qafzeh do represent a failed exodus, it was broader and more complex than previously thought.62
In addition to the evidence from the archeological and climatic record, genetic studies have also suggested some support for a Northern route. A study of Y chromosome haplogroup distributions together with 10 microsatellite loci and 45 binary markers in different African and Near Eastern populations found that the Levant was the most supported route for the primary migratory movements between Africa and Eurasia.63 In a more recent paper, Pagani et al sequenced the genomes of 100 Egyptians and 125 individuals from five Ethiopian ethnic groups (Amhara, Oromo, Ethiopian Somali, Wolayta, and Gumuz).64 After attempting to mask West Eurasian genetic components inherited via recent non-African admixture within the last 4 kya, they showed that modern non-African haplotypes were more similar to Egyptian haplotypes than to Ethiopian haplotypes, thus suggesting that Egypt was the more likely route in the exodus out of Africa migration, assuming the efficacy of their masking procedure. However, as noted earlier, one limitation of such studies that analyze modern DNA is that extant populations may not be good representatives of past populations due to factors such as population replacement, migrations, admixture, and drift.
Southern route
In contrast, mtDNA studies have traditionally favored a Southern route across the Bab el Mandeb strait at the mouth of the Red Sea.65–67 From there, modern humans are thought to have spread rapidly into regions of Southeast Asia and Oceania.68,69 For example, two studies have concluded that individuals assigned to haplogroup L3 migrated out of the continent via the Horn of Africa.67,70 Furthermore, Fernandes et al.71 analyzed three minor West-Eurasian haplogroups and found a relic distribution of these minor haplogroups suggestive of ancestry within the Arabian cradle, as expected under a Southern route. That being said, many mtDNA studies, including these, are based on the premise that haplogroup L3 represents a remnant Eastern African haplogroup. Groucutt et al have recently theorized that L3 does not provide conclusive evidence for a shared African ancestor, given human demographic history is likely to be less “tree-like” than has been consistently assumed by mtDNA analyses.72 As an example, they showed that L3 could have arisen inside or outside of Africa if gene flow occurred between the ancestors of Africans and non-Africans following their initial divergence.
Short Tandem Repeats (STR) and analysis of LD decay in combination with geographic data have also been used to support a Southern route via a single wave serial bottleneck model.73 Under this model, it is thought that a group crossed the mouth of the Red Sea and traveled along the Southern coast of the Arabian Peninsula toward India as “beachcombers,” exploiting shellfish and other marine products.74 Migrations then continued in an iterative wave as populations dispersed and expanded into uninhabited areas. This is consistent with a glacial maximum occurring during this time period, which caused sea levels to fall allowing potential passage across the mouth of the Red Sea.
From an archeological perspective, evidence indicative of maritime exploitation is extremely limited. The discovery of artifacts from the Abdur Reef Limestone in the Red Sea and archeological sites in the Gulf Basin that indicate long-standing human occupation earlier than 100 kya may offer some evidence; however, whether these represent the activities of the ancestors of modern-day human groups is still an open question.75,76 Furthermore, Boivin et al caution that while coastal regions may have been important, a coastal-focused dispersal would still have been problematic and not necessarily conducive to rapid out of Africa dispersal.77
The Timing of the out of Africa Event
Another key debate has focused on the precise timing of the exit of the first humans out of Africa (Table 1). Currently, there are two conflicting proposals that dominate the literature, each differing by several tens of thousands of years and not mutually exclusive. The first claims that the Eurasian dispersal took place around 50–60 kya, reaching Australia by 45–50 kya.19,67,71,78–83 The second posits that there was a much earlier exodus around 100–130 kya, prior to the eruption of Mount Toba (Northern Sumatra) dated to 74 kya.84
The Toba eruption is important in the context of dating the OoA event, as stone tools thought to be associated with AMH have been found embedded in the volcanic ash.85 If these tools are indeed from AMH, this provides clear evidence that modern humans had reached Southeast Asia before the Toba eruption more than 74 kya. The stone tools discovered in the archeological site Jebel Faya (present-day United Arab Emirates) and the Nubian complex of Dhofar (present-day Oman) have provided further support for an early migration via Arabia.86,87 More importantly, the recent discovery of 47 human teeth in the Fuyan Cave in Daoxian (Southern China), dated to 80–120 kya and unequivocally assigned to modern humans,88 supports the dispersal of AMH throughout Asia during the early Late Pleistocene. However, it remains to be seen if individuals among these groups contributed genetically to modern populations or represent another failed exodus similar to that proposed in the Levant, a question that DNA analyses are uniquely capable of shedding light on assuming genetic information can be interrogated reliably from these samples. Other pieces of archeological evidence that may help place AMH in the Far East over 70–120 kya are the human remains from Zhirendong (South China),89 the teeth from Late Pleistocene Luna Cave,90 the famous Southern Chinese Liujiang skeleton, which seems to have anatomically modern features,91 and the Callao man, a human foot bone, discovered in Philippines that dates to 67 kya (Fig. 1).92
Genetic studies have as yet been unable to settle the conflicting archeological evidence for these different dates, which can also be classified as pre-Toba (100–130 kya) or post-Toba (around 50–60 kya). Given the lack of ancient DNA (aDNA) data temporally and spatially, such genetic-based approaches have focused on inference using modern DNA. Studies based on reconstructing mtDNA phylogenies have suggested a date for modern humans leaving Africa between 60 and 40 kya,80 while dates inferred from STR analysis also fall within these estimates, positing an expansion date of around 50 kya for central African, European, and East Asian populations.82,93 Time estimates from whole-genome sequencing data have been more variable and depend largely on the choice of model used. For example, studies using the allele frequency spectrum, identity-by-state, or coalescent-based models suggest a divergence time of 60–50 kya,94–96 while analyses based on the pairwise sequencing Markovian coalescent (PSMC) model suggest that the divergence began 100–80 kya, with gene flow occurring until 20 kya.97
A key issue in the estimation of OoA dates using autosomal data is that the Yoruba of West Africa are commonly used as the reference point for AMH departure from East Africa, despite mtDNA and autosomal studies indicating a deep time separation of West and East African populations.98 Furthermore, many approaches assume that modern human groups are related via a simple bifurcating tree, which is likely an over simplistic view of human history. Another fundamental problem with many of the estimates used to date divergence times is that they are highly dependent on the choice of mutation rate, which can be estimated using a wide number of different approaches that often yield disparate values. The accumulation of heritable changes in the genome has traditionally been calculated from the divergence between humans and chimpanzees at pseudogenes, assuming a divergence time of around 6–7 million years ago (phylogenetic mutation rate 2.5 × 10−8/base/generation). With the advent of deep sequencing, it is now possible to directly calculate the mutation rate among present-day humans from parent-offspring trios. Using this method, the mutation rate has been estimated at 1.2 × 10−8/base/generation, half of the phylogenetic mutation rate, thus doubling the estimated divergence dates of Africans and suggesting that events in human evolution have occurred earlier than suggested previously.98–104 More recently, Harris reported that the rate of mutation has likely not been stable since the origin of modern humans, revealing higher mutation rates (particularly in the transition 5′-TCC-3′ to 5′-TTC-3′) in Europeans relative to African or Asian populations thus suggesting it may be too simplistic to assume the mutation rate is consistent across different populations.105 In addition to this, there is also considerable uncertainty in terms of the effect of paternal age at time of conception in the mutation rate with respect to ancestral populations.102 Recent work has attempted to mitigate some of these difficulties by instead calibrating estimates against fine-scale meiotic recombination maps. Using eight diploid genomes from modern non-Africans, Lipson et al calculated a mutation rate of 1.61 ± 0.13 × 10−8, which falls between phylogenetic and pedigree-based approaches.106
aDNA is becoming another major tool in appropriate calibration of mutation rate estimates and is likely to greatly refine our understanding of population divergence times, as it allows direct comparison of present-day and accurately dated ancient human DNA. For example, Fu et al used 10 whole mtDNA sequences from ancient AMHs spanning Europe and East Asia from 40 kya to directly estimate the mtDNA substitution rates based on a tip calibration approach. Using an amended mitochondrial substitution rate of 1.57 × 10−8, they dated the last major gene flow between Africans and non-Africans to 95 kya.107 Later work utilized high coverage aDNA from a 45,000-year-old western Siberian individual called Ust’-Ishim and a technique based on modeling the number of substitutions in relation to the PSMC inferred history, which led to slightly higher estimates of 1.3–1.8 × 10−8 per base per generation.108 It is likely that an increasing availability of ancient samples from different time periods will assist in further refining these estimates.
Intermixing with Neanderthals and Denisovans
On their migration out of Africa, AMH were not alone, with at least two distinct groups of archaic humans inhabiting Eurasia on their arrival: the Neanderthals and Denisovans (Table 1). Traditionally, the replacement or the OoA model assumed no intermixing between AMH leaving Africa and archaic hominins such as the Neanderthals. The revised replacement model, however, allows for gene flow with these archaic forms following the OoA dispersal, which is also consistent with the original multiregional model proposed by Weidenreich but still differs from the candelabra model.
Some of the most exciting outcomes of work on aDNA have been the publication of full Neanderthal and Denisovan genomes.109–111 Neanderthals, named after Neander valley in Germany where the species was discovered, are thought to have first appeared 250–200 kya,112 although the exact range is still under debate, and persisted, at least in regions of Southern Europe, until around 30 kya (Fig. 1).113 Initial genetic analyses focused on mtDNA, which is more easily extracted and amplified in ancient samples, and suggested no intermixing between Neanderthals and modern humans as they migrated into Eurasia.114–117 However, such analyses that rely on only single-locus data such as mtDNA can suffer from a lack of power. The first draft of the Neanderthal whole genome was published in 2010 where, in a landmark study, Green et al found that Neanderthals interbred with modern humans, contributing detectable segments of their genomes (1.5%–2.1%) to present-day populations outside of Africa.118 More recent estimates, based on mutation rate estimates refined by precise data for recombination rates as described previously, date this admixture to within 35–49 kya.106 These new dates, which are more recent than previously published estimates,119 are in good agreement with recent work that suggested Neanderthal admixture with modern humans was not restricted to the time period of the OoA migration in the Near East but instead may have also occurred more recently in Europe. Applying D statistics,120 which measure correlations in allele frequencies due to shared drift and/or admixture,121 to DNA extracted and enriched from the Oase 1 human mandible found in Romania and dated to 37–42 kya,122 Fu et al found that 6%–9% of the Oase 1 genome was derived from Neanderthals. Analyzing the lengths of segments in Oase 1 of likely Neanderthal ancestry, they inferred that Neanderthal introgression into this individual occurred in the previous four to six generations (Fig. 1).123 The authors caution that Oase 1 likely did not contribute to modern humans but nonetheless it provides a striking example of relatively recent Neanderthal introgression. Interestingly, Kuhlwilm et al have recently provided evidence for modern human introgression into the ancestors of Neanderthals from the Altai Mountains 100 kya in the Near East, although not in Europe, thus suggesting that a group of AMH might have left Africa before 100 kya.124 This is also in agreement with the Daoxian remains,88 and other fossil remains described previously that helped place the AMH in East Asia in the early Late Pleistocene.
Analysis of a hominin finger phalanx discovered at the Denisova Cave of the Altai Mountains in Southern Siberia confirmed the existence of a genetically distinct group of archaic humans related to Neanderthals, named the Denisovans (Fig. 1).110,111 The Denisovan lineage was classified based on genetic evidence and estimated to have diverged genetically from Neanderthals 381–473 kya assuming a simple bifurcating tree.109 One startling discovery was that despite being discovered and identified in Siberia, the Denisovan genome was found to share detectable segments of DNA (3%–5% of the genome) in common with modern-day Near Oceanians, including New Guineans, Australians, and Mamanwas (a Negrito group from the Philippines).110,111,125 Since then, Denisovan introgression has been detected in further populations, for example, those from East Eurasia and Native America,109,126 suggesting the extent of Denisovan ancestry in modern populations is likely more widespread than has been previously thought. There is some evidence that haplotypes associated with archaic human introgression have been maintained in modern groups as a result of them providing advantageous genetic variation. For example, a recent study proposes that a haplotype that is adaptive to hypoxic conditions in high altitude Tibetans has a structure consistent with introgression from a Denisovan or Denisovan-related individuals, suggesting that gene flow with other hominins may be important in human adaptation to local environments.127
Single or Multiple Waves of Migration?
An additional topic of ongoing controversy relates to whether the first modern humans to leave Africa were part of a unique dispersal event or whether in fact there were multiple waves of dispersal out of Africa to populate the rest of the world (Fig. 1). A variety of dispersal models exist, which all fall within these extremes, as addressed by Groucutt et al.72
Single dispersal
A single dispersal model states that AMH migrated out of Africa to the rest of Eurasia as a single wave exiting via a single route, whether North or South. As discussed previously, variants of the single dispersal model differ in their timing, for example, after 50 kya with complete population replacement under the upper Paleolithic model,128,129 versus those postulating as early as 100–125 kya based on the Skhul and Qafzeh remains and climatic data.130,131 They also differ on the exact route taken, for example, single coastal dispersal models 50–75 kya132 versus models governed by the location of discovered archeological artifacts, such as the Jebel Faya and Nubian models, based on artifacts found in present-day United Arab Emirates and North-East Africa, respectively.86,133
Under all of these models, genetic evidence suggests that migration out of Africa was accompanied by a severe bottleneck in the initial migrating group(s), drastically reducing the genetic diversity before a rapid expansion. The serial founder effect model is one possible description of a single dispersal model,134 by which the dispersal out of Africa occurred initially via a single migrant group and continued in an iterative way as individuals dispersed into unoccupied regions, expanded in population size, and gave rise to new groups of founder individuals who again expanded into unoccupied areas. This serial founder process could give rise to the patterns of increasing genetic drift and decreasing genetic diversity observed in groups as geographic distance from Africa increases.135
However, recent work by Pickrell and Reich has shown that the inverse correlation observed between heterozygosity and geographic distance can be generated under further historical models that all deviate in some way from the serial founder effect model and yet not as extreme as the multiple dispersals model. Primarily, they present models of dispersal characterized by fewer bottlenecks and more pervasive admixture that can also account for the patterns of heterozygosity we see in global populations.136
East Asia is a key region when considering the nature of the first AMH dispersals. Under a single dispersal model, the peopling of East Asia is hypothesized to have occurred via a single wave, with, for example, Aboriginal Australians surmised to have diversified from within this East Asian wave of individuals. Recent whole-genome studies reveal a split between Europeans and Asians dating to 17–43 kya,137,138 which seems in conflict with archeological evidence from greater Australia (Australia and Melanesia, including New Guinea), suggesting the presence of AMH occupation in this region around 50 kya.139 This evidence casts doubt on the single wave dispersal framework, as it has been suggested that this would allow insufficient time for AMH to have migrated to greater Australia. Addressing this idea, Wollstein et al investigated several dispersal hypotheses relating to the colonization and demographic history of Oceania.140 Using dense SNP array data and by simulating different demographic models for the occupation of this region, they found that the most parsimonious scenario involved a split of New Guineans from a common Eurasian ancestor population, rather than a separate early migration event.
Multiple dispersals
Lahr and Foley were the first to propose that AMH populated the world via multiple rather than a single wave of expansion from a morphologically variable population in Africa.141,142 Under their multiple dispersals model, there was an initial dispersal between 50 and 100 kya through South Arabia to Southeast Asia (the Southern route) and a second migration following the Northern route, through the Levant, that led to the colonization of the rest of Eurasia between 40 and 50 kya. They suggested that each of these dispersals was different in terms of the associated artifacts, with the first migration into the Arabian Peninsula affiliated with Middle Paleolithic stone tools and the later expansion through the Levantine corridor into Europe consistent with the appearance of upper Paleolithic tools. Later work by Field and Lahr used a geographical information system-based model, incorporating climatic data from 74 to 59 kya, to show that both a Northern and a Southern route out of Africa would have been possible given environmental barriers.143
More recently, McEvoy et al tested the multiple dispersals hypothesis by applying a LD-based approach to ∼200,000 SNPs in 17 global populations.144 They estimated the split time between these populations using mathematical models that relate the split time to FST between populations and the effective population size (Ne) of populations, assuming a simple bifurcating tree model. By testing divergence times in West and East Eurasian populations simultaneously, they found evidence for a more complex OoA scenario than that suggested by a single dispersal model. Namely, they demonstrated a significant difference in estimates of African/European and African/East Asian divergence times (40 and 36 kya, respectively). These dates are more recent than those previously proposed but nonetheless may indicate that Europe and East Asia were occupied by separate AMH dispersals. The same technique was used in recent work to explore divergence times between Europe and Africa and Australo-Melanesia and Africa.145 The split time for European and East African populations (57–76 kya) was again estimated to be somewhat more recent than that for East Asia and Africa (73–88 kya), and significantly more recent than that between Australo-Melanesians and Africa (87–119 kya) even after accounting for Denisovan introgression into the ancestors of Australo-Melanesians. According to the authors, this is again inconsistent with a single wave dispersal and suggests that Australo-Melanesian populations retain signals of an ancient divergence from Africa. This fits well with the discovery of artifacts associated with human colonization at the Huon Peninsula of Papuan New Guinea and the Mungo Man remains, the oldest AMH found in Australia, dated at 40 kya.146–148
Templeton stressed the importance of genetic interchange under a multiple dispersals scenario.149 His model suggests that ancestors of modern humans, some of which did not necessarily have AMH features, expanded out of Africa many times during the Pleistocene and that these expansions were never sufficient to remove the genetic contributions of previous dispersals, resulting in interbreeding between dispersing groups rather than population replacement. Further support for this model was recently provided by Reyes-Centeno et al who used morphological data from Holocene human cranial samples found in Asia, in conjunction with genetic data, to evaluate models of modern human dispersals out of Africa.150 They concluded that a single dispersal model was likely too simplistic and, as with other studies, found that modern humans first went south upon leaving African and only later took the Northern route in a second expansion wave. Recently published work providing evidence for modern human introgression into the ancestors of East Asian Neanderthals 100 kya also gives support for the multiple dispersals hypothesis.124
In the same vein, it has been suggested that some isolated Southeast Asian/Oceania populations, such as Papuans140,151 and Andamanese and Malaysian Negritos,125,152,153 represent relic populations of a first wave OoA. Some of this evidence is in light of autosomal DNA studies that have indicated Southeast Asia was settled by multiple waves of peoples, the first most related ancestrally to modern day groups such as the Onge and the second more closely related ancestrally to modern day East Asians.125 However, it remains challenging to disentangle whether differences in inferred ancestry are the result of long-term physical isolation and low population sizes rather than a separate wave of OoA migration.
Genetic evidence for an early Southern exit was presented by Rasmussen et al who analyzed a lock of hair from a 100-year-old Australian Aborigine.154 Applying D statistics to segregating sites in the genomes of modern Africans, Europeans, and Asians, they provided evidence for an early branching of Australian Aborigines 75–62 kya. These results suggest that the ancestors of Australian Aborigines are some of the best modern day representatives of a possible early human dispersal into East Asia, although such a conclusion is still a matter of controversy.
Opponents of an early migration into Australia and Oceania assert that if an early migration had taken place before AMH spread into Eurasia, then we would not expect to see evidence of Neanderthal admixture in these genomes given our current understanding of the Neanderthal geographic range.118,153 A conciliatory explanation for the fact that Australo-Melanesians have similar levels of Neanderthal admixture as other non-African populations has been proposed by Weaver,155 who has speculated that the Neanderthal genetic component present in Australasians may be the result of introgression from another group that was in direct contact with Neanderthals.109,125,156,157
Back into Africa Migrations
Another major difficulty in using DNA from modern individuals to the study of OoA migrations is the high proportion of non-African ancestry in modern-day Africans. The first indicators of what is termed the “back to Africa” migrations were obtained from phylogeny and phylogeography of mtDNA haplogroups U6 and M1, which have an origin outside Africa and are currently largely distributed within North and East Africa.158,159 There is now robust evidence for recent migrations back to Africa from non-African populations, as exemplified by high levels of non-African ancestry in present-day Horn of Africa (Ethiopia, Eritrea, Djbouti, and Somalia) and Southern Africa,160 where Eurasian ancestry proportions can be as high as 40%–50% in, for example, some Ethiopian populations.161,162 What is less clear, however, is both the number and the timing of these migrations, as there seems to be a lack of consensus among archeological, mtDNA, Y chromosome, and genomic studies. Using genome-wide data, back to Africa admixture into the Horn of Africa was dated using ROLLOFF,120 ALDER,162 and GLOBETROTTER163 software programs to around 3 kya, coinciding with the origin of the Ethiosemitic languages.109,160 Others have argued for older, perhaps additional, back to Africa migrations using both autosomal164,165 and mtDNA analyses.71,166–169 Any such admixture complicates the ability to study the OoA hypothesis using the genomes of modern day groups. For example, it is plausible that Eurasian gene flow into Africa, together with the high genetic drift reported for East Asians compared to Europeans,170 might explain the reduced African/European compared to African/East Asian divergence times mentioned in the previous work supporting multiple dispersals out of Africa.168,169
Intriguingly, the genome of a 4,500-year-old Ethiopian individual (thus predating the 3 kya Eurasian backflow) named Mota has recently been published (Fig. 1 and Table 1).171 Given that this specimen represents the first aDNA sample from Africa, it will likely become crucial for understanding African genetic diversity and accounting for confounding factors when using DNA from modern populations to reconstruct ancient events. However, the power to draw precise inference from a single such sample is likely limited, so that any resulting conclusions should be treated with caution.
Conclusions
Unraveling the origins and migratory movements of the first H. sapiens is key to understand the genetic diversity of modern-day populations. That said, the vast passage of time generates a level of complexity where conflicts are often encountered in the synthesis of paleontological, archeological, and climatic data.
Much of recent genetic work has moved away from analyses of uniparental systems and instead focuses on using genome-wide autosomal data to reconstruct past events. In light of a lack of high coverage aDNA samples from geographically and temporally representative regions, many studies have also focused on using modern populations to answer questions relating to OoA migration. This is extremely challenging, given that the majority of modern populations are admixed and many African populations have received non-African ancestry as a result of back to Africa migrations. Furthermore, population-specific drift effects are likely to be important, particularly when addressing the question of a single or multiple dispersals out of Africa, particularly given higher reported drift in East Asians compared to Europeans170 may distort estimates of split times in relation to Africa. Given extensive differential drift and admixture, it is likely that modern populations are not very accurate representatives of past populations. With this in mind, developing techniques to study ancient demography using present-day populations is likely to yield only limited success.
We are now entering an era where aDNA samples are increasingly available from different localities and time points. Given the development of new techniques in aDNA recovery and improvements in approaches to minimize contamination and account for postmortem damage, researchers now have access to DNA of both sufficient quality and coverage for the most sophisticated analytic approaches. Good examples are the publicly available high coverage Neanderthal and Denisovan genomes, which have revised our understanding of the OoA event through the identification and dating of admixture between these ancient hominins and AMH leaving Africa, something which was not possible using uniparental systems.
Ancient samples allow direct assessment of the genetic structures of past populations. Using these, in conjunction with carefully selected modern populations will likely allow far greater resolution when reconstructing the paths our ancestors took, as well as in the calibration of AMH mutation rates. However, obtaining samples from key geographical regions (eg, many parts of Africa) currently remains challenging due to poor preservation conditions in certain climates, particularly for older samples.172,173 Thus, it remains to be seen whether informative data relevant to the OoA question can be acquired. Future success in settling some of these long-standing debates will likely require a unified approach that integrates observed patterns in ancient and modern DNA with a growing body of archeological and anthropological data.
Acknowledgments
We would like to thank Prof. Dallas Swallow and Dr. Santos Alonso for their critical reading and helpful comments on the manuscript. We also thank the editor and eight anonymous reviewers for their constructive comments and suggestions.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
PEER REVIEW: Eight peer reviewers contributed to the peer review report. Reviewers’ reports totaled 6,345 words, excluding any confidential comments to the academic editor.
FUNDING: SL is supported by BBSRC (grant number BB/L009382/1). LvD is supported by CoMPLEX via EPSRC (grant number EP/F500351/1). GH is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (grant number 098386/Z/12/Z) and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the manuscript: SL, LvD. Made critical revisions and approved final version: GH. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Darwin C. The Descent of Man, and Selection in Relation to Sex. London: John Murray; 1871. [Google Scholar]
- 2.Huxley TH. Evidence as to Man’s Place in Nature. London: Williams & Norgate; 1863. pp. 114–5. [Google Scholar]
- 3.Weidenreich F. Some problems dealing with ancient man. Am Anthropol. 1940;42:380–2. [Google Scholar]
- 4.Templeton AR. Genetics and recent human evolution. Evolution. 2007;61:1507–19. doi: 10.1111/j.1558-5646.2007.00164.x. [DOI] [PubMed] [Google Scholar]
- 5.Coon CS. The Origin of Races. New York, NY: Knopf; 1962. [Google Scholar]
- 6.Wu X. The well preserved cranium of an early Homo sapiens from Dali, Shanxi. Sciencia Sinica. 1981;2:200–6. [PubMed] [Google Scholar]
- 7.Stringer CB, Andrews P. Genetic and fossil evidence for the origin of modern humans. Science. 1988;239:1263–8. doi: 10.1126/science.3125610. [DOI] [PubMed] [Google Scholar]
- 8.Stringer CB. Replacement, continuity and the origin of Homo sapiens. In: Brauer G, Smith FH, editors. Continuity or Replacement? Controversies in Homo sapiens Evolution. Rotterdam: Balkema; 1992. pp. 9–24. [Google Scholar]
- 9.Zietkiewicz E, Yotova V, Gehl D, et al. Haplotypes in the dystrophin DNA segment point to a mosaic origin of modern human diversity. Am J Hum Genet. 2003;73:994–1015. doi: 10.1086/378777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sankararaman S, Mallick S, Dannemann M, et al. The genomic landscape of neanderthal ancestry in present-day humans. Nature. 2014;507:354–7. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–13. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 12.Stringer C. Modern human origins: progress and prospects. Philos Trans R Soc Lond B Biol Sci. 2002;357:563–79. doi: 10.1098/rstb.2001.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavalli-Sforza L, Feldman MW. The application of molecular genetic approaches to the study of human evolution. Nat Genet. 2003;33:266–75. doi: 10.1038/ng1113. [DOI] [PubMed] [Google Scholar]
- 14.Relethford JH. Genetic evidence and the modern human origins debate. Heredity. 2008;100:555–63. doi: 10.1038/hdy.2008.14. [DOI] [PubMed] [Google Scholar]
- 15.Tattersall I. Human origins: out of Africa. Proc Natl Acad Sci U S A. 2009;106:16018–21. doi: 10.1073/pnas.0903207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature. 1987;325:31–6. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 17.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–7. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 18.Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc Natl Acad Sci U S A. 1995;92:532–6. doi: 10.1073/pnas.92.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macaulay V, Hill C, Achilli A, et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–6. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 20.Metspalu M, Kivisild T, Metspalu E, et al. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 2004;5:26. doi: 10.1186/1471-2156-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovchinnikov IV, Götherström A, Romanova GP, Kraritonov VM, Lidén K, Goodwin W. Molecular analysis of Neanderthal DNA from the northern caucasus. Nature. 2000;404:490–3. doi: 10.1038/35006625. [DOI] [PubMed] [Google Scholar]
- 22.Relethford JH. Absence of regional affinities of Neandertal DNA with living humans does not reject multiregional evolution. Am J Phys Anthropol. 2001;115:95–8. doi: 10.1002/ajpa.1060. [DOI] [PubMed] [Google Scholar]
- 23.Caramelli D, Lalueza-Fox C, Vernesi C, et al. Evidence for a genetic discontinuity between Neandertals and 24,000-year-old anatomically modern Europeans. Proc Natl Acad Sci U S A. 2003;100:6593–7. doi: 10.1073/pnas.1130343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson R, Pritchard JK, Shen P, Oefner PJ, Feldman MW. Recent common ancestry of human Y chromosomes: evidence from DNA sequence data. Proc Natl Acad Sci U S A. 2000;97:7360–5. doi: 10.1073/pnas.97.13.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Underhill PA, Shen P, Lin AA, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–61. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 26.Hawks J. The Y chromosome and the replacement hypothesis. Science. 2001;293:567. doi: 10.1126/science.293.5530.567a. [DOI] [PubMed] [Google Scholar]
- 27.Takahata N, Lee SH, Satta Y. Testing multiregionality of modern human origins. Mol Biol Evol. 2001;18:172–83. doi: 10.1093/oxfordjournals.molbev.a003791. [DOI] [PubMed] [Google Scholar]
- 28.Alonso S, Armour JA. A highly variable segment of human subterminal 16p reveals a history of population growth for modern humans outside Africa. Proc Natl Acad Sci U S A. 2001;98:864–9. doi: 10.1073/pnas.98.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–5. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 30.Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet. 2003;72:1171–86. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Curr Biol. 2005;15:159–60. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci U S A. 2005;102:15942–7. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harding RM, McVean G. A structured ancestral population for the evolution of modern humans. Curr Opin Genet Dev. 2004;14:1–8. doi: 10.1016/j.gde.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Goldstein DB, Chikhi L. Human migrations and population structure: what we know and why it matters. Annu Rev Genomics Hum Genet. 2002;3:129–52. doi: 10.1146/annurev.genom.3.022502.103200. [DOI] [PubMed] [Google Scholar]
- 35.Excoffier L. Human demographic history: refining the recent African origin model. Curr Opin Genet Dev. 2002;12:675–82. doi: 10.1016/s0959-437x(02)00350-7. [DOI] [PubMed] [Google Scholar]
- 36.Watkins WS, Rogers AR, Ostler CT, et al. Genetic variation among world populations: inferences from 100 Alu insertion polymorphisms. Genome Res. 2003;13:1607–18. doi: 10.1101/gr.894603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labuda D, Zietkiewicz E, Yotova V. Archaic lineages in the history of modern humans. Genetics. 2000;156:799–808. doi: 10.1093/genetics/156.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satta Y, Takahata N. The distribution of the ancestral haplotype in finite stepping-stone models with population expansion. Mol Ecol. 2004;13:877–86. doi: 10.1046/j.1365-294x.2003.02069.x. [DOI] [PubMed] [Google Scholar]
- 39.Eriksson A, Manica A. Effect of ancient population structure on the degree of polymorphism shared between modern human populations and ancient hominins. Proc Natl Acad Sci U S A. 2012;109:13956–60. doi: 10.1073/pnas.1200567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eriksson A, Manica A. The doubly conditioned frequency spectrum does not distinguish between ancient population structure and hybridisation. Mol Biol Evol. 2014;31:1618–21. doi: 10.1093/molbev/msu103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowery RK, Uribe G, Jimenez EB, et al. Neanderthal and Denisova genetic affinities with contemporary humans: introgression versus common ancestral polymorphisms. Gene. 2013;530:83–94. doi: 10.1016/j.gene.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Gunz P, Bookstein FL, Mitteroecker P, Stadlmayr A, Seidler H, Weber GW. Early modern human diversity suggests subdivided population structure and a complex out-of-Africa scenario. Proc Natl Acad Sci U S A. 2009;106:6094–8. doi: 10.1073/pnas.0808160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scerri EML, Drake NA, Jennings R, Groucutt HS. Earliest evidence for the structure of Homo sapiens populations in Africa. Quat Sci Rev. 2014;101:207–16. [Google Scholar]
- 44.McDougall I, Brown FH, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–6. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 45.Clark JD, Beyene Y, WoldeGabriel G, et al. Stratigraphic, chronological and behavioural contexts of Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:747–52. doi: 10.1038/nature01670. [DOI] [PubMed] [Google Scholar]
- 46.McCarthy RC, Lucas LA. Morphometric re-assessment of BOU-VP-16/1 from Herto, Ethiopia. J Hum Evol. 2014;74:114–7. doi: 10.1016/j.jhevol.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 47.Henn BM, Gignoux CR, Jobin M, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci U S A. 2011;108:5154–62. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlebusch CM, Skoglund P, Sjödin P, et al. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–9. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pickrell JK, Patterson N, Barbieri C, et al. The genetic prehistory of southern Africa. Nat Commun. 2012;3:1143. doi: 10.1038/ncomms2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HL, Ratan A, Perry GH, Montenegro A, Miller W, Schuster SC. Khoisan hunter-gatherers have been the largest population throughout most of modern-human demographic history. Nat Commun. 2014;5:5692. doi: 10.1038/ncomms6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blome MW, Cohen AS, Tryon CA, Brooks AS, Russell J. The environmental context for the origins of modern human diversity: a synthesis of regional variability in African climate 150,000–30,000 years ago. J Hum Evol. 2012;62:563–92. doi: 10.1016/j.jhevol.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Rito T, Richards MB, Fernandes V, et al. The first modern human dispersals across Africa. PLoS One. 2013;8:e80031. doi: 10.1371/journal.pone.0080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith TM, Tafforeau P, Reid DJ, et al. Earliest evidence of modern humans life history in North African early. Homo sapiens. Proc Natl Acad Sci U S A. 2007;104:6128–33. doi: 10.1073/pnas.0700747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cruciani F, Trombetta B, Massaia A, Destro-Bisol G, Sellitto D, Scozzari R. A revised root for the human Y chromosomal phylogenetic tree: the origin of patrilineal diversity in Africa. Am J Hum Genet. 2011;88(6):814–8. doi: 10.1016/j.ajhg.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fadhlaoui-Zid K, Haber M, Martínez-Cruz B, Zalloua P, Elgaaied AB, Comas D. Genome-wide and paternal diversity reveal a recent origin of human populations in North Africa. PLoS One. 2013;8:e80293. doi: 10.1371/journal.pone.0080293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hovers E. The Lithic Assemblages of Qafzeh Cave. New York, NY: Oxford University Press; 2009. [Google Scholar]
- 57.Grün R, Stringer C, McDermott F, et al. U-series and ESR analyses of bones and teeth relating to the human burials from Skhul. J Hum Evol. 2005;49:316–34. doi: 10.1016/j.jhevol.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 58.Oppenheimer S. Out of Eden: the Peopling of the World. London: Constable; 2003. [Google Scholar]
- 59.Hershkovitz I, Marder O, Ayalon A, et al. Levantine cranium from Manot Cave (Israel) foreshows the first European modern humans. Nature. 2015;520:216–9. doi: 10.1038/nature14134. [DOI] [PubMed] [Google Scholar]
- 60.Pope KO, Terrell JE. Environmental setting of human migrations in the circum-Pacific region. J Biogeogr. 2008;35:1–21. [Google Scholar]
- 61.Kingdom J. Self-Made Man and His Undoing: The Radical Reworking of Evolution Theory. New York, NY: Simon & Schuster; 1993. [Google Scholar]
- 62.Groucutt HS, White TM, Clark-Balzan L, et al. Human occupation of the Arabian Empty Qarter during MIS 5: evidence from Mundafan Al-Buhayrah, Saudi Arabia. Quat Sci Rev. 2015;119:116–35. [Google Scholar]
- 63.Luis JR, Rowold DJ, Regueiro M, et al. The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet. 2004;74:532–44. doi: 10.1086/382286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pagani L, Schiffels S, Gurdasani D, et al. Tracing the route of modern humans out of Africa by using 225 human genome sequences from Ethiopians and Egyptians. Am J Hum Genet. 2015;96:986–91. doi: 10.1016/j.ajhg.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cavalli-Sforza LL, Piazza A, Menozzi P, Mountain J. Reconstruction of human evolution: bringing together genetic, archaeological, and linguistic data. Proc Natl Acad Sci U S A. 1988;85:6002–6. doi: 10.1073/pnas.85.16.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet. 1999;23:437–41. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- 67.Soares P, Alshamali F, Pereira JB, et al. The expansion of mtDNA Haplogroup L3 within and out of Africa. Mol Biol Evol. 2012;29:915–27. doi: 10.1093/molbev/msr245. [DOI] [PubMed] [Google Scholar]
- 68.Forster P, Matsumura S. Did early humans go north or south? Science. 2005;308:965–6. doi: 10.1126/science.1113261. [DOI] [PubMed] [Google Scholar]
- 69.Reed FA, Tishkoff SA. African human diversity, origins and migrations. Curr Opin Genet Dev. 2006;16:597–605. doi: 10.1016/j.gde.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 70.Torroni A, Achilli A, Macaulay V, Richards M, Bandelt H. Harvesting the fruit of the human mtDNA tree. Trends in Genetics. 2006;22:339–45. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 71.Fernandes V, Alshamali F, Alves M, et al. The Arabian cradle: mitochondrial relicts of the first steps along the southern route out of Africa. Am J Hum Genet. 2012;90:347–55. doi: 10.1016/j.ajhg.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Groucutt HS, Petraglia MD, Bailey G, et al. Rethinking the dispersal of Homo sapiens out of Africa. Evol Anthropol. 2015;24:149–64. doi: 10.1002/evan.21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet. 2006;79:230–7. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oppenheimer S. Out-of-Africa, the peopling of continents and islands: tracing uniparental gene trees across the map. Proc Natl Acad Sci U S A. 2012;367:770–84. doi: 10.1098/rstb.2011.0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walter RC, Buffer RT, Bruggemann JH, et al. Early human occupation of the Red Sea Coast of Eritrea during the last interglacial. Nature. 2000;405:65–9. doi: 10.1038/35011048. [DOI] [PubMed] [Google Scholar]
- 76.Rose JI. New light on human prehistory in the Arabo-Persian Gulf Oasis. Curr Anthropol. 2010;51:849–83. [Google Scholar]
- 77.Boivin N, Fuller DQ, Dennell R, Allaby R, Petraglia MD. Human dispersal across diverse environments of Asia during the Upper Pleistocene. Quat Int. 2013;300:32–47. [Google Scholar]
- 78.Mellars P, Gori KC, Carr M, Soares PA, Richards MB. Genetic and archaeological perspectives on the initial modern human colonization of Southern Asia. Proc Natl Acad Sci U S A. 2013;110:10699–704. doi: 10.1073/pnas.1306043110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mellars P. Going east: new genetic and archaeological perspectives on the modern human colonization of Eurasia. Science. 2006;313:796–800. doi: 10.1126/science.1128402. [DOI] [PubMed] [Google Scholar]
- 80.Underhill PA, Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Ann Rev Genet. 2007;41:539–64. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- 81.O’Connell JF, Allen J. The restaurant at the end of the Universe: modelling the colonisation of Sahul. Aust Archaeol. 2012;74:5–31. [Google Scholar]
- 82.Shi W, Ayub Q, Vermeulen M, et al. A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP-CEPH populations. Mol Biol Evol. 2010;27:385–93. doi: 10.1093/molbev/msp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stringer C. Palaeoanthropology. Coasting out of Africa. Nature. 2000;405(24–5):27. doi: 10.1038/35011166. [DOI] [PubMed] [Google Scholar]
- 84.Rampino MR, Self S. Volcanic winter and accelerated glaciation following the Toba super-eruption. Nature. 1992;359:50–2. [Google Scholar]
- 85.Petraglia M, Korisettar R, Boivin N, et al. Middle paleolithic assemblages from the Indian subcontinent before and after the Toba Super-Eruption. Science. 2007;317:114–6. doi: 10.1126/science.1141564. [DOI] [PubMed] [Google Scholar]
- 86.Armitage SJ, Jasim SA, Marks AE, Parker AG, Usik VI, Uerpmann H. The southern route “Out of Africa”: evidence for an early expansion of modern humans into Arabia. Science. 2011;331:453–6. doi: 10.1126/science.1199113. [DOI] [PubMed] [Google Scholar]
- 87.Rose JI, Usik VI, Marks AE, et al. The Nubian complex of Dhofar, Oman: an African Middle Stone Age industry in Southern Arabia. PLoS One. 2011;6:e28239. doi: 10.1371/journal.pone.0028239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu W, Martinón-Torres M, Cai Y, et al. The earliest unequivocally modern humans in Southern China. Nature. 2015;526:696–9. doi: 10.1038/nature15696. [DOI] [PubMed] [Google Scholar]
- 89.Liu W, Jin CZ, Zhang YQ, et al. Human remains from Zhirendong, South China, and modern human emergence in East Asia. Proc Natl Acad Sci U S A. 2010;107:19201–6. doi: 10.1073/pnas.1014386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bae CJ, Wang W, Zhao J, Huang S, Tian F, Shen G. Modern human teeth from Late Pleistocene Luna Cave (Guangxi, China) Quat Int. 2014;354:169–83. [Google Scholar]
- 91.Rosenberg K. A late Pleistocene human skeleton from Liujiang, China suggests regional population variation in sexual dimorphism in the human pelvis. Var Evol. 2002;10:5–17. [Google Scholar]
- 92.Mijares AS, Détroit F, Piper P, et al. New evidence for a 67,000-year-old human presence at Callao Cave, Luzon, Philippines. J Hum Evol. 2010;59:123–32. doi: 10.1016/j.jhevol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 93.Zhivotovsky LA, Bennett L, Bowcock AM, Feldman MW. Human population expansion and microsatellite variation. Mol Biol Evol. 2000;17:757–67. doi: 10.1093/oxfordjournals.molbev.a026354. [DOI] [PubMed] [Google Scholar]
- 94.Gravel S, Henn BM, Gutenkunst RN, et al. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci U S A. 2011;108:11983–8. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harris K, Nielsen R. Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet. 2013;9:e1003521. doi: 10.1371/journal.pgen.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet. 2011;43:1031–4. doi: 10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;13:493–6. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scally A, Durbin R. Revisiting the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 2012;13:745–53. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 99.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 2014;46:919–25. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roach JC, Glusman G, Smit AF, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Conrad DF, Keebler JE, DePristo MA, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–4. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kong A, Frigge ML, Masson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–5. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Campbell CD, Chong JX, Malig M, et al. Estimating the human mutation rate using autozygosity in a founder population. Nat Genet. 2012;44:1277–81. doi: 10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Besenbacher S, Liu S, Izarzugaza JM, et al. Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nat Commun. 2015(6):5969. doi: 10.1038/ncomms6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harris K. Evidence for recent, population-specific evolution of the human mutation rate. Proc Natl Acad Sci U S A. 2015;112:3439–44. doi: 10.1073/pnas.1418652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lipson M, Loh P, Sankararaman S, Patterson N, Berger B, Reich D. Calibrating the human mutation rate via ancestral recombindation density in diploid genomes. PLoS Genet. 2015;11:e1005550. doi: 10.1371/journal.pgen.1005550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fu Q, Mittnik A, Johnson PLF, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23(7):553–9. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fu Q, Li H, Moorjani P, et al. Genome sequence of a 45,000-year-old modern human from Western Siberia. Nature. 2014;514:445–9. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prüfer K, Racimo F, Patterson N, et al. The complete genome sequence of a Neanderthal from the Altai mountains. Nature. 2014;505:43–9. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reich D, Green RE, Kircher M, et al. genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–60. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Meyer M, Kircher M, Gansauge MT, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–6. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hublin JJ. The origin of Neandertals. Proc Natl Acad Sci U S A. 2009;106:16022–7. doi: 10.1073/pnas.0904119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Finlayson C, Pacheco FG, Rodríguez-Vidal J, et al. Late survival of Neanderthals at the southernmost extreme of Europe. Nature. 2006;443:850–3. doi: 10.1038/nature05195. [DOI] [PubMed] [Google Scholar]
- 114.Krings M, Stone A, Schmitz RW, et al. Neanderthal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 115.Currat M, Excoffier L. Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol. 2004;2:e421. doi: 10.1371/journal.pbio.0020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Serre D, Langaney A, Chech M, et al. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2004;2:e57. doi: 10.1371/journal.pbio.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Green R, Malaspinas A-S, Krause J, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–26. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Green RE, Krause J, Briggs AW, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sankararaman S, Patterson N, Li H, Pääbo S, Reich D. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 2012;8:e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Patterson NJ, Moorjani O, Luo Y, et al. Ancent admixture in human history. Genetics. 2012;192:1065–93. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28:2239–52. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Trinkaus E, Moldovan O, Milota S, et al. An early modern human from the Pestera Cu Oase, Romania. Proc Natl Acad Sci U S A. 2003;110:11231–36. doi: 10.1073/pnas.2035108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fu Q, Hajdinjak M, Moldovan OT, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–9. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kuhlwilm M, Gronau I, Hubisz MJ, et al. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature. 2016;530(7591):429–33. doi: 10.1038/nature16544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reich D, Patterson N, Kircher M, et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89:516–28. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qin P, Stoneking M. Denisovan ancestry in East Eurasian and native American populations. Mol Biol Evol. 2015;32:2665–74. doi: 10.1093/molbev/msv141. [DOI] [PubMed] [Google Scholar]
- 127.Huerta-Sánchez E, Jin X, Asan, et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–7. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Klein RG. The Human Career: Human Biological and Cultural Origins. Chicago: Chicago University Press; 2009. [Google Scholar]
- 129.Shea JJ. The archaeology of an illusion: the middle-upper paleolithic transition in the Levant. In: Le Tenesor JM, Jagher R, Otte R, editors. The Lower and Middle Paleolithic in the Middle East and Neighbouring Regions. ERAUL 126. Liége: Université de Liége; 2011. pp. 169–82. [Google Scholar]
- 130.McDermott F, Grun R, Stringer GB, Hawkesworth CJ. Mass spectrometric dates for Israeli Neanderthal/early modern sites. Nature. 1993;363:252–5. doi: 10.1038/363252a0. [DOI] [PubMed] [Google Scholar]
- 131.Frumkin A, Bar-Yosef O, Schwarcz HP. Possibly paleohydrologic and paleo-climatic effects on hominin migration and occupation of the Levantine Middle Paleolithic. J Hum Evol. 2011;60:427–51. doi: 10.1016/j.jhevol.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 132.Oppenheimer S. A single southern exit of modern humans from Africa: before or after Toba? Quat Int. 2012;258:88–99. [Google Scholar]
- 133.Usik VI, Rose JI, Hilbert YH, et al. Nubian complex reduction strategies in Dhofar, Southern Oman. Quat Int. 2013;300:244–66. [Google Scholar]
- 134.Deshpande O, Batzoglo S, Feldman MW, Cavalli-Sforza LL. A serial founder effect model for human settlement out of Africa. Proc Biol Sci. 2009;276:291–300. doi: 10.1098/rspb.2008.0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pemberton TJ, DeGiorgio M, Rosenberg NA. Population structure in a comprehensive set on human microsatellite variation. G3 (Bethesda) 2013;3:891–907. doi: 10.1534/g3.113.005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pickrell J, Reich D. Towards a new history and geography of human genes informed by ancient DNA. Trends Genet. 2014;30:377–89. doi: 10.1016/j.tig.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat Genet. 2009;41:66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bowler JM, Jones R, Allen H, Thorne AG. Pleistocene remains from Australia: a living site and human cremation from Lake Mungo, Western New South Wales. World Archaeol. 1970;1:39–60. doi: 10.1080/00438243.1970.9979463. [DOI] [PubMed] [Google Scholar]
- 140.Wollstein A, Lao O, Becker C, et al. Demographic history of Oceania inferred from genome-wide data. Curr Biol. 2010;20:1983–92. doi: 10.1016/j.cub.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 141.Lahr M, Foley R. Multiple dispersals and modern human origins. Evol Anthropol. 1994;3:48–60. [Google Scholar]
- 142.Lahr M, Foley R. Towards a theory of modern human origins: geography, demography and diversity in recent human evolution. Am J Phys Anthropol. 1998;41:137–76. doi: 10.1002/(sici)1096-8644(1998)107:27+<137::aid-ajpa6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 143.Field JS, Lahr MM. Assessment of the southern dispersal: GIS-based analysis of potential routes at oxygen isotopic stage 4. J World Prehistory. 2006;19:1–45. [Google Scholar]
- 144.McEvoy BP, Powell JE, Goddard ME, Visscher PM. Human population dispersal “Out of Africa” estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 2011;21:821–9. doi: 10.1101/gr.119636.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tassi F, Ghirotto S, Mezzavilla M, Vilaça ST, de Santi L, Barbujani G. Early modern human dispersal from Africa: genomic evidence for multiple waves of migration. Investig Genet. 2015(6):13. doi: 10.1186/s13323-015-0030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Groube L, Chappell J, Muke J, Price D. A 40,000 year-old occupation site at Huon Peninsula, Papua New Guinea. Nature. 1986;324:453–5. doi: 10.1038/324453a0. [DOI] [PubMed] [Google Scholar]
- 147.O’Connell JF, Allen J. Dating the colonization of Sahul (Pleistocene Australia-New Guinea): a review of recent research. J Archaeol Sci. 2004;31:835–53. [Google Scholar]
- 148.Bowler JM, Johnston H, Olley JM, et al. New ages for human occupation and climatic change at Lake Mungo, Australia. Nature. 2003;421:837–40. doi: 10.1038/nature01383. [DOI] [PubMed] [Google Scholar]
- 149.Templeton A. Out of Africa again and again. Nature. 2002;416:45–51. doi: 10.1038/416045a. [DOI] [PubMed] [Google Scholar]
- 150.Reyes-Centeno H, Hubbe M, Hanihara T, Stringer C, Harvati K. Testing modern human out-of-Africa dispersal models and implications for modern human origins. J Hum Evol. 2015;87:95–106. doi: 10.1016/j.jhevol.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 151.Reyes-Centeno H, Ghirotto S, Detroit F, Grimaud-Hervé D, Barbujani G, Harvati K. Genomic and cranial phenotype data support multiple modern human dispersals from Africa and a Southern route into Asia. Proc Natl Acad Sci U S A. 2014;111:7248–53. doi: 10.1073/pnas.1323666111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461(489):494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Aghakhanian F, Yunus Y, Naidu R, et al. Unravelling the genetic history of Negritos and Indigenous populations of Southeast Asia. Genome Biol Evol. 2015;7:1206–15. doi: 10.1093/gbe/evv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rasmussen M, Guo X, Wang Y, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–8. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weaver TD. Tracing the paths of modern humans from Africa. Proc Natl Acad Sci U S A. 2014;111:7170–1. doi: 10.1073/pnas.1405852111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Skoglund P, Jakobsson M. Archaic human ancestry in East Asia. Proc Natl Acad Sci U S A. 2011;108:18301–6. doi: 10.1073/pnas.1108181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cooper A, Stringer CB. Did the Denisovans cross Wallace’s Line? Science. 2013;342:321–3. doi: 10.1126/science.1244869. [DOI] [PubMed] [Google Scholar]
- 158.Maca-Meyer N, Gonzalez AM, Larruga JM, Flores C, Cabrera VM. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet. 2001(2):13. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gonzalez AM, Larruga JM, Abu-Amero KK, Shi Y, Pestano J, Cabrera VM. Mitochondrial lineage M1 traces an early human backflow to Africa. BMC Genomics. 2007(8):223. doi: 10.1186/1471-2164-8-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pickrell JK, Patterson N, Loh PR, et al. Ancient west Eurasian ancestry in southern and eastern Africa. Proc Natl Acad Sci U S A. 2014;111:2632–7. doi: 10.1073/pnas.1313787111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pagani L, Kivisild T, Tarekegn A, et al. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet. 2012;91:83–96. doi: 10.1016/j.ajhg.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Loh P-R, Lipson M, Patterson N, et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–54. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hellenthal G, Busby GB, Band G, et al. A genetic Atlas of human admixture history. Science. 2014;343:747–51. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Henn BM, Botigué LR, Gravel S, et al. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hodgson JA, Mulligan CJ, Al-Meeri A, Raaum RL. Early back-to-Africa migration into the Horn of Africa. PLoS Genet. 2014;10:e1004393. doi: 10.1371/journal.pgen.1004393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cerný V, Mulligan CJ, Fernandes V, et al. Internal diversification of mitochondrial haplogroup R0a reveals post-last glacial maximum demographic expansions in South Arabia. Mol Biol Evol. 2011;28:71–8. doi: 10.1093/molbev/msq178. [DOI] [PubMed] [Google Scholar]
- 167.Musilová E, Fernandes V, Silva NM, et al. Population history of the Red Sea-genetic exchanges between the Arabian Peninsula and East Africa signaled in the mitochondrial DNA HV1 Haplogroup. Am J Phys Anthropol. 2011;145:592–8. doi: 10.1002/ajpa.21522. [DOI] [PubMed] [Google Scholar]
- 168.Non AL, Al-Meeri A, Raaum RL, Sanchez LF, Mulligan CJ. Mitochondrial DNA reveals distinct evolutionary histories for Jewish populations in Yemen and Ethiopia. Am J Phys Anthropol. 2011;144:1–10. doi: 10.1002/ajpa.21360. [DOI] [PubMed] [Google Scholar]
- 169.Pennarun E, Kivisild T, Metspalu E, et al. Divorcing the Late Upper Palaeolithic demographic histories of mtDNA haplogroups M1 and U6 in Africa. BMC Evol Biol. 2012(12):234. doi: 10.1186/1471-2148-12-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Keinan A, Mullikin J, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–5. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Llorente MG, Jones ER, Eriksson A, et al. Ancient Ethiopian genome reveals extensive Eurasian admixture throughout the African continent. Science. 2015;350(6262):820–2. doi: 10.1126/science.aad2879. [DOI] [PubMed] [Google Scholar]
- 172.Smith CI, Chamberlain AT, Riley MS, Stringer C, Collins MJ. The thermal history of human fossils and the likelihood of successful DNA amplification. J Hum Evol. 2003;45:203–17. doi: 10.1016/s0047-2484(03)00106-4. [DOI] [PubMed] [Google Scholar]
- 173.Allentoft ME, Collins M, Harker D, et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc Biol Sci. 2012;279:4724–33. doi: 10.1098/rspb.2012.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]