Abstract
Genetic factors appear to be highly relevant to predicting differential risk for the development of post-traumatic stress disorder (PTSD). In a discovery sample, we conducted a genome-wide association study (GWAS) for PTSD using a small military cohort (Systems Biology PTSD Biomarkers Consortium; SBPBC, N = 147) that was designed as a case-controlled sample of highly exposed, recently returning veterans with and without combat-related PTSD. A genome-wide significant single nucleotide polymorphism (SNP), rs717947, at chromosome 4p15 (N = 147, β = 31.34, P = 1.28×10−8) was found to associate with the gold-standard diagnostic measure for PTSD (the Clinician Administered PTSD Scale). We conducted replication and follow-up studies in an external sample, a larger urban community cohort (Grady Trauma Project, GTP, N = 2006), to determine the robustness and putative functionality of this risk variant. In the GTP replication sample, SNP rs717947 associated with PTSD diagnosis in females (N = 2006, P = 0.005), but not males. SNP rs717947 was also found to be a methylation quantitative trait locus (meQTL) in the GTP replication sample (N = 157, P = 0.002). Further, the risk allele of rs717947 was associated with decreased medial and dorsolateral cortical activation to fearful faces (N = 53, P < 0.05) in the GTP replication sample. These data identify a genome-wide significant polymorphism conferring risk for PTSD, which was associated with differential epigenetic regulation and with differential cortical responses to fear in a replication sample. These results may provide new insight into understanding genetic and epigenetic regulation of PTSD and intermediate phenotypes that contribute to this disorder.
Keywords: GWAS, PTSD, fMRI, meQTL, epigenetic
INTRODUCTION
Although post-traumatic stress disorder (PTSD) requires the experience of a life-threatening trauma to occur, twin and family studies have suggested that up to 30–40% of the risk for PTSD following significant trauma is genetically heritable (e.g., [True et al., 1993]). Genetic studies to date have been relatively small, but have yielded approximately 15–20 different genes that might associate with PTSD or PTSD risk. These candidate studies are well-discussed in several recent reviews [Pitman et al., 2012; Almli et al., 2014b]. There have been three published GWAS, at the time of this writing, focused on PTSD [Guffanti et al., 2013; Logue et al., 2013; Xie et al., 2013]. The genome-wide significant findings have yielded variants within or near potentially interesting candidate genes, including RORA, TLL, and COBL, as well as within a long noncoding RNA, lincRNA AC068718.1. The advantage of GWAS is that it is hypothesis neutral, and thus allows the identification of variants not selected for candidate gene/single nucleotide polymorphism (SNP)-based analysis. However, there is a stringent multiple test burden with GWAS, which is necessary to avoid attributing the finding to false discovery.
Since it is difficult to get very large sample sizes within a single cohort, and individual heterogeneity is introduced by them, another approach is to conduct GWAS in highly homogenous, well-characterized samples where exposure levels are controlled in both cases and controls. For instance, a study using very well-matched trauma control cohorts may provide statistical power that is not found in epidemiological samples of patient cases compared to non-patient controls. Alternatively, an extreme phenotype design, where there is enrichment of extreme cases and controls, can lead to increased odds ratios, and improved power to detect associations, especially with quantitative traits [Van Gestel et al., 2000]. An ideal solution from these methods would be a discovery sample in which the cases differ in having extremely severe PTSD compared to those similarly exposed to trauma, but with no PTSD at all.
In this study, we aimed to increase our understanding of genetic risk for PTSD by conducting a GWAS for PTSD in a discovery sample with an extreme phenotype design and cases and controls having similar exposures. Our discovery sample consisted of a deeply phenotyped, well-controlled, similarly exposed, but relatively small military sample (Systems Biology PTSD Biomarkers Consortium; SBPBC) [Yan et al., 2013; Lindqvist et al., 2014; Yehuda et al., 2015]. To validate GWAS findings, we conducted a replication study in a sample consisting of subjects from an independent cohort, the civilian Grady Trauma Project (GTP) [Binder et al., 2008; Almli et al., 2013]. The GTP is a large, primarily African-American highly-traumatized community cohort recruited from the general medical clinics of Grady Memorial Hospital, a publicly funded hospital that serves economically-disadvantaged individuals in Atlanta, Georgia. Using the replication cohort (GTP), we are able to expand our understanding of the GWAS findings through functional genomic and neuroimaging association analyses. Identification and replication of PTSD association across such divergent cohorts, combined with examination of GWAS findings at epigenetic and neural intermediate phenotype levels, has led to a novel and interesting potential PTSD-associated genomic locus.
METHODS
Participants in Discovery Cohort
The SBPBC cohort utilized for initial GWAS discovery in this report was recruited as part of a larger study that is designed to identify biomarkers for PTSD diagnosis in cross-sectional and longitudinal studies in male and female Iraq and Afghanistan veterans [Yan et al., 2013; Lindqvist et al., 2014; Yehuda et al., 2015]. Participants were recruited at New York University Langone Medical Center and Icahn School of Medicine at Mount Sinai and from New York City Veteran Affairs medical centers at Manhattan and Bronx, as well as other veteran service organizations and the community. All procedures were approved by the Institute Review Boards of NYU School of Medicine and Mount Sinai School of Medicine. Participants gave written informed consent after receiving a complete description of the study. General inclusion criteria include being a US veteran who served in Operation Enduring Freedom in Afghanistan and/or Operation Iraqi Freedom, between the age of 20 and 60 years, being able to understand the protocol and willing to provide written informed consent. Consistent with the goal of having extreme phenotypes, with PTSD being the primary difference between groups, exclusion criteria included loss of consciousness for more than 10 min, neurological disorders, current alcohol and substance abuse disorders, suicidality, lifetime history of any psychiatric disorder with psychotic features, bipolar disorder or obsessive-compulsive disorder.
Doctoral level clinical psychologists conducted structured diagnostic interviews. Criteria for the PTSD group include warzone exposure and related PTSD symptoms of at least three months duration as indexed by the clinician administered PTSD scale (CAPS) [Blake et al., 1995], consisting of the sum of symptom frequency and intensity. The criteria for control group also included warzone exposure, but subjects did not meet CAPS criteria for lifetime or current combat or civilian PTSD. Warzone exposure was defined as exposure to an OEF/OIF (GWOT) Criterion A event and was confirmed by clinical interview. The Structured Clinical Interview for DSM-IV Diagnosis [Ventura et al., 1998] was used to diagnose comorbid disorders and to assess for exclusion criteria. The military version of the PTSD Checklist (PCL-M) [Weathers et al., 2013], and the Beck depression index (BDI) [Beck et al., 1961] were also used as self-report questionnaires to assess symptom severity. After clinical screening, subjects who met the diagnosis criteria for PTSD with CAPS score >40 were included in the PTSD group and those with CAPS score <20 were included in the control group. These cut-offs were chosen to ensure clinically significant levels of symptomatology in the cases, and to exclude sub-threshold cases in the control group. Because of the nature of warzone trauma and the recruitment feasibility of combat veterans, PTSD subjects in the present study were chronic cases, and both groups had undergone prolonged warzone exposure. Demographic and psychiatric characteristics of the discovery sample are shown in Table I. We used t-tests to determine statistical differences between cases/controls with the quantitative data and chi-squared tests for categorical data.
TABLE I.
Demographic and Clinical Characteristics of the Discovery Sample (SBPBC Cohort) Shown as Means (SD) or Counts (Percent)
| Total (N = 147) | PTSD − (N = 63) | PTSD − (N = 84) | |
|---|---|---|---|
| Age | 32.8 (8.0) | 33.6 (8.1) | 32.1 (8.0) |
| Gender (% male) | 130 88.4% | 57 90.5% | 73 86.9% |
| Race/ethnicity | |||
| Hispanic | 57 (38.8%) | 32 (50.8%) | 25 (29.8%) |
| Non-hispanic Asian | 6 (4.1%) | 0 | 6 (7.1%) |
| Non-hispanic black | 35 (23.8%) | 16 (25.4%) | 19 (22.6%) |
| Non-hispanic white | 45 (30.6%) | 14 (22.2%) | 31 (36.9%) |
| Non-hispanic other | 4 (2.7%) | 1 (1.6%) | 3 (3.6%) |
| Education | |||
| Less than 12th grade | 3 (2.0%) | 1 (1.6%) | 2 (2.4%) |
| High school grad or GED | 39 (26.5%) | 21 (33.3%) | 18 (21.4%) |
| 2 years college, AA degree | 47 (32.0%) | 22 (34.9%) | 25 (29.8%) |
| 4 years college, BA degree | 43 (29.3%) | 17 (27.0%) | 26 (31.0%) |
| Masters degree | 14 (9.5%) | 2 (3.2%) | 12 (14.3%) |
| Doctoral degree | 1 (0.7%) | 0 (0%) | 1 (1.2%) |
| Relationship status | |||
| Single | 59 (40.1%) | 22 (34.9%) | 37 (44.0%) |
| Steady relationship or living together | 23 (15.6%) | 11 (17.5%) | 12 (14.3%) |
| Married | 39 (26.5%) | 16 (25.4%) | 23 (27.4%) |
| Divorced | 26 (17.1%) | 14 (22.2%) | 12 (14.3%) |
| Early trauma exposurea,* | 5.8 (4.9) | 6.9 (5.7) | 5.0 (4.2) |
| PTSD severity (clinician-rated)b,* | 31.7 (34.7) | 69.3 (17.0) | 3.5 (5.2) |
| PTSD severity (self-reported)c,* | 41.0 (20.3) | 60.9 (12.5) | 26.0 (8.9) |
| Depression severityd,* | 13.8 (13.0) | 24.7 (11.4) | 5.6 (6.3) |
| Military experiencee | 36.1 (14.2) | 52.2 (12.3) | 30.2 (9.6) |
| Negative life eventsf,* | 10.4 (10.9) | 17.8 (12.0) | 5.1 (5.9) |
AA, associate’s degree; BA, bachelor’s degree; GED, general educational development; PTSD, post-traumatic stress disorder.
ETISR total [Bremner et al., 2000].
CAPS [Blake et al., 1995].
PCL-M [Weathers et al., 2013].
BDI [Beck et al., 1961].
DRRI_D [Vogt et al., 2013].
Life events scale negative-past 12 months [Sarason et al., 1978].
P<0.05.
Genetic Data: Quality Control and Analyses
Genotyping
Using DNA extracted from blood samples, genome-wide SNP genotyping of 147 subjects (we note that one individual withdrew from the study) from the SBPBC discovery cohort was conducted using Illumina’s HumanOmniExpress BeadChip. After calls were made with GenomeStudio (Illumina Inc), there were 730,493 individual SNPs with genotype information. We performed quality control on the SNP data using PLINK [Purcell et al., 2007] (Supplementary Fig. 1). We first used crude quality control filters prior to performing principal-component analysis (PCA) to infer axes of ancestry (Supplementary Fig. 2A). We removed 3666 SNPs for low call rates (less than 95%) and 16,786 SNPs with a frequency of less than 0.01. All individuals had call rates of 98% or greater. Prior to PCA, we used PLINK to prune the autosome only data in windows of 50 base pairs, removing one SNP from each pair of SNPs with r2>0.05 to obtain a set of roughly independent SNPs. We further filtered SNPs for analysis with call rates of less than or equal to 98%, minor allele frequencies (MAF) of less than 0.1, and Hardy–Weinberg equilibrium (HWE) P-values of less than 1×10−5 (removing 13,935, 137,997, and 686 more SNPs, respectively). After quality control, there were 147 individuals (130 males and 17 females) that were genotyped on 557,423 SNPs.
Statistical analyses for GWAS
Using the statistical package PLINK, we regressed the outcome of current CAPS score on allele count assuming an additive model (0, 1, or 2 copies of the risk allele), including sex and the top three principal components of genome-wide data as covariates. We note that we used the first three principal components within the discovery sample to prevent possible model overfitting and loss of power. To verify the robustness of the top SNP, we used logistic regression to test for the association between PTSD diagnosis (using CAPS) and allele count, including the same covariates as above. Given the mixed ancestry of the discovery samples, we also tested significant variants using self-reported race instead of principal components, as a sensitivity analysis.
Replication Cohort
Participants
The replication cohort consisted of study participants from Grady Memorial Hospital (Atlanta, Georgia) as part of the GTP. As previously shown [Ressler et al., 2011], these participants were adult, primarily female, highly traumatized and impoverished, with high rates of PTSD. To minimize genetic differences due to ancestry, individuals within three standard deviations of the medians of the first and second principal components of self-reported African Americans, were selected for analysis [Almli et al., 2014a].
Genetic association analysis
Given that the discovery sample was primarily male, and prior findings of genomic differences in PTSD across the sexes [Ressler et al., 2011; Gillespie et al., 2013], we conducted an analysis stratified by sex. Using the categorical measure of PTSD, we conducted SNP-based association analysis in the GTP samples for the genome-wide significant SNP. Analyses were run parallel to those described in the SBPBC cohort. In males and females separately, we regressed PTSD diagnosis, based on DSM A-D criteria using responses to the modified PTSD Symptom Scale (PSS) [Falsetti et al., 1993], on allele count (0, 1, or 2 risk alleles) as above. Due to the larger sample size, we used the top 10 principal components [Price et al., 2006; Lin and Zhoa, 2009] and chip type (Illumina’s HumanOmniExpress or Omni1-Quad BeadChip) as covariates.
Follow-Up Analyses in Replication Cohort
Methylation quantitative trait locus analysis
DNA methylation was assessed as previously described [Mehta et al., 2013]. Briefly, 1 microgram of DNA was bisulfite-treated, and methylation at >485,000 CpG sites was interrogated using the Human-Methylation450 BeadChip (Illumina). Beta values were generated with BeadStudio and were set to missing (no call) if detection p-values exceeded .001. CpGassoc [Barfield et al., 2012] was used to remove samples with probe detection call rates <95% and those with an average intensity value of either <50% of the experiment-wide sample mean or <2,000 arbitrary units (AU). In addition, CpG sites with missing data for >10% of samples were excluded from analysis. Beta Mixture Quantile dilation (BMIQ) was used to normalize each dataset [Teschendorff et al., 2013]. Individual level data from the 11 CpG sites within 1 MB of rs717947 were examined for this study. For each CpG site, methylation is represented as the average of methylated and unmethylated DNA across all cells that make up an individual DNA sample.
Given that rs717947 is located in an intergenic region that appears to be epigenetically-regulated based on human postmortem prefrontal cortex methylation data [Maunakea et al., 2010], we identified methylation quantitative trait loci (meQTL) in the GTP cohort by applying the approach described previously [Smith et al., 2014] to the HumanMethylation450 data. Using R (www.R-project.org), the relationship between the proportion of methylation at the CpG site and SNP was examined via linear regression, where methylation was modeled as a linear function of the number of reference alleles (0, 1, or 2) with sex, age and the top three principal components from GWAS as covariates. Thus, an association, if found between genotype and methylation, represents genotype-dependent averages across the entire cohort. We also performed the same analysis in females only, consistent with the strategy described for SNP replication above. We examined 11 CpG sites and adjusted for each as an independent test; thus, P < 0.0045 (0.05/11 sites) was noted as statistically significant.
Neuroimaging
To determine the neural correlates of the PTSD risk allele, we examined fMRI data among a subset of traumatized women drawn from the GTP replication cohort. Participants completed a task designed to engage threat-processing networks, passively viewing static fearful and neutral face stimuli. Fearful and neutral face stimuli were presented in a block design. Trials included a face stimulus presented for 500 ms, followed by a 500 ms presentation of a fixation cross. Subjects were instructed to pay attention to the faces, and did not make any behavioral response, to minimize motion artifacts and neural activation unrelated to processing the visual stimulus. Detailed procedures and data processing methods are described elsewhere [Stevens et al., 2013].
RESULTS
The demographic characteristics of the SBPBC subjects are shown in Table I. The SBPBC cohort had an overall mean of 31.7 (SD = 34.7, range 0–136) for PTSD symptom scores; however, because the cohort was designed with extreme phenotypes, symptom severity was much higher in cases [PTSD+: mean = 69.4 (16.9)] than controls [PTSD−: 3.5 (5.2)]. We report that the demographic characteristics were similar between cases and controls (P > 0.05). The following psychiatric variables, Early Trauma Exposure, PTSD severity (clinician-rated), PTSD severity (self-reported), Depression severity, and Negative Life Events, showed differences between cases and controls, with cases endorsing higher symptoms.
GWAS
Using the military SBPBC cohort, we conducted a GWAS using a quantitative measure of PTSD symptoms as the outcome with sex, and the top three principal components as covariates. There was no evidence to suggest inflation of test statistics in our GWAS given a lambda of 1.00 (Supplementary Fig. 2B). The GWAS yielded one genome-wide significant SNP, rs717947, at chromosome 4p15 (N = 147, β (SE) = 31.34 (5.19), P = 1.28×10−8) that associated with current CAPS score (Fig. 1A). SNP rs717947 had a MAF of 0.18, and a HWE p-value of 1 (4 TT/45 TC/98 CC) in the whole sample and 0.18 in controls. Although most of the top SNPs for the GWAS were intragenic, with the exception of Collagen, Type IV, Alpha 2 (COL4A2), several loci are represented 4p15.1, 12q15, 20p12.1, with SNPs in likely linkage disequilibrium (Table II). Sensitivity analysis covarying for self-reported race instead of the top three principal components showed similar results for the association between rs717947 and PTSD symptoms (data not shown). The risk allele (T) carriers consistently show higher CAPS scores compared to individuals with the CC genotype [mean (SD): TT/TC = 52.7 (34.7), CC = 21.5 (29.8)] (Supplementary Fig. 3A). Follow-up analysis of SNP rs717947 showed that this SNP is robust to different measures of PTSD, both categorical and self-reported measures. Using the categorical PTSD diagnosis measure (based on CAPS) as an outcome revealed an odds ratio (OR) of 8.6 (N = 147, 95% confidence interval (CI) = 3.5–21.6, P = 3.84×10−6) (Fig. 2A). Given that the discovery sample was an extreme phenotype design based on CAPS, we verified that self-reported PTSD symptoms (not used for initial sample selection) produced similar results (N = 147, β (SE) = 17.37 (3.10), P = 1.203×10−7). Plots of self-reported PTSD symptoms are similar to symptoms using CAPS (Supplementary Fig. 3B).
FIG. 1.
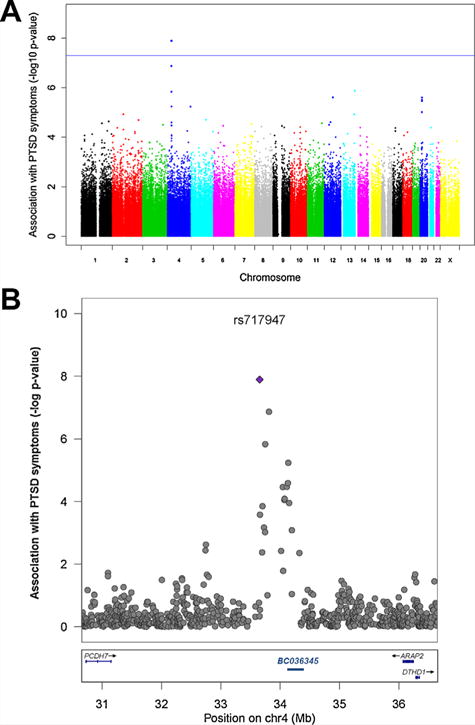
Chromosome 4 SNP is Associated with PTSD at a Genome-Wide Level. (A) Manhattan plot for PTSD symptoms as measured by the CAPS in the SBPBC cohort (N = 147) showing a peak on chromosome 4, with top SNP rs717947 (β (SE) = 31.34 (5.19), P = 1.28×10−8). (B) Regional plot of chromosome 4 showing a peak of SNPs with low p-values near the genome-wide significant hit (rs717947 shown as a blue diamond) and non-coding RNA BC036345 (chr4: 33,897,961–34,041,515). Plot generated using LocusZoom (http://csg.sph.umich.edu/locuszoom/).
TABLE II.
Top 10 Variants From GWAS of PTSD Symptoms as Measured by the CAPS
| Chr | Gene | Approx distance to downstream gene | SNP | BP location | Allele | β (SE) | P-value |
|---|---|---|---|---|---|---|---|
| 4 | intergenic | 2629 kb, DTHD1 | rs717947 | 33653757 | T | 31.34 (5.19) | 1.28×10−8 |
| 4 | intergenic | 2472 kb, DTHD1 | rs13104973 | 33810391 | C | 29.04 (5.23) | 1.35×10−7 |
| 13 | COL4A2 | intragenic | rs4773155 | 110971066 | A | 20.03 (3.97) | 1.35×10−6 |
| 4 | intergenic | 2536 kb, DTHD1 | rs7684329 | 33747424 | A | 27.04 (5.38) | 1.47×10−6 |
| 12 | intergenic | 473 kb, RAP1B | rs17104851 | 68531344 | A | −23.74 (4.83) | 2.47×10−6 |
| 12 | intergenic | 458 kb, RAP1B | rs3181035 | 68546396 | A | −23.74 (4.83) | 2.47×10−6 |
| 20 | intergenic | 1069 kb, PCSK2 | rs4239715 | 16138249 | G | 18.00 (3.67) | 2.51×10−6 |
| 20 | intergenic | 1065 kb, PCSK2 | rs6080152 | 16141991 | C | −18.95 (3.90) | 3.14×10−6 |
| 20 | intergenic | 1101 kb, PCSK2 | rs6080131 | 16106040 | G | 18.59 (3.85) | 3.48×10−6 |
| 4 | intergenic | 2148 kb, DTHD1 | rs13113036 | 34135538 | C | 23.19 (4.92) | 5.75×10−6 |
SNPs on chromosome 4 are likely in linkage disequilibrium (shown in Fig. 1b).
FIG. 2.
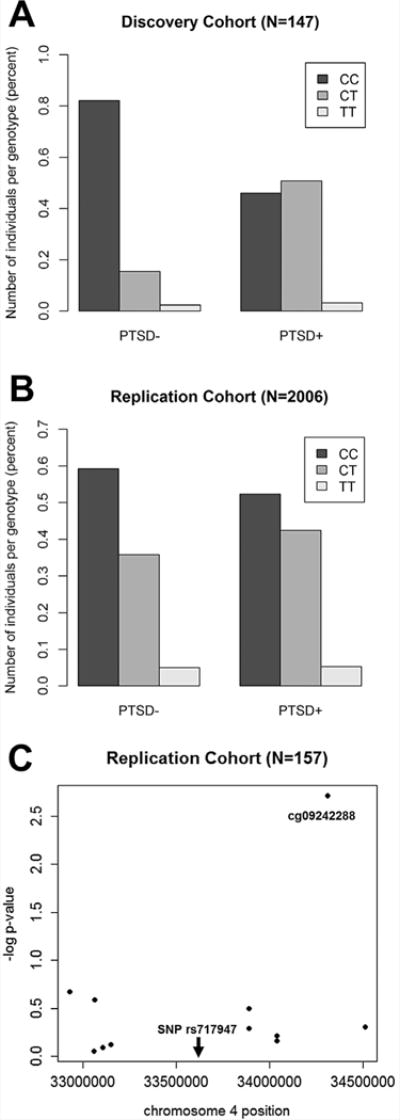
Replication of Genome-wide PTSD-associated SNP and Evidence for an meQTL. Replication results for associations of genome-wide significant SNP, rs717947, with PTSD diagnosis in the (A) discovery cohort (N = 147, OR = 8.6 CI = 3.5–21.6, P = 3.84×10−6). and (B) replication cohort (females only, N = 2006, OR = 1.25, CI = 1.1–1.5, P = 0.005). (C) Of the 11 CpG within 1 MB of the genome-wide SNP, rs717947 affects expression levels (probe cg09242288) with the risk genotype (TT) showing decreased proportion of methylation (N = 157, β (SE) = −0.02 (0.007), P = 0.002].
Given uncertainty on whether trauma exposure should be considered as a covariate in GWAS of PTSD, we next examined whether the main effect of rs717947 would be significant for CAPS scores after controlling for early childhood trauma exposure or overall negative life events. In both cases, the SNP remained genome-wide significant [Covariate: Early Trauma Inventory, N = 146, β (SE) = 31.5 (5.2), P = 1.13×10−8; Negative life events, N = 146, β (SE) = 24.9 (4.3), P = 3.29×10−8].
Replication of rs717947
Using the civilian GTP cohort, we attempted to replicate the association between SNP rs717947 and PTSD diagnosis. Due to our past findings of SNPs differentially associating with PTSD in males and females (e.g., Ressler et al., 2011) in addition to the male majority of the discovery sample, we stratified our analyses by sex. In females, we found that carriers of the risk allele (T) had increased odds of PTSD diagnosis (N=2006, OR=1.25, CI=1.1–1.5, p=0.005) (Fig. 2B); in contrast, no association was found in males (N=862, p=0.37). Follow-up analyses showed that 6 of the 9 SNPs in the regional peak on chromosome 4 identified from the discovery sample also associate with PTSD diagnosis in the same direction in the GTP replication cohort (nominal p<0.1; Supplementary Table I). We also note that in the GTP sample, this SNP, rs717947, was not associated with comorbid disorders, such as depression as measured by BDI (p=0.8), alcohol abuse by AUDIT (p=0.4), or drug abuse by DAST (p=0.6).
rs717947 is a Methylation QTL
Although the gene structure and function of this region of chromosome 4 is unclear, bioinformatics analyses suggest that this peak of association is within an intergenic region that appears to be epigenetically-regulated based on postmortem prefrontal cortex methylation data (Supplementary Fig 4). Therefore, we examined whether the GWAS-associated SNP is potentially a functional SNP, based on its association with DNA methylation in a subset of the full GTP cohort with methylation data. Of the 11 CpG sites within 1 MB of the SNP, one probe was found to be significant after multiple test correction (Supplementary Table II). The number of risk alleles of rs717947 predicted the proportion of methylation from probe cg09242288 [N = 157, β (SE) = −0.02 (0.007), P = 0.002] in the overall cohort (Fig. 2C), indicating that the SNP is an meQTL. Though power to detect associations was reduced when the analysis was restricted to females (N = 99), the effect sizes was equivalent β = −0.02. Methylation of cg09242288 did not predict PTSD independent of genotype.
Neuroimaging Correlates of rs717947
We next wished to examine whether this SNP may be associated with an intermediate neural phenotype related to fear or threat, using prior data from fMRI with a fearful faces task in a subset of the civilian GTP cohort. The subset of females with fMRI data did not appear to significantly differ on the clinical or demographic variables from the full genetic sample of GTP females (Supplementary Table III). To determine brain regions whose response to threat stimuli was influenced by genotype, we conducted a whole-brain analysis of the response to fearful relative to neutral pictures (Fig. 3, Supplementary Table IV). We observed a linear association between the number of risk alleles and decreasing activation in bilateral dorsolateral prefrontal cortex (dlPFC) and dorsomedial prefrontal cortex (dmPFC). These regions have previously been shown to play key roles in emotion regulation (e.g., Ochsner et al., 2002), and decreased dlPFC activation has been linked with PTSD symptoms [Aupperle et al., 2012; Fani et al., 2012].
FIG. 3.

Dorsomedial and Dorsolateral PFC Differentially Respond to Fearful Faces as a function of Genotype. Functional MRI was used to examine whether the PTSD-associated SNP was related to differential brain regional activation with fearful cues. (A) Example of neutral and fearful face stimuli from the fearful faces task. (B) Axial section showing medial and lateral prefrontal signal associated with genotype. Significant clusters associated with genotype are shown at a whole-brain corrected threshold of P < 0.05, N = 53 (33 CC, 16 TC, 4 TT). (C) Dorsolateral and (D) Dorsomedial regions surviving whole brain correction. Images are shown in neurological orientation, on template slices in Montreal Neurological Institute (MNI) space. Correction for multiple comparisons used a combined height-extent threshold calculated using monte carlo simulation in Alphasim, with 1,000 iterations and a cluster-forming threshold of P < 0.01.
DISCUSSION
Identified in a small military cohort designed based on extreme phenotypes, we found a genome-wide significant SNP that associates with PTSD that replicates in a much larger civilian community cohort. In the discovery sample (SBPBC), the genome-wide significant SNP, rs717947, at chromosome 4p15 associated with PTSD symptoms from the CAPS, as well as the PTSD diagnostic measure from the CAPS. Subsequently, we replicated the top SNP, as well as several others within the association peak, with PSS-determined PTSD diagnosis from the much larger traumatized civilian cohort. Using the GTP replication cohort, our data also suggest that rs717947 may be functionally significant as the SNP was found to be an meQTL, and the risk allele of rs717947 was associated with altered medial and dorsolateral prefrontal activation to fearful faces.
These findings are notable as very few GWAS of PTSD have yet been published, and of those, replication has been limited. Furthermore, functional status of the prior GWAS SNPs has been unclear at the genomic or neural levels. Our data add to this literature in identifying a novel candidate region in chromosome 4 that appears to be a region of moderate epigenetic regulation. Using human tissue derived from postmortem brain, a research team from the Department of Neurosurgery, UC San Francisco, investigated DNA methylation at 24 out of 28 million CpG sites across the haploid genome and found that a majority of cytosine methylation (5mC) occurs in intronic and intergenic regions [Maunakea et al., 2010]. In fact, less than 3% of CpG islands located in 5′ promoter regions are methylated. While a lot of emphasis has historically been focused on methylation at the 5′ promoter region for its role in regulation of gene expression, intergenic DNA methylation may also play a key role in gene regulation, cell-type specificity, and epigenetic response to environment. Li et al have recently shown that distinct populations of neurons have differential methylation at intergenic loci [Li et al., 2014]. Furthermore, they suggest that methylation differences within these intergenic regions may be a result of experience-induced methylation that contributes to the development of neuropsychiatric disorders. Interestingly, in silico analysis of the genomic region surrounding rs717947 (200 bp) reveals a strong signal for DNA methylation (MeDIP) within the brain, suggesting that this intergenic region can be methylated (Supplementary Fig. 4). In fact, rs717947 may also be part of a CpG dinucleotide. Given this scenario, it is reasonable to assume that if this SNP is indeed a CpG, a T allele, in place of the ancestral C allele would eliminate the potential for DNA methylation at this site. This is highly speculative but could be tested further by methylation specific PCR or pyro sequencing of bisulfite treated DNA at this particular SNP.
The region at 4p15 may be a distal regulatory region in cis of one or more of the neighboring genes, PCDH7, ARAP2, and DTHD1, or that this region of DNA may harbor yet unknown functions such as noncoding RNA (ncRNA) or chromatin regulatory regions. Indeed, the peak of SNPs likely in linkage disequilibrium with the genome-wide significant SNP on chromosome 4 are in, or very close to, the ncRNA BC036345 (chr4: 33,897,961–34,041,515) of unknown function. However, further work needs to be performed to identify the effect of the ncRNA within this region. Related to its possible roles in these neighboring genes, of note PCDH7 (protocadherin 7) is expressed within the brain [Hertel et al., 2012], and it has recently been associated with sleep regulation, epilepsy, and neurocognitive function (e.g., Ollila et al., 2014). ARAP2 is also expressed in cortex and hippocampus, and appears to be involved in cellular cytoskeletal dynamics and endosome regulation (e.g., Chen et al., 2013). Less is known about DTHD1 functioning, but it has been associated with retinal dystrophy [Abu-Safieh et al., 2013]. Thus all of the genes surrounding this SNP have potentially interesting characteristics as candidates expressed within the central nervous system (CNS) and putatively associated with other forms of CNS pathology.
To further examine whether there were any associations between rs717947 and intermediate phenotypes associated with PTSD, we examined brain activation during a fear and threat processing task—passive viewing of fearful faces, which we and others have previously associated with differential responses to PTSD and PTSD-related genetic pathways [Andero et al., 2013; Stevens et al., 2013; Stevens et al., 2014]. We found that when confronted with threat stimuli, individuals carrying the rs717947 risk allele showed decreased activation among prefrontal regions that regulate negative emotion (dlPFC and dmPFC). Dysregulation of negative emotion, whether related to inability to extinguish fear, hypervigilance to threatening cues, or generalization of fearful stimuli, are all important intermediate phenotypes related to PTSD. These data suggest that SNPs within this region of chromosome 4 associated with PTSD and differential genomic methylation, are also associated with differential intermediate fear-related neural phenotypes.
There are several limitations of this study. The sample size of the discovery cohort (SBPBC) was very small; however, we believe that the extreme phenotype design and well-controlled nature of the study enhance its power to determine genetic differences between cases and controls. While the extreme phenotype design may yield interesting information, traditional modeling of the phenotype is challenging. Follow-up studies using all continuous values of this phenotype are essential for confirmation of rs717947 as a risk variant for PTSD. We have used alternative PTSD phenotypes, not based on extreme phenotype thresholds, to assess the robustness of the association. The cohort is also of mixed race/ethnicity. Although we used principal components to control for ancestry differences in the samples, it is possible that the susceptibility for disease in risk allele carriers is actually driven by subtle differences in the genetic backgrounds of the different races. Additionally, the discovery and replication cohorts are quite different, particularly in sex and trauma type. Finally, almost half of the cases in the discovery cohort had comorbid depression, a level of comorbidity common across PTSD patients and seen within our civilian cohort as well. Further research is necessary to determine whether SNP rs717947 is specific to PTSD or to comorbid PTSD and depression.
In summary, we have identified a novel genomic locus associated with PTSD symptoms and diagnosis across two very different cohorts—one military, primarily of mixed race and male, and one civilian, African American, and primarily female. The observation will need further replication across other cohorts. However, the combination of significance across mixed samples, association with differential regional DNA methylation, and association with neural intermediate phenotypes related to PTSD are all compelling, and suggest this region as a potentially important genomic region for further study. Further genome wide association studies for PTSD, such as those currently planned within the Psychiatric Genomics Consortium PTSD workgroup [Koenen et al., 2013], will continue to dissect the genomic architecture underlying PTSD, with hope for new biomarkers and therapeutic targets.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Mental Health (MH071537 and MH096764 to K.J.R.; T32 MH87977-5 to L.M.A.) and the Department of Defense (DoD: W81XWH-09-2-0044 to CRM and W911NF-09-1-0298 to R.Y.). Support was also received from Emory and Grady Memorial Hospital General Clinical Research Center, NIH National Centers for Research Resources (M01RR00039), Howard Hughes Medical Institute (K.J.R.), and Steven and Alexandra Cohen Foundation (CRM). The project described was also supported by Grant Number #UL1TR000067 from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH). It’s contents are solely the responsibility of the authors and do not necessarily represent the official views of NCATS or NIH. We are grateful for the staff and participants from the Grady Trauma Project for their time and effort in supporting this research.
Grant sponsor: National Institutes of Mental Health; Grant numbers: MH071537, MH096764; Grant sponsor: Department of Defense; Grant numbers: W81XWH-09-2-0044, W911NF-09-1-0298; Grant sponsor: Emory and Grady Memorial Hospital General Clinical Research Center; Grant sponsor: NIH National Centers for Research Resources; Grant number: M01RR00039; Grant sponsor: Howard Hughes Medical Institute; Grant sponsor: Steven and Alexandra Cohen Foundation; Grant sponsor: National Center for Advancing Translational Sciences; Grant number: #UL1TR000067.
Footnotes
Conflict of interest: None.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Abu-Safieh L, Alrashed M, Anazi S, Alkuraya H, Khan AO, Al-Owain M, Al-Zahrani J, Al-Abdi L, Hashem M, Al-Tarimi S, Sebai MA, Shamia A, Ray-Zack MD, Nassan M, Al-Hassnan ZN, Rahbeeni Z, Waheeb S, Alkharashi A, Abboud E, Al-Hazzaa SA, Alkuraya FS. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res. 2013;23:236–247. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almli LM, Duncan R, Feng H, Ghosh D, Binder EB, Bradley B, Ressler KJ, Conneely KN, Epstein MP. Correcting systematic inflation in genetic association tests that consider interaction effects: Application to a genome-wide association study of posttraumatic stress disorder. JAMA Psychiatry. 2014a;71:1392–1399. doi: 10.1001/jamapsychiatry.2014.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almli LM, Fani N, Smith AK, Ressler KJ. Genetic approaches to understanding post-traumatic stress disorder. Int J Neuropsychopharmacol. 2014b;17:355–370. doi: 10.1017/S1461145713001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almli LM, Mercer KB, Kerley K, Feng H, Bradley B, Conneely KN, Ressler KJ. ADCYAP1R1 genotype associates with post-traumatic stress symptoms in highly traumatized African-American females. Am J Med Genet Part B Neuropsychiatr Genet. 2013;162B:262–272. doi: 10.1002/ajmg.b.32145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andero R, Brothers SP, Jovanovic T, Chen YT, Salah-Uddin H, Cameron M, Bannister TD, Almli L, Stevens JS, Bradley B, Binder EB, Wahlestedt C, Ressler KJ. Amygdala-dependent fear is regulated by Oprl1 in mice and humans with PTSD. Sci Transl Med. 2013;5:188ra173. doi: 10.1126/scitranslmed.3005656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aupperle RL, Allard CB, Grimes EM, Simmons AN, Flagan T, Behrooznia M, Cissell SH, Twamley EW, Thorp SR, Norman SB, Paulus MP, Stein MB. Dorsolateral prefrontal cortex activation during emotional anticipation and neuropsychological performance in posttraumatic stress disorder. Arch Gen Psychiatry. 2012;69:360–371. doi: 10.1001/archgenpsychiatry.2011.1539. [DOI] [PubMed] [Google Scholar]
- Barfield R, Kilaru V, Smith A, Conneely K. CpGassoc: An R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28:1280–1281. doi: 10.1093/bioinformatics/bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Arch Gen Psychiatry. 1961;4:561–571. doi: 10.1001/archpsyc.1961.01710120031004. [DOI] [PubMed] [Google Scholar]
- Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, Heim CM, Nemeroff CB, Schwartz AC, Cubells JF, Ressler KJ. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299:1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, Keane TM. The development of a Clinician-Administered PTSD Scale. J Trauma Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- Bremner JD, Vermetten E, Mazure CM. Development and preliminary psychometric properties of an instrument for the measurement of childhood trauma: The Early Trauma Inventory. Depress Anxiety. 2000;12:1–12. doi: 10.1002/1520-6394(2000)12:1<1::AID-DA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Chen PW, Jian X, Yoon HY, Randazzo PA. ARAP2 signals through Arf6 and Rac1 to control focal adhesion morphology. J Biol Chem. 2013;288:5849–5860. doi: 10.1074/jbc.M112.415778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetti SA, Resnick HS, Resick PA, Kilpatrick D. The Modified PTSD Symptom Scale: A brief self-report measure of postraumatic stress disorder. The Behavior Therapist. 1993;16:161–162. [Google Scholar]
- Fani N, Tone EB, Phifer J, Norrholm SD, Bradley B, Ressler KJ, Kamkwalala A, Jovanovic T. Attention bias toward threat is associated with exaggerated fear expression and impaired extinction in PTSD. Psychol Med. 2012;42:533–543. doi: 10.1017/S0033291711001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CF, Almli LM, Smith AK, Bradley B, Kerley K, Crain DF, Mercer KB, Weiss T, Phifer J, Tang Y, Cubells JF, Binder EB, Conneely KN, Ressler KJ. Sex dependent influence of a functional polymorphism in steroid 5-alpha-reductase type 2 (SRD5A2) on post-traumatic stress symptoms. Am J Med Genet Part B Neuropsychiatr Genet. 2013;162B:283–292. doi: 10.1002/ajmg.b.32147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti G, Galea S, Yan L, Roberts AL, Solovieff N, Aiello AE, Smoller JW, De Vivo I, Ranu H, Uddin M, Wildman DE, Purcell S, Koenen KC. Genome-wide association study implicates a novel RNA gene, the lincRNA AC068718.1, as a risk factor for post-traumatic stress disorder in women. Psychoneuroendocrinology. 2013;38:3029–3038. doi: 10.1016/j.psyneuen.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel N, Redies C, Medina L. Cadherin expression delineates the divisions of the postnatal and adult mouse amygdala. J Comp Neurol. 2012;520:3982–4012. doi: 10.1002/cne.23140. [DOI] [PubMed] [Google Scholar]
- Koenen KC, Duncan LE, Liberzon I, Ressler KJ. From candidate genes to genome-wide association: The challenges and promise of posttraumatic stress disorder genetic studies. Biol Psychiatry. 2013;74:634–636. doi: 10.1016/j.biopsych.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Baker-Andresen D, Zhao Q, Marshall V, Bredy TW. Methyl CpG Binding Domain Ultra-Sequencing: A novel method for identifying inter-individual and cell-type-specific variation in DNA methylation. Genes Brain Behav. 2014;13:721–731. doi: 10.1111/gbb.12150. [DOI] [PubMed] [Google Scholar]
- Lin S, Zhoa H. Handbook on analyzing human genetic data: Computational approaches and software. New York: Springer; 2009. [Google Scholar]
- Lindqvist D, Wolkowitz OM, Mellon S, Yehuda R, Flory JD, Henn-Haase C, Bierer LM, Abu-Amara D, Coy M, Neylan TC, Makotkine I, Reus VI, Yan X, Taylor NM, Marmar CR, Dhabhar FS. Proinflammatory milieu in combat-related PTSD is independent of depression and early life stress. Brain, Behavior, and Immunity. 2014 doi: 10.1016/j.bbi.2014.06.003. in press. [DOI] [PubMed] [Google Scholar]
- Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, Uddin M, Wildman D, Galea S, Koenen KC, Miller MW. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2013;18:937–942. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, Rex-Haffner M, Loeschner A, Gonik M, Mercer KB, Bradley B, Müller-Myhsok B, Ressler KJ, Binder EB. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013;110:8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner KN, Bunge SA, Gross JJ, Gabrieli JD. Rethinking feelings: An FMRI study of the cognitive regulation of emotion. J Cogn Neurosci. 2002;14:1215–1229. doi: 10.1162/089892902760807212. [DOI] [PubMed] [Google Scholar]
- Ollila HM, Kettunen J, Pietilainen O, Aho V, Silander K, Kronholm E, Perola M, Lahti J, Raikkonen K, Widen E, Palotie A, Eriksson JG, Partonen T, Kaprio J, Salomaa V, Raitakari O, Lehtimäki T, Sallinen M, Härmä M, Porkka-Heiskanen T, Paunio T. Genome-wide association study of sleep duration in the Finnish population. J Sleep Res. 2014;23:609–618. doi: 10.1111/jsr.12175. [DOI] [PubMed] [Google Scholar]
- Pitman RK, Rasmusson AM, Koenen KC, Shin LM, Orr SP, Gilbertson MW, Milad MR, Liberzon I. Biological studies of post-traumatic stress disorder. Nat Rev Neurosci. 2012;13:769–787. doi: 10.1038/nrn3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, Norrholm SD, Kilaru V, Smith AK, Myers AJ, Ramirez M, Engel A, Hammack SE, Toufexis D, Braas KM, Binder EB, May V. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarason IG, Johnson JH, Siegel JM. Assessing the impact of life changes: Development of the life experiences survey. J Consult Clin Psychol. 1978;46:932–946. doi: 10.1037//0022-006x.46.5.932. [DOI] [PubMed] [Google Scholar]
- Smith AK, Kilaru V, Kocak M, Almli LM, Mercer KB, Ressler KJ, Tylavsky FA, Conneely KN. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics. 2014;15:145. doi: 10.1186/1471-2164-15-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JS, Almli LM, Fani N, Gutman DA, Bradley B, Norrholm SD, Reiser E, Ely TD, Dhanani R, Glover EM, Jovanovic T, Ressler KJ. PACAP receptor gene polymorphism impacts fear responses in the amygdala and hippocampus. Proc Natl Acad Sci USA. 2014;111:3158–3163. doi: 10.1073/pnas.1318954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JS, Jovanovic T, Fani N, Ely TD, Glover EM, Bradley B, Ressler KJ. Disrupted amygdala-prefrontal functional connectivity in civilian women with posttraumatic stress disorder. J Psychiatr Res. 2013;47:1469–1478. doi: 10.1016/j.jpsychires.2013.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True WR, Rice J, Eisen SA, Heath AC, Goldberg J, Lyons MJ, Nowak J. A twin study of genetic and environmental contributions to liability for posttraumatic stress symptoms. Arch Gen Psychiatry. 1993;50:257–264. doi: 10.1001/archpsyc.1993.01820160019002. [DOI] [PubMed] [Google Scholar]
- Van Gestel S, Houwing-Duistermaat JJ, Adolfsson R, van Duijn CM, Van Broeckhoven C. Power of selective genotyping in genetic association analyses of quantitative traits. Behav Genet. 2000;30:141–146. doi: 10.1023/a:1001907321955. [DOI] [PubMed] [Google Scholar]
- Ventura J, Liberman RP, Green MF, Shaner A, Mintz J. Training and quality assurance with the Structured Clinical Interview for DSM-IV (SCID-I/P) Psychiatry Res. 1998;79:163–173. doi: 10.1016/s0165-1781(98)00038-9. [DOI] [PubMed] [Google Scholar]
- Vogt DS, Smith BN, King LA, King DW, Knight JA, Vasterling JJ. Deployment Risk and Resilience Inventory-2 (DRRI-2): An updated tool for assessing psychosocial risk and resilience factors among service members and veterans. J Trauma Stress. 2013;26:710–717. doi: 10.1002/jts.21868. [DOI] [PubMed] [Google Scholar]
- Weathers FW, Litz BT, Keane TM, Palmieri PA, Marx BP, Schnurr PP. The PTSD Checklist for DSM-5 (PCL-5) 2013 Scale available from the National Center for PTSD at www.ptsd.va.gov.
- Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013;74:656–663. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Brown AD, Lazar M, Cressman VL, Henn-Haase C, Neylan TC, Shalev A, Wolkowitz OM, Hamilton SP, Yehuda R, Sodickson DK, Weiner MW, Marmar CR. Spontaneous brain activity in combat related PTSD. Neurosci Lett. 2013;547:1–5. doi: 10.1016/j.neulet.2013.04.032. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F, Makotkine I, Daskalakis NP, Marmar CR, Meaney MJ. Lower methylation of glucocorticoid receptor gene promoter 1 in peripheral blood of veterans with posttraumatic stress disorder. Biol Psychiatry. 2015;77:356–364. doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


