Abstract
The objective of this study was to investigate an apparent increase in linezolid-nonsusceptible staphylococci and enterococci following a laboratory change in antimicrobial susceptibility testing from disk diffusion to an automated susceptibility testing system. Isolates with nonsusceptible results (n = 27) from Vitek2 were subjected to a battery of confirmatory testing which included disk diffusion, Microscan broth microdilution, Clinical and Laboratory Standards Institute (CLSI) reference broth microdilution, gradient diffusion (Etest), 23S rRNA gene sequencing, and cfr PCR. Our results show that there is poor correlation between methods and that only 70 to 75% of isolates were confirmed as linezolid resistant with alternative phenotypic testing methods (disk diffusion, Microscan broth microdilution, CLSI broth microdilution, and Etest). 23S rRNA gene sequencing identified mutations previously associated with linezolid resistance in 16 (59.3%) isolates, and the cfr gene was detected in 3 (11.1%) isolates. Mutations located at positions 2576 and 2534 of the 23S rRNA gene were most common. In addition, two previously undescribed variants (at positions 2083 and 2345 of the 23S rRNA gene) were also identified and may contribute to linezolid resistance.
INTRODUCTION
Linezolid is a synthetic oxazolidinone antibiotic that inhibits bacterial protein synthesis by blocking the first step in ribosome assembly (1). This mechanism of action is unique to the oxazolidinone class of antibiotics, and thus cross-resistance from other antibiotics with protein synthesis targets is unlikely. The spectrum of activity of linezolid includes most Gram-positive organisms and mycobacteria but not Gram-negative organisms. Since its introduction for clinical use in 2000, linezolid has remained nearly uniformly active against all Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE), and has been an important therapeutic agent for treating infections with these multidrug-resistant organisms (2). Recent data from global surveillance programs suggest that >99% of staphylococci (including MRSA and coagulase-negative staphylococci [CoNS]) and enterococci (including VRE) are susceptible to linezolid (2, 3). However, nonsusceptibility rates can be higher within individual medical centers and when resistant clones disseminate within a specific geographic area or health care institution (4).
Antimicrobial susceptibility testing results are important in the process of deciding whether to use linezolid as therapeutic agent, especially when using linezolid to treat infections due to multidrug-resistant organisms. There are several potential mechanisms that can mediate linezolid resistance. The most common mechanism of resistance is mutation of the 23S rRNA subunit, leading to alteration of the peptidyltransferase center (PTC) where conserved residues interact directly with linezolid (5). Gram-positive organisms possess multiple 23S rRNA alleles (4 to 6 copies), and development of clinically significant resistance requires that more than one allele be mutated (6). This likely explains the low frequency of linezolid resistance.
Mutation of linezolid binding sites is the most common mechanisms of resistance; however, acquired resistance mechanisms have now been described (7). One example of nonmutational resistance is acquisition of the natural cfr (chloramphenicol-florfenicol resistance) gene, which is a plasmid-carried gene encoding a protein which catalyzes the posttranscriptional methylation of the C-8 atom of a key residue (A2503) in the 23S rRNA (8). cfr is a highly mobile genetic element that facilitates interspecies spread, and to date, cfr has been identified in staphylococci (both S. aureus and CoNS), enterococci, streptococci, and other, less common Gram-positive pathogens (5). The result of methylation by the cfr product is a multidrug resistance phenotype that includes linezolid, lincosamides, and streptogramins (9).
Given the critical nature of infections that are treated with linezolid, it is imperative that laboratory methods accurately detect resistance. The objective of this study was to investigate an apparent increase in linezolid-nonsusceptible staphylococci and enterococci following a laboratory change in antimicrobial susceptibility testing from disk diffusion to an automated susceptibility testing system.
MATERIALS AND METHODS
Bacterial strains.
Isolates were recovered from clinical specimens at Barnes-Jewish Hospital (St. Louis, MO) as part of routine clinical care from April to November 2012. The inclusion criterion was any Gram-positive isolate testing intermediate or resistant to linezolid using the Vitek2 system.
Antimicrobial susceptibility testing.
An initial finding of linezolid nonsusceptibility on the Vitek2 was confirmed with repeat testing. Kirby-Bauer disk diffusion using a 30-μg disk (Becton, Dickinson and Company) was performed on Mueller-Hinton agar (Thermofisher, Lenexa, KS); testing was performed and interpreted according to CLSI standards (10). Per CLSI recommendations, linezolid zone sizes for staphylococci were interpreted using transmitted light, and isolates were evaluated for visible growth detected with the unaided eye. Isolates with any discernible growth located inside the zone were classified as resistant. Gradient diffusion testing using Etest (bioMérieux, Durham, NC) on Mueller-Hinton agar (Thermofisher) was performed and interpreted according to the manufacturer's recommendations, and thus the Etest was read at 90% inhibition. Testing with the Vitek2 GP70 card and Microscan positive MIC panel PM29 was performed according to the manufacturers' recommendations. The PM29 was inoculated using the Prompt method and interpreted visually. If trailing endpoints were observed on the PM29, the MIC was interpreted as the first well in which trailing was observed. It should be noted that bioMérieux lists linezolid as a card limitation for the testing of Enterococcus species isolates. For disk diffusion, gradient diffusion, and Microscan testing, quality control was conducted each day that testing was performed; for Vitek2 analysis, quality control was performed weekly.
At the Centers for Disease Control and Prevention (CDC), 96-well MIC trays were prepared using 100 μl of cation-adjusted Mueller-Hinton broth (CAMHB) (BBL, Sparks, MD) per well; trays were kept frozen at 70°C and then thawed when ready for use. The linezolid broth microdilution (BMD) testing range was 1 to 16 μg/ml. BMD MIC testing was performed by CLSI standard methods (11, 12). A sterile 95-pin inoculator and reservoir combination system (Evergreen Scientific, Los Angeles, CA) was used to deliver 10 μl of inoculum per well. Quality control was performed with Enterococcus faecalis ATCC 29212 and Staphylococcus aureus ATCC 29213 on the MIC panels containing linezolid; results were in acceptable ranges.
23S rRNA gene sequencing.
Bacterial DNA was extracted from isolated colonies or broth using 30 μl of the PrepMan Ultra reagent (Applied Biosystems). Extraction supernatant (1 μl) was used directly for PCR amplification using the Easy-A PCR enzyme (Stratagene) and then sequenced bidirectionally using the same PCR primers on the 3130xl genetic analyzer (Life Technologies, Foster City, CA, USA). The PCR primers (forward, 5′-AACGATTTGGGCACTGTCTCAACG-3′; reverse, 5′-AATTTCCTACGCCCACGACGGATA-3′) were designed for targeted amplification of a 660-bp region of the 23S rRNA gene domain V that is highly conserved between Staphylococcus spp. and Enterococcus spp. This targeted region includes the well-characterized linezolid resistance mutations G2576T, T2500A, and G2234A.
Variants in the 23S rRNA gene were determined by comparing the generated sequence to the reference sequence from GenBank (accession number X68425). Heterozygous sequencing chromatogram peaks were confirmed in the corresponding nucleotide position in the complementary sequence reaction.
cfr PCR.
PCR for detection of cfr was performed using a modification of a previously published method (13). In brief, DNA was extracted from bacterial isolates grown on sheep blood agar (Remel) using the BiOstic bacteremia DNA isolation kit (MoBio, Carlsbad, CA). Approximately 100 ng of DNA was subjected to PCR using primers cfr-fw (5′-TGA AGT ATA AAG CAG GTT GGG AG) and cfr-rev (5′-ACC ATA TAA TTG AC CACA AGC AGC) and a PuReTaq Ready-To-Go PCR bead (GE Healthcare, Buckinghamshire, United Kingdom) (13). PCR cycling conditions were as follows: 94°C for 10 min; 35 cycles of 94°C for 30 s, 57°C for 30 s, and 74°C for 30 s; and a final elongation step at 72°C for 10 min. Products were visualized using agarose gel electrophoresis. Quality control was performed with each PCR run; S. aureus ATCC 25923 was used as a negative control, and S. aureus 004-737X 2007 was used as a positive cfr control strain (this strain was kindly provided by JMI Laboratories).
Data analysis.
Because intermethod correlation was poor, two gold standards were used in this study to assess the performance of each method. Gold standard 1 (GS1) was defined as any molecular result (23S rRNA gene sequencing or cfr PCR) that identified an established resistance determinant and/or yielded 100% concordance between phenotypic test results. Gold standard 2 (GS2) was determined by reference broth microdilution, which was performed by the CDC.
RESULTS
Isolates.
Twenty-seven isolates (11 Enterococcus [71% vancomycin resistant], eight S. aureus [88% methicillin resistant], and eight S. epidermidis [100% methicillin resistant]) were categorized as either intermediate or resistant to linezolid by Vitek2 (Table 1). All isolates were recovered from clinical specimens, including blood (n = 10), urine (n = 5), cystic fibrosis sputum (n = 5), abscess (n = 2), wound (n = 1), thoracentesis fluid (n = 1), bone marrow (n = 1), bone (n = 1), and otherwise undefined biopsy (n = 1) specimens (Table 1). In total, 23 isolates tested as resistant by Vitek2 with MICs of >8 μg/ml, and 4 enterococcal isolates tested as intermediate with MICs of 4 μg/ml.
TABLE 1.
Summary of test results for isolates with reduced susceptibility to linezolida
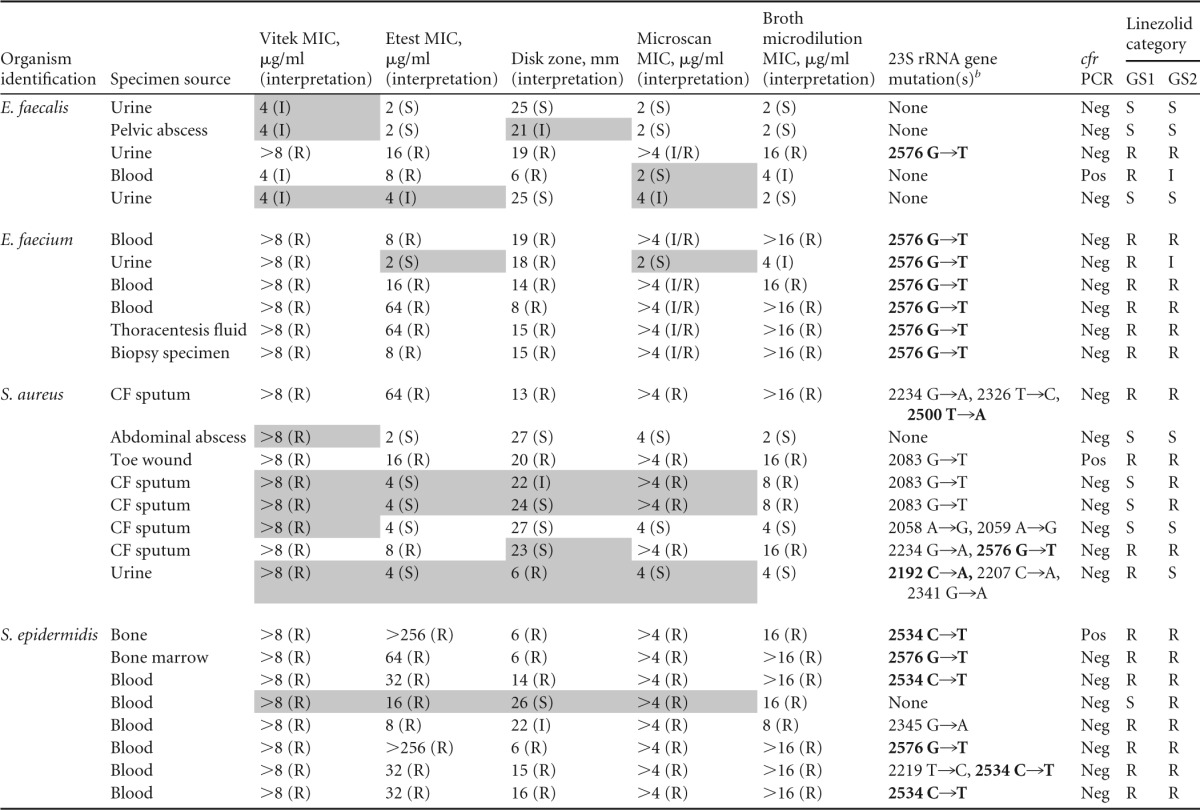
Shading indicates an error with either gold standard 1 (GS1) or gold standard 2 (GS2). GS1 is defined by a positive result for a molecular marker of linezolid resistance and/or complete agreement amongst phenotypic tests. GS2 is defined by the CLSI broth microdilution result. CF, cystic fibrosis; R, resistant; I, intermediate; S, susceptible; Neg, negative; Pos, positive.
b Bold indicates a mutation of known significance.
23S rRNA gene sequencing.
To interrogate for mutations in the 23S rRNA gene that can confer resistance to linezolid, the 23S rRNA gene was sequenced. Of the 27 isolates, 16 (59%) had mutations that have been previously reported to confer linezolid resistance. Mutations which have not been previously associated with linezolid resistance were identified in 9 (33%) of the isolates. Consistent with previous literature, the most common mutation identified was a guanine-to-thymidine transition at nucleotide 2576 (n = 10). The G2576T mutation was identified most commonly in the enterococci (n = 7) but was also seen in S. aureus (n = 1) and S. epidermidis (n = 2) (Table 1). Additional mutations previously associated with linezolid resistance were also identified but at a lower frequency, including T2500A (S. aureus [n = 1]), C2192A (S. aureus [n = 1]), and C2534T (S. epidermidis [n = 4]). Interestingly, several previously undescribed variants were also detected (Table 2). The combination of ambiguous phenotypic profiles and overlapping mechanisms of resistance makes it difficult to establish the significance of some of these novel variants. However, our data suggest that G2083T, which was identified in two S. aureus isolates, and G2345A, which was identified from a single S. epidermidis isolate, may be the sole cause of linezolid resistance in those isolates. Both of these variants were present in isolates with no other identified mechanisms of resistance and with phenotypic resistance profiles consistent with linezolid resistance. Lastly, 6 isolates had no identified 23S rRNA gene variants.
TABLE 2.
Characterization of 23S rRNA variants detected in this study but not previously characterized
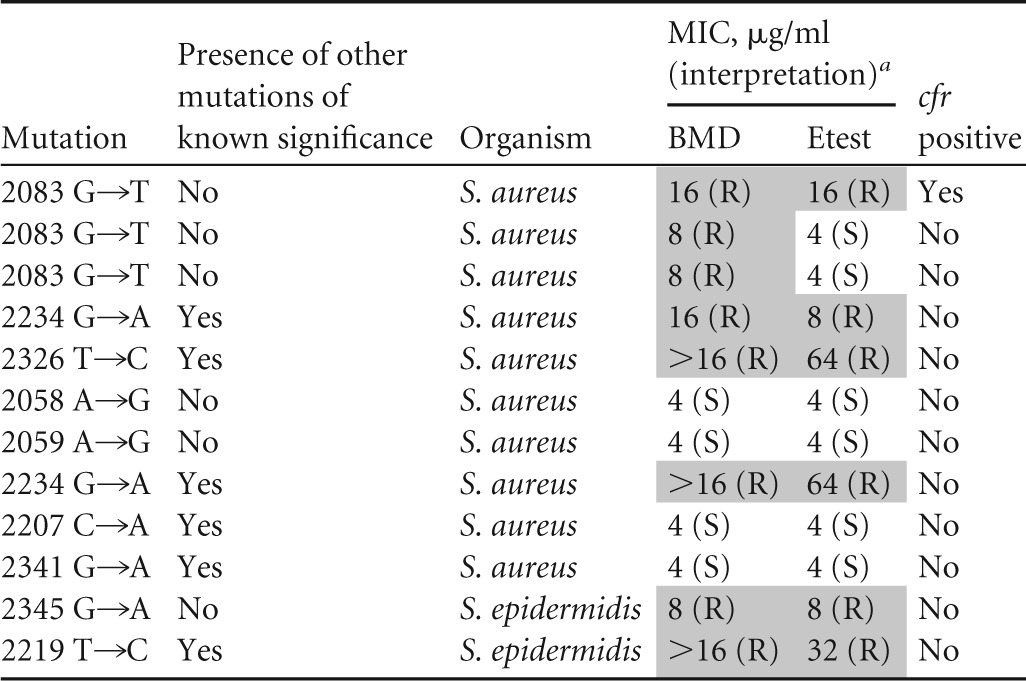
a Shading indicates reduced susceptibility. BMD, broth microdilution; R, resistant; S, susceptible.
Assessment of cfr.
Three isolates were positive for cfr by PCR. One isolate of Enterococcus faecalis was cfr positive and had no mutation in the 23S rRNA gene. This isolate tested as intermediate (MIC = 4 μg/ml) by the Vitek2 and broth microdilution, resistant by Etest (MIC = 8 μg/ml) and disk diffusion (zone diameter = 6 mm), and susceptible by Microscan broth microdilution (MIC = 2 μg/ml). This is in contrast to the case for the S. aureus and S. epidermidis isolates containing cfr; these isolates were resistant to linezolid by all testing methods employed. However, these isolates also possessed variants in the 23S rRNA gene, and it may be that the combination of the two concomitant mechanisms contributed to a more resistant phenotype.
Antimicrobial susceptibility testing.
Etest, disk diffusion, broth microdilution, and Microscan broth microdilution results confirmed reduced susceptibility in 19 (70%), 21 (78%), 20 (75%), and 20 (75%) of isolates tested, respectively (Table 1). In an effort to evaluate the accuracy of Vitek2, Etest, disk diffusion, and Microscan broth microdilution detection of reduced linezolid susceptibility, two gold standards were established. The first gold standard (GS1) defined linezolid resistance as an isolate with a positive molecular assay for linezolid resistance (that is, a previously described 23S rRNA gene mutation associated with linezolid resistance or presence of cfr) or complete categorical agreement among phenotypic methods. Using this strict gold standard, eight (30%), two (7.5%), two (7.5%), and four (15%) of Vitek2, Etest, disk diffusion, and Microscan broth microdilution results were falsely resistant, respectively. Similarly, Etest, disk diffusion, and Microscan broth microdilution each generated falsely susceptible results for two (7.4%), two (7.4%), and three (11%) isolates, respectively. Of note, these methods did not correlate well, and the isolates that were falsely classified as susceptible differed between methods.
A second gold standard (GS2) defined linezolid resistance according to reference broth microdilution testing performed by the CDC. By this measure, 5 (19%), one (3.7%), two (7.4%), and one (3.7%) of Vitek2, Etest, disk diffusion, and Microscan broth microdilution results were falsely resistant, respectively. In addition, Etest and disk diffusion each generated falsely susceptible results for three (11%) isolates, while Microscan broth microdilution generated two (7.4%) falsely susceptible results. As a result of the fact that these isolates were selected on the basis of linezolid nonsusceptibility using Vitek2, the analytical performance characteristics of that method cannot be accurately characterized.
Regardless of the organism being tested, these data show that there is poor correlation between susceptibility testing methods when it comes to detecting linezolid resistance. This is a significant finding, as current CLSI standards suggest that laboratories should confirm reduced linezolid susceptibility in staphylococci and enterococci using a second method (14). Of the 27 isolates included in this study, only 14 (51.9%) produced concordant results across all methods (Vitek2, Etest, disk diffusion, Microscan broth microdilution, CDC broth microdilution, and molecular [23S rRNA gene and cfr PCR considered together]), suggesting that confirmatory methods readily available to clinical laboratories are likely to produce conflicting results. Most laboratories do not have access to the molecular methods used in this study and must therefore rely on phenotypic confirmatory methods. If molecular methods are excluded, only 15 (55.6%) of the 27 isolates produced concordant results for all phenotypic methods (Table 1).
DISCUSSION
This is one of the first studies to perform a systematic comparison of susceptibility testing methods for the detection of linezolid resistance. However, Qi et al. did evaluate the ability of Etest, agar dilution, broth microdilution, disk diffusion, and Vitek2 to detect decreased linezolid susceptibility in enterococci with G2576T mutations (15). The purpose of the study by Qi et al. was to assess the impact of G2576T mutation on phenotypic susceptibility. Despite testing a relatively small sample size of linezolid-resistant isolates and only two species of Enterococcus (E. faecium [n = 14] and E. faecalis [n = 5]), the authors were able to conclude that for enterococci with the G2576T mutation, the gradient diffusion method was the least reliable method and overcalled resistance more than any other method. One limitation of the study by Qi et al. is that it was conducted between 2004 and 2005, and the only mutation they assessed was the G2576T mutation. Since that time, numerous other variants, some of which have been described in our study, have been identified, in addition to the emergence of cfr-mediated resistance. In addition, our investigation includes Staphylococcus spp., which were not tested in the study by Qi et al.
In another investigation, Tenover et al. evaluated enterococci (n = 50) and staphylococci (n = 50), of which 17 and 15 were nonsusceptible, respectively. They compared seven methods, the Vitek, Vitek2, Microscan WalkAway, BD Phoenix (BD Diagnostic Systems, Sparks, MD), Etest, disk diffusion, and broth microdilution (16). As in our study, they found that there was poor categorical agreement between methods. When testing linezolid-nonsusceptible staphylococci with Etest, disk diffusion, Vitek2, and Microscan, very major errors were generated for 40%, 53.3%, 6.7%, and 6.7% of isolates, respectively. Performance was better with enterococci, but very major errors were still generated for 6% (Etest), 4% (disk diffusion), 10% (Vitek2), and 6% (Microscan) of isolates.
Our findings illustrate the difficulty in establishing breakpoints for antibiotics to which resistance is rare. In addition, the issue can be exacerbated and categorical interpretation can be a challenge when the wild-type MIC distribution hovers around or approaches that of the susceptible/resistant breakpoint, as is the case for linezolid and the staphylococci and enterococci (2). Further, in the absence of an intermediate category (staphylococci), it is possible that the allowable error for any susceptibly testing method (i.e., plus or minus one doubling dilution) contributed to the lack of category agreement observed in some instances in our investigation.
Linezolid is a bacteriostatic antibiotic and as such can generate susceptibility patterns that can be challenging to interpret (16, 17). Etest, for example, requires that 90% inhibition be considered when determining the MIC, while disk diffusion requires the use of transmitted light. In both cases there is some subjectivity in reading zone sizes, which may lead to poor test correlation as demonstrated here and by others (16). Broth microdilution also poses some interpretive difficulty, as trailing endpoints can be encountered and may explain MIC variation between studies (17).
Our study identified several previously unreported 23S rRNA gene variants in linezolid-resistant isolates which may represent novel linezolid resistance determinants. The linezolid binding site is surrounded by eight 23S rRNA gene nucleotides (18). Resistance to linezolid has primarily been characterized by mutations found in close proximity to the peptidyltransferase center (PTC). However, it has been postulated that binding specificity may be determined by nucleotides that are distal to the primary binding site (18). Assessment of the protein structure published by Wilson et al. suggests that several of the variants identified in our study are in close proximity to the linezolid binding site and could possibly lead to resistance (19). Nucleotides 2061 and 2062 lie at the heart of the linezolid binding site and have been shown to confer resistance. The variant A2058G, A2059G, and G2083T mutations are close to nucleotides 2061 and 2062. In addition, nucleotide 2058 has been shown to confer linezolid resistance in Mycobacterium smegmatis, which strongly suggests that it may have the same effect in Gram-positive bacteria. Unfortunately, our phenotypic results were ambiguous for the isolate harboring 2058 and 2059 variants, and we cannot make a definitive conclusion about their significance.
Two isolates harboring a variant at nucleotide 2083 demonstrated phenotypic resistance and were cfr PCR negative. Interestingly, these isolates were phenotypically resistant when tested by broth microdilution and Vitek2 but were susceptible by gradient diffusion. One of the two isolates demonstrated intermediate resistance by disk diffusion. Despite the lack of agreement between phenotypic results for isolates harboring a variant at nucleotide 2083, these isolates are clearly not exhibiting wild-type susceptibility to linezolid. However, the 2083 variant was also found in a third isolate that was cfr positive, and therefore, at this time, it does remain a variant of unknown significance.
Lastly, a single isolate of S. epidermidis was cfr negative and found to harbor a 23S rRNA gene variant at nucleotide 2345. This isolate was linezolid nonsusceptible by all phenotypic methods. Interestingly, the location of this variant is distal to the linezolid binding pocket, so the mechanism by which it confers resistance remains to be determined.
This study has several limitations. First, all isolates were selected for inclusion in this study by testing intermediate or resistant to linezolid on the Vitek2. This selection bias does not permit any meaningful conclusions regarding the rate at which the Vitek2 generates falsely resistant or susceptible results. Establishing a false susceptibility rate for Vitek2 would have required screening for linezolid resistance using another method, which was outside the scope of this study. Second, the genes encoding the L3 and L4 ribosomal proteins were not sequenced. Mutations in these proteins have only recently been described, and their association with resistance is less certain than that of mutations found in the 23S rRNA gene. Initially, in vitro studies suggested that these mutations conferred reduced susceptibility to linezolid (20). Since that time, though, few studies have confirmed the association between these mutations and linezolid resistance, making this only a minor limitation of our study. Third, this study was based on isolates yielding reduced susceptibility when tested with the Vitek2. Given that linezolid testing for enterococci is listed as a card limitation on the Vitek2, it is not surprising that resistance was not always confirmed in these isolates. Lastly, because reduced susceptibility to linezolid is a rare occurrence, this study is limited by small numbers of isolates representing each species.
Here we demonstrate poor intermethod correlation for linezolid susceptibility testing; this result raises the question of how laboratories should confirm linezolid resistance. Laboratories are likely to encounter discrepant results with different methods for testing linezolid. Our data suggest that Etest and disk diffusion were the least likely to generate falsely resistant results and may therefore represent specific confirmatory methods. However, both methods did yield falsely susceptible results (very major errors), which is of concern and must be considered when deciding how to handle discrepant confirmatory methods. Emerging molecular biomarkers of resistance such as cfr and the 23S rRNA gene are promising, but as demonstrated by this study, they cannot be used in isolation and furthermore are not readily available for many clinical laboratories.
There are multiple mechanisms that can confer linezolid resistance in Gram-positive bacteria, and the MICs associated with resistance can be variable, depending on species and genotype. Laboratories should be aware that overall, linezolid resistance is an uncommon finding, and confirmatory testing should be performed when resistance is identified.
In conclusion, we agree that laboratories should continue to follow the confirmatory algorithm outlined in appendix A of the CLSI M100-S25 document (11). While following CLSI guidance, laboratories should be reminded that reading endpoints for linezolid susceptibility testing can be challenging and that testing is best performed by experienced laboratory personnel. These data suggest that Etest and disk diffusion demonstrate sufficiently high specificity for resistance and are therefore acceptable for use in this capacity. Reference broth microdilution is considered the gold standard and is encouraged when possible; however, few laboratories have access to this method, necessitating that isolates be sent to reference laboratories for confirmatory testing. The results from 23S rRNA gene and cfr PCR are useful when they are positive; however, when they are negative, the possibility of resistance is not excluded. These data indicate that in the absence of broth microdilution, laboratories will fail to confirm resistance in a high percentage of isolates. Given the significance of falsely reporting an isolate as susceptible, many laboratories will elect to report the isolate as resistant regardless of the confirmatory test result. However, this decision is one that should be made on a case-by-case basis and in consultation with the laboratory medical director and infectious disease practitioners. Once decisions about how to handle discrepant results are made, laboratories can standardize the timing and manner in which these susceptibility results are communicated.
ACKNOWLEDGMENTS
We are grateful to the technologists of the Barnes Jewish Hospital Clinical Microbiology Laboratory for their ongoing efforts for the patients of Barnes Jewish Hospital. We thank Carol Weber and Meghan Wallace for technical assistance with susceptibility testing and banking of bacterial isolates. The reference broth microdilution testing was performed at the Antimicrobial Susceptibility Testing Laboratory, Centers for Disease Control and Prevention, Atlanta, GA, by David Lonsway and Karen Anderson.
C.-A.D.B has received research support from bioMérieux, Accelerate Diagnostics, and Cepheid and personal fees from bioMérieux and Thermofisher Scientific. C.D.D. has received research support from Cepheid and personal fees from Nanosphere and Thermofisher Scientific.
REFERENCES
- 1.Zurenko GE, Ford CW, Hutchinson DK, Brickner SJ, Barbachyn MR. 1997. Oxazolidinone antibacterial agents: development of the clinical candidates eperezolid and linezolid. Expert Opin Invest Drugs 6:151–158. doi: 10.1517/13543784.6.2.151. [DOI] [PubMed] [Google Scholar]
- 2.Jones RN, Sader HS, Flamm RK. 2013. Update of dalbavancin spectrum and potency in the USA: report from the SENTRY Antimicrobial Surveillance Program (2011). Diagn Microbiol Infect Dis 75:304–307. doi: 10.1016/j.diagmicrobio.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Mendes RE, Farrell DJ, Sader HS, Streit JM, Jones RN. 2015. Update of the telavancin activity in vitro tested against a worldwide collection of Gram-positive clinical isolates (2013), when applying the revised susceptibility testing method. Diagn Microbiol Infect Dis 81:275–279. doi: 10.1016/j.diagmicrobio.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 4.McLaughlin M, Malczynski M, Qi C, Barajas G, Radetski J, Zembower T, Scheetz MH. 2013. Virulence of vancomycin-resistant Enterococcus faecium according to linezolid resistance and clinical outbreak status. Antimicrob Agents Chemother 57:3923–3927. doi: 10.1128/AAC.00192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendes RE, Deshpande LM, Jones RN. 2014. Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist Updat 17:1–12. doi: 10.1016/j.drup.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Marshall SH, Donskey CJ, Hutton-Thomas R, Salata RA, Rice LB. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother 46:3334–3336. doi: 10.1128/AAC.46.10.3334-3336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arias CA, Vallejo M, Reyes J, Panesso D, Moreno J, Castaneda E, Villegas MV, Murray BE, Quinn JP. 2008. Clinical and microbiological aspects of linezolid resistance mediated by the cfr gene encoding a 23S rRNA methyltransferase. J Clin Microbiol 46:892–896. doi: 10.1128/JCM.01886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grove TL, Livada J, Schwalm EL, Green MT, Booker SJ, Silakov A. 2013. A substrate radical intermediate in catalysis by the antibiotic resistance protein Cfr. Nat Chem Biol 9:422–427. doi: 10.1038/nchembio.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol 64:1506–1514. doi: 10.1111/j.1365-2958.2007.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. 2012. Performance standards for antimicrobial susceptibility testing; M100-S22. CLSI, Wayne, PA. [Google Scholar]
- 11.CLSI. 2015. Performance standards for antimicrobial susceptibility testing: twenty-fifth informational supplement. M100–S25. CLSI, Wayne, PA. [Google Scholar]
- 12.CLSI. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 10th ed M07–A10. CLSI, Wayne, PA. [Google Scholar]
- 13.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol 57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 14.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; M100-S24. CLSI, Wayne, PA. [Google Scholar]
- 15.Qi C, Zheng X, Obias A, Scheetz MH, Malczynski M, Warren JR. 2006. Comparison of testing methods for detection of decreased linezolid susceptibility due to G2576T mutation of the 23S rRNA gene in Enterococcus faecium and Enterococcus faecalis. J Clin Microbiol 44:1098–1100. doi: 10.1128/JCM.44.3.1098-1100.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover FC, Williams PP, Stocker S, Thompson A, Clark LA, Limbago B, Carey RB, Poppe SM, Shinabarger D, McGowan JE Jr. 2007. Accuracy of six antimicrobial susceptibility methods for testing linezolid against staphylococci and enterococci. J Clin Microbiol 45:2917–2922. doi: 10.1128/JCM.00913-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livermore DM. 2003. Linezolid in vitro: mechanism and antibacterial spectrum. J Antimicrob Chemother 51(Suppl 2):ii9–ii16. [DOI] [PubMed] [Google Scholar]
- 18.Long KS, Munck C, Andersen TM, Schaub MA, Hobbie SN, Bottger EC, Vester B. 2010. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross-resistance. Antimicrob Agents Chemother 54:4705–4713. doi: 10.1128/AAC.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci U S A 105:13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locke JB, Hilgers M, Shaw KJ. 2009. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob Agents Chemother 53:5275–5278. doi: 10.1128/AAC.01032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


