Abstract
Background Most, if not all, organisms possess the ability to alter their phenotype in direct response to changes in their environment, a phenomenon known as phenotypic plasticity. Selection can break this environmental sensitivity, however, and cause a formerly environmentally induced trait to evolve to become fixed through a process called genetic assimilation. Essentially, genetic assimilation can be viewed as the evolution of environmental robustness in what was formerly an environmentally sensitive trait. Because genetic assimilation has long been suggested to play a key role in the origins of phenotypic novelty and possibly even new species, identifying and characterizing the proximate mechanisms that underlie genetic assimilation may advance our basic understanding of how novel traits and species evolve.
Scope This review begins by discussing how the evolution of phenotypic plasticity, followed by genetic assimilation, might promote the origins of new traits and possibly fuel speciation and adaptive radiation. The evidence implicating genetic assimilation in evolutionary innovation and diversification is then briefly considered. Next, the potential causes of phenotypic plasticity generally and genetic assimilation specifically are examined at the genetic, molecular and physiological levels and approaches that can improve our understanding of these mechanisms are described. The review concludes by outlining major challenges for future work.
Conclusions Identifying and characterizing the proximate mechanisms involved in phenotypic plasticity and genetic assimilation promises to help advance our basic understanding of evolutionary innovation and diversification.
Keywords: Genetic accommodation, genetic assimilation, phenotypic plasticity, cis and trans regulatory evolution, canalization, developmental robustness, species diversity
INTRODUCTION
Phenotypic plasticity – the ability of an individual organism to change its phenotype in direct response to stimuli or inputs from its environment (Nijhout, 2003; West-Eberhard, 2003) – is commonplace. Yet the evolutionary significance of such developmental flexibility remains controversial (Laland et al., 2014; Wray et al., 2014). Although many evolutionary biologists have long held that plasticity has no relevance for evolution (other than to perhaps act as a stabilizing force by dampening any diversifying effects of selection; Huey et al., 2003; Price et al., 2003; de Jong, 2005), a growing number of researchers have suggested that plasticity can actually act as a diversifying force in evolution. Indeed, plasticity might play a key role in fostering new traits and species, and may even fuel adaptive radiation (West-Eberhard, 1986, 1989, 2003; Pigliucci and Murren, 2003; Schlichting, 2004; Pfennig et al., 2010; Moczek et al., 2011).
Although several routes have been proposed for how plasticity might impact evolutionary innovation and diversification (Bradshaw, 1965; Stearns, 1989; West-Eberhard, 1989, 2003; Pigliucci and Murren, 2003; Price et al., 2003; Schlichting, 2004; Pfennig and McGee, 2010; Pfennig et al., 2010; Moczek et al., 2011; Thibert-Plante and Hendry, 2011; Fitzpatrick, 2012), one widely cited pathway involves an evolutionary process known as ‘genetic assimilation’ (sensu Waddington, 1952, 1953). Genetic assimilation occurs when a trait that was originally triggered by the environment loses this environmental sensitivity (i.e. plasticity) and ultimately becomes ‘fixed’ or expressed constitutively in a population. Another way of wording this phenomenon is that an induced trait loses its environmental sensitivity and thereby becomes robust to the environment. Not only might this process represent a common way for new traits to arise (reviewed in West-Eberhard, 2003; Pfennig et al., 2010; Moczek et al., 2011), but if some populations undergo genetic assimilation and others do not – and if the affected traits influence the likelihood that these populations can exchange genes – then the differential loss of plasticity might represent a crucial step in the formation of new species (West-Eberhard, 1986, 1989, 2003, 2005; Pigliucci and Murren, 2003; Pfennig et al., 2010; Schwander and Leimar, 2011).
In this review, we explore this potential pathway to innovation and diversification. We begin by examining how the evolution of phenotypic plasticity, followed by its loss via genetic assimilation, might facilitate genetic evolution and thereby promote the origins of new traits and even new species. We then discuss the various proximate (i.e. genetic, molecular and physiological) mechanisms that potentially underpin both phenotypic plasticity and genetic assimilation. We conclude by outlining future directions that promise to help illuminate plasticity’s role in the origins of biodiversity.
THE FLEXIBLE – AND INFLEXIBLE – ORGANISM
All organisms can alter some aspect of their phenotype in response to changes in their environment; if not their morphology, then their physiology, behaviour and/or gene expression (Nijhout, 2003; Sultan, 2007; Gilbert and Epel, 2009). For example, temperature often influences phenotype because nearly all enzyme activity is temperature-dependent; food contains potent chemical signals that can dramatically alter phenotypes; light can stimulate plants to produce different-shaped leaves and shoots; pressure can cause plant stems and roots and animal muscles and bones to grow differently; and other organisms (be they pathogens, predators or competitors) can cause an individual plant or animal to release hormones, which can alter the phenotype that the individual produces (reviewed in Gilbert and Epel, 2009). Such phenotypic plasticity is so prevalent that it can be considered a defining feature of living things (Pfennig, 2004).
To understand why plasticity is ubiquitous, consider that all organisms encounter variation in their environment, and these fluctuations can destabilize development and thereby disrupt the match between the organism’s phenotype and its environment (Whitman and Agrawal, 2009). Phenotypic plasticity can lessen such mismatches and thereby enhance fitness (Ghalambor et al., 2007). Generally, phenotypic plasticity is favoured when organisms confront environmental variation, when no fixed trait is best suited for all environmental conditions, when cues are available that reliably signal change in local conditions, and when the fitness benefits outweigh the costs of expressing plasticity (Berrigan and Scheiner, 2004; Travis, 2009; Whitman and Agrawal, 2009).
When the above conditions favouring plasticity are met, plasticity is generally thought to evolve because selection favours genotypes that are more responsive to the changes in the environment (Schlichting and Pigliucci, 1998). Indeed, in nearly every natural population surveyed, different genotypes are typically found to vary not only in whether they respond to a particular change in their environment but also in the manner in which they respond – in other words, different genotypes typically express different environmentally contingent phenotypic responses (Gupta and Lewontin, 1982; Sultan and Bazzaz, 1993; Kingsolver et al., 2004). Such ‘reaction norms’ provide the heritable variation on which selection can act to promote an evolutionary change in plasticity (Schlichting and Pigliucci, 1998; Windig et al., 2004).
Once it evolves, however, plasticity can subsequently be reduced or even lost evolutionarily, and this loss can have profound evolutionary consequences. Indeed, for more than a century various researchers have hypothesized that the gain and subsequent loss of plasticity can facilitate genetic evolution and thereby fuel the origins of new, ecologically relevant traits (Baldwin, 1902; Schmalhausen, 1949 [1986]; Waddington, 1953; West-Eberhard, 2003). According to one widely cited model for how this process might unfold (West-Eberhard, 2003), when selection acts on quantitative genetic variation regulating the expression of an initially environmentally induced trait, it can promote the evolution of either increased or decreased plasticity through the process known as ‘genetic accommodation’ (Fig. 1). Formally, genetic accommodation is defined as a mechanism of evolution wherein a novel phenotype, generated by either a mutation or an environmental perturbation, is refined into an adaptive phenotype through a series of quantitative genetic changes (West-Eberhard, 2003). In this sense, genetic assimilation represents a specific form of genetic accommodation in which plasticity decreases to the point that a trait becomes constitutively expressed (Waddington, 1953) (Fig. 1).
Fig. 1.
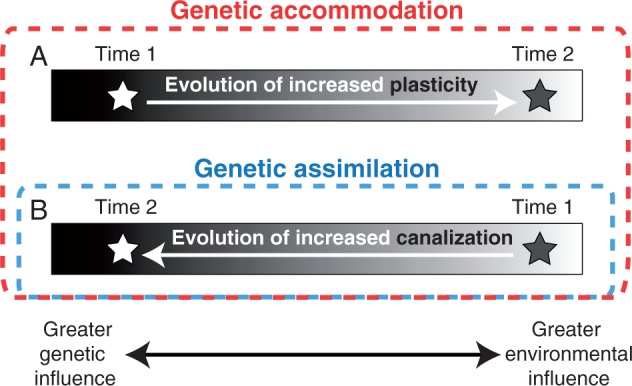
A diagram illustrating the distinction between genetic accommodation and genetic assimilation. Genetic accommodation is any adaptive genetic change in the environmental regulation of a phenotype. For example, a trait may evolve either (A) increased or (B) decreased environmental sensitivity (i.e. phenotypic plasticity). The complete loss of phenotypic plasticity (i.e. increased canalization) is an extreme form of genetic accommodation known as genetic assimilation. Reproduced with permission from Pfennig (2015).
Genetic accommodation/assimilation can occur because most (if not all) traits are influenced by both genes and the environment (Gilbert and Epel, 2009). These two influences on phenotype are potentially evolutionarily interchangeable, meaning that selection can slide trait regulation anywhere along a continuum from total environmental control to total genetic control (Fig. 1). Thus, when genetic variation for the degree of environmental influence is present (as is nearly always the case; see above), selection can act on this variation and promote the evolution of either increased or decreased environmental sensitivity (West-Eberhard, 2003). If selection favours the elimination of all environmental influences – i.e. genetic assimilation – the end result is a genetically ‘fixed’ or ‘canalized’ trait (Waddington, 1942); i.e. a trait that is invariably produced regardless of normal changes in the environment (Fig. 2).
Fig. 2.
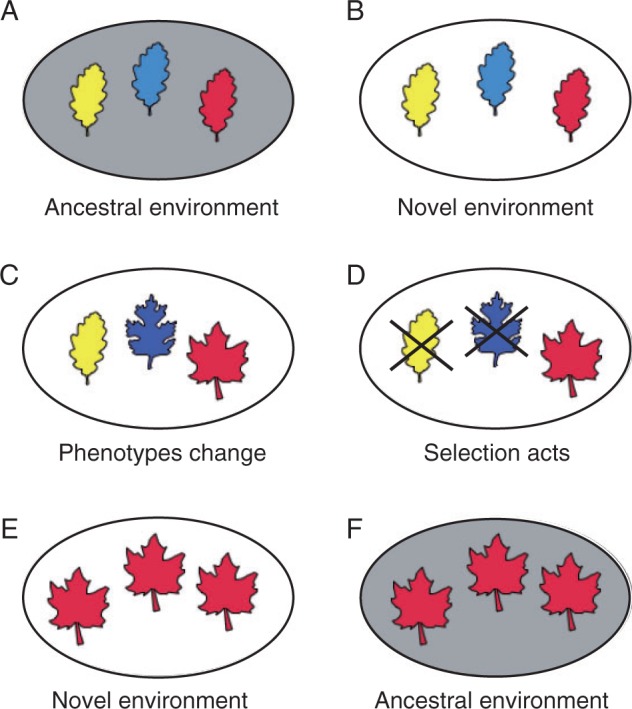
Phenotypic plasticity, followed by genetic assimilation, may facilitate the evolution of a new, canalized trait regardless of the environment through the following steps (here, the trait is a new leaf shape; different colours represent different genotypes). (A) A genetically variable population (B) experiences a novel environment (indicated here as a change from a shaded to an unshaded background). (C) Consequently, the environment induces novel phenotypes (different leaf shapes), but different genotypes respond differently (by producing different-shaped leaves). (D) Selection disfavours those genotypes that produce maladaptive phenotypes (leaf shapes) in the novel environment (indicated here by an X). (E) Such selection may result in the evolution of a novel, canalized trait (a novel leaf shape) that is expressed regardless of the environment. (F) That is, the novel trait is produced even when the environment changes back to the original, ancestral state.
Although genetic accommodation can occur whether a novel trait is mutationally or environmentally induced (West-Eberhard, 2003), environmentally triggered novelties are likely to have greater evolutionary potential than mutationally induced ones, for at least three reasons (West-Eberhard, 2003). First, changes in the environment often impact many individuals simultaneously, in contrast to genetic mutations, which initially affect only one individual and its immediate descendants (not to mention that the vast majority of mutations are deleterious; Kassen and Bataillon, 2006; Halligan and Keightley, 2009). This widespread impact of environmental change enables a newly induced trait to be tested among diverse genotypes, thereby providing fertile ground for selection to act and increasing the chances that genetic accommodation will occur (West-Eberhard, 2003).
Second, although the chance that a particular mutation will occur is not influenced by whether or not the organism is in an environment where that mutation would be advantageous – i.e. adaptively directed mutation does not occur (Sniegowski and Lenski, 1995) – the situation is quite different for an environmentally triggered novel trait. Such a trait is always associated with a particular environment – the one that triggered it. Therefore, environmentally induced traits are more likely than mutationally induced novelties to experience consistent selection and directional modification (West-Eberhard, 2003). This allows new environments to immediately produce and select among new phenotypes and rapidly refine their expression (Badyaev, 2005).
Third, plasticity promotes the storage and release of ‘cryptic genetic variation’, i.e. variation that is expressed only under atypical conditions (Gibson and Dworkin, 2004; Hermisson and Wagner, 2004; Dworkin, 2005a; Schlichting, 2008; Ledón-Rettig et al., 2010; Paaby and Rockman, 2014). The release of such variation ultimately makes genetic accommodation possible (Moczek, 2007; Moczek et al., 2011). Phenotypic plasticity facilitates the build-up of cryptic genetic variation, both because the effects of novel genetic variants are buffered by compensatory plastic responses (Moczek, 2008) and because environment-specific genes experience relaxed selection in the non-inducing environment (Lahti et al., 2009; but see Hunt et al., 2011; Leichty et al., 2012). Essentially, because living systems are robust to mutations and minor environmental perturbation (Masel and Siegal, 2009), they can accumulate genetic variation without that variation having any phenotypic effects and thereby being removed by selection. However, if the system is disturbed (e.g. following exposure to a novel environment and the stress that often accompanies such situations; Badyaev, 2005), robustness breaks down, and the formerly cryptic genetic variation is revealed phenotypically and exposed to selection. Because this storage and release of cryptic genetic variation is the biological analogue of an electric capacitor (which stores and later releases electric charge), it has been dubbed ‘evolutionary capacitance’ (Masel, 2013). Evolutionary capacitors – which have been implicated in the heat shock protein Hsp90 (Rutherford and Lindquist, 1998), yeast prions (True and Lindquist, 2000) and complex gene networks (Bergman and Siegal, 2003) – represent molecular switch mechanisms that can shift genetic variation between cryptic and exposed states (Masel, 2013).
In sum, there are multiple reasons to expect environmentally triggered novelties to have greater evolutionary potential than mutationally induced ones. Indeed, genetic mutations might contribute not so much to the initial origins of phenotypic novelties as to the pool of genetic variation that ultimately makes genetic accommodation/assimilation possible. Or, as West-Eberhard (2003, p. 158) put it, ‘genes are followers, not necessarily leaders, in phenotypic evolution’.
GENETIC ASSIMILATION’S ROLE IN DIVERSIFYING EVOLUTION
Genetic assimilation has long been regarded as potentially crucial in the origins of novel traits (Waddington, 1952, 1953, 2003; Pigliucci and Murren, 2003; Aubret and Shine, 2009; Lande, 2009; Moczek et al., 2011). With genetic assimilation, the origin of a new, canalized trait does not require new genes; instead, selection can promote the origins of a novel trait by acting on existing genetic and epigenetic variation in a population (Schlichting and Pigliucci, 1998; e.g. see Emlen et al., 2007; Ledón-Rettig et al., 2008; Aubret and Shine, 2009; Pfennig and Martin, 2009, 2010; Scoville and Pfrender, 2010). In other words, a plastic trait can be converted into a canalized trait through evolutionary adjustments in the regulation of a trait’s expression.
Although laboratory studies have demonstrated that genetic assimilation can promote novelty (Waddington, 1952, 1953), and there are several suggestive field studies (e.g. Losos et al., 2000; Pfennig and Murphy, 2000; Wund et al., 2008; Scoville and Pfrender, 2010; Robinson, 2013; reviewed in Schlichting and Wund, 2014), ascertaining whether or not genetic assimilation actually has contributed to the evolution of any complex, novel trait in any natural population has long been questioned (Simpson, 1953; Williams, 1966; Orr, 1999; de Jong, 2005; Futuyma, 2013; Wray et al., 2014). A chief difficulty with establishing whether genetic assimilation has promoted novelty is that, once a novel trait has evolved, its evolution cannot be studied in situ (Hall, 1999). One way around this problem is to study lineages (i.e. populations or species) that are thought to be ancestral to the lineage possessing the novel trait in question (West-Eberhard, 2003; Badyaev, 2005; Ghalambor et al., 2007; Ledón-Rettig et al., 2008). Using such an approach, one can then test whether: (1) ancestral species express the trait only through plasticity; (2) novel environments uncover cryptic genetic variation; and (3) trait expression has been refined in derived species. This approach has recently been applied in, among other systems, spadefoot toads and has revealed that a novel ecomorph (the ‘carnivore’ morph) has likely arisen through a ‘plasticity-first’ scenario (Ledón-Rettig et al., 2008, 2010). Moreover, a recent meta-analysis has uncovered several convincing cases in which genes appear to be ‘followers’ in the origins of novel traits (Schwander and Leimar, 2011). For other possible examples in which genetic assimilation might have promoted novelty, including in plants, see the recent review by Schlichting and Wund (2014).
Genetic assimilation might even play a role in speciation. When an induced phenotype becomes expressed constitutively, environmentally induced variation within populations or species can be translated into diverse phenotypes between populations and species. Thus, genetic assimilation generates diversity because it produces fixed (genetic) differences among populations due specifically to the shift from a plastic to a non-plastic phenotype. As noted in the Introduction, if a trait that is involved in mating choice, habitat use or reproductive mode undergoes genetic assimilation in some populations but not in others (e.g. Diggle and Miller, 2013), then this differential loss of plasticity might lead to the evolution of reproductive isolation between these populations and, possibly, speciation (West-Eberhard, 1986, 1989, 2003; Pfennig et al., 2010). Theory has demonstrated genetic assimilation’s capability for promoting diversification (Lande, 2009), and empirical studies find that phenotypic plasticity produces intraspecific variation that parallels interspecific variation within the same clade, suggesting that the former might often form the basis for the latter (Badyaev and Foresman, 2000; Losos et al., 2000; Pfennig and Murphy, 2000; Gomez-Mestre and Buchholz, 2006; Bull-Herenu and Arroyo, 2009). Moreover, there are numerous examples in which a formerly plastic trait has undergone canalization (i.e. lost its plasticity) in a particular lineage, and such shifts are typically accompanied by speciation (Schwander and Leimar, 2011). However, further study is needed to ascertain what role, if any, genetic assimilation plays in speciation (Pfennig et al., 2010; Nosil, 2012).
Phenotypic plasticity, followed by genetic assimilation, might also promote adaptive radiation, influencing both the likelihood of occurrence and the patterns of diversity that emerge (reviewed in West-Eberhard, 2003; Wund et al., 2008; Pfennig et al., 2010). In adaptive radiation, a single ancestral lineage diversifies rapidly in response to divergent selection pressures across numerous environments (Schluter, 2000). According to the ‘flexible stem’ hypothesis (West-Eberhard, 2003), an adaptive radiation arises when ecological circumstances favour diversification in an ancestral taxon that expresses phenotypic plasticity in the types of traits that characterize the adaptive radiation. Under such circumstances, when individuals are exposed to the same selective environments, plasticity in the ancestral lineage repeatedly reveals the same sets of phenotypes. This model might explain ‘replicate’ adaptive radiations (i.e. the situation in which many descendant species evolve parallel ecotypic variation in response to similar selection pressures) in a number of systems (Meyer, 1987; Losos et al., 2000; Wund et al., 2008). However, further study is needed to test this model more rigorously.
Having discussed the potential evolutionary consequences of phenotypic plasticity and genetic assimilation, we now turn to the important issue of the possible proximate mechanisms that underpin both processes.
POTENTIAL PROXIMATE MECHANISMS OF PHENOTYPIC PLASTICITY AND GENETIC ASSIMILATION
Mechanisms of phenotypic plasticity
Given the potential importance of genetic assimilation in evolutionary innovation and diversification (as outlined above), understanding the molecular mechanisms involved in this process might improve our knowledge of how novel traits evolve; however, such an understanding can also help in assessing whether genetic assimilation is likely to be common or rare in nature. Given that genetic assimilation represents a loss of phenotypic plasticity, a natural starting point for thinking about the mechanisms that might underlie genetic assimilation is the molecular causes of phenotypic plasticity.
Phenotypic plasticity typically involves the following three general steps. First, an individual’s sensory system detects and transduces information about its external environment. In some cases, the environmental stimulus and any response that it elicits may be very general, as with phenotypic changes wrought by changes in an individual’s temperature or nutrition (reviewed in Gilbert and Epel, 2009). In other cases, the requisite stimuli and responses are highly specific. For example, certain plants possess receptor proteins that detect only the plant’s most common natural enemies (Zhao et al., 2005). Second, the signal detected by the sensory system is transduced into a molecular response at the biochemical level that alters the activities within cells. In the case of multicellular organisms, this information may be conveyed elsewhere in the organism’s body via hormonally mediated signals. Indeed, hormones underlie nearly all instances of plasticity in animals and likely also play an important role in plasticity in plants (Gilbert and Epel, 2009). Third, the target cells, organs or tissues respond accordingly by altering phenotype.
Ultimately, phenotypic plasticity is nearly always accompanied by changes in gene expression (Aubin-Horth and Renn, 2009), an observation that has provided key insight into the molecular mechanisms of phenotypic plasticity (Gilbert and Epel, 2009). In particular, differential gene expression (and, hence, phenotypic plasticity) often involves alterations in the binding of transcription factors to a gene’s promoter or other regulatory elements. Transcription factors differ in the sequences that they recognize, their abilities to activate or repress transcription and their responsiveness to external signals. Thus, one proposed mechanism of phenotypic plasticity is that the individual’s sensory system transduces information from its external environment by recruiting different transcription factors, thereby activating different genes. These newly activated genes may then trigger different hormones, which ultimately cause alternative phenotypes to be produced (Nijhout, 2003).
Any particular plastic response may involve many steps, potentially encompassing numerous genes and environmental and physiological factors. This complexity provides copious targets on which selection can act, from the types of signals that an individual’s sensory system can detect to the threshold amount of a particular hormone needed to trigger a phenotypic response (Pfennig et al., 2010; Moczek et al., 2011).
Potential mechanisms of genetic assimilation
For genetic assimilation to take place, the reaction norms that underlie a phenotype’s expression in a population must undergo an evolutionary shift, such that genotypes that express a phenotype robustly across a range of different environments become fixed in the population (Pigliucci et al., 2006). Experimental evidence indicates that such changes in reaction norms may be driven by variation in signalling pathways that mediate the relationship between genotype, environment and phenotype. One way such variants might act is by changing developmental thresholds for environmentally influenced hormones and other key signalling molecules (Moczek and Nijhout, 2002; Suzuki and Nijhout, 2006).
Although there have yet to be case studies reported in which genetic assimilation has been linked to specific genetic changes (but see Scoville and Pfrender, 2010), work on related topics – including the molecular basis of gene expression variation, genotype–environment interaction, and robustness – provides valuable insights into possible causes of genetic assimilation. As with phenotypic plasticity in general, genetic variants that alter gene regulation are likely important contributors to genetic assimilation (Pfennig and Ehrenreich, 2014). Populations commonly harbour large numbers of genetic variants that affect gene expression (e.g. Brem et al., 2002; Schadt et al., 2003; Morley et al., 2004). Additionally, many new mutations that arise within populations impact gene regulation (Landry et al., 2007). Thus, ample genetic diversity in gene expression exists within populations that affects gene regulation and may serve as a reservoir of cryptic phenotypic effects (Gibson and Dworkin, 2004; Moczek, 2007; Le Rouzic and Carlborg, 2008; Paaby and Rockman, 2014).
There are multiple plausible ways in which changes in gene regulation might facilitate canalization of a plastic trait (Fig. 3). For example, the regulation of genes that control a plastic trait might become decoupled from their environmental cue (Fig. 3A) or might evolve regulation by a secondary pathway that makes their expression robust to the environment (Fig. 3B; e.g. Matsui et al., 2015). Describing such changes in gene regulation in a binary manner is helpful in considering possible models for genetic assimilation, but it is important to note that real cases of genetic assimilation are likely to be caused by quantitative genetic changes in gene regulation, i.e. changes involving one or more loci with quantitative effects on transcript levels (Ledón-Rettig et al., 2014). These genetic changes that facilitate genetic assimilation may occur in phenotypically important genes themselves or, alternatively, they may occur in the upstream regulators of these genes. These two classes of variants are referred to as cis and trans regulatory polymorphisms, respectively (Albert and Kruglyak, 2015).
Fig. 3.
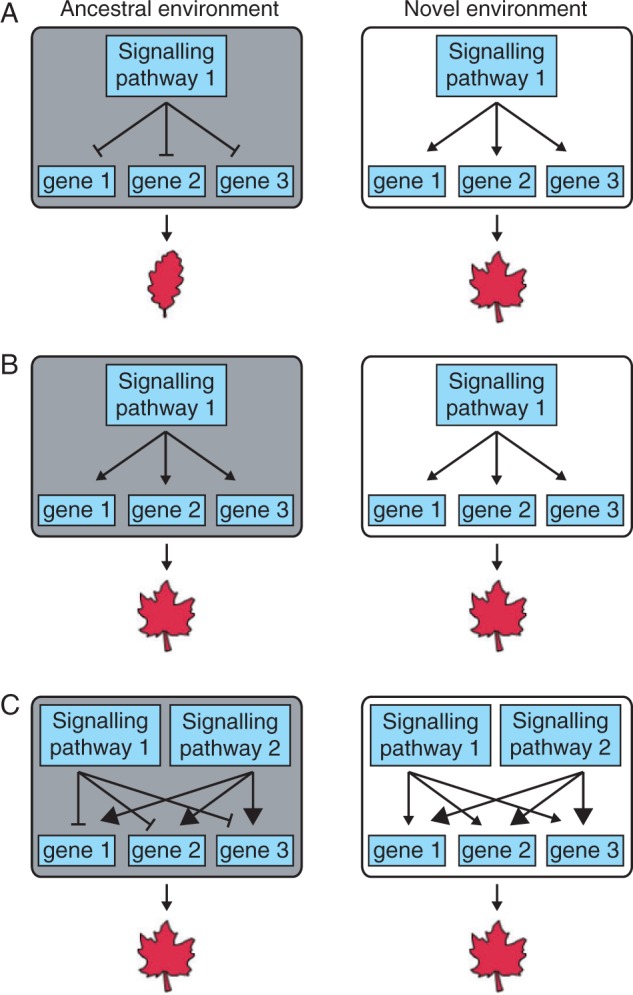
Potential causes of phenotypic plasticity and genetic assimilation. (A) A new trait arises due to a change in gene regulation triggered by a novel environment. Two plausible mechanisms for genetic assimilation of the novel trait are illustrated: (B) evolution of the pathway underlying the novel trait such that it is no longer environmentally responsive; and (C) rewiring of development such that a new, environment-independent pathway causes the novel phenotype to be constitutively expressed. Note that genes 1, 2 and 3 represent genes that must be transcribed for the novel phenotype to be expressed, while signalling pathways 1 and 2 are pathways that can activate these genes. The thicker arrows in (C) indicate that signalling pathway 2 has a stronger effect on genes 1, 2 and 3 than signalling pathway 1.
Strong arguments can be made for both cis and trans regulatory evolution contributing to genetic assimilation. With regard to cis regulatory polymorphisms, because they generally exhibit lower sensitivity to the environment than trans regulatory polymorphisms (Smith and Kruglyak, 2008; Cubillos et al., 2014), cis regulatory variants have the potential to rapidly canalize the expression of individual genes. The most important types of cis regulatory variants within the context of genetic assimilation are likely to be those that create or disrupt transcription factor binding sites (Wittkopp and Kalay, 2012). For example, loss of a binding site for a conditionally active transcription factor might eliminate a gene’s sensitivity to the environment. Alternatively, gain of a binding site for a constitutively active transcription factor might also result in the decoupling of a gene’s expression from the environment by leading to increased redundancy in a gene’s regulation and a corresponding higher robustness in the gene’s expression across conditions.
Although the rewiring of gene regulation by cis regulatory polymorphism is a plausible mechanism for genetic assimilation (as described above), variants that influence gene expression in trans, or through a combination of cis and trans effects, might also be capable of causing genetic assimilation. This is particularly true when considering novel traits that depend on many genes having their expression altered, as cis variants only affect their cognate gene (and potentially its downstream targets if it is a transcription factor), while trans variants can potentially impact the regulation of many genes (e.g. Brem et al., 2002; Yvert et al., 2003).
There is at least one additional important difference between cis and trans regulatory variants with respect to their potential to contribute to genetic assimilation: their mutational target spaces. While genes usually have one promoter, their expression is often influenced by a large number of trans factors. Despite transcription factors being the most direct regulators of gene expression, many other factors can influence the abundances of transcripts and proteins within a cell (Yvert et al., 2003; Albert and Kruglyak, 2015). These include, but are not limited to, environmental sensors and other components of signalling cascades that modulate the activities of transcription factors in response to environmental cues, as well as proteins and non-coding RNAs that directly determine transcript stability and translation. Because so many factors affect the expression of a gene in trans, the mutational target space for trans effects is often one or more orders of magnitude larger than for cis effects (Denver et al., 2005; Landry et al., 2007; Gruber et al., 2012).
The larger mutational target space for trans regulatory variants than cis regulatory variants may suggest there is a greater chance for polymorphisms that occur in trans to decouple a trait from its environmental cue. In particular, trans regulatory variants that cause signalling activity in conditionally active pathways, even in the absence of their inductive cues, might result in genetic assimilation. Examples of such environmentally insensitive variants have been described. For instance, laboratory strains of Saccharomyces cerevisiae possess an allele of GPA1, a component of the mating pheromone responsive mitogen activated protein kinase (MAPK) pathway, that shows high activity even in the absence of mating pheromone (Yvert et al., 2003). MAPK pathways are evolutionarily conserved across eukaryotes and play diverse roles in development and physiology (Seger and Krebs, 1995; Schaeffer and Weber, 1999; Hamel et al., 2006), suggesting that similar phenomena could occur in other species.
Our description of the role of changes in gene regulation in cis and trans in genetic assimilation is an oversimplification in that we have largely focused on the mechanisms by which individual genetic polymorphisms might contribute. However, it is well known that heritable changes in most traits are influenced by numerous genetic variants (Mackay et al., 2009) and that these variants can show complicated non-additive effects on gene expression due to their collective influence on gene regulatory networks (Omholt et al., 2000; Gjuvsland et al., 2007; Nuzhdin et al., 2012). Furthermore, genetic variation in other molecular processes, such as post-translational regulation of proteins or changes in protein–protein interactions, might contribute to genetic assimilation. Moving forward, it will be important to develop case studies in which genetic assimilation has been conclusively shown and the involved genetic variants identified and functionally characterized. In the following section we discuss approaches that can be used to explore this problem.
APPROACHES FOR INVESTIGATING THE MOLECULAR MECHANISMS THAT UNDERLIE GENETIC ASSIMILATION
Current genetic and genomic approaches provide powerful tools for characterizing the mechanisms underlying genetic assimilation. Admittedly, these techniques will work best for traits that have well-understood evolutionary histories and can be studied in model organisms, which typically have high-quality genomes and transcriptomes. However, as new sequencing technologies make it easier to generate high-quality genomes and transcriptomes in other species, these approaches should increase in their applicability to non-model organisms. Here, we first describe methods for examining how canalization of a phenotype might arise due to changes in gene regulation across genotypes and environments. We then discuss mapping techniques that can be used to identify genes and genetic variants that cause regulatory changes that result in genetic assimilation. We also mention how systematic mutagenesis screens in model organisms might improve understanding of genetic assimilation.
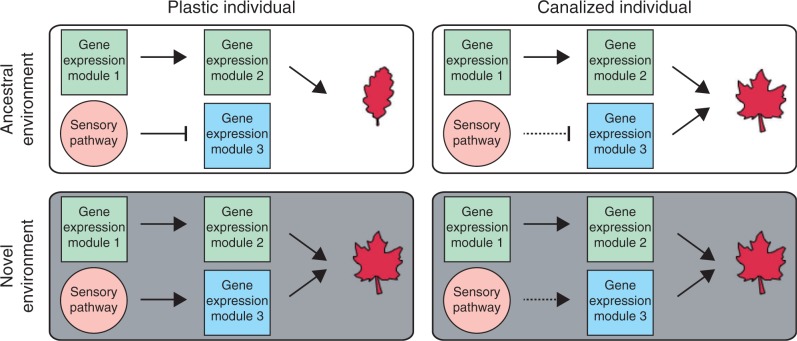
All organisms exhibit modular gene expression throughout development (e.g. Arbeitman et al., 2002; Schmid et al., 2005). Alterations in how these gene modules are expressed across developmental stages and tissues is known to play an important role in phenotypic evolution (as discussed in Peter and Davidson, 2011; Ichihashi et al., 2014 and elsewhere), and likely also contributes to phenotypic plasticity (Schneider et al., 2014) and genetic assimilation (Pfennig and Ehrenreich, 2014) (Fig. 4). RNA-Seq, at multiple stages of development, can be used to quantify the abundances of all transcripts in an organism’s genome, even for non-model species (Wang et al., 2009), providing a foundation for defining modules of co-expressed genes and examining the regulatory relationships among these modules (Nuzhdin et al., 2012; Schneider et al., 2014).
Fig. 4.
Examining how changes in gene regulatory networks contribute to genetic assimilation. Expression analysis across development can be employed to determine transcriptional changes that enable constitutive expression of a phenotype. Comparison of plastic and canalized genotypes in the ancestral and novel environment can be used to identify gene modules that underlie canalization. Here, we have shown a hypothetical example of what might be found in such a study. In the example, the sensitivity of gene expression module 3 to regulation by an environmentally responsive sensory pathway has been reduced in canalized individuals (indicated by a dotted line). Due to this reduction, gene expression module 3 is expressed when individuals are not reared in the novel, inductive environment. This higher expression leads to constitutive expression of the new trait across conditions.
A strategy that will likely help in characterizing the molecular basis of genetic assimilation is comparing expression patterns among canalized and plastic genotypes in both their ancestral and novel environments (Fig. 4). Of course, this approach requires knowledge of the evolutionary history of plasticity in the study organisms, and the number of systems in which this is known may be limited (but see examples in Schwander and Leimar, 2011; Schlichting and Wund, 2014). Nevertheless, through such an experiment, gene modules that are differentially expressed across conditions in plastic individuals but expressed at similar levels across conditions in canalized individuals can be identified. Bioinformatic analysis of genes in these modules (Schneider et al., 2014), potentially in combination with experimental techniques for studying the binding of proteins to DNA, such as ChIP-Seq (Kidder et al., 2011) and DNase-FLASH (Vierstra et al., 2014), can then be used to determine the changes in transcription factor activity that have resulted in canalization. Recent work has shown that these approaches for studying how protein–DNA interactions regulate chromatin structure, transcription and phenotypic outcome can be applied in non-model organisms (Simola et al., 2013).
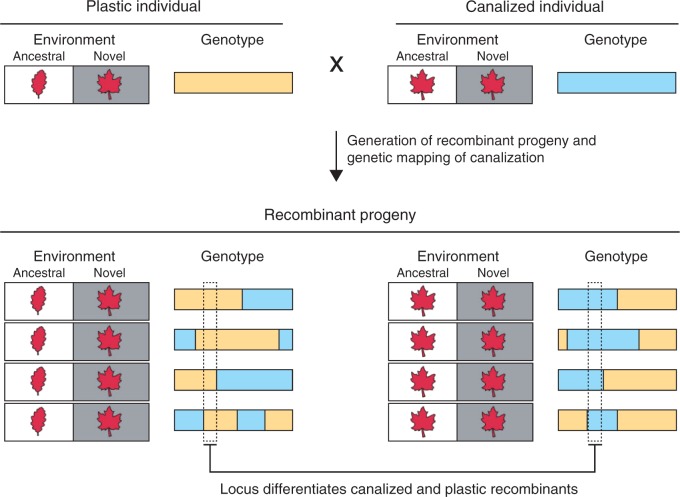
Given that canalization requires the fixation of genetic variants that make a trait robust to the environment, this robustness itself can be viewed as a quantitative trait (e.g. de Visser et al., 2003; Dworkin, 2005b; Flatt, 2005; Levy and Siegal, 2012; Queitsch et al., 2012) and subjected to linkage mapping (Lander and Botstein, 1989) (Fig. 5). For this approach to work, it must be possible to mate individuals from a canalized population or species to individuals from an ancestral population or species that has remained plastic (Fig. 5). Furthermore, recombinants from these crosses must be viable.
Fig. 5.
Genetic mapping of loci that underlie genetic assimilation. Crosses between canalized and plastic genotypes can be used to determine the loci that underlie canalization of a novel trait.
Although historically it might have been difficult to use genetic mapping to identify loci that canalize a trait, next-generation sequencing has revolutionized genetic mapping by crosses (e.g. Ehrenreich et al., 2010; Bloom et al., 2013). Indeed, new sequencing technologies have made it cheaper and easier to identify markers for conducting linkage mapping, and have also simplified the process of genotyping cross progeny (Andolfatto et al., 2011). Recent studies have shown that by analysing large genetic mapping populations, high statistical power and precise mapping resolution can be achieved, facilitating detection and cloning of most, if not all, of the loci involved in a trait (Ehrenreich et al., 2010; Bloom et al., 2013; Taylor and Ehrenreich, 2014). Within the context of genetic assimilation, cloning of the genes and genetic variants underlying these loci can help to shed light on the molecular mechanisms that cause canalization.
Finally, much of what we know about genetic assimilation comes from model organisms. Indeed, Waddington’s original work on genetic assimilation was conducted in Drosophila melanogaster (Waddington, 1952, 1953), and in recent years work in multiple model systems, including Arabidopsis (Queitsch et al., 2002; Sangster et al., 2008a, b), Caenorhabditis (Felix, 2007; Milloz et al., 2008; Duveau and Felix, 2012), Drosophila (Gibson and Hogness, 1996; Rutherford and Lindquist, 1998; Gibson et al., 1999; Dworkin et al., 2003) and yeast (Jarosz and Lindquist, 2010; Tirosh et al., 2010; Halfmann et al., 2012), has been used to explore the mechanisms that reveal cryptic genetic variation and may give rise to genetic assimilation. However, such work is still in its infancy, meaning that these model systems have great potential to advance our understanding of genetic assimilation in ways that would be very difficult in non-model systems. For example, saturating screens can be conducted in these organisms to identify mutations that decouple plastic traits from their environmental stimuli. When layered onto our knowledge of the gene regulatory networks of these species, general rules about how to manipulate the genotype–environment–phenotype relationship may emerge. As new genetic engineering approaches, such as CRISPR/Cas9 (Mali et al., 2013), gain increasing usage across species, comparable analysis techniques may be possible in non-model organisms as well.
CONCLUSIONS
Improving our understanding of genetic assimilation may provide valuable new insights into the origins of novel traits and species. Because the number of examples in which genetic assimilation has been convincingly demonstrated is small, efforts to develop more case studies of genetic assimilation, especially in natural populations, may facilitate major advances in this research area. Identifying and cloning the genes and genetic variants that underlie these examples can shed crucial new light onto the mechanisms underlying genetic assimilation. However, to fully explain the mechanisms that cause genetic assimilation, it will likely be necessary to examine how the genetic variants underlying genetic assimilation alter developmental gene regulatory programmes across genotypes and environments. Examples in which genetic assimilation has been shown and characterized at the molecular level will facilitate new insights into the processes that shape diversity in nature. Such work may also have impacts outside of evolutionary biology by expanding our basic knowledge of how to modify biological systems to produce new traits.
ACKNOWLEDGEMENTS
We thank two anonymous referees for their comments, Rainer Melzer and Günter Theissen for inviting us to prepare this review, and Takeshi Matsui for helping us to design figures. I.M.E. was supported by grants from the National Institutes of Health (R01GM110255 and R21AI108939), National Science Foundation (MCB1330874), the Army Research Office (W911NF-14-1-0318) and Rose Hills Foundation, as well as a Sloan Research Fellowship. D.W.P. was supported by a grant from the NSF (DEB1019479).
LITERATURE CITED
- Albert FW, Kruglyak L. 2015. The role of regulatory variation in complex traits and disease. Nat Rev Genet 16: 197–212. [DOI] [PubMed] [Google Scholar]
- Andolfatto P, Davison D, Erezyilmaz D, et al. 2011. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res 21: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EE, Imam F, et al. 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270–2275. [DOI] [PubMed] [Google Scholar]
- Aubin-Horth N, Renn SCP. 2009. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Molecular Ecology 18: 3763–3780. [DOI] [PubMed] [Google Scholar]
- Aubret F, Shine R. 2009. Genetic assimilation and the postcolonization erosion of phenotypic plasticity in island tiger snakes. Current Biology 19: 1932–1936. [DOI] [PubMed] [Google Scholar]
- Badyaev AV. 2005. Stress-induced variation in evolution: from behavioural plasticity to genetic assimilation. Proceedings of the Royal Society of London Series B: Biological Sciences 272: 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badyaev AV, Foresman KR. 2000. Extreme environmental change and evolution: stress-induced morphological variation is strongly concordant with patterns of evolutionary divergence in shrew mandibles. Proceedings of the Royal Society of London Series B: Biological Sciences 267: 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JM. 1902. Development and evolution. New York: Macmillan. [Google Scholar]
- Bergman A, Siegal ML. 2003. Evolutionary capacitance as a general feature of complex gene networks. Nature 424: 549–552. [DOI] [PubMed] [Google Scholar]
- Berrigan D, Scheiner SM. 2004. Modeling the evolution of phenotypic plasticity. In: DeWitt TJ, Scheiner SM, eds. Phenotypic plasticity: functional and conceptual approaches. New York: Oxford University Press. [Google Scholar]
- Bloom JS, Ehrenreich IM, Loo WT, Lite TL, Kruglyak L. 2013. Finding the sources of missing heritability in a yeast cross. Nature 494: 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw AD. 1965. Evolutionary significance of phenotypic plasticity in plants. Advances in Genetics 13: 115–155. [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–725. [DOI] [PubMed] [Google Scholar]
- Bull-Herenu K, Arroyo MTK. 2009. Phenological and morphological differentiation in annual Chaetanthera moenchioides (Asteraceae) over an aridity gradient. Plant Systematics and Evolution 278: 159–167. [Google Scholar]
- Cubillos FA, Stegle O, Grondin C, et al. 2014. Extensive cis-regulatory variation robust to environmental perturbation in Arabidopsis. Plant Cell 26: 4298–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nature Genetics 37: 544–548. [DOI] [PubMed] [Google Scholar]
- Diggle PK, Miller JS. 2013. Developmental plasticity, genetic assimilation, and the evolutionary diversification of sexual expression in Solanum. American Journal of Botany 100: 1050–1060. [DOI] [PubMed] [Google Scholar]
- Duveau F, Felix MA. 2012. Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans. PLoS Biology 10: e1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin I. 2005a Canalization, cryptic variation, and developmental buffering: a critical examination and analytical perspective. In: Hallgrímsson B, Hall BK, eds. Variation: a central concept in biology. New York: Elsevier/Academic Press, 131–158. [Google Scholar]
- Dworkin I. 2005b Towards a genetic architecture of cryptic genetic variation and genetic assimilation: the contribution of K. G. Bateman. Journal of Genetics 84: 223–226. [DOI] [PubMed] [Google Scholar]
- Dworkin I, Palsson A, Birdsall K, Gibson G. 2003. Evidence that Egfr contributes to cryptic genetic variation for photoreceptor determination in natural populations of Drosophila melanogaster. Current Biology 13: 1888–1893. [DOI] [PubMed] [Google Scholar]
- Ehrenreich IM, Torabi N, Jia Y, et al. 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464: 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlen DJ, Lavine LC, Ewen-Campen B. 2007. On the origin and evolutionary diversification of beetle horns. Proceedings of the National Academy of Sciences of the USA 104: 8661–8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA. 2007. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Current Biology 17: 103–114. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick BM. 2012. Underappreciated consequences of phenotypic plasticity for ecological speciation. International Journal of Ecology 2012: 256017. [Google Scholar]
- Flatt T. 2005. The evolutionary genetics of canalization. Quarterly Review of Biology 80: 287–316. [DOI] [PubMed] [Google Scholar]
- Futuyma DJ. 2013. Evolution. Sunderland, MA: Sinauer. [Google Scholar]
- Ghalambor CK, McKay JK, Carroll S, Reznick DN. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation to new environments. Functional Ecology 21: 394–407. [Google Scholar]
- Gibson G, Dworkin I. 2004. Uncovering cryptic genetic variation. Nature Reviews. Genetics 5: 1199–1212. [DOI] [PubMed] [Google Scholar]
- Gibson G, Hogness DS. 1996. Effect of polymorphism in the Drosophila regulatory gene Ultrabithorax on homeotic stability. Science 271: 200–203. [DOI] [PubMed] [Google Scholar]
- Gibson G, Wemple M, van Helden S. 1999. Potential variance affecting homeotic Ultrabithorax and Antennapedia phenotypes in Drosophila melanogaster. Genetics 151: 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF, Epel D. 2009. Ecological developmental biology: integrating epigenetics, medicine, and evolution. Sunderland, MA: Sinauer. [Google Scholar]
- Gjuvsland AB, Hayes BJ, Omholt SW, Carlborg O. 2007. Statistical epistasis is a generic feature of gene regulatory networks. Genetics 175: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mestre I, Buchholz DR. 2006. Developmental plasticity mirrors differences among taxa in spadefoot toads linking plasticity and diversity. Proceedings of the National Academy of Sciences of the USA 103: 19021–19026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber JD, Vogel K, Kalay G, Wittkopp PJ. 2012. Contrasting properties of gene-specific regulatory, coding, and copy number mutations in Saccharomyces cerevisiae: frequency, effects, and dominance. PLoS Genetics 8: e1002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AP, Lewontin RC. 1982. A study of reaction norms in natural populations of Drosophila pseudoobscura. Evolution 36: 934–948. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, et al. 2012. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482: 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BK. 1999. Evolutionary developmental biology. Dordrecht: Kluwer. [Google Scholar]
- Halligan DL, Keightley PD. 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annual Review of Ecology, Evolution, and Systematics 40: 151–172. [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, et al. 2006. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends in Plant Science 11: 192–198. [DOI] [PubMed] [Google Scholar]
- Hermisson J, Wagner GP. 2004. The population genetic theory of hidden variation and genetic robustness. Genetics 168: 2271–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey RB, Hertz PE, Sinervo B. 2003. Behavioral drive versus behavioral inertia in evolution: a null model approach. American Naturalist 161: 357–366. [DOI] [PubMed] [Google Scholar]
- Hunt BG, Ometto L, Wurmb Y, et al. 2011. Relaxed selection is a precursor to the evolution of phenotypic plasticity. Proceedings of the National Academy of Sciences of the USA 108: 15936–15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Aguilar-Martinez JA, Farhi M, et al. 2014. Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proceedings of the National Academy of Sciences of the USA 111: E2616–E2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Lindquist S. 2010. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 330: 1820–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong G. 2005. Evolution of phenotypic plasticity: patterns of plasticity and the emergence of ecotypes. New Phytologist 166: 101–117. [DOI] [PubMed] [Google Scholar]
- Kassen R, Bataillon TM. 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nature Genetics 38: 484–488. [DOI] [PubMed] [Google Scholar]
- Kidder BL, Hu G, Zhao K. 2011. ChIP-Seq: technical considerations for obtaining high-quality data. Nature Immunology 12: 918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsolver JG, Ragland GJ, Schlichta JG. 2004. Quantitative genetics of continuous reaction norms: thermal sensitivity of caterpillar growth rates. Evolution 58: 1521–1529. [DOI] [PubMed] [Google Scholar]
- Lahti DC, Johnson NA, Ajie BC, et al. 2009. Relaxed selection in the wild. Trends in Ecology & Evolution 24: 487–496. [DOI] [PubMed] [Google Scholar]
- Laland KN, Uller T, Feldman MW, et al. 2014. Does evolutionary theory need a rethink? Yes, urgently. Nature 514: 161–164. [DOI] [PubMed] [Google Scholar]
- Lande R. 2009. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. Journal of Evolutionary Biology 22: 1435–1446. [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D. 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. 2007. Genetic properties influencing the evolvability of gene expression. Science 317: 118–121. [DOI] [PubMed] [Google Scholar]
- Le Rouzic A, Carlborg O. 2008. Evolutionary potential of hidden genetic variation. Trends in Ecology & Evolution 23: 33–37. [DOI] [PubMed] [Google Scholar]
- Ledón-Rettig CC, Pfennig DW, Chunco AJ, Dworkin I. 2014. Cryptic genetic variation in natural populations: a predictive framework. Integrative and Comparative Biology 54: 783–793. [DOI] [PubMed] [Google Scholar]
- Ledón-Rettig CC, Pfennig DW, Nascone-Yoder N. 2008. Ancestral variation and the potential for genetic accommodation in larval amphibians: implications for the evolution of novel feeding strategies. Evolution and Development, 10: 316–325. [DOI] [PubMed] [Google Scholar]
- Ledón-Rettig CC, Pfennig DW, Crespi EJ. 2010. Diet and hormone manipulations reveal cryptic genetic variation: implications for the evolution of novel feeding strategies. Proceedings of the Royal Society of London Series B: Biological Sciences 277: 3569–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichty AR, Pfennig DW, Jones CD, Pfennig KS. 2012. Relaxed genetic constraint is ancestral to the evolution of phenotypic plasticity. Intergrative and Comparative Biology 52: 16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML. 2012. The robustness continuum. Advances in Experimental Medicine and Biology 751: 431–452. [DOI] [PubMed] [Google Scholar]
- Losos JB, Creer DA, Glossip D, et al. 2000. Evolutionary implications of phenotypic plasticity in the hindlimb of the lizard Anolis sagrei. Evolution 54: 301–305. [DOI] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. 2009. The genetics of quantitative traits: challenges and prospects. Nature Reviews Genetics 10: 565–577. [DOI] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. 2013. Cas9 as a versatile tool for engineering biology. Nature Methods 10: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J. 2013. Q&A: evolutionary capacitance. BMC Biology 11: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Siegal ML. 2009. Robustness: mechanisms and consequences. Trends in Genetics 25: 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Linder R, Phan J, Seidl F, Ehrenreich IM. 2015. Regulatory rewiring in a cross causes extensive genetic heterogeneity. Genetics. doi:10.1534/genetics.115.180661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A. 1987. Phenotypic plasticity and heterochrony in Ciclasoma managuense (Pisces, Cichlidae) and their implications for speciation in cichlid fishes. Evolution 41: 1357–1369. [DOI] [PubMed] [Google Scholar]
- Milloz J, Duveau F, Nuez I, Felix MA. 2008. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes & Development 22: 3064–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek AP. 2007. Developmental capacitance, genetic accommodation, and adaptive evolution. Evolution and Development 9: 299–305. [DOI] [PubMed] [Google Scholar]
- Moczek AP. 2008. On the origins of novelty in development and evolution. Bioessays 30: 432–447. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Nijhout HF. 2002. Developmental mechanisms of threshold evolution in a polyphenic beetle. Evolution and Development 4: 252–264. [DOI] [PubMed] [Google Scholar]
- Moczek AP, Sultan SE, Foster S, et al. 2011. The role of developmental plasticity in evolutionary innovation. Proceedings of the Royal Society of London Series B: Biological Sciences 278: 2705–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, et al. 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430: 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout HF. 2003. Development and evolution of adaptive polyphenisms. Evolution and Development 5: 9–18. [DOI] [PubMed] [Google Scholar]
- Nosil P. 2012. Ecological speciation. New York: Oxford University Press. [Google Scholar]
- Nuzhdin SV, Friesen ML, McIntyre LM. 2012. Genotype-phenotype mapping in a post-GWAS world. Trends in Genetics 28: 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omholt SW, Plahte E, Oyehaug L, Xiang K. 2000. Gene regulatory networks generating the phenomena of additivity, dominance and epistasis. Genetics 155: 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. 1999. An evolutionary dead end? Science 285: 343–344. [Google Scholar]
- Paaby AB, Rockman MV. 2014. Cyptic genetic variation: evolution’s hidden substrate. Nature Reviews. Genetics 15: 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144: 970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig DW. 2004. Putting genes in perspective. American Scientist 92: 84–86. [Google Scholar]
- Pfennig DW. 2015. Ecological evolutionary developmental biology. In: Kliman R, ed. Encyclopedia of evolutionary biology. Amsterdam: Elsevier, in press. [Google Scholar]
- Pfennig DW, Ehrenreich IM. 2014. Towards a gene regulatory network perspective on phenotypic plasticity, genetic accommodation and genetic assimilation. Molecular Ecology 23: 4438–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig DW, Martin RA. 2009. A maternal effect mediates rapid population divergence and character displacement in spadefoot toads. Evolution 63: 898–909. [DOI] [PubMed] [Google Scholar]
- Pfennig DW, Martin RA. 2010. Evolution of character displacement in spadefoot toads: different proximate mechanisms in different species. Evolution 64: 2331–2341. [DOI] [PubMed] [Google Scholar]
- Pfennig DW, McGee M. 2010. Resource polyphenism increases species richness: a test of the hypothesis. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences 365: 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig DW, Murphy PJ. 2000. Character displacement in polyphenic tadpoles. Evolution 54: 1738–1749. [DOI] [PubMed] [Google Scholar]
- Pfennig DW, Wund MA, Snell-Rood EC, Cruickshank T, Schlichting CD, Moczek AP. 2010. Phenotypic plasticity’s impacts on diversification and speciation. Trends in Ecology & Evolution 25: 459–467. [DOI] [PubMed] [Google Scholar]
- Pigliucci M, Murren CJ. 2003. Genetic assimilation and a possible evolutionary paradox: can macroevolution sometimes be so fast as to pass us by? Evolution 57: 1455–1464. [DOI] [PubMed] [Google Scholar]
- Pigliucci M, Murren CJ, Schlichting CD. 2006. Phenotypic plasticity and evolution by genetic assimilation. Journal of Experimental Biology 209: 2362–2367. [DOI] [PubMed] [Google Scholar]
- Price TD, Qvarnstrom A, Irwin DE. 2003. The role of phenotypic plasticity in driving genetic evolution. Proceedings of the Royal Society of London Series B: Biological Sciences 270: 1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Carlson KD, Girirajan S. 2012. Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS Genetics 8: e1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624. [DOI] [PubMed] [Google Scholar]
- Robinson BW. 2013. Evolution of growth by genetic accommodation in Icelandic freshwater stickleback. Proceedings of the Royal Society of London Series B: Biological Sciences 280: 2013.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342. [DOI] [PubMed] [Google Scholar]
- Sangster TA, Salathia N, Undurraga S, et al. 2008a HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proceedings of the National Acadamy of Sciences of the USA 105: 2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster TA, Salathia N, Lee HN, Watanabe E, Schellenberg K, et al. 2008b HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the USA 105: 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, et al. 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. [DOI] [PubMed] [Google Scholar]
- Schaeffer HJ, Weber MJ. 1999. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Molecular and Cellular Biology 19: 2435–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting CD. 2004. The role of phenotypic plasticity in diversification. In: DeWitt TJ, Scheiner SM, eds. Phenotypic plasticity: functional and conceptual approaches. New York: Oxford University Press, 191–200. [Google Scholar]
- Schlichting CD. 2008. Hidden reaction norms, cryptic genetic variation, and evolvability. Annals of the New York Academy of Sciences 1133: 187–203. [DOI] [PubMed] [Google Scholar]
- Schlichting CD, Pigliucci M. 1998. Phenotypic evolution: a reaction norm perspective. Sunderland, MA: Sinauer. [Google Scholar]
- Schlichting CD, Wund MA. 2014. Phenotypic plasticity and epigenetic marking: an assessment of evidence for genetic accommodation. Evolution 68: 656–672. [DOI] [PubMed] [Google Scholar]
- Schluter D. 2000. The ecology of adaptive radiation. Oxford: Oxford University Press. [Google Scholar]
- Schmalhausen II. 1949. [1986]. Factors of evolution: the theory of stabilizing selection. Chicago: University of Chicago Press. [Google Scholar]
- Schmid M, Davison TS, Henz SR, et al. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Schneider RF, Li Y, Meyer A, Gunter HM. 2014. Regulatory gene networks that shape the development of adaptive phenotypic plasticity in a cichlid fish. Molecular Ecology 23: 4511–4526. [DOI] [PubMed] [Google Scholar]
- Schwander T, Leimar O. 2011. Genes as leaders and followers in evolution. Trends in Ecology & Evolution 26: 143–151. [DOI] [PubMed] [Google Scholar]
- Scoville AG, Pfrender ME. 2010. Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. Proceedings of the National Academy of Sciences of the USA 107: 4260–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger R, Krebs EG. 1995. The MAPK signaling cascade. FASEB Journal 9: 726–735. [PubMed] [Google Scholar]
- Simola DF, Ye C, Mutti NS, et al. 2013. A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Research 23: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. 1953. The major features of evolution. New York: Columbia University Press. [Google Scholar]
- Smith EN, Kruglyak L. 2008. Gene-environment interaction in yeast gene expression. PLoS Biology 6: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Lenski RE. 1995. Mutation and adaptation: the directed mutation controversy in evolutionary perspective. Annual Review of Ecology and Systematics 26: 553–578. [Google Scholar]
- Stearns SC. 1989. The evolutionary significance of phenotypic plasticity. Bioscience 39: 436–445. [Google Scholar]
- Sultan SE. 2007. Development in context: the timely emergence of eco-devo. Trends in Ecology & Evolution 22: 575–582. [DOI] [PubMed] [Google Scholar]
- Sultan SE, Bazzaz FA. 1993. Phenotypic plasticity in Polygonum persicaria. I. Diversity and uniformity in genotypic norms of reaction to light. Evolution 47: 1009–1031. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nijhout HF. 2006. Evolution of a polyphenism by genetic accommodation. Science 311: 650–652. [DOI] [PubMed] [Google Scholar]
- Taylor MB, Ehrenreich IM. 2014. Genetic interactions involving five or more genes contribute to a complex trait in yeast. PLoS Genetics 10: e1004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Sigal N, Assia Y, Barkai N. 2010. Chromatin regulators as capacitors of interspecies variations in gene expression. Molecular Systematic Biology 6: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibert-Plante X, Hendry AP. 2011. The consequences of phenotypic plasticity for ecological speciation. Journal of Evolutionary Biology 24: 326–342. [DOI] [PubMed] [Google Scholar]
- Travis J. 2009. Phenotypic plasticity. In: Levin SA, ed. The Princeton guide to ecology. Princeton: Princeton University Press. [Google Scholar]
- True HL, Lindquist SL. 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483. [DOI] [PubMed] [Google Scholar]
- Vierstra J, Wang H, John S, Sandstrom R, Stamatoyannopoulos JA. 2014. Coupling transcription factor occupancy to nucleosome architecture with DNase-FLASH. Nature Methods 11: 66–72. [DOI] [PubMed] [Google Scholar]
- de Visser JA, Hermisson J, Wagner GP, et al. 2003. Perspective: evolution and detection of genetic robustness. Evolution 57: 1959–1972. [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565. [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1952. Selection of the genetic basis for an acquired character. Nature 169: 278. [DOI] [PubMed] [Google Scholar]
- Waddington CH. 1953. Genetic assimilation of an acquired character. Evolution 7: 118–126. [Google Scholar]
- Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews. Genetics 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ. 1986. Alternative adaptations, speciation, and phylogeny. Proceedings of the National Academy of Sciences of teh USA 83: 1388–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ. 1989. Phenotypic plasticity and the origins of diversity. Annual Review of Ecology and Systematics 20: 249–278. [Google Scholar]
- West-Eberhard MJ. 2003. Developmental plasticity and evolution. New York: Oxford University Press. [Google Scholar]
- West-Eberhard MJ. 2005. Developmental plasticity and the origin of species differences. Proceedings of the National Academy of Sciences of the USA 102: 6543–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman DW, Agrawal AA. 2009. What is phenotypic plasticity and why is it important? In: Whitman DW, Ananthakrishnan TN, eds. Phenotypic plasticity of insects. Enfield, NH: Science Publishers. [Google Scholar]
- Williams GC. 1966. Adaptation and natural selection. Princeton: Princeton University Press. [Google Scholar]
- Windig JJ, De Kovel CGF, De Jong G. 2004. Genetics and mechanics of plasticity. In: DeWitt TJ, Scheiner SM, eds. Phenotypic plasticity. Oxford: Oxford University Press. [Google Scholar]
- Wittkopp PJ, Kalay G. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nature Reviews. Genetics 13: 59–69. [DOI] [PubMed] [Google Scholar]
- Wray GA, Hoekstra HE, Futuyma DJ, et al. 2014. Does evolutionary theory need a rethink? No, all is well. Nature 514: 161–164. [DOI] [PubMed] [Google Scholar]
- Wund MA, Baker JA, Clancy B, Golub JL, Foster SA. 2008. A test of the “flexible stem” model of evolution: ancestral plasticity, genetic accommodation, and morphological divergence in the threespine stickleback radiation. American Naturalist 172: 449–462. [DOI] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, et al. 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nature Genetics 35: 57–64. [DOI] [PubMed] [Google Scholar]
- Zhao J, Davis LC, Verpoorte R. 2005. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnology Advances 23: 283–333. [DOI] [PubMed] [Google Scholar]