Abstract
Background Discussions of phenotypic robustness often consider scenarios where invariant phenotypes are optimal and assume that developmental mechanisms have evolved to buffer the phenotypes of specific traits against stochastic and environmental perturbations. However, plastic plant phenotypes that vary between environments or variable phenotypes that vary stochastically within an environment may also be advantageous in some scenarios.
Scope Here the conditions under which invariant, plastic and variable phenotypes of specific traits may confer a selective advantage in plants are examined. Drawing on work from microbes and multicellular organisms, the mechanisms that may give rise to each type of phenotype are discussed.
Conclusion In contrast to the view of robustness as being the ability of a genotype to produce a single, invariant phenotype, changes in a phenotype in response to the environment, or phenotypic variability within an environment, may also be delivered consistently (i.e. robustly). Thus, for some plant traits, mechanisms have probably evolved to produce plasticity or variability in a reliable manner.
Keywords: Noise, stochasticity, canalization, plasticity, instability, variability, robustness.
INTRODUCTION
Robustness is often defined as the ability of an organism to produce a constant phenotype in the face of environmental, genetic and stochastic perturbations (Debat and David, 2001; Lempe et al., 2013). Waddington coined a similar term, canalization, to describe how members of a population may have the same phenotype despite environmental and genetic variation (Waddington, 1942). Plastic traits, which vary with the environment, are sometimes described as uncanalized and may be considered to result from a lack of robustness (Wagner et al., 1997; Debat and David, 2001). Likewise, traits that are highly variable within an environment are often considered to be a result of poorly constrained development (Debat and David, 2001). However, as sessile organisms, sensitivity to the environment is an essential feature of plant development, and a number of well-characterized plastic responses allow plants to tune their phenotype reliably to prevailing environmental conditions. For example, in response to shading from neighbours, some species undergo increased stem elongation in a well-characterized shade avoidance response (Ballaré et al., 1987). Additionally, there are scenarios where increased phenotypic variability within a single environment may be advantageous. For instance, in environments with unpredictable weather patterns, there is evidence that increased variability in the timing of germination is adaptive in some species (Venable, 2007; Simons, 2009). Therefore, mechanisms that reproducibly (i.e. robustly) deliver increased phenotypic variance between or within environments may be under selection. Here we argue that it is useful to categorize phenotypes based on their variability within and between environments, rather than based on the narrower concept of robustness.
We use three terms to describe different types of phenotype: invariant, plastic and variable (Table 1). Throughout the text we use the term ‘phenotype’ to refer to characteristics of specific traits (e.g. stem length, number of branches and flower colour) rather than to describe the composite of a plant’s characteristics.
Table 1.
Working definitions used throughout the text
| Phenotype: the result of development of a specific trait, encoded by a particular genotype, which may be influenced by the environment. |
| Invariant phenotype: the phenotype produced by a genotype is relatively constant (low variance), and insensitive to differences in the environment. |
| Phenotypic plasticity: the phenotype produced by a genotype changes reproducibly in response to a difference in the environment [see Gianoli and Valladares (2012) for a discussion of a broader definition relevant for ecological studies]. |
| Variable phenotype: the phenotype produced by a genotype differs between individuals in the same environment. |
| Bet-hedging: A strategy that reduces the variance in fitness over time and comes at a cost of a reduced arithmetic mean fitness. A fraction of individuals of a particular genotype have a non-optimal phenotype in the mean environment, and the presence of this phenotype allows survival of at least some of the population in unpredictable and extreme conditions. |
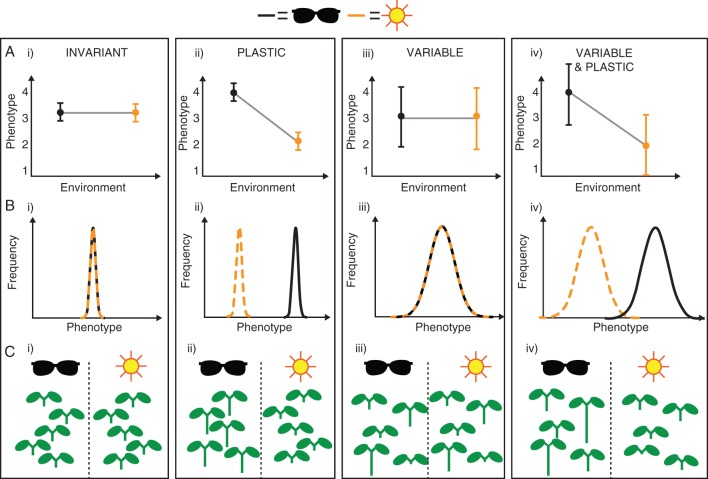
For a given genotype, an invariant phenotype is relatively constant and insensitive to differences in the environment (Table 1; Fig. 1). On the other hand, a plastic phenotype changes reproducibly in response to a change in the environment (Fig. 1). A variable phenotype varies substantially even between genetically identical individuals within the same environment (Fig. 1). This may be due to properties of regulatory networks underlying development of the trait which amplify stochasticity (see text below and Fig. 2). Within a given environment, plastic phenotypes may be more or less variable (Fig. 1).
Fig. 1.
—Types of phenotypic variation. (A) Expected change in the mean and variance between environments for the different types of phenotypic variation. Black dots and error bars indicate a shaded environment; orange indicates full sunlight. Error bars represent the true variance, as if the whole population was sampled. (B) Frequency distributions of phenotypes in the two environments. (C) Illustration of the four types of phenotypic variation, using hypocotyl length as an example phenotype. (i) An invariant response to the presence of shade/sunlight, such as that shown by shade-avoiding species which exhibit little phenotypic plasticity in response to shading (Gommers et al., 2013). (ii) A plastic shade avoidance response where hypocotyl elongation is triggered upon perception of shading (Pierik and de Wit, 2014). (iii) A variable hypocotyl length phenotype that varies within each environment but is not affected by shading. (iv) A variable and plastic hypocotyl length phenotype, which varies both within and between environments.
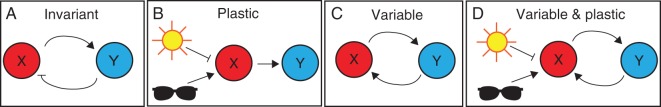
Fig. 2.
Network motifs underlying different types of phenotypic variation. (A) A negative feedback loop may restrict fluctuations in a protein’s levels and thereby contribute towards the development of an invariant phenotype. For example, protein X may cause the production of protein Y, which downregulates production of X, restricting fluctuations in X’s levels. (B) Phenotypic plasticity may be generated when environmental signals (here sunlight and shade) feed into gene-regulatory networks. In the scenario of shade avoidance, X represents phytochrome-interacting factors (PIFs), while Y represents their downstream transcriptional targets. Perception of full light by activation of phytochrome B (PhyB) causes inactivation or degradation of PIFs (X) (inhibitory arrow), preventing their downstream targets (Y) being activated. In shade, PhyB is inactive, allowing PIF (X) activity (arrow promoting X) and activation of their downstream targets (arrow promoting Y), leading to a change in phenotype. (C) A positive feedback loop can generate a bistable system where stochastic fluctuations in one protein’s levels can cause the noisy transition to a new state of gene expression, leading to intercellular variability in the timing of the transition. (D) If an environmental signal regulates the expression of a component of a positive feedback motif, the phenotype may be both variable within an environment and environmentally sensitive.
For each of these phenotypic categories, we discuss the ecological scenarios under which it might be advantageous. In addition, drawing from a large body of work on unicellular organisms and from work on variability and plasticity in multicellular systems, we review the mechanistic principles that could underlie the development of each type of phenotype.
In addition to developmental mechanisms of plasticity and variability, the ability of a given genotype to produce multiple phenotypes may be attributed to heritable epigenetic changes which can be influenced by the environment and cause transgenerational changes in gene expression (Richards, 2006; Bossdorf et al., 2008). By mediating maternal effects on offspring gene expression, such transgenerational epigenetic mechanisms are likely to cause differences between families of individuals. An in-depth discussion of specifically epigenetic mechanisms underlying phenotypic variation is beyond the scope of this review. Here we focus on mechanisms that could generate phenotypic variability even between genetically identical individuals descended from the same parents.
INVARIANT PHENOTYPES
Although plant development is remarkably plastic, some phenotypes are relatively constant, even between different environments. Early on in arabidopsis development, embryogenesis consistently generates mature embryos with the same basic body plan including correctly positioned shoot and root apices and two cotyledons (Park and Harada, 2008). This sets the foundation for growth and patterning at later stages of development which are often more environmentally sensitive. Flower patterning and morphology are also often relatively invariant. For example, the numbers of each type of floral organ (sepals, petals, stamens and carpels) are remarkably constant between individuals (Baker et al., 2005) and environments. The relative invariance of flower development may help to ensure that the different organs of the flower remain co-ordinated in shape and size throughout flower development, allowing protection of the reproductive organs and successful pollination. Invariance in flower morphology is also likely to be important from an ecological perspective. For example, within species that rely on a specific animal pollinator, individuals consistently generate specific flower morphologies that are recognized by specific pollinators, and, in some cases, restrict entry to the desired pollinator (Whitney and Glover, 2007). In this scenario, invariance in flower morphology and patterning is likely to be important for successful attraction, entry and pollination by insects. Invariant phenotypes may also be selected for in extreme and relatively invariant environments where a consistent and environmentally insensitive phenotype is likely to be favourable. For example, in contrast to shade-avoiding species that exhibit plasticity in stem length in response to shading, shade-tolerant species have relatively invariant stem lengths, regardless of light quality. A short, invariant stem length is hypothesized to reduce carbon wastage due to futile stem elongation and maintenance in scenarios where stem elongation will not enable light capture (Givnish, 1988; Valladares and Niinemets, 2008). Such invariant phenotypes are likely to have evolved under stabilizing selection that removes genotypes giving rise to deviations away from an optimum phenotype (Gibson and Wagner, 2000).
Mechanisms of minimizing variability
The development of invariant phenotypes is probably controlled by mechanisms that limit the effects of environmental variation and stochasticity on the final phenotype. As most of plant development is post-embryonic, developing organs are often exposed to environmental fluctuations much larger than those that normally occur during the development of many animal species (Lempe et al., 2013). Additionally, the molecular processes of transcription and translation are inherently noisy and can generate random differences in protein levels between cells (Raj and van Oudenaarden, 2008). Stochastic differences between cells can also be generated by spontaneous epi-mutations that alter the epigenetic regulation of gene expression (Blewitt et al., 2004). Noise may also arise independently of differences in gene expression during the allocation of cellular components to daughter cells during cell division. If components such as proteins and organelles are present at low copy numbers per cell, their stochastic allocation to daughters will lead to variability in a cell’s offspring (Berg, 1978; Oates, 2011). When all these processes are considered, the development of an invariant trait is clearly non-trivial. Such processes also have the potential to disrupt the generation of consistent phenotypic changes in response to specific environmental cues during the development of plastic phenotypes.
The extent of variability in development is under genetic control, and, in arabidopsis and maize, quantitative trait loci (QTL) controlling the extent of variability within individual environments have been mapped (Hall et al., 2007; Ordas et al., 2008; Jimenez-Gomez et al., 2011; Shen et al., 2012; Joseph et al., 2015). These studies have shown that some loci influence phenotypic variance without affecting the mean phenotype, and that distinct loci influence the variance of different phenotypes (Joseph et al., 2015). This suggests that specific molecular mechanisms exist to regulate the variability of particular traits.
Developmental studies addressing how phenotypes can be buffered against environmental and stochastic influences are beginning to emerge, and a large body of work on microbes has elucidated mechanisms that can promote cellular homogeneity by limiting the effects of noise in gene expression. Below, we discuss sources of noise in gene expression and how the architecture of gene regulatory networks, and the regulatory environment in which they operate, may buffer against stochasticity and environmental variability. Such mechanisms are likely to be important in the development of both invariant and plastic phenotypes. In some cases, the mechanisms discussed might also maintain invariant phenotypes in the face of genetic variation, although this is not covered here (Stearns et al., 1995; Rutherford and Lindquist, 1998; Gibson and Wagner, 2000; Meiklejohn and Hartl, 2002; Queitsch et al., 2002; Fraser and Schadt, 2010; Lempe et al., 2013).
Sources of noise in gene expression
Changes in gene expression that occur in response to spatial and temporal cues form the core of most developmental mechanisms. However, the small numbers of molecules that are involved in these processes make them inherently noisy (Li and Xie, 2011). Transcription initiation occurs stochastically due to a low copy number of genes per cell, and the random arrival of transcription factors at promoters (Blake et al., 2003; Raser and O’Shea, 2004; Newman et al., 2006). Following initiation of transcription, a burst of mRNA production may occur (Golding et al., 2005; Blake et al., 2006; Chubb et al., 2006; Raj et al., 2006; Raj and van Oudenaarden, 2008). Each mRNA molecule can then be translated multiple times, resulting in a burst of protein production following the stochastic initiation of transcription (Ozbudak et al., 2002; Fraser et al., 2004; Yu et al., 2006).
Stochasticity in protein production not only means that an individual cell will vary over time, but also that at a given time in development, cells within a population will be heterogeneous in their protein levels (Lander, 2011). This could disrupt developmental processes when groups of cells are required to respond to upstream signals. Additional variability can arise since, when a protein regulates the expression of downstream genes, fluctuations in its levels can propagate through the gene-regulatory network (Thattai and van Oudenaarden, 2001; Becskei et al., 2005; Hooshangi et al., 2005). Stochasticity may even be amplified in gene expression cascades where multiple noisy components are placed in series (Lander, 2011). Noise can also disrupt the cellular memory of previously received cues (Acar et al., 2005). Thus, stochastic fluctuations in gene expression have the potential to disrupt developmental signalling and increase phenotypic variability between individuals.
These stochastic fluctuations in protein levels can be minimized if rates of transcription are relatively high and the rates of translation per mRNA are low (e.g. due to elevated mRNA degradation) since this prevents bursts of proteins being generated from rarely produced mRNAs (Ozbudak et al., 2002; Fraser et al., 2004). Consistent with the relative rates of transcription and translation being regulated to limit stochastic effects, in yeast, essential genes tend to have higher rates of transcription and lower rates of translation than non-essential genes which produce proteins at similar rates (Fraser et al., 2004). Protein turnover rates may also be regulated to reduce stochastic fluctuations. If a protein decays relatively slowly, its total level in the cell will be governed by the integral of its synthesis over time, allowing the effects of stochastic and environmental fluctuations on production rates to be averaged out (Raj and van Oudenaarden, 2008).
Buffering variability through regulatory architecture
The interactions between genes in a cell, or between gene-regulatory networks in different cells, may be structured to minimize variation in development despite stochastic or environmentally induced fluctuations in protein production.
Negative feedback
Negative autoregulation is a common motif in genetic networks and involves a protein feeding back to downregulate its own expression, either directly or through the production of a second regulatory protein (Fig. 2A) (Alon, 2007). This feedback may help to restrict fluctuations in a protein’s levels and reduce noise propagation through a network: if the protein’s levels become elevated, its production will decrease; but, if its levels drop, its production rate will increase (Becskei and Serrano, 2000; Thattai and van Oudenaarden, 2001; Nevozhay et al., 2009). However, due to the noise inherent in gene expression, the ability to suppress noise through negative feedback involving regulatory proteins is likely to be limited to cases where large numbers of regulatory molecules are involved (Lestas et al., 2010). This is because, in a system where protein X promotes the production of a regulatory protein (Y) which in turn downregulates X (Fig. 2A), the production of Y in response to X will be a stochastic event. Unless large numbers of Y molecules are made in response to X (minimizing stochastic effects), the total levels of Y are unlikely accurately to reflect the amount of X, leading to imperfect control of X by Y (Lestas et al., 2010). Also, if there is a time delay in the production of Y and its effect on X's expression, this could lead to oscillations in the levels of both proteins (Novák and Tyson, 2008).
Network redundancy
Many flowering plants have large genomes as a result of both small- and large-scale genome duplications, with two whole-genome duplications believed to have occurred during the evolution of angiosperms (Cui et al., 2006; Vanneste et al., 2014). As a consequence, many genes have multiple paralogues, which, in some cases, have at least partly overlapping functions (Veitia, 2005). Such redundancy can make networks resistant to mutation. For example, a number of families of transcription factors that each have multiple redundant members contribute to the development of distinct upper (adaxial) and lower (abaxial) surfaces of leaves and other plant lateral organs (McConnell and Barton, 1998; Eshed et al., 2001; McConnell et al., 2001; Emery et al., 2003; Yamaguchi et al., 2012). Adaxial and abaxial identity determinants mutually inhibit each other to specify precisely an adaxial–abaxial boundary along the mid-plane of the leaf. Loss-of-function mutations in single components rarely cause a complete loss of patterning, indicating that the network is robust to changes in the levels of its components (Eshed et al., 2001; Emery et al., 2003; Husbands et al., 2009). The presence of redundant copies of the transcription factors involved could also reduce variability due to stochastic effects. If multiple transcription factors perform a common function, stochastic bursts in the production of each individual protein would be buffered at the level of downstream gene expression. This is because this output would be proportional to the total level of all the transcription factors at a given time, allowing the effects of stochastic bursts in the production of each one to be averaged out across the different proteins.
Intercellular signalling
The development and correct behaviour of multicellular structures requires that the gene regulatory networks operating in individual cells can be co-ordinated and regulated to achieve a reliable tissue-scale output. This can be achieved through intercellular signalling which links the behaviour of the gene-regulatory networks operating in each cell to its environment within the tissue.
Co-ordination of individual cells to achieve a reliable tissue-scale output
Intercellular signalling may allow the dynamics of intracellular processes to be synchronized across a tissue. This occurs in the mammalian suprachiasmatic nucleus (part of the hypothalamus) where neurons show circadian oscillations in firing rates. If these cells are dissociated, their circadian rhythms become desynchronized (Welsh et al., 1995). Somewhat similarly, in arabidopsis leaves, the presence of spatio-temporal waves of circadian clock gene expression suggests that intercellular coupling helps to co-ordinate the circadian clocks of individual cells (Wenden et al., 2012). A similar phenomenon of intracellular coupling was observed in the context of somite development in the vertebrate mesoderm. The segmentation of the mesoderm into somites is controlled by oscillations in gene expression that provide temporal cues for somite formation, referred to as the segmentation clock (Palmeirim et al., 1997). If mesodermal cells are dissociated from each other, the oscillations within individual cells become unstable, with highly variable amplitudes (Masamizu et al., 2006). These examples suggest that signalling between cells synchronizes and stabilizes their oscillations, which would otherwise be noisy.
In contrast to the generation of tissue-scale oscillations, the maintenance of a relatively constant average level of gene expression within a tissue may sometimes be best achieved by regulatory networks that prevent synchronization of groups of cells. This has been proposed to be the case for nuclear factor-κB (NF-κB) signalling, which plays an important role in inflammation (Paszek et al., 2010). Upon perception of cytokines (paracrine signalling factors), the NF-κB transcription factor oscillates between being located in the cytoplasm and in the nucleus (Nelson et al., 2004) where it influences the expression of downstream genes, including those encoding cytokines (Tian et al., 2005; Ashall et al., 2009; Paszek et al., 2010). If neighbouring cells were co-ordinated in their perception and production of cytokines, this system would provide the potential for positive feedback (between the production of cytokines and NF-κB expression), leading to a strong inflammatory response. However, through computational modelling, it was shown that the signalling network involved appears to be optimal for giving rise to NF-κB oscillations that are unco-ordinated between cells (Paszek et al., 2010). It has been proposed that by desynchronizing the oscillations of individual cells, the gene-regulatory network restricts the extent of positive feedback and prevents large fluctuations in amounts of cytokines at the tissue level (Paszek et al., 2010).
Generation of long-range patterns in the face of noise
Intercellular signalling via morphogen gradients plays an important role in large-scale developmental patterning by generating reproducible differences in gene expression across a tissue (Lander, 2011). The classical model of a morphogen gradient involves a graded signal inducing separate domains of gene expression in a field of cells that respond to threshold concentrations of the signal (Wolpert, 1969). To generate boundaries of gene expression that are reproducible between individuals, neighbouring cells must consistently be able to perceive differences in morphogen concentration and induce different gene expression states (Gregor et al., 2007). Therefore, stochastic fluctuations in morphogen concentrations and in the ability of cells to perceive the morphogen are likely to impact on the precision of this process. However, a number of different strategies appear to be employed in plant and animal development to generate invariant outcomes from graded intercellular signals.
Early in Drosophila embryogenesis, a maternally controlled anterior–posterior gradient in Bicoid (Bcd) protein activates zygotic expression of hunchback (hb) in a concentration-dependent manner (Driever and Nusslein-Volhard, 1988a, b; Struhl et al., 1989). hb expression is activated in the anterior half of the embryo, where the Bcd concentration is highest. The relationship between Bcd and Hb protein concentrations is remarkably precise despite a relatively high degree of stochasticity in the transcription of hb (Gregor et al., 2007; Little et al., 2013). This has been proposed to be due to spatial and temporal averaging of Bcd concentrations during the control of hb expression (Gregor et al., 2007; Little et al., 2013). hb transcripts have a relatively long lifetime and therefore Hb protein levels are a read-out of the cumulative production of hb mRNAs over time. As described above, time averaging of hb transcription in this way has the potential to reduce the effects of stochastic transcription initiation and noise in Bcd concentration (Little et al., 2013). Also, as the embryo is a syncitium at this stage of development, averaging may occur through short-range diffusion of Hb between neighbouring nuclei, which can smooth out fluctuations in Bcd and Hb levels and yield more precise boundaries (Erdmann et al., 2009). Boundaries specified by the Hb morphogen gradient appear then to be further refined by cross-regulation between gap genes induced in neighbouring tissue domains (Jaeger, 2011; Gursky et al., 2012).
Interestingly, plants appear to employ a unique strategy to overcome the disruptive effects of noise on the sensing of gradients: they maximize the steepness of the gradient over the whole region of the tissue in which patterning occurs. This allows them to solve the problem of generating large-scale positional information in the face of stochastic perturbations, since the difference in signal between adjacent cells will be larger than the noise. This appears to be the case in the arabidopsis root, where an auxin concentration gradient, with its maximum at the root tip, influences specification of the boundaries between different zones of cellular behaviour (Aida et al., 2004; Blilou et al., 2005; Galinha et al., 2007; Grieneisen et al., 2007; Petersson et al., 2009). Rather than being generated through a localized source combined with global decay (as is the case for many morphogen gradients in animals), the auxin gradient is generated through a reflux loop of auxin transport created by the pattern of polarly localized PIN proteins (auxin efflux carriers) in the root (Blilou et al., 2005; Grieneisen et al., 2007). In the internal tissue of the root, auxin is transported towards its tip, while, in outer tissue, transport is directed back up towards the shoot. In intermediate cell layers, PIN proteins are localized inwards, returning auxin back to inner tissue from the outer cell layers (Blilou et al., 2005). Taking the known biophysical parameters involved in auxin transport into account, the shape and steepness of the auxin gradient generated through the reflux loop mechanism was compared with other possible mechanisms, such as source decay or unidirectional transport towards the root tip (Grieneisen et al., 2012). This revealed that the auxin transport reflux loop is probably the most effective mechanism for generating the observed steep auxin gradient over the large spatial range of the root.
Parallel patterning systems
Another approach to achieve invariant tissue patterning is to use two or more signalling systems in parallel to provide positional information. Indeed, recent work on phyllotaxis revealed that plants achieve relatively reproducible spacing of successive organs through the action of two interacting signalling systems, one involving polarized transport of auxin, and the other involving diffusion of a cytokinin signalling inhibitor (Besnard et al., 2014). The auxin efflux carrier, PIN1, forms polarity convergence points with high intracellular auxin that specify the position of each new organ in the shoot apical meristem (Reinhardt et al., 2000, 2003). The polarized transport of auxin towards existing primordia is predicted to cause depletion of auxin in surrounding tissue, meaning that new primordia can only be initiated at a distance from existing ones where the auxin concentration becomes sufficiently high (Jönsson et al., 2006; Smith et al., 2006; Stoma et al., 2008). However, in the presence of variability in auxin concentrations and primordium size and upon increases in meristem size, the positional information generated by this mechanism may be ambiguous, causing the next two sites for organ initiation to be indistinguishable (Mirabet et al., 2012; Landrein et al., 2014). This can cause pairs of organs to initiate simultaneously or the order of organ initiation to be swapped. Mutations in AHP6, a cytokinin signalling inhibitor, cause an increased frequency of these aberrations, suggesting that this second signal contributes to the invariance of phyllotaxis (Besnard et al., 2014). Indeed, accumulation of auxin in new primordia activates AHP6 expression, and intercellular movement of AHP6 generates a graded distribution around each young primordium. This additional layer of information can be used to distinguish between the next two sites for primordium initiation, decreasing the variability of phyllotactic patterning.
Regulating effects of intercellular variability on organ growth
In the context of growth, intercellular signalling may ensure that a precise final state is reached despite stochasticity throughout development. For example, a signal that terminates growth may be increasingly produced in a tissue as it approaches its final state, ensuring an accurate final outcome. Such a mechanism functions in the mouse olfactory epithelium, where stem cells divide to produce progenitors that pass through distinct lineage stages before differentiating into mature neurons (Lander et al., 2009; Lander, 2011). Signalling molecules are produced by differentiated cells that feed-back to inhibit the self-renewal of progenitor cells, terminating proliferation at the correct time. A model of such a system was shown to produce a reproducible final outcome even in the face of variation in cell cycle speeds and initial stem cell number (Lander et al., 2009). A role for non-cell-autonomous signals in regulating the final size of determinate plant organs is indicated by the phenomenon of compensation, whereby mutants with reduced cell division still reach a similar final organ size by adjusting the rates or duration of organ expansion (Ferjani et al., 2007; Powell and Lenhard, 2012). The mechanism underlying compensation could contribute to the consistent generation of similar organ sizes in the face of stochastic variations in individual cell behaviour.
A role for tissue stresses in regulating growth heterogeneity has also been proposed. Heterogeneous growth of neighbouring regions of tissues is predicted to create local stresses. If these were sensed and used to influence cellular behaviour, this could in turn influence growth variability (Shraiman, 2005). Such feedback appears to occur in plants where cells orient their microtubules parallel to the local principle direction of stress (Hamant et al., 2008). As a consequence, cell walls become reinforced along the principle direction of stress as microtubules guide the synthesis of cellulose microfibrils (Lloyd and Chan, 2008). Thus, growth in this direction is expected to be restricted (Hamant et al., 2008). Via computational modelling, it was shown that when cell-autonomous growth rates (the rate at which a cell would grow if removed from the tissue) are variable, such mechanical feedback can reduce overall growth rate variability in the tissue (Uyttewaal et al., 2012). However, if the strength of feedback is increased, it can effectively overshoot and amplify differences in growth rate between regions. Surprisingly, this latter scenario seems to be the case in the shoot apical meristem, where an apparent reduction in mechanical feedback (in a mutant with a reduced ability to align its microtubules with the principle direction of stress) causes reduced growth rate variability (Uyttewaal et al., 2012). In the meristem, amplified growth variability due to mechanical feedback was proposed to promote organ emergence (Uyttewaal et al., 2012). However, little is known about the effects of mechanical feedback on growth heterogeneity in other tissues.
Regulatory environment
The examples discussed above illustrate how changes in the properties of specific gene-regulatory networks can influence the variability of their output on a multicellular scale. However, there are other more general processes that may reduce variability across a large number of different gene-regulatory networks in parallel.
Chaperone proteins
Chaperone proteins bind to a number of target proteins and influence their stability. The HSP90 chaperone binds to developmental regulators and other signalling proteins when they are in unstable states. When bound, the chaperone prevents its target protein from misfolding until a correct signalling event occurs and triggers a conformational change (Zuehlke and Johnson, 2010). Disruption of HSP90 function in Drosophila or arabidopsis leads to increased variability in many phenotypes, suggesting that its role in protein stability contributes to invariance in a number of developmental processes (Rutherford and Lindquist, 1998; Queitsch et al., 2002).
Recent work on yeast suggests that, within highly connected networks, there are many other genes that play a similar role to HSP90 in reducing variability. When thousands of haploid yeast single-gene knockout strains were screened for their phenotypic variance, about 5 % of genes were required for achievement of a wild-type level of phenotypic variability, causing increased variability when knocked out (Levy and Siegal, 2008). A large proportion of these genes was involved in core cellular processes such as chromosome organization, mRNA processing and protein modification. Also, their products tended to be highly connected within protein–protein interaction networks, suggesting that they may have general functions with many client proteins, as is the case with HSP90.
Physical constraints and mechanical feedback
In plant tissues, neighbouring cells are connected via cell walls and therefore cannot slide relative to each other. This connectedness of tissues constrains the growth of each local region so that each cell or local region of the tissue may not grow at the same rate as it would if it were in isolation (Coen et al., 2004). The constraints imposed by such mechanical connectivity can restrict the overall shape changes occurring in a tissue as a result of locally altered growth (Boudon et al., 2015) and therefore could result in locally variable growth being smoothed out on the tissue scale, increasing homogeneity. In contrast to the mechanism of mechanical feedback discussed above, this could occur without mechanical forces being sensed by cells to alter directly specific aspects of their behaviour. This mechanism would therefore act as a more general buffer for growth variability.
Slowing of growth in adverse conditions
An important factor influencing whether plant phenotypes can be buffered against environmental variability appears to be the effect of stress on growth rate (Achard et al., 2006). The perception of multiple types of stress can lead to a generally reduced growth rate throughout the plant via increased levels of DELLA proteins, which repress the activity of growth-promoting transcription factors (Achard et al., 2006; Harberd et al., 2009). This DELLA-dependent mechanism of stress-induced growth reduction was shown to increase survival in stressful environments (Achard et al., 2006). There are several ways in which slow growth might confer stress resistance. One possibility is that by slowing down growth in adverse environmental conditions, resource consumption by growth is temporarily reduced, allowing resources to be diverted towards homoeostatic mechanisms that counteract the effect of stress (Harberd et al., 2009). Alternatively, slow growth may be inherently less sensitive to stress and stochasticity because of the buffering provided by integrating regulatory processes over longer periods of time. Slow growth could therefore contribute to the development of invariant phenotypes by reducing the phenotypic effects of potentially detrimental environmental fluctuations.
In summary, there are many mechanisms that might contribute to the development of invariant phenotypes by generating reproducible developmental outputs despite stochastic effects. Since environmental changes probably cause changes in gene expression, developmental mechanisms that confer robustness to stochastic changes in gene expression may also mitigate the effects of environmentally induced changes (Meiklejohn and Hartl, 2002). A number of these mechanisms relate to properties of specific gene-regulatory networks controlling developmental processes. Such networks may include negative feedback to reduce stochastic bursts in protein levels. The presence of paralogous genes with similar functions may also reduce stochasticity at the level of particular protein functions. At the tissue level, the gene-regulatory networks of individual cells may be coupled via intercellular signalling or maintained in a decoupled state to reduce variability across the tissue. During patterning, noise in developmental signals and their perception may be compensated for by employment of spatio-temporal averaging. The generation of steep signal gradients and the use of parallel patterning systems may also help to provide unambiguous positional information. Additionally, the regulatory context may act to reduce variability by providing a more generalized buffering of a number of different developmental processes. Chaperone proteins, physical constraints of growing tissues and the regulated repression of growth to aid the maintenance of homeostasis may each restrict the phenotypic variability arising from a number of different developmental pathways. These aspects of the regulatory environment might be seen as imposing a 30 mph speed limit on a number of roads in parallel to prevent accidents.
PHENOTYPIC PLASTICITY
The development of many plant phenotypes changes in response to the environment. For some traits, plasticity may be an adaptive response and involve signalling pathways that were selected to cause a specific change in development in response to the environment. However, not all plastic responses are adaptive, and some may be due to the inevitable consequences of resource limitation or stress on growth and physiology (Sultan, 2000; van Kleunen and Fischer, 2005). Many instances of phenotypic plasticity appear to be functionally appropriate and therefore examples of the adaptive scenario (Sultan, 2000). Here we illustrate types of molecular mechanisms that can underlie adaptive plasticity using two well-characterized examples in plants: shade avoidance and the control of flowering time by the perception of winter. We then review some empirical evidence for the adaptive significance of plastic phenotypes. Finally, we explore the relationship between plasticity and variability and discuss the benefits, limitations, and costs involved in the evolution of phenotypic plasticity, illustrating that under certain conditions, invariant or variable phenotypes may be favoured over plastic ones.
Developmental mechanisms of plasticity
Plants often change their development in response to cues that are likely to vary spatially in their natural environment and impact upon fitness. One well-studied example is the avoidance of shading. When plants are present at high densities, competition for light can reduce their growth and fitness (Schmitt and Wulff, 1993; Schmitt et al., 1999). Plants are able to use the red to far-red light ratio to predict future shading, as nearby neighbours (which do not yet cause shading) absorb red light but reflect far-red light (Ballaré et al., 1987). In response, early flowering and a range of morphological changes can be triggered that increase chances of light capture, including increased elongation of stems and reduced branching (Schmitt and Wulff, 1993; Ruberti et al., 2012).
Plants employ a number of different photoreceptors to perceive their light environment and control development accordingly (Fig. 2B). The phytochrome B clade of photoreceptors is the most important in perception of the red to far-red light ratio, which is indicative of the degree of shading (Casal, 2013). Active phytochrome B physically interacts with the PHYTOCHROME INTERACTING FACTOR (PIF) family of transcription factors, causing either their degradation or inactivation. Increased far-red light promotes the conversion of phytochrome B to an inactive form, which allows PIF levels to increase rapidly and enables their binding to target promoters. PIFs control the expression of a number of genes required for shade avoidance, including cell wall-modifying enzymes and auxin biosynthetic enzymes. Indeed, many of the phenotypes associated with shade avoidance, including reduced branching, stem elongation and upwards bending of leaves, are characteristic of elevated auxin levels. Phytochrome B also feeds more directly into the pathways influencing flowering and branching (Jang et al., 2008; Finlayson et al., 2010). Thus, in the case of shade avoidance, phenotypic plasticity is caused by a specific mechanism of environmental perception that feeds into core developmental pathways.
As well as changing development in response to cues that may be spatially heterogeneous, plants use environmental cues to control the timing of key life history events such as flowering time and germination. Flowering time is influenced by a number of seasonal cues including daylength, temperature and prolonged periods of cold, as well as responding to indicators of unfavourable conditions such as salinity, shading and a lack of nutrient availability (de Jong and Leyser, 2012; Kim and Sung, 2014). Following their perception, these cues converge to influence the expression of three floral integrator genes. These integrators control flowering time by regulating expression of floral meristem identity genes, which when expressed result in a switch from vegetative to reproductive development (de Jong and Leyser, 2012; Kim and Sung, 2014).
One of the best characterized pathways influencing flowering time is the vernalization pathway, which promotes flowering in response to prolonged periods of cold (Ietswaart et al., 2012; Song et al., 2012; Kim and Sung, 2014). Arabidopsis accessions differ in their requirement for vernalization, and this largely depends on genetic variation at the FRIGIDA (FRI) locus. FRI promotes the expression of a floral repressor, FLOWERING LOCUS C (FLC), which becomes stably downregulated in the cold. Accessions from northern Europe tend to have functional alleles of FRI, and therefore require vernalization to flower. These winter annual accessions tend to over-winter in a vegetative state (when vernalization occurs) and then flower the following year in response to increasing daylength. For this to occur, FLC downregulation must be mitotically stable for several months, providing a memory of perceived winter.
Recent work has shown that FLC expression is modulated by multiple regulators to ensure stable and robust downregulation following winter (Ietswaart et al., 2012; Song et al., 2012). After around 2 weeks of cold treatment, FLC undergoes a decrease in expression levels. This appears to involve a number of overlapping pathways that recruit Polycomb repressive complex 2 (PRC2), a highly conserved chromatin-modifying complex, to the FLC locus (Heo and Sung, 2011; Song et al., 2012). Through the action of PRC2, continued cold causes a gradual build-up of repressive histone modifications at a particular region (termed the nucleation region) of the FLC locus. Upon return to warm conditions, repressive modifications can spread throughout the rest of the locus, and this spreading is required for the maintenance of FLC repression and memory of vernalization (Angel et al., 2011).
The stable silencing of FLC upon return to warm appears to be a probabilistic event in each cell: following a moderate duration of cold, only a small proportion of nuclei stably silence FLC, whereas a longer duration of cold allows a higher proportion of nuclei to establish stable silencing (Angel et al., 2011). The floral integrator FLOWERING-LOCUS T (FT ) is repressed by FLC and can act as a long-range signal. FT levels are therefore thought to provide an average read-out of FLC expression across a number of cells (Corbesier et al., 2007; Angel et al., 2011). Thus, the extent of flowering acceleration is proportional to the time spent in the cold and the proportion of nuclei that stably repress FLC expression. The slow and quantitative response of the FLC locus to cold means that several weeks of cold are needed for flowering to be significantly accelerated. This is likely to remove sensitivity to fluctuations in temperature occurring at short time scales (e.g. a one-off frost in autumn) but allows a robust plastic response upon perception of meaningful environmental cues.
Evidence for adaptive plasticity
As mentioned above, not all instances of plasticity will be adaptive, and, in fact, evidence for plasticity conferring a fitness advantage is relatively difficult to acquire. This is because it is necessary to show that the plastic phenotype confers an increased fitness across relevant environments relative to a non-plastic phenotype (Schmitt et al., 1999; Sultan, 2000). One way to do this is to compare the fitness of a wild-type plant, which shows a plastic response, with that of mutants or transgenic plants that lack the plasticity. This was successfully achieved for the shade avoidance response (Schmitt et al., 1995, 1999). Wild-type tobacco plants were sown at high and low density alongside transgenic tobacco plants overexpressing PHYTOCHROME A (PHYA) which are defective in the shade avoidance response. At low density, PHYA overexpressor and wild-type plants had comparable fitness, indicating that PHYA overexpression does not cause deleterious pleiotropic effects. However, at high density, wild-type plants outcompeted the PHYA overexpressors, indicating that shade avoidance at high density confers an advantage. In a complementary experiment, wild-type plants of Brassica rapa were grown alongside mutants deficient in phytochrome B, which have a constitutive shade avoidance response. These mutants were less fit than the wild type when grown at low densities. Thus, the plasticity of shade avoidance-related phenotypes in wild-type plants confers an advantage at both low and high densities.
In the case of plasticity in flowering time, the requirement for vernalization in winter annuals from northern Europe is likely to be adaptive. Evidence for this comes from a study of 360 arabidopsis accessions that were sown in a field site in Rhode Island (northern USA) in autumn or spring (Korves et al., 2007). When sown in autumn, accessions carrying a functional allele of FRI (conferring a vernalization requirement) had better survival over winter (due to delayed flowering) and therefore increased fitness compared with accessions with a non-functional FRI allele (which flowered too early and thus had reduced fitness). Therefore, for plants native to northern latitudes, which will experience cold winters, acceleration of flowering following a period of cold is likely to be adaptive. However, in this case, it is likely to be environmental sensitivity rather than plasticity per se which is adaptive. Any one plant germinating close to its parent plant is unlikely to be required to respond appropriately to diverse lengths and severities of winter. Thus the delay in flowering of vernalization-requiring accessions in the absence of winter may not be advantageous (Korves et al., 2007). Indeed, there is evidence that non-functional alleles of FRI have undergone multiple selective sweeps in central Europe, indicating that a lack of vernalization requirement (and thus a lack of plasticity) might be advantageous in the absence of very cold winters (Le Corre et al., 2002; Le Corre, 2005).
The relationship between plasticity and variability
Plastic phenotypes have sometimes been described as uncanalized against environmental variation, implying that plasticity results as a consequence of a lack of mechanisms conferring reliable developmental outcomes (Debat and David, 2001; Meiklejohn and Hartl, 2002). However, the shade avoidance and vernalization examples show that specific mechanisms have evolved that generate consistent and pre-emptive responses to the environment. In some cases, plasticity may therefore be considered to be robust.
However, sometimes plasticity of a phenotype may be correlated with the variability of this phenotype within an environment. A positive correlation was found within a set of arabidopsis recombinant inbred lines, where lines that showed more plastic responses to nitrate availability were also more variable within an environment (Tonsor et al., 2013). Consistent with these findings, in yeast, genes with highly plastic expression often show highly variable expression within a constant environment (Landry et al., 2007; Choi and Kim, 2009; Lehner, 2010). Genes with high expression variability both within and between environments tend to have specific mechanisms of gene expression regulation including the presence of a nucleosome in a specific regulatory region of the promoter, higher rates of histone exchange and the presence of TATA box elements in promoters (Choi and Kim, 2009; Lehner, 2010). However, at the gene expression level, plasticity and variability are not always correlated as essential genes tend to lack these mechanisms of gene expression regulation, but can still show plasticity whilst maintaining low variability (Lehner, 2010). Additionally, studies on Drosophila have not found a clear relationship between phenotypic plasticity and variability (Scheiner et al., 1991). In a study on seed germination in populations of Lobelia inflata, it was found that plasticity of germination timing in response to temperature was negatively correlated with the variability in germination time (Simons, 2014).
There are several mechanistic explanations for why plasticity and variability might sometimes be positively correlated. It may be the case that the development of some phenotypes is poorly constrained due to a lack of mechanisms buffering against environmental or stochastic perturbations. These phenotypes may therefore be variable within an environment and change in response to environmental fluctuations, showing plastic responses that are not necessarily adaptive. In other cases, the evolution of adaptive plasticity might lead to increases in variability. This could occur if the evolution of plasticity involves changes in regulatory architecture that allow an increased range of phenotypes to be produced in response to the environment. As a consequence of these changes, the phenotype may be less buffered against stochastic effects, leading to increased variability. Another possibility is that the evolution of sensitivity to meaningful environmental cues allowing a plastic response may, as a side effect, lead to increased sensitivity to microenvironmental cues, giving rise to increased variability within an apparently homogeneous environment. Alternatively, in some cases, separable mechanisms may give rise to increased plasticity and variability, and both may be favoured by selection in particular environments (see the section on variable phenotypes). In the case of the negative correlation between plasticity and variability seen in L. inflata seed germination, it has been proposed that this may be due to a constraint on the total amount of phenotypic variability for a specific trait that can be produced through the combination of plasticity or variability (Simons, 2014). To test these different possibilities, it is necessary to improve our understanding of the developmental mechanisms involved in phenotypic plasticity and variability and how these mechanisms change during the evolution of increased plasticity.
Evolution of adaptive plasticity
If plants could instantly change their development to suit best the prevailing environment this would appear to be an ideal strategy. Indeed, theoretical work shows that phenotypic plasticity increases fitness in spatially and temporally varying environments where environmental cues are good predictors of the optimal strategies in the future (Van Tienderen, 1997; Tufto, 2000; Givnish, 2002). Plasticity may also allow successful colonization of new or changed environments by allowing the persistence of a genotype in the altered conditions (Crispo, 2008). At early stages of colonization of a new environment, an increase in plasticity may be selected for and the plastic phenotype may become fixed (Ghalambor et al., 2007; Lande, 2009). If the plastic phenotype is sub-optimal, it may then be refined by natural selection operating in the new habitat (Pigliucci et al., 2006; Crispo, 2008; Lande, 2009). Consistent with a role for plasticity in successful colonization of new habitats, more invasive species tend to occupy a greater range of habitats, and have relatively high phenotypic plasticity (Baker, 1974; Williams et al., 1995; Sultan, 2003). However, despite the advantages of plasticity, many factors can constrain its evolution and influence the likelihood that a plastic phenotype is favoured over invariant or variable phenotypes in a given environment (DeWitt et al., 1998; Alpert and Simms, 2002; Givnish, 2002).
An inherent constraint to the evolution of plasticity is that genes conferring phenotypic plasticity, which are usually expressed only in certain environmental conditions, are likely to be under relaxed selection compared with genes expressed in all environments. This is because only the copies of the gene that are expressed will experience selection, whereas, if the inductive environment is never encountered, the gene will not be expressed, and its effect will be neutral. This means that advantageous alleles conferring a plastic phenotype will take longer to be fixed than those involved in the generation of an adaptive invariant phenotype (Snell-Rood et al., 2010). Thus, in environments where spatial or temporal variability occurs rarely (causing plasticity genes to be rarely expressed), non-plastic specialists may be favoured over generalists with relatively high plasticity in a number of traits.
Plasticity of a trait may be considered to have a cost if it causes a reduced mean fitness in a given environment compared with an invariant phenotype with the same mean trait value (DeWitt et al., 1998). Costs of plasticity could be related to detrimental pleiotropic or epistatic effects when a developmental process comes under environmental control. Because of this, increased modularity of developmental processes has been proposed to enable evolution of plasticity (Snell-Rood et al., 2010). If the development of different traits is controlled by relatively independent genetic networks, then a developmental module could evolve to become environmentally sensitive without disrupting other developmental processes. It has also been proposed that energetic costs associated with machinery that allows perception, integration and response to environmental cues could provide a cost to plasticity (DeWitt et al., 1998; Givnish, 2002), although it has been argued that these costs would probably be minimized by natural selection (Schlichting and Pigliucci, 1998; Givnish, 2002).
Another potential cost of plasticity is that the change in phenotype produced in response to a specific environmental cue might be detrimental in the presence of other co-occurring abiotic and biotic variables (Valladares et al., 2007). For example, tree seedlings that exhibit a strong stem elongation response to shading are more susceptible to cold temperatures than seedlings with a less plastic stem elongation phenotype (Valladares et al., 2007).
A limitation to the benefits of plasticity arises if, due to developmental constraints, a plastic phenotype cannot achieve as extreme a trait value in a given environment as an invariant phenotype can (DeWitt et al., 1998; Alpert and Simms, 2002). In severe environmental conditions where extreme trait values confer a selective advantage, invariant phenotypes may therefore be more advantageous. This is consistent with the observation that generalist species that inhabit a wide variety of relatively moderate environment conditions frequently exhibit developmental plasticity, while specialists that inhabit more extreme environments tend to have invariant phenotypes (Wilson and Yoshimura, 1994; Sultan, 2000, 2003).
Another limit to the effectiveness of plasticity comes from how well temporal environmental variability can be matched with changes in development. The lag time between the perception of an environmental change and a phenotypic outcome may be large compared with the time scale on which the environment changes. In this case, by the time a plastic phenotype is produced, it may be too late to prevent detrimental effects on fitness or it may no longer be appropriate for the current environment (DeWitt et al., 1998; Alpert and Simms, 2002). One solution to this problem is to use cues to influence development that predict future environmental changes (Alpert and Simms, 2002). The two instances of plasticity focused on here provide examples of such a solution. In the case of shade avoidance, morphological changes can be triggered before shading occurs through changes in the red:far-red light ratio due to reflection from neighbours (Ballaré et al., 1987). Flowering time is also influenced by a number of cues (such as cold) that indirectly indicate future optimal conditions for flowering (de Jong and Leyser, 2012).
However, in environments that vary stochastically, predictable cues about the environment to be encountered might not exist and plasticity is unlikely to be advantageous (Leimar et al., 2006). Indeed, using stochastic simulations of populations of individuals in variable environments, Reed et al. (2010) showed that strong plasticity is favoured when environmental cues provide good predictors of future selective pressures, but increases the risk of extinction when cues are unreliable. Low predictability of the optimum strategy for the future environment may have especially important implications for irreversible developmental events duch as the transition to flowering or seed germination (Cohen, 1966; Simons and Johnston, 2003). In the absence of reliable cues, variable phenotypes, where an individual genotype produces a range of different phenotypes relatively independently of environmental cues, have been proposed to be favourable over phenotypic plasticity (Simons, 2011).
VARIABLE PHENOTYPES
Variability as a bet-hedging strategy
Variable phenotypes may be produced as bet-hedging strategies in unpredictable environments where fluctuating natural selection favours different phenotypes across generations (Cohen, 1966; Lewontin and Cohen, 1969; Simons, 2011). As discussed above, in such an environment, the advantages of phenotypic plasticity may be limited due to the lack of reliable cues that can be used to provide information about the optimal future phenotype. An invariant phenotype may also be disadvantageous. This is because, even if it was optimal for the mean environment across generations, an invariant phenotype could result in extinction if extreme variations in the environment occurred. In contrast, a variable phenotype, produced as a bet-hedging strategy (see Table 1), would allow continued survival of a fraction of the population, even though the increased phenotypic variability would probably reduce population fitness in the mean environment. Thus, in unpredictable environments, variance in a phenotype may confer a selective advantage by reducing variance in fitness from generation to generation. This reduction in fitness variance across generations can have a large positive effect on long-term population fitness, which is severely reduced by occasional drops in population size (Gillespie, 1974; Seger and Brockmann, 1987; Philippi and Seger, 1989; Simons, 2011).
As discussed previously, phenotypic variability is under genetic control, and molecular mechanisms exist that can modulate its levels (Queitsch et al., 2002; Hall et al., 2007; Ansel et al., 2008; Viney and Reece, 2013). Thus, it is possible that increased variability as a bet-hedging strategy could be under positive selection in unpredictable environments. Indeed, there is empirical evidence that this has occurred in a number of systems (Simons, 2011). One of the best studied examples is the phenomenon of bacterial persistence, whereby in stressful conditions (such as in the presence of antibiotics) a slow-growing fraction of cells in a clonal population can survive and therefore later regenerate the population. Even in the absence of stress, bacteria can switch stochastically into a slow-growing mode, which can persist if conditions become stressful (Balaban et al., 2004; Maisonneuve et al., 2013). The slow-growing phenotype decreases the mean fitness of the genotype in favourable conditions, but reduces the risk of extinction should stressful conditions arise (Martins and Locke, 2015).
In plants, seedling mortality is often very high, and ecological theory suggests that bet-hedging germination strategies are likely to be beneficial in temporally variable and unpredictable environments (e.g. deserts with sporadic rainfall patterns) (Cohen et al., 1966; Simons and Johnston, 1997). Thus, species in unpredictable environments are expected to have increased variances in germination time. This hypothesis has been tested in desert annuals, which spread germination between seasons by producing a fraction of seeds that is dormant and resists germination in the season of seed production. Consistent with bet-hedging theory, a study of ten desert annual species over 22 years revealed that species experiencing more unpredictable reproductive success upon germination had higher fractions of dormant seeds (Venable, 2007). The species with increased variability in germination time tended to be those most sensitive to variation in precipitation and thus with greater variability in survival.
Spreading offspring germination times within a season to cope with an unpredictable environment can also constitute a bet-hedging strategy. This phenomenon has been studied in L. inflata, an annual species from a temperate habitat in which most seeds are non-dormant and show relatively high variation in time to germination within a season (Simons and Johnston, 2006). Naturally inbred lines of L .inflata show high variance in germination time even when grown under controlled laboratory conditions (Simons and Johnston, 2006). Simons et al. (2009) estimated the extent to which selection on L. inflata germination time fluctuates between years by germinating seeds in the lab and transferring the seedlings to field sites at different times of the year, for 5 years. The survival of plants transferred at different times of the year was measured so that for each year the optimum timing of germination could be estimated. This revealed that the optimal germination time varied widely between the 5 years. Simulations to determine the optimum germination strategy in these conditions revealed that variability in germination time, rather than synchronous germination at the mean optimum time across years, maximized long-term fitness (Simons, 2009). The extent of variability predicted to be optimal was in fact similar to the observed variability, suggesting that the observed germination strategy is consistent with bet-hedging (Simons and Johnston, 2006; Simons, 2009).
Selection for variable germination rates may also minimize intersibling competition for resources. Selection for increased variance in germination rates is expected to be particularly strong for selfing species that produce large numbers of seeds simultaneously and have short seed dispersal distances (Cheplick, 1992; Hyatt and Evans, 1998; Kobayashi and Yamamura, 2000). In such a species, genetically identical offspring are likely to undergo competition, which could reduce fitness of the maternal genotype. Spreading seed germination over time would be likely to reduce this competition (Ellner, 1986; Cheplick, 1992). There is some experimental evidence that plants may be able to regulate the dormancy of their seeds (and therefore the extent to which germination is spread over several seasons) according to the likelihood that sibling competition will occur, with larger families and seeds with lower dispersal having increased variability in germination rate (Cheplick, 1996; Hyatt and Evans, 1998).
It should be noted that the timing of germination is sensitive to a number of environmental cues experienced by the mother plant and by the seed, and thus is likely to be both plastic and variable in a number of species (Fenner, 1991; Finch-Savage and Leubner-Metzger, 2006; Graeber et al., 2012; Footitt et al., 2014). Indeed, in the presence of low environmental predictability, but where some cues about the future are available, a plastic and variable phenotype is predicted to be optimal (Wong and Ackerly, 2005). In such a scenario, a bet-hedging strategy may evolve as an increase in the variance around reaction norms for a plastic phenotype (Simons and Johnston, 1997; Simons, 2011).
In addition to germination timing, other plant phenotypes may also have been selected to have high variability within an environment. This has been proposed to be the case for metabolic pathways that produce toxins to defend against herbivory. A prolonged period of exposure of an insect population to a particular toxin can encourage the evolution of toxin resistance. It may therefore be beneficial for plants to prevent prolonged exposure of herbivores to a particular toxin by stochastically varying toxin production (Shelton, 2004). Indeed, variability in the levels of defence compounds produced by arabidopsis plants in a common environment is genetically controlled and is greater than the variability in production of primary metabolites (Joseph et al., 2015). Variability in lateral root emergence has also been hypothesized to be advantageous (Forde, 2009). However, in many cases, phenotypes might be variable between individuals because the system is poorly constrained and mechanisms to buffer against noise in development have not been selected.
Molecular mechanisms underlying variability
Currently, little is known about the mechanisms that underlie functional variability between genetically identical plants within an environment. One possibility is that what appears to be variability within a homogeneous environment is actually due to plasticity in response to unknown microenvironmental cues (Bradshaw, 1965). Indeed, in a study where almost genetically identical L. inflata seeds were grown under growth chamber conditions, the position of seeds within Petri dishes could account for around a third of the variance in the time to germination, indicating that microenvironmental plasticity can account for at least some of the variability (Simons and Johnston, 2006). However, it is also possible that interindividual variation exists due to a lack of noise-buffering mechanisms in development, as discussed in the first section. Molecular mechanisms may also exist that amplify stochastic fluctuations to generate increased variability within a homogeneous environment. Although little is known about whether such noise-harnessing mechanisms exist in plants, they have been described for microbes and during the development of some multicellular organisms (Raj and van Oudenaarden, 2008; Eldar and Elowitz, 2010; Martins and Locke, 2015).
In microbes, it has been shown that gene-regulatory networks can amplify stochastic fluctuations to generate sub-populations of cells with distinct gene expression states (Alon, 2007; Raj and van Oudenaarden, 2008; Eldar and Elowitz, 2010; Viney and Reece, 2013; Martins and Locke, 2015). Bistability (the existence of two stable states) is often generated by positive feedback acting upon gene expression, i.e. an increase in a protein’s levels causes an increase in the production rate of that protein (Fig. 2C). In this case, the presence of low protein levels will be stable in a fraction of cells. However, stochastic fluctuations may increase protein levels above a threshold, causing a transition to a new stable state of gene expression (Choi et al., 2008). Thus, two sub-populations of cells may be generated, one with high and one with low expression levels of the protein. More complex network architectures which combine a number of interlinked network motifs can give rise to a greater number of possible stable states, allowing increased heterogeneity in the cell population (Alon, 2007). As well as possibly playing a role in creating functional interindividual differences, stochastically induced gene expression states may be used to generate differences between equivalent cells during tissue patterning or cellular differentiation. In this scenario, the realizations of the patterns produced may be variable between individuals, but functionally equivalent (Meyer and Roeder, 2014).
These examples can produce alternative stable states of gene expression; however, genetic networks also exist that can cause unstable, temporary transitions into alternative cellular states. These networks are called excitable circuits and combine a fast-acting positive feedback loop with slower acting negative feedback (Süel et al., 2006; Eldar and Elowitz, 2010; Martins and Locke, 2015). Stochastic fluctuations may trigger the positive feedback, causing a transition to an alternative state. The activation of negative feedback may then bring the system back to the original state. Since this mechanism allows independent stochastic transitions between states in each cell, it provides a means for generating variable phenotypes within a population of genetically identical cells at a given time.
Mechanisms may also exist to create cell heterogeneity in response to environmental cues, thus generating variability in a plastic phenotype. For example, in bacteria, stressful conditions such as starvation cause a large fraction of cells to undergo sporulation, in which highly resistant, dormant spores are produced (Phillips and Strauch, 2002; Schultz et al., 2009). The rest of the population may remain in an active state, allowing them to continue to grow if environmental conditions improve. In Bacillus subtilis, sporulation is thought to be triggered when the levels of a phosphorylated protein, Spo0A, exceed a particular threshold (Chastanet et al., 2010). Phosphorylation of Spo0A involves a four-step phosphorelay mechanism that is activated upon nutrient deprivation (Phillips and Strauch, 2002). Upon induction of sporulation, the activity of phosphorylated Spo0A is heterogeneous amongst cells within a population, which is proposed to explain why only a fraction of cells undergo sporulation (Chastanet et al., 2010). This heterogeneity has been proposed to be due to noise in the transfer of phosphate along the phosphorelay pathway. The presence of four steps in the phosphorelay (rather than a lower number) has been hypothesized to increase noise accumulation along the signal transduction pathway, maximizing cellular heterogeneity, and ensuring that only a fraction of the population undergoes sporulation (Chastanet et al., 2010). Although unknown, one might speculate that in plants, such a mechanism of differential regulator accumulation could underlie the discussed variability in germination timing upon the perception of permissive environmental cues by a population of seeds.
Variability can also be generated through a deterministic mechanism of differential inheritance. Again, yeast provides an example, concerning its dependence on metal ions for survival. If a cell experiences a depletion of metal ions when it undergoes budding (which generates a smaller daughter cell), most of the vacuole containing essential ions is retained by the mother (Avraham et al., 2013). In this situation, the mother cell continues to divide, whereas the daughter’s cell cycle arrests in G1 phase. This has been proposed to prevent the dilution of ions by cell division (Avraham et al., 2013). In this case, differential inheritance occurs deterministically (the vacuole is always inherited by the older mother cell) and generates two populations of cells with different behaviours. Cell fate diversity is also frequently generated by asymmetric cell divisions in the development of multicellular organisms (Macara and Mili, 2008; De Smet and Beeckman, 2011). One hypothesis to explain variability in germination time is that differential inheritance by a mother’s offspring creates inter-seed differences in a deterministic manner.
SUMMARY AND CONCLUSION
In contrast to traditional views that consider plasticity and variable phenotypes as resulting from poorly buffered development, we emphasize that specific genetic mechanisms are likely to be involved in reproducibly (i.e. robustly) delivering invariant, plastic and variable phenotypes. Each of these types of phenotype may be favoured in different ecological scenarios. Invariant phenotypes are likely to have evolved in environments where a constant phenotypic optimum exists, whereas plastic phenotypes are expected to evolve in the presence of predictable changes in the environment. When the environment varies unpredictably, variable phenotypes probably reduce the risk of extinction and promote the long-term survival of the population.
Plants of the same genotype in a given environment are likely to show more variability or plasticity for some traits than others. For example, for a hypothetical plant with a specific insect pollinator, living in an environment where the timing of rainfall is highly unpredictable and neighbouring plants often shade each other, flower morphology may be predicted to be relatively invariant, while germination might be expected to be variable and stem length plastic.
Due to their ecological significance, we argue that developmental mechanisms have probably evolved to allow the robust formation of each of the three types of phenotype. We have illustrated how a number of mechanisms can buffer development against stochastic and environmental perturbations, allowing invariant phenotypes to be produced. There are also well-characterized mechanisms that alter development in response to environmental cues. Finally, the developmental mechanisms involved in producing variable plant phenotypes are the least well understood, although QTL mapping has shown that variance in a number of phenotypes in arabidopsis is under genetic control (Hall et al., 2007). An important future step is to extend this work towards gaining a mechanistic understanding of phenotypic variability in plants.
Another important question is the extent to which plastic, variable and invariable phenotypes are beneficial in the face of climate change. This has been discussed for plasticity (Jump and Penuelas, 2005; Ghalambor et al., 2007; Chevin et al., 2010; Nicotra et al., 2010) which, as mentioned previously, may facilitate survival in a new environment, allowing the phenotype then to be optimized by natural selection (Ghalambor et al., 2007). This is likely to be the case if the phenotypic change that is induced in the new environment changes in the direction of the optimum phenotype in that environment (Ghalambor et al., 2007). However, in a changing climate, the environmental cues that trigger a plastic response may no longer be good indicators of the future environment, and this could lead to the induction of inappropriate phenotypes, which could be detrimental for survival (Ghalambor et al., 2007; Nicotra et al., 2010). In the case of changes in flowering time in response to climate change in the last century, there is evidence for the former scenario. Plant species with flowering times that are plastic with respect to temperature have undergone less rapid declines in abundance than species in which flowering time is less plastic (Willis et al., 2008). This may be due to a better ability of such species to co-ordinate their flowering time with the availability of insect pollinators or appropriate growing conditions. Phenotypic variability could also enable survival in future climates in which weather patterns are expected to become more extreme and unpredictable (IPCC, 2012). More variable phenotypes, produced as bet-hedging strategies, are likely to allow at least some individuals of a population to survive in unpredictable and extreme conditions (Cohen, 1966; Lewontin and Cohen, 1969; Simons, 2011).
ACKNOWLEDGEMENTS
We thank Hugo Tavares for useful suggestions and critical reading of the manuscript. This work was supported by the Gatsby Foundation.
LITERATURE CITED
- Acar M, Becskei A, van Oudenaarden A. 2005. Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232. [DOI] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, et al. 2006. Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94. [DOI] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, et al. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Alon U. 2007. Network motifs: theory and experimental approaches. Nature Reviews Genetics 8: 450–461. [DOI] [PubMed] [Google Scholar]
- Alpert P, Simms E. 2002. The relative advantages of plasticity and fixity in different environments: when is it good for a plant to adjust? Evolutionary Ecology 16: 285–297. [Google Scholar]
- Angel A, Song J, Dean C, Howard M. 2011. A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476: 105–108. [DOI] [PubMed] [Google Scholar]
- Ansel J, Bottin H, Rodriguez-Beltran C, et al. 2008. Cell-to-cell stochastic variation in gene expression is a complex genetic trait. PLoS Genetics 4: e1000049. doi:10.1371/journal.pgen.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashall L, Horton CA, Nelson DE, et al. 2009. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science 324: 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham N, Soifer I, Carmi M, Barkai N. 2013. Increasing population growth by asymmetric segregation of a limiting resource during cell division. Molecular Systems Biology 9: 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CC, Sieber P, Wellmer F, Meyerowitz EM. 2005. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Current Biology 15: 303–315. [DOI] [PubMed] [Google Scholar]
- Baker HG. 1974. The evolution of weeds. Annual Review of Ecology and Systematics 5: 1–24. [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305: 1622–1625. [DOI] [PubMed] [Google Scholar]
- Ballaré C, Sanchez R, Scopel AL, Casal J, Ghersa C. 1987. Early detection of neighbour plants by phytochrome perception of spectral changes in reflected sunlight. Plant, Cell and Environment 10: 551–557. [Google Scholar]
- Becskei A, Kaufmann BB, van Oudenaarden A. 2005. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nature Genetics 37: 937–44. [DOI] [PubMed] [Google Scholar]
- Becskei A, Serrano L. 2000. Engineering stability in gene networks by autoregulation. Nature 405: 590–593. [DOI] [PubMed] [Google Scholar]
- Berg OG. 1978. A model for the statistical fluctuations of protein numbers in a microbial population. Journal of Theoretical Biology 71: 587–603. [DOI] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, et al. 2014. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505: 417–21. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, et al. 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Molecular Cell 24: 853–865. [DOI] [PubMed] [Google Scholar]
- Blake WJ, KÆrn M, Cantor CR, Collins JJ. 2003. Noise in eukaryotic gene expression. Nature 422: 633–637. [DOI] [PubMed] [Google Scholar]
- Blewitt ME, Chong S, Whitelaw E. 2004. How the mouse got its spots. Trends in Genetics 20: 550–554. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, et al. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Richards CL, Pigliucci M. 2008. Epigenetics for ecologists. Ecology Letters 11: 106–115. [DOI] [PubMed] [Google Scholar]
- Boudon F, Chopard J, Ali O, et al. 2015. A computational framework for 3D mechanical modeling of plant morphogenesis with cellular resolution. PLoS Computational Biology 11: e1003950. doi:10.1371/journal.pcbi.1003950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw AD. 1965. Evolutionary significance of phenotypic plasticity in plants. Advances in Genetics 13: 115–155. [Google Scholar]
- Casal JJ. 2013. Photoreceptor signaling networks in plant responses to shade. Annual Review of Plant Biology 64: 403–427. [DOI] [PubMed] [Google Scholar]
- Chastanet A, Vitkup D, Yuan G-C, Norman TM, Liu JS, Losick RM. 2010. Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proceedings of the National Academy of Sciences, USA 107: 8486–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheplick G. 1992. Sibling competition in plants. Journal of Ecology 80: 567–575. [Google Scholar]
- Cheplick G. 1996. Do seed germination patterns in cleistogamous annual grasses reduce the risk of sibling competition? Journal of Ecology 84: 247–255. [Google Scholar]
- Chevin L-M, Lande R, Mace GM. 2010. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biology 8: e1000357. doi:10.1371/journal.pbio.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Kim YJ. 2009. Intrinsic variability of gene expression encoded in nucleosome positioning sequences. Nature Genetics 41: 498–503. [DOI] [PubMed] [Google Scholar]
- Choi PJ, Cai L, Frieda K, Xie XS. 2008. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science 322: 442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH. 2006. Transcriptional pulsing of a developmental gene. Current Biology 16: 1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E, Rolland-Lagan A-G, Matthews M, Bangham JA, Prusinkiewicz P. 2004. The genetics of geometry. Proceedings of the National Academy of Sciences, USA 101: 4728–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D. 1966. Optimizing reproduction in a randomly varying environment. Journal of Theoretical Biology 12: 119–129. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Crispo E. 2008. Modifying effects of phenotypic plasticity on interactions among natural selection, adaptation and gene flow. Journal of Evolutionary Biology 21: 1460–1469. [DOI] [PubMed] [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, et al. 2006. Widespread genome duplications throughout the history of flowering plants. Genome Research 16: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debat V, David P. 2001. Mapping phenotypes: canalization, plasticity and developmental stability. Trends in Ecology and Evolution 16: 555–561. [Google Scholar]
- De Smet I, Beeckman T. 2011. Asymmetric cell division in land plants and algae: the driving force for differentiation. Nature Reviews Molecular Cell Biology 12: 177–88. [DOI] [PubMed] [Google Scholar]
- DeWitt TJ, Sih A, Wilson DS. 1998. Costs and limits of phenotypic plasticity. Trends in Ecology and Evolution 13: 77–81. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. 1988a. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54: 95–104. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. 1988b. A gradient of bicoid protein in Drosophila embryos. Cell 54: 83–93. [DOI] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB. 2010. Functional roles for noise in genetic circuits. Nature 467: 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellner S. 1986. Germination dimorphisms and parent–offspring conflict in seed germination. Journal of Theoretical Biology 123: 173–185. [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, et al. 2003. Radial patterning of Arabidopsis shoots by class IIIHD-ZIP and KANADI genes. Current Biology 13: 1768–1774. [DOI] [PubMed] [Google Scholar]
- Erdmann T, Howard M, ten Wolde PR. 2009. Role of spatial averaging in the precision of gene expression patterns. Physical Review Letters 103: 258101. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. 2001. Establishment of polarity in lateral organs of plants. Current Biology 11: 1251–1260. [DOI] [PubMed] [Google Scholar]
- Fenner M. 1991. The effects of the parent environment on seed germinability. Seed Science Research 1: 75–84. [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H. 2007. Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiology 144: 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. 2006. Seed dormancy and the control of germination. New Phytologist 171: 501–523. [DOI] [PubMed] [Google Scholar]
- Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ. 2010. Phytochrome regulation of branching in Arabidopsis. Plant Physiology 152: 1914–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Clay HA, Dent K, Finch-Savage WE. 2014. Environment sensing in spring-dispersed seeds of a winter annual Arabidopsis influences the regulation of dormancy to align germination potential with seasonal changes. New Phytologist 202: 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG. 2009. Is it good noise? The role of developmental instability in the shaping of a root system. Journal of Experimental Botany 60: 3989–4002. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Schadt EE. 2010. The quantitative genetics of phenotypic robustness. PLoS One 5: e8635. doi:10.1371/journal.pone.0008635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. 2004. Noise minimization in eukaryotic gene expression. PLoS Biology 2: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, et al. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Ghalambor CK, McKay JK, Carroll SP, Reznick DN. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Functional Ecology 21: 394–407. [Google Scholar]
- Gianoli E, Valladares F. 2012. Studying phenotypic plasticity: the advantages of a broad approach. Biological Journal of the Linnean Society 105: 1–7. [Google Scholar]
- Gibson G, Wagner G. 2000. Canalization in evolutionary genetics: a stabilizing theory? Bioessays 22: 372–380. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. 1974. Natural selection for within-generation variance in offspring number. Genetics 76: 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ. 1988. Adaptation to sun and shade: a whole-plant perspective. Functional Plant Biology 15: 63–92. [Google Scholar]
- Givnish TJ. 2002. Ecological constraints on the evolution of plasticity in plants. Evolutionary Ecology 16: 213–242. [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. 2005. Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036. [DOI] [PubMed] [Google Scholar]
- Gommers CM, Visser EJ, St Onge KR, Voesenek LA, Pierik R. 2013. Shade tolerance: when growing tall is not an option. Trends in Plant Science 18: 65–71. [DOI] [PubMed] [Google Scholar]
- Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJ. 2012. Molecular mechanisms of seed dormancy. Plant, Cell and Environment 35: 1769–1786. [DOI] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W. 2007. Probing the limits to positional information. Cell 130: 153–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AFM, Hogeweg P, Scheres B. 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Scheres B, Hogeweg P, Maree AFM. 2012. Morphogengineering roots: comparing mechanisms of morphogen gradient formation. BMC Systems Biology 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursky VV, Surkova SY, Samsonova MG. 2012. Mechanisms of developmental robustness. Biosystems 109: 329–335. [DOI] [PubMed] [Google Scholar]
- Hall MC, Dworkin I, Ungerer MC, Purugganan M. 2007. Genetics of microenvironmental canalization in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 104: 13717–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jönsson H, et al. 2008. Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655. [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. 2009. The angiosperm gibberellin–GID1–DELLA growth regulatory mechanism: how an ‘inhibitor of an inhibitor’ enables flexible response to fluctuating environments. The Plant Cell 21: 1328–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S. 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79. [DOI] [PubMed] [Google Scholar]
- Hooshangi S, Thiberge S, Weiss R. 2005. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proceedings of the National Academy of Sciences, USA 102: 3581–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands AY, Chitwood DH, Plavskin Y, Timmermans MCP. 2009. Signals and prepatterns: new insights into organ polarity in plants. Genes and Development 23: 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt LA, Evans AS. 1998. Is decreased germination fraction associated with risk of sibling competition? Oikos 83: 29–35. [Google Scholar]
- Ietswaart R, Wu Z, Dean C. 2012. Flowering time control: another window to the connection between antisense RNA and chromatin. Trends in Genetics 28: 445–453. [DOI] [PubMed] [Google Scholar]
- IPCC. 2012. Summary for policymakers. Managing the risks of extreme events and disasters to advance climate change adaptation. A Special Report of Working Groups I and II of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press, 1–19. [Google Scholar]
- Jaeger J. 2011. The gap gene network. Cellular and Molecular Life Sciences 68: 243–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, et al. 2008. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO Journal 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Gomez JM, Corwin JA, Joseph B, Maloof JN, Kliebenstein DJ. 2011. Genomic analysis of QTLs and genes altering natural variation in stochastic noise. PLoS Genetics 7: e1002295. doi:10.1371/journal.pgen.1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M, Leyser O. 2012. Developmental plasticity in plants. Cold Spring Harbor Symposia on Quantitative Biology 77: 63–73. [DOI] [PubMed] [Google Scholar]
- Jönsson H, Heisler MG, Shapiro BE, Meyerowitz EM, Mjolsness E. 2006. An auxin-driven polarized transport model for phyllotaxis. Proceedings of the National Academy of Sciences, USA 103: 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Corwin JA, Kliebenstein DJ. 2015. Genetic variation in the nuclear and organellar genomes modulates stochastic variation in the metabolome, growth, and defense. PLoS Genetics 11: e1004779. doi:10.1371/journal.pgen.1004779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump AS, Penuelas J. 2005. Running to stand still: adaptation and the response of plants to rapid climate change. Ecology Letters 8: 1010–1020. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sung S. 2014. Genetic and epigenetic mechanisms underlying vernalization. Arabidopsis Book 12: e0171. doi:10.1199/tab.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kleunen M, Fischer M. 2005. Constraints on the evolution of adaptive phenotypic plasticity in plants. New Phytologist 166: 49–60. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamura N. 2000. Evolution of seed dormancy due to sib competition: effect of dispersal and inbreeding. Journal of Theoretical Biology 202: 11–24. [DOI] [PubMed] [Google Scholar]
- Korves TM, Schmid KJ, Caicedo AL, et al. 2007. Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. American Naturalist 169: E141–E157. [DOI] [PubMed] [Google Scholar]
- Lande R. 2009. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. Journal of Evolutionary Biology 22: 1435–1446. [DOI] [PubMed] [Google Scholar]
- Lander AD. 2011. Pattern, growth, and control. Cell 144: 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander AD, Gokoffski KK, Wan FY, Nie Q, Calof AL. 2009. Cell lineages and the logic of proliferative control. PLoS Biology 7: e15. doi:10.1371/journal.pbio.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Refahi Y, Besnard F, et al. 2014. Meristem size contributes to the robustness of phyllotaxis in Arabidopsis. Journal of Experimental Botany 66: 1317–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. 2007. Genetic properties influencing the evolvability of gene expression. Science 317: 118–121. [DOI] [PubMed] [Google Scholar]
- Le Corre V. 2005. Variation at two flowering time genes within and among populations of Arabidopsis thaliana: comparison with markers and traits. Molecular Ecology 14: 4181–4192. [DOI] [PubMed] [Google Scholar]
- Le Corre V, Roux F, Reboud X. 2002. DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Molecular Biology and Evolution 19: 1261–1271. [DOI] [PubMed] [Google Scholar]
- Lehner B. 2010. Conflict between noise and plasticity in yeast. PLoS Genetics 6: e1001185. doi:10.1371/journal.pgen.1001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimar O, Hammerstein P, Van Dooren TJ. 2006. A new perspective on developmental plasticity and the principles of adaptive morph determination. American Naturalist 167: 367–376. [DOI] [PubMed] [Google Scholar]
- Lempe J, Lachowiec J, Sullivan AM, Queitsch C. 2013. Molecular mechanisms of robustness in plants. Current Opinion in Plant Biology 16: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestas I, Vinnicombe G, Paulsson J. 2010. Fundamental limits on the suppression of molecular fluctuations. Nature 467: 174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML. 2008. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biology 6: e264. doi:10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Cohen D. 1969. On population growth in a randomly varying environment. Proceedings of the National Academy of Sciences, USA 62: 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Xie XS. 2011. Central dogma at the single-molecule level in living cells. Nature 475: 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Tikhonov M, Gregor T. 2013. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 154: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C, Chan J. 2008. The parallel lives of microtubules and cellulose microfibrils. Current Opinion in Plant Biology 11: 641–646. [DOI] [PubMed] [Google Scholar]
- Macara IG, Mili S. 2008. Polarity and differential inheritance – universal attributes of life? Cell 135: 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin–antitoxin activity. Cell 154: 1140–1150. [DOI] [PubMed] [Google Scholar]
- Martins BM, Locke JC. 2015. Microbial individuality: how single-cell heterogeneity enables population level strategies. Current Opinion in Microbiology 24C: 104–112. [DOI] [PubMed] [Google Scholar]
- Masamizu Y, Ohtsuka T, Takashima Y, et al. 2006. Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proceedings of the National Academy of Sciences, USA 103: 1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. 1998. Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Hartl DL. 2002. A single mode of canalization. Trends in Ecology and Evolution 17: 468–473. [Google Scholar]
- Meyer HM, Roeder AH. 2014. Stochasticity in plant cellular growth and patterning. Frontiers in Plant Science 5: 420. doi:10.3389/fpls.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabet V, Besnard F, Vernoux T, Boudaoud A. 2012. Noise and robustness in phyllotaxis. PLoS Computational Biology, 8: e1002389. doi:10.1371/journal.pcbi.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D, Ihekwaba A, Elliott M, et al. 2004. Oscillations in NF-κB signaling control the dynamics of gene expression. Science 306: 704–708. [DOI] [PubMed] [Google Scholar]
- Nevozhay D, Adams RM, Murphy KF, Josic K, Balazsi G. 2009. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proceedings of the National Academy of Sciences, USA 106: 5123–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, et al. 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846. [DOI] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, et al. 2010. Plant phenotypic plasticity in a changing climate. Trends in Plant Science 15: 684–692. [DOI] [PubMed] [Google Scholar]
- Novák B, Tyson JJ. 2008. Design principles of biochemical oscillators. Nature Reviews Molecular Cell Biology 9: 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates AC. 2011. What’s all the noise about developmental stochasticity? Development 138: 601–607. [DOI] [PubMed] [Google Scholar]
- Ordas B, Malvar RA, Hill WG. 2008. Genetic variation and quantitative trait loci associated with developmental stability and the environmental correlation between traits in maize. Genetics Research 90: 385–395. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. 2002. Regulation of noise in the expression of a single gene. Nature Genetics 31: 69–73. [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. 1997. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91: 639–648. [DOI] [PubMed] [Google Scholar]
- Park S, Harada J. 2008. Arabidopsis embryogenesis. Methods in Molecular Biology 427: 3–16. [DOI] [PubMed] [Google Scholar]
- Paszek P, Ryan S, Ashall L, et al. 2010. Population robustness arising from cellular heterogeneity. Proceedings of the National Academy of Sciences, USA 107: 11644–11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, et al. 2009. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. The Plant Cell 21: 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippi T, Seger J. 1989. Hedging one’s evolutionary bets, revisited. Trends in Ecology and Evolution 4: 41–44. [DOI] [PubMed] [Google Scholar]
- Phillips Z, Strauch M. 2002. Bacillus subtilis sporulation and stationary phase gene expression. Cellular and Molecular Life Sciences 59: 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, de Wit M. 2014. Shade avoidance: phytochrome signalling and other aboveground neighbour detection cues. Journal of Experimental Botany 65: 2815–2824. [DOI] [PubMed] [Google Scholar]
- Pigliucci M, Murren CJ, Schlichting CD. 2006. Phenotypic plasticity and evolution by genetic assimilation. Journal of Experimental Biology 209: 2362–2367. [DOI] [PubMed] [Google Scholar]
- Powell AE, Lenhard M. 2012. Control of organ size in plants. Current Biology 22: R360–R367. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. 2006. Stochastic mRNA synthesis in mammalian cells. PLoS Biology 4: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. 2008. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135: 216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed TE, Waples RS, Schindler DE, Hard JJ, Kinnison MT. 2010. Phenotypic plasticity and population viability: the importance of environmental predictability. Proceedings, Biological Sciences 277: 3391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C. 2000. Auxin regulates the initiation and radial position of plant lateral organs. The Plant Cell 12: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, et al. 2003. Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Richards EJ. 2006. Inherited epigenetic variation – revisiting soft inheritance. Nature Reviews Genetics 7: 395–401. [DOI] [PubMed] [Google Scholar]
- Ruberti I, Sessa G, Ciolfi A, Possenti M, Carabelli M, Morelli G. 2012. Plant adaptation to dynamically changing environment: the shade avoidance response. Biotechnology Advances 30: 1047–1058. [DOI] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342. [DOI] [PubMed] [Google Scholar]
- Scheiner SM, Caplan RL, Lyman RF. 1991. The genetics of phenotypic plasticity. III. Genetic correlations and fluctuating asymmetries. Journal of Evolutionary Biology 4: 51–68. [Google Scholar]
- Schlichting CD, Pigliucci M. 1998. Phenotypic evolution: a reaction norm perspective. Sunderland, MA: Sinauer Associates.
- Schmitt J, Dudley SA, Pigliucci M. 1999. Manipulative approaches to testing adaptive plasticity: phytochrome-mediated shade-avoidance responses in plants. American Naturalist 154: S43–S54. [DOI] [PubMed] [Google Scholar]
- Schmitt J, McCormac AC, Smith H. 1995. A test of the adaptive plasticity hypothesis using transgenic and mutant plants disabled in phytochrome-mediated elongation responses to neighbors. American Naturalist 146: 937–953. [Google Scholar]
- Schmitt J, Wulff RD. 1993. Light spectral quality, phytochrome and plant competition. Trends in Ecology and Evolution 8: 47–51. [DOI] [PubMed] [Google Scholar]
- Schultz D, Wolynes PG, Jacob EB, Onuchic JN. 2009. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proceedings of the National Academy of Sciences, USA 106: 21027–21034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger J, Brockmann J. 1987. What is bet-hedging? Oxford Surveys in Evolutionary Biology 4: 182–211.
- Shelton AL. 2004. Variation in chemical defences of plants may improve the effectiveness of defence. Evolutionary Ecology Research 6: 709–726. [Google Scholar]
- Shelton AL. 2005. Within-plant variation in glucosinolate concentrations of Raphanus sativus across multiple scales. Journal of Chemical Ecology 31: 1711–1732. [DOI] [PubMed] [Google Scholar]
- Shen X, Pettersson M, Rönnegård L, Carlborg Ö. 2012. Inheritance beyond plain heritability: variance-controlling genes in Arabidopsis thaliana. PLoS Genetics 8: e1002839. doi:10.1371/journal.pgen.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shraiman BI. 2005. Mechanical feedback as a possible regulator of tissue growth. Proceedings of the National Academy of Sciences, USA 102: 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons A. 2014. Playing smart vs. playing safe: the joint expression of phenotypic plasticity and potential bet hedging across and within thermal environments. Journal of Evolutionary Biology 27: 1047–1056. [DOI] [PubMed] [Google Scholar]
- Simons AM. 2009. Fluctuating natural selection accounts for the evolution of diversification bet hedging. Proceedings, Biological Sciences 276: 1987–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons AM. 2011. Modes of response to environmental change and the elusive empirical evidence for bet hedging. Proceedings of the Royal Society B: Biological Sciences 278: 1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons AM, Johnston MO. 1997. Developmental instability as a bet-hedging strategy. Oikos 80: 401–406. [Google Scholar]
- Simons AM, Johnston MO. 2003. Suboptimal timing of reproduction in Lobelia inflata may be a conservative bet-hedging strategy. Journal of Evolutionary Biology 16: 233–243. [DOI] [PubMed] [Google Scholar]
- Simons AM, Johnston MO. 2006. Environmental and genetic sources of diversification in the timing of seed germination: implications for the evolution of bet hedging. Evolution 60: 2280–92. [PubMed] [Google Scholar]
- Smith RS, Guyomarc'h S, Mandel T, Reinhardt D, Kuhlemeier C, Prusinkiewicz P. 2006. A plausible model of phyllotaxis. Proceedings of the National Academy of Sciences, USA 103: 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell-Rood EC, Van Dyken JD, Cruickshank T, Wade MJ, Moczek AP. 2010. Toward a population genetic framework of developmental evolution: the costs, limits, and consequences of phenotypic plasticity. Bioessays 32: 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Angel A, Howard M, Dean C. 2012. Vernalization – a cold-induced epigenetic switch. Journal of Cell Science 125: 3723–3731. [DOI] [PubMed] [Google Scholar]
- Stearns SC, Kaiser M, Kawecki TJ. 1995. The differential genetic and environmental canalization of fitness components in Drosophila melanogaster. Journal of Evolutionary Biology 8: 539–557. [Google Scholar]
- Stoma S, Lucas M, Chopard J, Schaedel M, Traas J, Godin C. 2008. Flux-based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLoS Computational Biology 4: e1000207. doi:10.1371/journal.pcbi.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Struhl K, Macdonald PM. 1989. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57: 1259–1273. [DOI] [PubMed] [Google Scholar]
- Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. 2006. An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440: 545–550. [DOI] [PubMed] [Google Scholar]
- Sultan SE. 2000. Phenotypic plasticity for plant development, function and life history. Trends in Plant Science 5: 537–542. [DOI] [PubMed] [Google Scholar]
- Sultan SE. 2003. Phenotypic plasticity in plants: a case study in ecological development. Evolution and Development 5: 25–33. [DOI] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A. 2001. Intrinsic noise in gene regulatory networks. Proceedings of the National Academy of Sciences, USA 98: 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Nowak DE, Brasier AR. 2005. A TNF-induced gene expression program under oscillatory NF-κB control. BMC Genomics 6: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonsor SJ, Elnaccash TW, Scheiner SM. 2013. Developmental instability is genetically correlated with phenotypic plasticity, constraining heritability, and fitness. Evolution 67: 2923–2935. [DOI] [PubMed] [Google Scholar]
- Tufto J. 2000. The evolution of plasticity and nonplastic spatial and temporal adaptations in the presence of imperfect environmental cues. American Naturalist 156: 121–130. [DOI] [PubMed] [Google Scholar]
- Uyttewaal M, Burian A, Alim K, et al. 2012. Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149: 439–451. [DOI] [PubMed] [Google Scholar]
- Valladares F, Gianoli E, Gómez JM. 2007. Ecological limits to plant phenotypic plasticity. New Phytologist 176: 749–763. [DOI] [PubMed] [Google Scholar]
- Valladares F, Niinemets Ü. 2008. Shade tolerance, a key plant feature of complex nature and consequences. Annual Review of Ecology, Evolution, and Systematics 39: 237–257. [Google Scholar]
- Van Tienderen PH. 1997. Generalists, specialists, and the evolution of phenotypic plasticity in sympatric populations of distinct species. Evolution 51: 1372–1380. [DOI] [PubMed] [Google Scholar]
- Vanneste K, Maere S, Van de Peer Y. 2014. Tangled up in two: a burst of genome duplications at the end of the Cretaceous and the consequences for plant evolution. Philosophical Transactions of the Royal Society B: Biological Sciences 369: 20130353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA. 2005. Paralogs in polyploids: one for all and all for one? The Plant Cell 17: 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venable DL. 2007. Bet hedging in a guild of desert annuals. Ecology 88: 1086–1090. [DOI] [PubMed] [Google Scholar]
- Viney M, Reece SE. 2013. Adaptive noise. Proceedings of the Royal Society of London B: Biological Sciences 280: 20131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. 1942. Canalization of development and the inheritance of acquired characters Nature 150: 563–565. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Booth G, Bagheri-Chaichian H. 1997. A population genetic theory of canalization. Evolution 51: 329–347. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. 1995. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14: 697–706. [DOI] [PubMed] [Google Scholar]
- Wenden B, Toner DL, Hodge SK, Grima R, Millar AJ. 2012. Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proceedings of the National Academy of Sciences, USA 109: 6757–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney H, Glover B. 2007. Morphology and development of floral features recognised by pollinators. Arthropod-Plant Interactions 1: 147–158. [Google Scholar]
- Williams DG, Mack RN, Black RA. 1995. Ecophysiology of introduced Pennisetum setaceum on Hawaii: the role of phenotypic plasticity. Ecology 76: 1569–1580. [Google Scholar]
- Willis CG, Ruhfel B, Primack RB, Miller-Rushing AJ, Davis CC. 2008. Phylogenetic patterns of species loss in Thoreau’s woods are driven by climate change. Proceedings of the National Academy of Sciences, USA 105: 17029–17033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DS, Yoshimura J. 1994. On the coexistence of specialists and generalists. American Naturalist 144: 692–707. [Google Scholar]
- Wolpert L. 1969. Positional information and the spatial pattern of cellular differentiation. Journal of Theoretical Biology 25: 1–47. [DOI] [PubMed] [Google Scholar]
- Wong TG, Ackerly DD. 2005. Optimal reproductive allocation in annuals and an informational constraint on plasticity. New Phytologist 166: 159–71. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Nukazuka A, Tsukaya H. 2012. Leaf adaxial–abaxial polarity specification and lamina outgrowth: evolution and development. Plant and Cell Physiology 53: 1180–1194. [DOI] [PubMed] [Google Scholar]
- Yu J, Xiao J, Ren X, Lao K, Xie XS. 2006. Probing gene expression in live cells, one protein molecule at a time. Science 311: 1600–1603. [DOI] [PubMed] [Google Scholar]
- Zuehlke A, Johnson JL. 2010. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers 93: 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]