Article title: “Size and positional effects of promoter RNA segments on virus-induced RNA-directed DNA methylation and transcriptional gene silencing”
Authors: Shungo Otagaki, Miou Kawai, Chikara Masuta, and Akira Kanazawa
Journal: Epigenetics
Bibliometrics: Volume 6, Issue 6, pages 681–691
DOI: 10.4161/epi.6.6.16214
An incorrect version of Figure 6 was published in the original online and print publications. The correct version of Figure 6 is published below:
Figure 6.
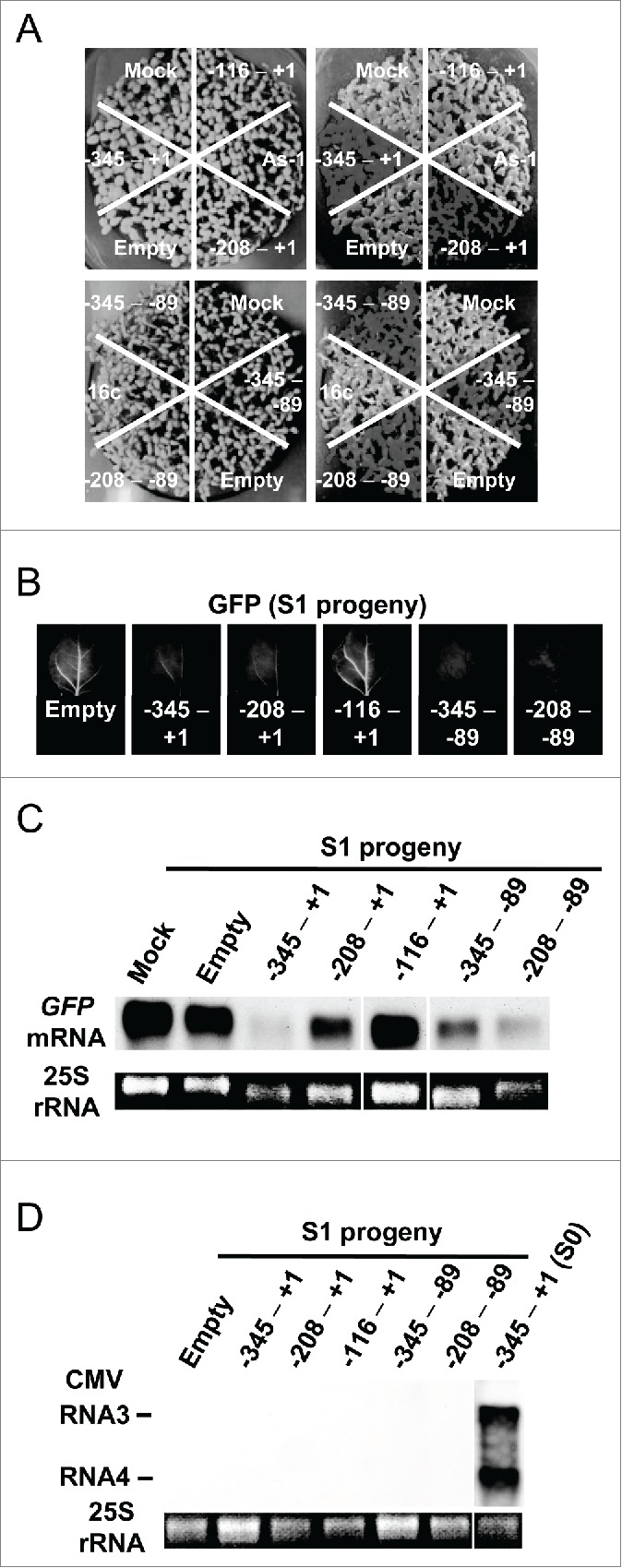
Inheritance of TGS in the self-reproductive plants (S1 generation). (A) GFP fluorescence of the progeny plants infected with CMV-A1 vectors. Fifty-five seeds were sown in one block on 1/4 MS medium and photographed under white (left part) or blue light (right part) 1 week post-germination. 16c and mock indicate the progeny of non-inoculated and mock-inoculated 16c plant, respectively. Others indicate the progeny of plants infected with the recombinant CMVs. (B) Upper leaves of S1 plants photographed under UV illumination. Fluorescence emission was collected using the long-pass filter. (C) Level of mRNA of the GFP gene in S1 plants. RNA isolated from the progeny of a mock-inoculated plant was hybridized as a negative control. (D) Northern blot analysis of RNA isolated from S1 plants indicating the absence of recombinant CMV. The blots were hybridized with a GFP-specific (C) or the CMV-specific (D) probe, respectively. RNA isolated from a primary infected plant (S0 plant) at 18 DPI was hybridized as a positive control. Ethidium bromide stained 25S rRNA bands below the part demonstrate that an equal amount of RNA was loaded. The images of RNA samples that were not adjacent to each other in the original gel were separated by a line.
The authors apologize for any inconvenience caused.


