Abstract
Amyloid Precursor Protein (APP) and its metabolites play a critical role in Alzheimer’s disease pathogenesis. The idea that APP may function as a receptor has gained momentum based on its structural similarities to type I transmembrane receptors, and the identification of putative APP ligands. Here we review the recent experimental evidence in support of this notion and discuss how this concept is viewed in the field. Specifically, we focus on the structural and functional characteristics of APP as a cell surface receptor, and its interaction with adaptors and signaling proteins. We also address the importance of APP's function as a receptor in Alzheimer’s disease etiology and discuss how this function might be potentially important for the development of novel therapeutic approaches.
Keywords: Amyloid precursor protein, Alzheimer’s disease, cell surface receptors, signaling
The amyloid precursor protein
The amyloid precursor protein (APP) is a type-1 transmembrane protein that plays an essential role in Alzheimer’s disease (AD). It is the source of cerebral accumulation of β– amyloid peptides (Aβ), which accumulate in brain senile plaques. Cleavage of full-length APP by α- and β-secretases releases the large soluble ectodomain (sAPPα and sAPPβ, respectively), leaving behind membrane-bound C-terminal fragments (CTF), comprised of the transmembrane and short cytoplasmic domain (named C83 and C99, respectively) [1, 2]. Subsequent cleavage of APP-CTF by γ-secretase releases the APP intracellular domain (AICD) from the membrane and generates extracellular p3 or Aβ peptides (of 38–43 amino acids length) through α-secretase-mediated non-amyloidogenic pathway or β-secretase-mediated amyloidogenic pathway, respectively (Figure 1). In addition to the processing pathways described above, APP ectodomain can be cleaved by other secretases as recently described [3, 4]. Caspase cleavages of APP-CTF could also contribute to accumulating various APP intracellular fragments [5–8]. Because of its multiple cleavage sites and the production of several metabolites, numerous roles in the nervous system have been ascribed to APP, making it difficult to understand its true function(s) [2, 9–14].
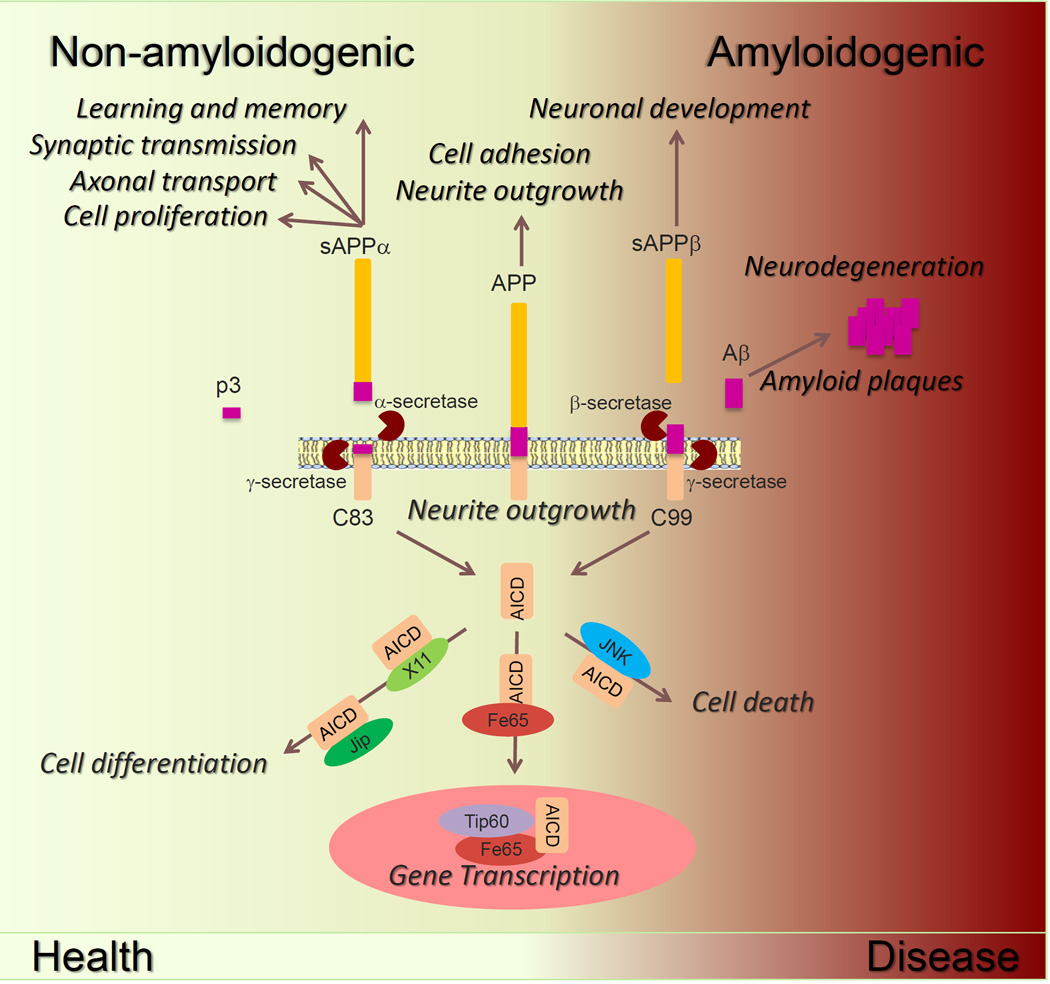
Figure 1. Variation on a theme – APP functions in health and disease.
APP is a versatile biologically active protein, with its N-terminal domains as well as its shorter C-terminal fragments involved in a variety of cellular functions [2, 9–14]. APP-FL possesses cell-adhesive properties and can modulate neurite outgrowth. Non-amyloidogenic soluble APPα fragment (sAPPα), resulting from α-secretase cleavage, displays neuroprotective and trophic properties. Several studies suggest that soluble APPβ (sAPPβ) has a role in neuronal development. Amyloidogenic Aβ deposition in senile plaques correlates with pathological AD-related neuronal dysfunction. Accumulation of APP-CTF at the membrane as C99 (β-CTF) or C83 (α-CTF) stimulates neurite outgrowth. Processing of APP by γ-secretase releases the intracellular AICD fragment from the membrane, which could interact with JNK to activate cell death, X11/JIP to stimulate cell differentiation, or with Fe65 to modulate gene transcription. Proper accumulation of APP and its fragments lead to healthy biological outcomes (depicted as yellow zone), whereas over accumulation of APP fragments (including Aβ and AICD) would lead to neurodegeneration and cell death (depicted as the red transitioning zone) [15–17].
Indisputably, Aβ peptide production and accumulation have been a focus of intense research in the field of AD [15–17]. The neurotoxic effects of Aβ have been well-documented using in vitro and in vivo model systems [18–20]. The “amyloid cascade hypothesis” was formulated to propose that excessive production of Aβ activates the neurodegenerative sequela of synaptic dysfunction, synapse loss and ultimately neuronal death [21]. The higher Aβ42/Aβ40 ratio is associated with familial AD-linked point mutations in the genes encoding APP and presenilin proteins. Considering, some have proposed that it is not Aβ peptide accumulation per se that causes AD [22]. But others debate that Aβ peptide appears to be necessary, but not sufficient, to cause AD [23]. Accordingly, it has been proposed that elevated Aβ levels may act as a trigger of other downstream pathogenic processes. Moreover, it appears that accumulation of AICD is detrimental to cellular function [24–26]. In contrast, low levels of Aβ have been reported to enhance neurotransmission and memory [27–29]; likewise membrane-targeted AICD favors neurite outgrowth [30, 31], supporting physiologic functions for Aβ and AICD. Finally, non-amyloidogenic p3 fragments have been shown to form calcium permeable channels at the plasma membrane and cause neurite degeneration [32]. Accordingly, physiological levels of APP contributes to overall cellular health, whereas excess APP levels lead to overproduction of extracellular Aβ and intracellular AICD, and consequently it becomes detrimental to cellular function (see schematic in Figure 1).
It has been proposed in the literature that APP can modulate synaptic function and neurite outgrowth throughout its cell-adhesion properties or through putative receptor-like intracellular signaling mechanism [2, 9, 33–38]. The idea that APP may act as a cell surface receptor was proposed several years ago, based mainly on structural similarities to type I membrane receptors [39]. Since then, it has been shown that APP may act as a receptor entity through functional similarities with growth factors and cell-adhesion molecules. This review will focus on several of these aspects, and discuss how the current evidence supports the idea that APP may indeed fall in the category of receptor-like molecules.
Proteolytic processing of APP
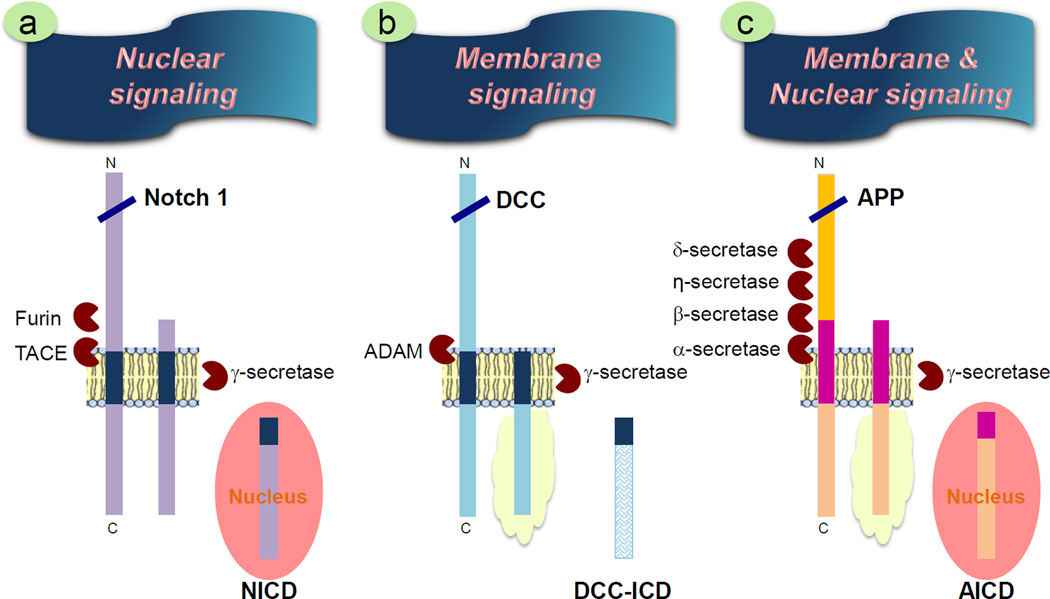
APP shares similarities in membrane topology and proteolytic processing with several γ-secretase substrates (including Notch and deleted in colorectal carcinomas / DCC) that function as cell surface receptors (Figure 2) [2, 40–42]. The majority of γ-secretase substrates are type-I transmembrane proteins, harboring a large ectodomain, a single-pass transmembrane domain, and a cytoplasmic C-terminal domain capable of mediating intracellular signaling. In addition, these substrates are well known for their diverse functions in neuronal outgrowth, synaptogenesis and axon guidance [41, 42]. As an example, following maturation through the secretory pathway, the Notch1 receptor is constitutively cleaved by a convertase of the furin family generating an extracellular and a transmembrane fragment, which remain associated with each other at the cell surface. Upon interaction with a ligand, Notch becomes susceptible to a second extracellular cleavage by A Disintegrin And Metalloprotease/TNF-α-Converting Enzyme (ADAM/TACE), which releases the large extracellular domain. The ectodomain shedding is followed by the intramembrane cleavage mediated by γ–secretase, which releases the Notch intracellular domain (NICD). This fragment rapidly translocates to the nucleus and modulates transcription [43, 44] (Figure 2a). In a similar way, DCC is exposed to metalloprotease-dependent proteolytic processing after ligand binding that results in the shedding of its ectodomain. The resulting membrane-tethered DCC-CTF is subject to γ-secretase processing. Following inhibition of γ-secretase cleavage, membrane-tethered DCC-CTF accumulates and induces signaling cascade that regulates neurite outgrowth and axon guidance [45, 46] (Figure 2b). The similarities between the processing of Notch, DCC and APP have prompted the speculation that membrane-tethered APP-CTF and AICD may play analogous signaling roles as described for membrane-tethered DCC-CTF and NICD, respectively (Figure 2c). Indeed, as discussed below, accumulation of membrane-associated APP-CTF could initiate an intracellular signaling cascade, whereas AICD released from the membrane would reach the nucleus to engage in transcription activities [30, 47–53]. Owing to the similarity with Notch and DCC processing, we propose a model in which APP’s (or APP-CTF’s) sojourn at the plasma membrane would facilitate membrane signaling associated with its CTF tail, until a time when γ-secretase cleavage will release the cytosolic tail for potential nuclear translocation. However, unlike the case of Notch or DCC receptors, specific ligand-induced α-secretase cleavage prior of APP-like receptor has not been well characterized. One would also consider that perhaps a variety of ligands could interact with APP to achieve “constitutive” ectodomain shedding of APP.
Figure 2. APP signaling: homology with other γ–secretase substrates.
γ-secretase is known to catalyze intramembranous cleavage of several type-1 membrane proteins, including DCC, Notch and APP. (a) Cleavage of Notch by γ–secretase releases Notch intracellular domain (NICD) which translocates to the nucleus to activate nuclear signaling [2, 40–44]. (b) Ligand-activated cleavage of DCC by α-secretase generates transmembrane DCC-CTF, which activates intracellular signaling through interaction with various signaling partners (depicted as yellow cloud). Intramembranous cleavage by γ-secretases releases the intracellular fragment (DCC-ICD) that is subsequently degraded, thus terminating DCC signaling [45]. (c) APP is sequentially cleaved by α- or β-secretases, and also recently-described η-secretase [3] and 5-secretase [4], generating various transmembrane APP-CTFs. The accumulation of APP-CTF at the plasma membrane facilitates intracellular signaling, which is terminated upon γ-secretase cleavage [30]. The resulting AICD fragment translocates to the nucleus to activate nuclear signaling [24, 25, 120, 121]. Thus, APP processing and signaling mediated by APP is analogous to both Notch and DCC signaling.
Structural properties of APP as a receptor-like entity
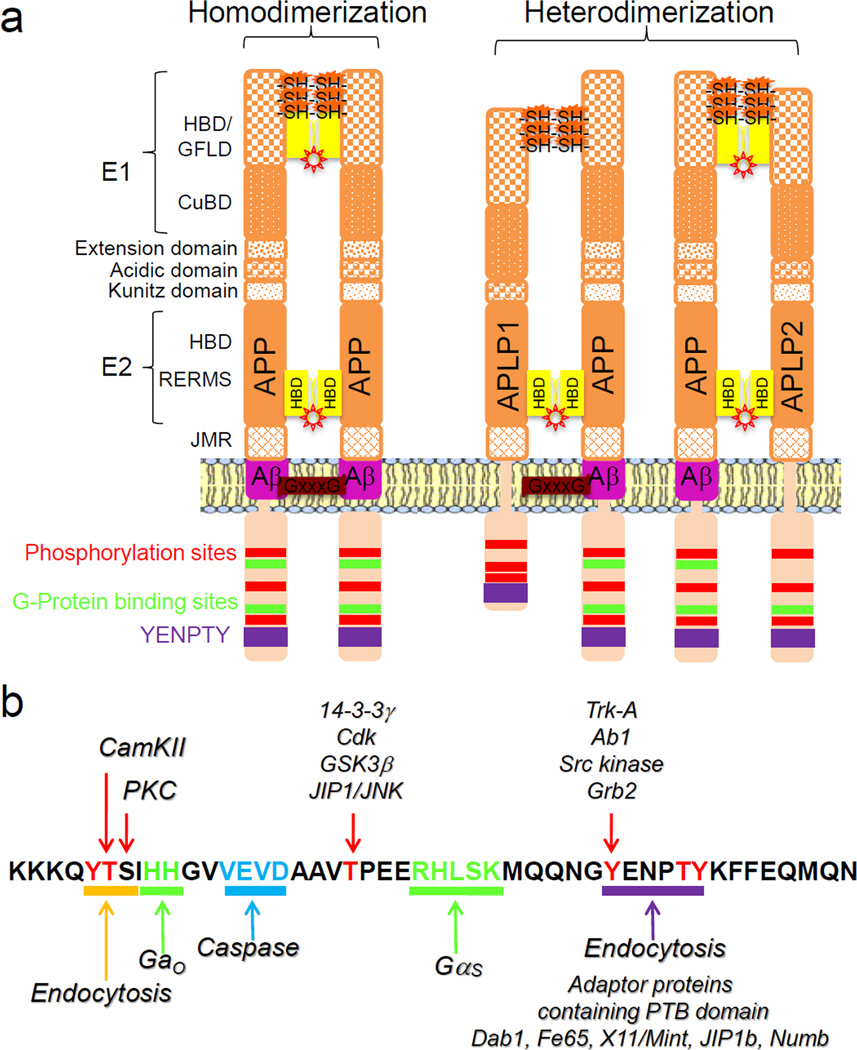
APP is expressed as three isoforms attributable to tissue-specific alternative splicing (Figure 3a) (reviewed in ref [13]). The APP695 isoform is predominantly expressed in neurons, whereas the APP751 and APP770 isoforms are expressed in non-neuronal populations. Two mammalian APP homologues, named amyloid precursor-like protein 1 and 2 (APLP-1 and APLP-2), share primary amino acid sequence, structure and conserved domains with APP (reviewed in refs. [38, 54]) (Figure 3a). Although APLP1 and APLP2 undergo proteolytic processing similar to APP, they do not contribute directly to AD neuropathology, as their sequences considerably diverge in the first half of Aβ domain. The extension domain, located at the end of the extracellular E1 domain, is also exclusive to APP. All APP members comprise E1 and E2 domains in the extracellular region that contains heparin-binding domains (HBD). The HBD exhibits an extremely positively charged surface, which would allow the binding of negatively charged molecules, such as heparin. The first HBD (also named growth factor-like domain, GFLD) at the N-terminus of APP contains three disulfide bridges that form between β-sheets; the structure of this domain resembles ligand recognition sites found in growth factors and receptor-like proteins [38, 55]. This high-affinity HBD was identified within the E1 HBD (Lys99-His110) using synthetic peptides, and mutagenesis of three charged residues within this APP domain reduced heparin-binding capacity [56]. Formation of disulfide bridges appears to stabilize APP structure at the cell surface [57–59], thus potentially favoring ligand induction of signaling cascade. APP also contains a second HBD within the membrane-proximal E2 domain, which can bind albeit with lower affinity to membrane-anchored heparan sulfate proteoglycans (HSPGs) [60]. It is known that HSPGs could regulate low-affinity binding sites to allow putative ligand(s) to bind with higher affinity to receptors [61, 62]. Accordingly, it has been suggested that membrane-anchored HSPGs may function as low-affinity co-receptors for soluble APP, and perhaps enhance its affinity to the putative sAPP receptor(s) including APP [60, 63–65]. Accordingly, an interaction of membrane-bound APP with HSPG may allow the formation of a more stable tertiary signaling complex that promotes growth or cell-adhesion properties of APP, as it is the case for many growth factors or cell-adhesion receptor-like proteins [66, 67]. Indeed, it has been shown that the HSPG interaction with the E1 HBD of APP is required to induce neurite outgrowth [56]. In addition, the penta-peptide sequence RERMS in the E2 domain was identified as an active site that promotes cell growth and neurite extension [68]. The presence of two HBD with different binding affinities is fascinating because many receptors possess high and low affinity ligand binding sites within the molecule to allow the receptor to initiate more rapid and sustained intracellular signaling cascade. This dual affinity HBD of APP might be critical for tight temporal regulation of its receptor-like properties, especially at the synapse. Therefore, APP structure may include a high-affinity ligand binding domain (the GFLD within E1 domain) and a low-affinity interaction site domain (the HSPG binding domain within the E2 domain). Altogether, APP structural properties support the idea that APP may act in several aspects as a receptor-like entity.
Figure 3. APP receptor-like architecture, its intracellular motifs and signaling partners.
(a) APP and its two mammalian homologues, APLP-1 and APLP-2, share similar primary structure and conserved extracellular domains (E1, E2, acidic, and Kunitz), juxtamembrane region (JMR), and intracellular phosphorylation, G-protein binding, and YENPTY internalization sites [38, 54]. APP can bind putative ligands with high affinity through disulfide bridge (SH-SH)-stabilized E1 domain and low affinity co-receptor proteins through the heparin recognition domains (E1 and E2, as shown as yellow HBD box and heparin ligand as a sun). Using these interacting domains, APP and its homologues can form homo or heteromeric complexes through cis formation within cellular compartments or transdimerization across cells, including synaptic structures [37, 58, 72, 76, 77, 80]. The Aβ domain of APP possesses three GxxxG motif sequences that allow tighter association to itself and perhaps to other proteins [80, 84]. (b) APP cytosolic domain possesses conserved sequence motifs responsible for a complex network of protein-protein interactions, which can account for a variety of cellular functions mediated by APP and its metabolites [25, 119–121, 123]. The main signaling domains include: two conserved trafficking internalization/endocytic signal (purple and yellow underlines), whose one is responsible for the binding of many adaptors proteins (purple underline), a caspase cleavage motif (blue), two G-proteins binding sites (green) and several phosphorylation sites (red).
The role of APP subdomains in dimerization process
In a manner similar to many type-1 transmembrane domain receptors [44, 69–71], APP can form homodimers as well as heterodimers by interacting with its homologues or putative ligands. Dimerization of APP is mediated by motifs present in the extracellular and the transmembrane domains of the protein. The extracellular E1 domain at the N-terminal region of APP has been characterized as the major interacting interface for homo- as well as heterodimerization of APP and APLPs at the cell surface [34, 37, 72]. According to structural analysis, dimerization of APP could be initiated first through dimerization of the E1 domain, the putative ligand recognition domain (see above; Figure 3a). The E2 domain is also capable of dimerizing, but with lower affinity. Nevertheless, heparin-induced dimerization of the E1 domain will lead to a local increase of APP concentration and the proximity of E2 domains at the plasma membrane, which will promote dimerization of this domain as well. However, changes in APP conformation is thought to prevent dimerization (summarized in [72]).
Further structural analysis of the extracellular E2 domain of APP supports the idea that APP can reversibly dimerize in an antiparallel orientation at least in vitro [73]. This very important concept may indeed account for the described cell-cell-adhesion properties of APP and subsequent signaling through homophilic interactions (reviewed in ref [37]). This effect can be exacerbated in a dose-dependent manner during initiation of axonal growth pathfinding [74, 75]. More recent studies showed clear indication that APP dimerization would facilitate synapse formation [58, 76, 77]. This structural feature may enable membrane-tethered APP to interact directly with soluble APP ectodomain (or the ectodomains of APP homologues) [60, 65]. As reported for lingo-1, a type-1 transmembrane protein, APP could function both as a ligand and a receptor through trans self-dimerization [78, 79]. Indeed, it has been shown that the disulfide bond in the E1 domain might be the main contributor for trans-dimerization between APP molecules and its homologues [34, 58, 59, 72], although the E2 domain could still account partially to enhance synaptic connections in the absence of E1 [77]. The importance of E1 and E2 domain interactions with APP and APP homologues has been confirmed [80, 81]. Based on the findings that endogenous APP and APLPs are rapidly processed by γ-secretase [82] and also the APP homologues differ in their extent to which they are cleaved by β-site APP cleaving enzyme 1 (BACE1) at the steady state in neurons (APLP1 is more readily cleaved relative to APP [83]), only a subset of neuronal APP or APLPs may be available to transdimerize with adjacent cells or form cell surface receptor-like complexes.
The transmembrane domain of APP also harbors three consecutive GxxxG motifs, which are known to mediate helix-helix association of transmembrane domains by direct glycine-glycine contacts [80, 84]. Despite the high degree of homology between APP and its homologues, the GxxxG motifs are not conserved. Only one GxxxG motif is found in APLP1, and this motif is absent in APLP2. Thus, it is unlikely that the GxxxG motifs could contribute to heterodimerization between APP and APLP2. The British dementia protein-2 (BRI2 or integral membrane protein 2B/ITM2B), a protein associated with familial Danish and British dementia, possesses a GxxxG motif that could form dimerization complexes through helix-helix interactions with APP [85, 86]. In addition, the GxxxG motifs in APP transmembrane domain could serve as signals for guided intracellular transport from endoplasmic reticulum compartment to cell surface [86–88]. Upon interaction with mature BRI2 polypeptide at the plasma membrane and in endocytic compartments, APP could no longer be cleaved efficiently by secretases, as a consequence APP processing and Aβ production are reduced [86, 89–91]. In addition, ErbB family of growth factor receptor tyrosine kinases possesses two conserved GxxxG dimerization motifs that are perhaps important for association with soluble APPα [92–94] (see below).
The APP ectodomain has also the property to adopt different conformations depending on the cellular pH [95]. A pH-dependent interaction between the GFLD subdomain interaction with the copper binding domain (CuBD) within the E1 domain will allow APP to adopt a closed conformation in more acidic compartment, whereas it will adopt a more opened conformation at neutral pH. Consequently, APP at the cell surface (pH ~7.4) – in its open conformation - would be poised to expose the E1 domain and favor association with other proteins or putative ligands, which is consistent with noticeable higher molecular weight complex formation and its dimerization pattern [95] (see Figure 4b). Thus, the pH-dependent molecular conformation switch may provide APP the ability to fulfill different physiological functions in different cellular localizations.
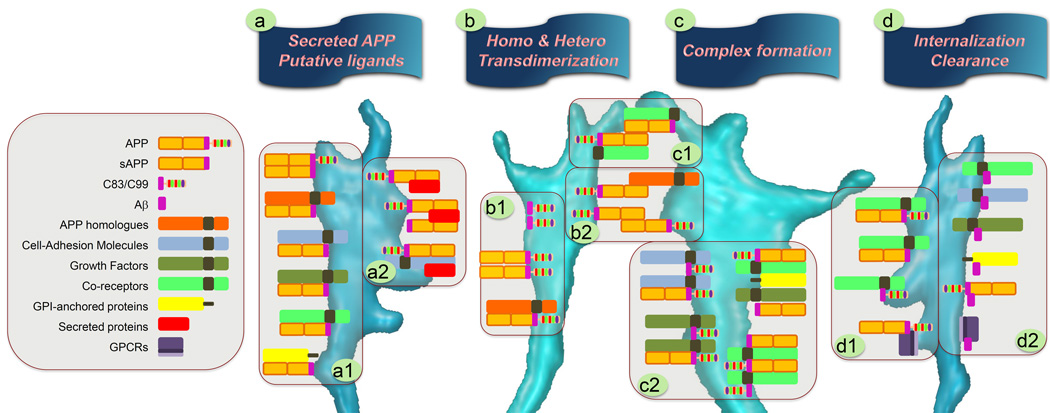
Figure 4. Model of APP and its metabolite cross-interactions with cell surface proteins.
The complexity of interactions between APP and APP metabolites and a variety of proteins is portrayed in this illustration. Neuronal growth cones are shown as a model to depict the multiple sites of interaction of APP, sAPP, AICD and its Aβ fragment (fuchsia) with a variety of proteins including: APP and its own homologues (orange), cell-adhesion molecule receptors (blue), growth factor receptors (green), other receptor-like proteins (fluo-green), G-protein-coupled receptors (GPCRs, purple), GPI-anchored proteins (yellow), and secreted ligand proteins (red). (a1) The first cartoon box depicts interactions of secreted soluble APP (sAPP) with a variety of cell surface receptors or proteins. Accordingly, sAPP has the ability to initiate signaling upon binding with a variety of type-1 transmembrane molecules including cell-adhesion molecules [151, 174, 180–187] and growth factor receptors [94, 156, 162, 188–191, 193]. (a2) Interactions of APP with putative ligands, through direct or indirect association, are illustrated on the right. (b) The schematics of several dimerization partners are shown. APP and its homologues can make homo- or heterodimers and form cell surface receptor-like complexes capable of modulating signaling events [56, 64, 151, 165, 167, 168, 170, 173, 175, 176, 226]. The extracellular region of APP-like receptor possesses the structural features to homodimerize or heterodimerize with its homologues in (b1) cis- and (b2) trans- cellular manner [65, 80, 151–153, 171, 172]. (c1) Cell-adhesive property of APP with adjacent neurons is shown in the left side of the schematic diagram. (c2) APP and its ectodomain associate with other cell surface receptors to form complexes that render them capable of mediating signaling through crosstalk interactions. (d1) APP, as a receptor entity, has the capability to internalize through interaction with endocytic partners, glycoproteins, and GPCRs. (d2) Receptor proteins that could conceptually affect Aβ clearance are also presented in the schematic.
Regulation of APP cell surface localization
Cell surface localization of APP would be a requisite for APP to fall under the category of receptor-like proteins and molecules involved in cell-cell trans-dimerization interaction across the membrane. Indeed, a subset of full-length APP (APP-FL) in all cells, including neurons, can be found at the cell surface, where it can interact with putative ligands (below). APP cell surface localization is regulated by a balance between the efficiency of APP secretory trafficking and internalization, and the efficiency of secretase processing [96]. Indeed, reduced ectodomain shedding increases surface residence, and there is evidence in the literature that APP family members may have different propensity for residence at the cell surface, at least in transfected non-neuronal cells [58, 59, 97]. Surface localization of APP family members correlates with higher stability of the proteins and their potential to dimerize at the cell surface, which appear to facilitate synapse formation [58, 76, 77, 97]. Residence of APP at the cell surface seems to be a common denominator of non-amyloidogenic α-secretase cleavage of APP. Accumulation of APP at the cell surface, following inhibition of its endocytic transit, favors α-secretase cleavage leading to a reduction of Aβ production while diminution of APP at the cell surface will increase BACE1-dependent amyloidogenic processing and secretion of Aβ [2, 96]. In support of this concept, it has been shown that APP interaction with the Nogo receptor (NgR) via a core region spanning residues 558 to 599 (APP695 numbering) prevents accumulation of APP at the cell surface and regulates Aβ production and amyloid deposition [98, 99].
Inhibition of γ-secretase activity causes an increase of cell surface accumulation of APP, although the underlying mechanism(s) are poorly understood [58, 100]. Moreover, it has been demonstrated that APP-CTF interaction with Dab1 results in an increase of cell surface APP [101]. Extracellular application of reelin, a cell surface matrix glycoprotein, promotes this effect, suggesting that extracellular factors could facilitate APP association with intracellular partners [101, 102] Other intracellular partners have been shown as well to regulate cell surface APP trafficking [96]. As an example, it has been shown that adaptor protein Mint interaction with APP favors endocytosis of APP, whereas deletion of Mint promotes cell surface accumulation of APP [103]. Increased interaction of APP with microtubule interacting proteins PAT1 and PAT1a has been reported to favor surface expression of APP in transfected cell lines, while PAT1a knockdown reduces the levels of APP at the plasma membrane [104, 105]. However, a recent study reported an inverse correlation between PAT1 expression and accumulation of endogenous cell surface APP in primary neurons, suggesting that PAT 1-APP interaction and APP trafficking might involve additional neuron-specific mechanisms [106].
APP plasma membrane localization can be regulated by post-translational glycosylation. Alteration in the expression of type I transmembrane protein 59 (TMEM59), a brain-specific membrane-associated protein, inhibited complex glycosylation of APP, reduced secretory APP trafficking to the cell surface, and reduced APP cleavage by both α- and β-secretase [107]. In contrast, knockdown of p23, a member of the p24 family protein that regulates vesicular trafficking in the early secretory pathway, promoted efficient glycosylation and surface accumulation of APP [108]. Both APP-FL and APP-CTF have been reported to undergo ubiquitination. The C-terminus Hsp70 interacting protein CHIP [109] and the F-box and leucine-rich repeat protein 2 (FBL2), a component of the E3 ubiquitin ligase complex [110], have been shown to promote APP ubiquitination and stabilization. In the case of FBL2 overexpression, there is an increase of APP at the cell surface due to a reduction of endocytosis [110].
Naturally, the presence of APP or APP homologues at the cell surface would facilitate the cell-adhesion properties of the molecule, especially in synaptic structures [58]. In addition, accumulation of APP at the cell surface will facilitate cis / trans-interactions with cell surface signaling molecules (including APP itself and its homologues) and putative soluble ligands to initiate co-receptor complex formation and subsequent signaling (Figure 4). It is appealing to think that these latter interactions may regulate secretase-dependent shedding of APP ectodomain as suggested for epidermal growth factor / epidermal growth factor receptors (EGFR) [111, 112]. EGFR family possesses more than a dozen of interacting ligands that could provide bidirectional signaling, with a forward signal mediated by the receptors and a reverse signal mediated by the ectodomain of membrane-anchor ligands. Likewise, shedding of the APP ectodomain by α-secretase at the cell surface might allow soluble APP to participate in paracrine signaling.
APP-mediated intracellular signaling
Cell surface receptors initiate intracellular signal transduction in response to extracellular signaling molecules. The idea that APP may behave as a receptor is supported by evidence of signaling associated with APP-CTF or membrane-tethered APP. APP and especially APP-CTF reside in membrane microdomains enriched in cholesterol and sphingolipids (called lipid rafts) [49, 113, 114], populated by several cell surface receptors and signaling molecules [115, 116]. Raft association of APP would position it in an ideal environment for interaction with signaling molecules. Data from our lab illustrated this point by revealing enhanced APP-induced signaling when γ-secretase cleavage of the APP-CTF was inhibited or when AICD was tethered to the membrane via raft-targeting lipid modifications [30]. Specifically, membrane-tethered AICD activates adenylate cyclase/cAMP/PKA-dependent intracellular signaling cascade. This signaling involves an interaction between AICD and the heterotrimeric G-protein subunit Gαs via the APP cytoplasmic sequence 672RHLSK676, which is a characteristic BBxxB motif found in G-protein binding segments of G-protein-coupled receptors (GPCR) and type-1 receptor proteins [117, 118]. Mutating this motif in AICD abolishes the interaction with Gαs and associated signaling [30]. Moreover, APP-CTF colocalizes and interacts with the Gαo subunit at the plasma membrane in insect and mammalian neurons [47–50]. Physical interaction with the Gαo subunit appears to involve a histidine doublet in the cytoplasmic tail of APP [49]. These studies provide clear evidence that APP could function analogous to a GPCR, where membrane-bound or -tethered AICD facilitates interactions and recruitment of cytosolic adaptors/effectors to exacerbate intracellular signaling.
APP contains at least eight conserved serine/threonine residues that undergo potential phosphorylation within its cytoplasmic domain. It is clear that phosphorylation on these residues can modulate APP trafficking and signal transduction (Figure 3b) [119–121]. In vitro studies showed that Ser655 on the APP cytosolic domain can be phosphorylated by protein kinase C (PKC) or Thr654 can be phosphorylated by calcium/calmodulin-dependent protein kinase II (CaMKII) [122]. In addition, APP-Thr668 (located within the motif 667VTPEER672) can be phosphorylated by c-Jun N-terminal kinases (JNK1/2/3), cyclin-dependent protein kinases (Cdk1 and Cdk5) and GSK3β. The phosphorylation of Thr668 residue emerges as the most critical phosphorylation site that may determine AICD subcellular compartment localization and impact its functions. Phosphorylation of Thr668 induces a significant conformational change in 682YENPTY687 endocytic motif, therefore affecting the binding specificity and affinity of APP cytoplasmic domain to other cytosolic partners [119, 121, 123].
Phosphorylation of APP at Thr668 renders AICD less vulnerable to degradation and cleavage by caspases, and interaction with Fe65 protects AICD from proteasome degradation [119, 123]. Indeed, APP phosphorylation at Thr668 site induces a conformational change that regulates interaction with Fe65 and subsequent translocation of AICD to the nucleus [51]. Binding of adaptor protein 14-3-3γ on the 667VTPEER672 motif further stabilizes AICD/Fe65 interaction. Consequently, this will contribute to enhance Fe65-dependent gene transactivation [124] as it has been described as well for JIP1 interaction, a scaffolding protein for the JNK kinase cascade [125]. JIP1 can facilitate JNK-dependent phosphorylation of Thr668 although its interaction is not necessary [125]. Down-regulation of JIP1 expression selectively impairs the trafficking of APP phosphorylated at the Thr668 residue [126]. APP-Thr668Glu, which mimics phosphorylated APP, was found to reside predominantly in neuritic tips/growth cones and promotes neurite outgrowth through a JIP1-dependent pathway in a neuronal cell line [126], whereas APP-Thr668Ala, a non-phosphorylatable mutant, failed to be efficiently transported [127]. Interestingly, JIP1 has been shown to mediate anterograde axonal transport of APP through the molecular motor kinesin-1 [128]. As a consequence of JIP1 phosphorylation states and association with APP, phosphorylated JIP1 could favor anterograde transported of APP while retrograde transport of APP is prominent in nonphosphorylated JIP1 expressing neurons [128] or neurons lacking JIP1 expression or interaction with APP [127] (but see [129]). Altogether, these findings emphasis the importance of the APP phosphorylation state in mediating axonal transport through protein interactions. As a consequence, these phosphorylation events would facilitate the delivery of APP receptor to its cellular targets and allow receptor signaling and function to occur in a spatially and temporally regulated manner.
APP cytosolic domain possesses additional conserved sequence motifs responsible for a complex network of protein-protein interactions that regulate APP trafficking [25, 119–121, 123] (Figure 3b). Selective phosphorylation of APP-CTF regulates the interaction with several cytosolic adaptor proteins. A number of these protein interactions are known to regulate APP trafficking and processing, as well as APP-mediated intracellular signaling cascades. As an example, APP-CTF contains two highly conserved motifs responsible for clathrin-mediated internalization, 682YENPTY687 and 653YTSI656 motifs [130] The 682YENPTY687 motif has been described to serve as a docking site for adaptor proteins that possess a phosphotyrosine-binding (PTB) domain, such as Dab1, Fe65, X11/MINT, JIP1b and Numb. Such adaptor proteins play critical roles in tyrosine kinase-mediated signal transduction, APP trafficking and localization [120, 121] Tyr682 phosphorylation of APP-CTF has been reported to be necessary for the binding adaptor proteins ShcA (Src and collagen homologue) and Grb2 (Growth factor receptor bound) leading to an activation of MAP kinase transduction pathways [131–133]. These membrane events could contribute to regulating proliferation, differentiation, mitosis, cell survival, apoptosis, as well as APP processing [120].
In addition to the adaptor protein interactions described above, the APP intracellular domain appears to initiate intricate crosstalk with several transmembrane receptor proteins (see below). Of interest, Bradykinin receptor-induced phosphoinositide-mediated Ca2+ signaling was diminished in cells lacking APP expression [134]. Although a direct site of interaction between bradykinin and APP was not determined, this inhibition was reversed by selective expression of APP-CTF. APP interaction with TrkA/NGF receptor can induce phosphorylation of APP-CTF Tyr682 residue [135, 136]. APP can interact as well with alcadein/calsyntenin, a type-1 transmembrane protein that contains cadherin repeat motifs [137]. Following sequential α-secretase and γ-secretase cleavages of alcadein, alcadein-ICD interacts with X11 and forms a ternary complex with APP-CTF [138]. Tripartite interaction of Dab1 and DISC1 (disrupted-in-schizophrenia-1, a protein with multiple coiled coil motifs) with APP through distinct binding sites in APP-CTF has been described [139]. APP-CTF also interacts with the type-1 transmembrane BRI2 protein, which is mutated in patients with familial British dementia [89]. Accordingly, APP is capable of initiating intricate crosstalk with other proteins through its cytosolic domain.
AICD has a short half-life and is rapidly degraded, making it an APP proteolytic fragment that is present at extremely low levels at the steady state [52, 97]. Despite being subject to efficient degradation, small amounts of AICD could translocate to the nucleus, which is similar to the fate of the Notch intracellular domain released from the Notch receptor by γ-secretase cleavage. Analogous to Notch receptor signaling, AICD engages in the regulation of gene expression (Figure 2a). AICD interaction with transcription factors, such as CP2/LSF/LBP1, Tip60 and the nucleosome assembly factor SET, is consistent with AICD’s participation in gene expression [24, 25, 120, 121]. Following intramembranous γ-secretase cleavage, phosphorylated AICD-Fe65 complex could be released in the cytosol and translocate to the nucleus [51]. Although, APP/Fe65 interaction is very important for AICD-mediated transcription, it does not appear to be necessary for its nuclear translocation [140]. AICD could enter the nucleus through mechanisms that are independent of Fe65 interaction, such as by interacting with JIP1 [125]. In addition, JIP1 has been reported to promote the transcriptional activity of the AICD [141]. Together with Fe65 and Tip60, AICD forms a transcriptionally active protein complex that has been reported to promote glycogen synthase kinase 3beta (GSK3β) gene expression [51, 53, 142]. On the other hand, interaction with X11/Mint would favor AICD cytoplasmic localization, therefore inhibiting AICD-mediated gene transactivation [143]. Interestingly, while signaling associated with AICD transcription appears to favor GSK3β activation [51, 144, 145], retaining AICD at the membrane would favor inhibition of GSK3β cascade [30]. It has been proposed that AICD generated through preferential endosomal BACE1 cleavage can be translocated to the nucleus to induce signaling, while production of AICD through sequential cleavage by α-secretase at the plasma membrane would favor perhaps membrane-induced signaling [30], and subsequent proteasomal degradation [97, 146]. Notably, comprehensive proteomic studies have revealed evidence of interactions of APP intracellular domain with the exocytic machinery at the synapse as well [147–151]. Clearly, these findings indicate that subcellular localization of AICD could have differing effects on cellular signaling and its potential involvement in AD pathogenicity.
Secreted APP and putative ligand-induced signaling
As described above, the N-terminal ectodomain of APP has the structural features and molecular domains that enable it to interact with a variety of receptor proteins (Figure 4a). Thus, soluble APP could initiate subsequent signaling upon binding with receptor molecules [2, 12, 65] (Table 1). This concept has emerged from several studies, which demonstrated that accumulation or overexpression of APP-NTF (and in some instances Aβ itself [17]) induced or exacerbated signaling associated with several classes of receptors as described above (see also Table 1). Recently, it has been shown that soluble APP and the APP-E1 domain could activate the Akt survival pathway following trophic factor depletion and that this activation involves an interaction of APP or APP-E1 domain with APP-FL [152]. In a similar fashion, Aβ interaction with APP induces G-protein-dependent enhancement of synaptic activity that involves the APP-E1 domain [153]. As previously discussed in section 1, APP-FL possesses the structural ability to bind ligand to its own E1 domain. This association could initiate subsequent G-protein coupled signaling events by engaging motifs within the cytosolic C-terminal tail of APP [152, 153]. This scenario is reminiscent of a complex formation between a ligand and an autoreceptor. Soluble APP can modulate several signaling pathways involved in neuroprotection and axonal outgrowth, including the PI3K/Akt pathway [154–156], the NF-kB pathway [154, 157], and the MAPK /Erk/Egr1 pathway [154, 158–161]. However, it has been shown that Aβ could repress CaMKII, Erk/MAPK and PI3K/Akt pathways through an interaction with insulin receptor tyrosine kinase autophosphorylation process [162], suggesting that soluble APP and Aβ might differentially influence signaling cascades.
Table I.
APP-mediated signaling and APP receptor-like interactions
| RECEPTOR-LIKE INTERACTION |
APP FRAGMENT INTERACTION |
SIGNALING | REFERENCES |
|---|---|---|---|
| APP/APP homologues | APP-FL | Erk | [80, 151, 153, 160] |
| sAPP | PI3K/Akt, GSK3β, Erk/MAPK/Egr1, NF-kB, | [65, 68, 151, 152, 154–161] | |
| Aβ | G-protein | [80, 151, 153, 171, 172] | |
| AICD, APP-CTF | cAMP/PKA, GSK3β, Wnt, IP3, Erk, nuclear | [30, 47–50, 119, 134, 151, 159] | |
| Ligands | |||
| Collagen I | APP-FL, sAPP | - | [167] |
| F-spondin | APP-FL, sAPP | - | [151, 170, 175] |
| Heparin | APP-FL, sAPP | - | [56] |
| Integrin | APP-FL, sAPP | - | [64, 173] |
| Laminin | APP-FL, sAPP, Aβ, AICD | - | [168] |
| Pancortin | APP-FL, sAPP | - | [175, 176] |
| Reelin | APP-FL, sAPP | - | [173, 175] |
| TSH | sAPP | MAPK (Elk-1) | [165] |
| Cell-adhesion molecules | |||
| EphB2 | Aβ | - | [180, 181] |
| FcγRIIb | Aβ | JNK | [187] |
| L1/NgCAM | sAPP | - | [174] |
| N-Cadherin | APP-FL, C99 | - | [186] |
| NCAM1 | APP-FL, sAPP | - | [151] |
| DCC | APP-FL, sAPPα | Erk1/2 | [182, 183] |
| Notch | APP-FL, sAPP | - | [184, 185] |
| Growth factor receptors | |||
| DR6 | sAPPβ | Caspase | [190, 197] |
| EGF receptor | sAPP | - | [94] |
| IGF/insulin receptor | sAPPα, Aβ | PI3K/PKB/Akt/GSK3, Erk/MAPK, CaMKII | [156, 162, 193] |
| LilrB2/PirB | Aβ | Cofilin | [188] |
| NGF receptor | APP-FL, AICD | TrkA | [135, 136, 191] |
| p75 | sAPP | cAMP/PKA | [189] |
| GPL-anchors | |||
| Contactins | APP-FL, sAPP | Nuclear | [151, 174, 175, 198] |
| Glypican | sAPP | - | [60, 169] |
| NgR | APP-FL, sAPP, Aβ | - | [98, 99] |
| PrP | APP-FL, sAPP, Aβ | Fyn kinase | [151, 199, 204] |
| GPCRs | |||
| Amylin-3 | Aβ | cAMP/PKA, Erk/MAPK, cFos, Akt | [210, 211] |
| B2-ADR | Aβ | cAMP/PKA | [209] |
| Frizzled | Aβ | Wnt/β-Catenin | [163] |
| GPR3 | APP-FL | - | [212] |
| mGluR5 | Aβ | Fyn kinase | [199, 204] |
| PTGER2 | APP-FL | - | [212] |
| Receotor-like proteins | |||
| Alcadein | APP-FL, C99 | - | [137] |
| BRI2 | APP-FL, C99 | - | [85, 86, 88–91] |
| Lingo-1 | APP-FL, sAPP | - | [151, 175, 243] |
| LRP family receptors | APP-FL, sAPP, Aβ, AICD, APP-CTF |
Nuclear, Wnt |
[91, 192, 223–225, 227–232, 234, 235] |
| Syndecan | sAPP | - | [60] |
A more complex effect on APP-induced signaling has been described for the GSK3β signaling cascade. Aβ can inhibit canonical Wnt/β-catenin signaling, which is linked to the inhibition of GSK3β activity, through a dose-dependent interaction with Frizzled family receptors and in competition with Wnt ligand [163]. Similarly, Aβ oligomers could prevent inhibition of GSK3β activity preferentially through competition with various neurotrophic factors albeit at different magnitudes (NGF > IGF1 = insulin > soluble APPa > BDNF) [156, 164]. Studies in cultured cells indicate that soluble APPα (with no apparent effect of soluble APPβ) leads to increased Ser-9 GSK3β phosphorylation, and concurrent loss of GSK3β activity, through signaling mechanisms involving IGF-1 and insulin receptors [156]. In addition, it was observed that an accumulation of soluble APPα parallels an Akt-dependent decrease of GSK3β activity whereas accumulation of Aβ parallels an increase of GSK3β activity in brains of young as compared to aged PS1/APP transgenic mice, respectively [156]. We have previously shown that APP-CTF targeted in the membrane could account for an adenylate cyclase-dependent inhibition of GSK3β [30]. In this latter case, we speculate that inhibition of GSK3β activity might occur through activation of APP receptor although this remains to be experimentally tested. The convergence of signaling events mediated by soluble APP and Aβ fragments highlight the fact that APP ectodomain may serve as a ligand for a range of receptor molecules that share similar intracellular signaling cascades, in the nervous system and peripheral tissue. This notion is exemplified by the involvement of soluble APP in the thyrotropin-mediated proliferation of thyroid follicle cells and a potential role for soluble APP in signaling associated with thyroid carcinogenesis [165, 166].
Interestingly, E1 and/or E2 domains in the extracellular region of APP have been reported to mediate substrate-specific interaction between neurons and extracellular matrix components that include collagen I [167], heparin [56], laminin [168], glypican [169], F-spondin [170], and β1 integrin [64]. Based on these interactions, some of these matrix proteins have been considered as candidate ligands for APP including soluble APP [65] and Aβ per se [153, 171, 172], F-spondin [170], reelin [173], lingo-1 [151], contactins [151, 174, 175], β1 integrin [64, 173], and pancortin [175, 176]. Among these, reelin, lingo-1, and pancortin-1 were found to significantly inhibit APP ectodomain shedding, suggesting their roles as physiological modulators of APP processing or APP secretase activity per se [175]. Interestingly, it has been shown that cell surface interaction with soluble reelin with APP resides within the E1 domain of APP - the putative ligand domain [173]. This association favors the accumulation of cell surface APP and further increases of neurite outgrowth in cell culture system, which is consistent with ligand/receptor-mediated signaling.
A recent study reported that two cysteine residues (Cys186 and Cys187) within the APP-E1 domain could be modified by S-palmitoylation and that palmitoylated APP was preferentially targeted to the lipid raft microdomains [177]. How APP undergoes palmitoylation within the luminal domain, and how palmitoylation of APP ectodomain contributes to lipid raft targeting are not clearly understood. Nevertheless, by analogy to Wnt and particularly Hedgehog proteins [178, 179], palmitoylated secreted APP may serve as an anchor to recruit additional proteins to engage in signaling. Further studies are needed to understand the role of ectodomain palmitoylation in APP’s function as a ligand in cellular processes.
Cross-interactions of APP and APP metabolites with receptor-like proteins
APP has been reported to interact with a variety of receptor-like proteins (see Table 1). The structural features of APP, which confer its adhesive properties, support the notion that APP and its proteolytic products are capable of initiating molecular interactions that could affect its own signaling or signaling through its interacting partners. Notably, APP (especially its extracellular domain that contains the Aβ sequence) has a preference for interaction with type-1 transmembrane molecules including well-described cell-adhesion and growth factor receptors (Figure 4). The following interactions with cell-adhesion receptor molecules have been described: EphB2 [180, 181], netrin-1 [182], DCC [183], Notch [184, 185], N-cadherin [186], NCAM1 [151], L1/NgCAM [174] and immunoglobulin G Fcγ receptor II-b / FcγRIIb [187]. APP and its metabolites can interact with many growth factor receptors including leukocyte immunoglobulin-like receptor B2 / LilrB2 / PirB [188], epidermal growth factor / EGF receptor [94], p75 / TrkA / NGF receptor [189–192], insulin growth factor / IGF [156], and insulin receptor [156, 162, 193]. Insulin receptor subunits, which are enriched in lipid rafts, were found to act together with Aβ and soluble APP and consequently perturb insulin-dependent phosphorylation signaling cascade in neurons [156, 162, 193] (for review on GSK3β see [164, 194]). It has been shown as well that interaction of Aβ with PiRB receptor enhances phosphatase / cofilin cascade signaling events [188].
Another commonality among the group of receptors discussed above is that intramembranous cleavage by γ-secretase regulates several of these molecules. Interestingly, it has been shown that soluble APPα has also the ability to bind BACE1, a type-1 membrane protein [195]. This contributed to restraining subsequent interaction with holo-APP that would normally favor amyloidogenic pathway. This finding demonstrates the possibility of APP N-terminal fragment to attenuate excess secretase activity and offer neuroprotection. Conversely, an interaction of APP ectodomain with death receptor 6 / DR6 (also named tumor necrosis factor receptor family member 21 and a BACE1 substrate [196]) has been described to induce axonal degeneration through caspase-6 activation [190, 197]. This exemplifies again the capacity of soluble APP to exert a ligand-receptor like interaction (Figure 4a). The divergence in APP function strengthens the idea that cellular localization of APP and its selective interaction with protein partners may impact in various ways the cellular destiny. How soluble APP could interplay with BACE1 and be influential on other receptor signaling such as DR6 or putative APP receptor remains to be determined.
In addition to cell-adhesion receptor molecules and growth factor receptors, the soluble APP fragment and Aβ have been described to interact as well with cell surface glycosyl phosphatidylinositol / GPI-anchor proteins and GPCRs (Figure 4c). The interaction of APP N-terminal fragment with a variety of GPI-anchor protein family has been described for glypican [60, 169], syndecan [60], Nogo receptor / NgR [98, 99], transient axonal glycoprotein 1 / TAG-1 [198], and prion protein / PrP [151, 199]. The interest of a complex formation with GPI molecules resides in the ability of GPI-anchored proteins to act as co-receptor for extracellular ligands as described in other systems [116, 200]. Per their preferential localization in lipid rafts, GPI-APP association would promote complex formation with signaling molecules and mediate signal transduction. Moreover, ligands captured by readily-diffusible GPI-anchored protein (perhaps in this scenario, the soluble APP) could be efficiently presented to less mobile signaling receptors, thus increasing the sensitivity of the ligand recognition and signaling [200].
PrP interaction with Aβ has received a lot of attention because of the propensity of PrP to induce neurodegeneration and associated memory-like dysfunction [201]. It has been proposed that cell surface PrP can act as a receptor for Aβ [202]. Through a series of studies, Stephen Strittmatter’s research team reported that Aβ could function as a link between multiple cellular components to alter synaptic function and cognition. More specifically, they found that mGluR5 could facilitate PrP-bound Aβ oligomers to activate Fyn kinase cascade that leads to dendritic spine retraction [199, 203, 204]. The presence of Aβ might stabilize mGluR5 receptor at the synapse in a manner that interferes with receptor mobility and clustering. This succession of events enables mGluR5 receptor to induce signaling cascade [205]. Moreover, Aβ peptide has been reported to facilitate endocytosis of other receptors including ionotropic N-methyl-D-aspartic acid (NMDA) and alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors that will affect greatly spine formation and synaptic efficacy [206–208] (reviewed in ref [16])
An interaction of APP with GPCRs seems more elusive. The molecular structure of the GPCR renders them less likely to physically interact with type-1 transmembrane proteins like APP. However, the small, soluble Aβ peptides with charged residues might possess this property (Figure 4d). Indeed, it has been reported that Aβ associates with a variety of GPCRs including metabotropic glutamate receptor/mGluR5 [204], beta-2 adrenergic receptor/β2-ADR [209], amylin-3 [210, 211], frizzled [163], G-protein coupled receptor 3/GPR3 [212], and protaglandin2 receptor/PTGER2 [212]. Interestingly, association with Aβ mediates internalization of β2-ADR, GPR3, and PTGER2 through β-arrestin [212, 213]. Strikingly, β-arrestin is a common denominator for GPCR endocytosis, as well as internalization of a variety of type-1 transmembrane receptors [214, 215] Thus, APP and its ectodomain (including soluble APP and Aβ fragments) are capable of mediating signaling through crosstalk with other cell surface receptors and perhaps their endocytic molecular components. A similar complexity of receptor cross-interactions has been described for a variety of growth factor receptors and receptor tyrosine kinases [71, 216].
Functional consequence of APP internalization on Aβ clearance
Receptors are internalized upon ligand binding, which allows the ligand-activated receptor to recruit signaling molecules and efficiently transduce intracellular signaling. In addition, receptor internalization is fundamental to initiate receptor down-regulation and desensitization, to promote its clearance from the cell surface, and to further target its own degradation [216–218]. It is a common understanding that GPCRs (and non-GPCRs as well) are rapidly desensitized through receptor phosphorylation and β-arrestin coupling, which promote receptor internalization from the plasma membrane [219]. Binding with β-arrestin results in a partial uncoupling of G-proteins, and subsequent receptor targeting to clathrin coated pits for endocytosis. As discussed above, the APP cytoplasmic tail can complex with trimeric G-proteins [30, 47–50], as well as bind to clathrin-dependent sorting molecules [14, 96, 220]. Although the consequential partnerships associated with both occurrences have not been truly investigated, it is tempting to speculate that APP may go through these regulatory steps to sustain its receptorlike function. What is clear at present is that an association between APP and clathrin partners (such as AP-2, PICALM, AP-4) could profoundly influence APP internalization from the surface as well as its journey through the intracellular compartments [1, 221, 222]. Interestingly, Aβ interaction with β2-adrenergic receptor through β-arrestin has been reported to facilitate internalization of Aβ - adrenergic receptor complex, leading to receptor degradation [213]. Similarly, Aβ interaction with several other GPCRs has been documented [163, 165, 204, 210–212] (see below and Figure 4d). Whether in these instances Aβ is co-internalized with the receptors in a manner that contributes to Aβ clearance remains to be determined.
The extent to which APP undergoes internalization and clearance has direct consequences on the efficiency on proteolytic processing of APP. In this regard, interaction of APP with very low-density lipoprotein receptor (VLDLR) [223] and low-density lipoprotein receptor-related proteins (LRPs) [224, 225], including megalin (also named LRP2) [226], apolipoprotein E receptor 2 (ApoER2, also named LRP8) [227, 228], or sortilin-related receptor containing LDLR class-A repeats (SORLA, also named LRP9) [229–232] are notable. It has been shown that the interaction of APP with LRP1 at the cell surface accelerates APP endocytosis, and enhances Aβ production [233]. This process involves the cytoplasmic adaptor protein Fe65, which simultaneously binds to NPxY (682YENPTY687) motif in the cytoplasmic tails of APP and LRP1 (or LRP family members) forming a trimeric complex [223, 234]. In contrast, a trimeric association of APP with F-spondin and ApoER2 favors cell surface accumulation of APP and consequential processing through α-secretase and a decrease of Aβ production [227, 228]. Thus, APP interaction with endocytic binding partners has significant effects on APP trafficking, amyloidogenic processing of APP and Aβ clearance [96, 220, 229, 233] (Figure 4d).
LRP family members can also influence APP metabolism through complex formation with other transmembrane receptor partners. For example, in cultured cells and in mouse brain LRP6-mediated Wnt signaling protects against Aβ production, perhaps by promoting a longer residence of APP at the cell surface and consequently reducing the extent of amyloidogenic processing in endocytic organelles [235]. LRP family members can interact with HSPG and serve as a co-receptor for Wnt and sonic hedgehog signaling [236]. As highlighted in previous studies, receptor-protein complex formation with HSPGs is necessary for receptor internalization and receptor clearance [61, 62, 237]. Indeed, it has been proposed that HSPG may serve as a critical cellular component to promote Aβ clearance [233, 238]. The association of LRP with APP through HSPG has been described [239]. More recently, it has been shown that the close interaction of APP with syndecan [60], a transmembrane cell surface HSPG known to be associated with internalization of several receptors including LRP and VLDL [61, 62, 237], might promote the internalization of APP or APP-CTF. Complex formation with LRP, BACE1 and Ran-binding protein 9 (RanBP9), a scaffolding protein involved in trafficking of membrane receptors [240], reduces APP cell surface localization, and favors internalization of APP and generate Aβ [241, 242]. Similar functional outcome has been proposed for the interaction of APP with lingo proteins [151, 243], which can form a complex with NgR and p75 [244, 245]. Lingo family of proteins has been associated with growth factor receptor internalization and subsequent proteasomal degradation in other systems [245, 246]. Knockdown of lingo expression reduces Aβ production, apparently by favoring α-secretase processing of APP. However, overexpression of lingo results in increased lysosomal degradation of APP [243].
In general, receptor function and associated signaling would be determined through desensitization and receptor endocytosis, mediated by phosphorylation and β-arrestin binding. In addition, ubiquitination is emerging as an important post-translational modification that regulates trafficking and signaling processes to govern receptor functions [247]. More recently, APP ubiquitination has been proposed as a regulatory mechanism to influence APP internalization, and possibly clearance of its CTF. It has been shown that ubiquitination of APP at the juxtamembrane lysine residues is important for PIP3-dependent endosomal sorting of APP, and interfering with this process can lead to increase Aβ generation [248]. APP ubiquitination by FBL2 promotes proteasome-mediated degradation of APP and affects cerebral amyloid burden in transgenic mice [110] (see above).
Concluding Remarks
Accumulating evidence strongly supports the idea that APP is not only a molecule that produces the toxic Aβ peptides, but is also a cell-adhesion molecule and a growth factor ligand that can (i) interact with various binding protein partners; (ii) result in activation of signaling pathways; and (iii) elicit physiological responses. Thus, secretase-dependent processing of holo-APP is an excellent way to modulate the levels of cell surface APP receptor and also confer additional cellular roles for APP through the generation of APP metabolites. In agreement with this concept, soluble APP can modulate several signaling pathways involved in neuroprotection and axonal outgrowth whereas Aβ could repress the same signaling pathways, suggesting that soluble APP and Aβ possess antagonistic roles. The diverse APP interactions with a variety of proteins make it a target for tight regulation. Dysregulation of APP interactions certainly would shift the balance in its proteolytic processing from non-amyloidogenic to amyloidogenic processing, with an outcome that contributes to the cerebral amyloid burden. Consequentially, loss of APP homeostatic regulation could shift the balance toward more pathological outcomes. Aberrant modulation of its signaling partners, its protein interaction network, its cell surface receptor connectivity, could influence drastically APP’s innate physiological functions as a signaling growth factor or as a cell-adhesion molecule in the central nervous system and peripheral tissue. Despite the identification of numerous APP-interacting proteins, the quest for the discovery of credible ligand(s) for putative APP receptor remains unfulfilled (see Outstanding Questions). On the other hand, it is plausible that the APP ectodomain itself might serve as a ligand for APP and possibly other receptors as it has been proposed for lingo-1 [78, 79]. In all, cell surface recycling of putative APP receptor, either through endocytic trafficking, glycosylation, ubiquitination or kinase-dependent processes, would modulate Aβ production. Portraying APP as a receptor entity may provide us a different or even more meaningful understanding of how APP could play an important role in health and disease.
Outstanding Questions.
-
□
Does highly regulated APP processing account for homeostatic responses of APP receptor and consequently its functions?
-
□
Would wild-type and pathogenic APP adopt different pH-dependent conformations in a manner that prevents or facilitates protein dimerization or putative ligand interactions?
-
□
What would be the purpose of having APP and its metabolites in different subcellular compartments?
-
□
How would APP's multiple interactions with extracellular and intracellular binding partners regulate the activation of signaling cascades to influence pathophysiological outcomes?
-
□
Would APP dimerization or complex formation with other receptors affect their function and vice versa?
-
□
Rigorous research on APP over the past decades, stimulated by APP’s central role in AD pathogenesis, has led to the identification of a large number of APP interaction partners. The sheer number of APP interacting proteins gives the impression that APP is a major hub in cellular protein-protein interaction network. Nevertheless, it is a daunting task to determine which of these diverse interactions contributes to the multiple functions of APP under physiological or pathological conditions. Could the knowledge on APP interactors lead to a better understanding of those critical for the pathological outcome, and thus contribute to the disease process? Would it be possible to target APP interactions to modulate disease pathogenesis?
-
□
Dysregulation of APP interactions with endocytic partners is known to affect APP internalization and consequently APP processing and Aβ accumulation. Would it be possible to therapeutically modulate APP processing by targeting APP interactions with endocytic partners?
Trends.
-
□
The difficulty in defining the precise physiological and pathological function(s) of APP lies primarily in its complex proteolytic processing that generates various metabolites.
-
□
Based on its structural properties and its diverse functions, APP is not only a molecule that serves as the substrate for Alzheimer’s disease-associated Aβ peptides, but is also a receptor, a cell-adhesion molecule, and a growth factor ligand capable of activating signaling pathways that elicit physiological responses.
-
□
Portraying APP and its metabolites as ligand/receptor entity may provide us a more meaningful understanding of how APP could play an important role in health and disease.
Acknowledgments
Research in the authors’ labs were supported by grants from the National Institutes of Health (NS055223, AG046710, and AG042762 to ATP; and AG019070 and AG051230 to GT), BrightFocus Foundation (ATP), Cure Alzheimer's Fund (GT), Illinois Department of Public Health (CD and ATP), and Alzheimer’s Association (CD and ATP). We thank the members of our labs for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no competing financial interests.
References
- 1.Haass C, et al. Trafficking and Proteolytic Processing of APP. Cold Spring Harbor perspectives in medicine. 2012;2:a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willem M, et al. eta-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature. 2015 doi: 10.1038/nature14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z, et al. Delta-secretase cleaves amyloid precursor protein and regulates the pathogenesis in Alzheimer's disease. Nature communications. 2015;6:8762. doi: 10.1038/ncomms9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pellegrini L, et al. Alternative, non-secretase processing of Alzheimer's beta-amyloid precursor protein during apoptosis by caspase-6 and −8. J Biol Chem. 1999;274:21011–21016. doi: 10.1074/jbc.274.30.21011. [DOI] [PubMed] [Google Scholar]
- 6.Gervais FG, et al. Involvement of caspases in proteolytic cleavage of Alzheimer's amyloid-beta precursor protein and amyloidogenic A beta peptide formation. Cell. 1999;97:395–406. doi: 10.1016/s0092-8674(00)80748-5. [DOI] [PubMed] [Google Scholar]
- 7.Weidemann A, et al. Proteolytic processing of the Alzheimer's disease amyloid precursor protein within its cytoplasmic domain by caspase-like proteases. J Biol Chem. 1999;274:5823–5829. doi: 10.1074/jbc.274.9.5823. [DOI] [PubMed] [Google Scholar]
- 8.Lu DC, et al. A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor. Nat Med. 2000;6:397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- 9.Turner PR, et al. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70:1–32. doi: 10.1016/s0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, et al. Proteolytic processing of Alzheimer's beta-amyloid precursor protein. J Neurochem. 2012;120(Suppl 1):9–21. doi: 10.1111/j.1471-4159.2011.07519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Q, et al. APP physiological and pathophysiological functions: insights from animal models. Cell Res. 2012:1–12. doi: 10.1038/cr.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe MS, Guenette SY. APP at a glance. J Cell Sci. 2007;120:3157–3161. doi: 10.1242/jcs.03481. [DOI] [PubMed] [Google Scholar]
- 13.Zheng H, Koo EH. Biology and pathophysiology of the amyloid precursor protein. Mol Neurodegener. 2011;6:27. doi: 10.1186/1750-1326-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nhan HS, et al. The multifaceted nature of amyloid precursor protein and its proteolytic fragments: friends and foes. Acta neuropathologica. 2014 doi: 10.1007/s00401-014-1347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masters CL, Selkoe DJ. Biochemistry of amyloid beta-protein and amyloid deposits in Alzheimer disease. Cold Spring Harbor perspectives in medicine. 2012;2:a006262. doi: 10.1101/cshperspect.a006262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Querfurth HW, LaFerla FM. Alzheimer's disease. The New England journal of medicine. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 17.Benilova I, et al. The toxic Abeta oligomer and Alzheimer's disease: an emperor in need of clothes. Nat Neurosci. 2012;15:349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 18.Tu S, et al. Oligomeric Abeta-induced synaptic dysfunction in Alzheimer's disease. Mol Neurodegener. 2014;9:48. doi: 10.1186/1750-1326-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mucke L, Selkoe DJ. Neurotoxicity of amyloid beta-protein: synaptic and network dysfunction. Cold Spring Harbor perspectives in medicine. 2012;2:a006338. doi: 10.1101/cshperspect.a006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shankar GM, Walsh DM. Alzheimer's disease: synaptic dysfunction and Abeta. Mol Neurodegener. 2009;4:48. doi: 10.1186/1750-1326-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 22.Herrup K. The case for rejecting the amyloid cascade hypothesis. Nat Neurosci. 2015;18:794–799. doi: 10.1038/nn.4017. [DOI] [PubMed] [Google Scholar]
- 23.Musiek ES, Holtzman DM. Three dimensions of the amyloid hypothesis: time, space and 'wingmen'. Nat Neurosci. 2015;18:800–806. doi: 10.1038/nn.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckett C, et al. Nuclear signalling by membrane protein intracellular domains: the AICD enigma. Cell Signal. 2012;24:402–409. doi: 10.1016/j.cellsig.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Pardossi-Piquard R, Checler F. The physiology of the beta-amyloid precursor protein intracellular domain AICD. J Neurochem. 2012;120(Suppl 1):109–124. doi: 10.1111/j.1471-4159.2011.07475.x. [DOI] [PubMed] [Google Scholar]
- 26.Passer B, et al. Generation of an apoptotic intracellular peptide by gamma-secretase cleavage of Alzheimer's amyloid beta protein precursor. J Alzheimers Dis. 2000;2:289–301. doi: 10.3233/jad-2000-23-408. [DOI] [PubMed] [Google Scholar]
- 27.Puzzo D, et al. Picomolar amyloid-beta positively modulates synaptic plasticity and memory in hippocampus. J Neurosci. 2008;28:14537–14545. doi: 10.1523/JNEUROSCI.2692-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morley JE, et al. A physiological role for amyloid-beta protein:enhancement of learning and memory. J Alzheimers Dis. 2010;19:441–449. doi: 10.3233/JAD-2009-1230. [DOI] [PubMed] [Google Scholar]
- 29.Abramov E, et al. Amyloid-beta as a positive endogenous regulator of release probability at hippocampal synapses. Nat Neurosci. 2009;12:1567–1576. doi: 10.1038/nn.2433. [DOI] [PubMed] [Google Scholar]
- 30.Deyts C, et al. Novel GαS-protein signaling associated with membrane-tethered APP intracellular domain. J Neurosci. 2012;32:1714–1729. doi: 10.1523/JNEUROSCI.5433-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyssen M, et al. Amyloid precursor protein promotes post-developmental neurite arborization in the Drosophila brain. Embo J. 2005;24:2944–2955. doi: 10.1038/sj.emboj.7600757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang H, et al. Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer's Disease and Down syndrome. Proc Natl Acad Sci U S A. 2010;107:6538–6543. doi: 10.1073/pnas.0914251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ando K, et al. Role of phosphorylation of Alzheimer's amyloid precursor protein during neuronal differentiation. J Neurosci. 1999;19:4421–4427. doi: 10.1523/JNEUROSCI.19-11-04421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soba P, et al. Homo- and heterodimerization of APP family members promotes intercellular adhesion. Embo J. 2005;24:3624–3634. doi: 10.1038/sj.emboj.7600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu WQ, et al. Cell-surface beta-amyloid precursor protein stimulates neurite outgrowth of hippocampal neurons in an isoform-dependent manner. J Neurosci. 1995;15:2157–2167. doi: 10.1523/JNEUROSCI.15-03-02157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller UC, Zheng H. Physiological functions of APP family proteins. Cold Spring Harbor perspectives in medicine. 2012;2:a006288. doi: 10.1101/cshperspect.a006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumkotter F, et al. Structural aspects and physiological consequences of APP/APLP trans-dimerization. Exp Brain Res. 2012;217:389–395. doi: 10.1007/s00221-011-2878-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coburger I, et al. The structural biology of the amyloid precursor protein APP - a complex puzzle reveals its multi-domain architecture. Biological chemistry. 2014;395:485–498. doi: 10.1515/hsz-2013-0280. [DOI] [PubMed] [Google Scholar]
- 39.Kang J, et al. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 40.Parks AL, Curtis D. Presenilin diversifies its portfolio. Trends Genet. 2007;23:140–150. doi: 10.1016/j.tig.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Haapasalo A, Kovacs DM. The many substrates of presenilin/gamma-secretase. J Alzheimers Dis. 2011;25:3–28. doi: 10.3233/JAD-2011-101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarthy JV, et al. Presenilin-dependent regulated intramembrane proteolysis and gamma-secretase activity. Cell Mol Life Sci. 2009;66:1534–1555. doi: 10.1007/s00018-009-8435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ables JL, et al. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parent AT, et al. Presenilin attenuates receptor-mediated signaling and synaptic function. J Neurosci. 2005;25:1540–1549. doi: 10.1523/JNEUROSCI.3850-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai G, et al. Presenilin-dependent receptor processing is required for axon guidance. Cell. 2011;144:106–118. doi: 10.1016/j.cell.2010.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimoto I, et al. Alzheimer amyloid protein precursor complexes with brain GTP-binding protein G(o) Nature. 1993;362:75–79. doi: 10.1038/362075a0. [DOI] [PubMed] [Google Scholar]
- 48.Ramaker JM, et al. Amyloid precursor proteins interact with the heterotrimeric G protein Go in the control of neuronal migration. J Neurosci. 2013;33:10165–10181. doi: 10.1523/JNEUROSCI.1146-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brouillet E, et al. The amyloid precursor protein interacts with Go heterotrimeric protein within a cell compartment specialized in signal transduction. J Neurosci. 1999;19:1717–1727. doi: 10.1523/JNEUROSCI.19-05-01717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaked GM, et al. Interactions between the amyloid precursor protein C-terminal domain and G proteins mediate calcium dysregulation and amyloid beta toxicity in Alzheimer's disease. FEBS J. 2009;276:2736–2751. doi: 10.1111/j.1742-4658.2009.06997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang KA, et al. Phosphorylation of amyloid precursor protein (APP) at Thr668 regulates the nuclear translocation of the APP intracellular domain and induces neurodegeneration. Mol Cell Biol. 2006;26:4327–4338. doi: 10.1128/MCB.02393-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cupers P, et al. The amyloid precursor protein (APP)-cytoplasmic fragment generated by gamma-secretase is rapidly degraded but distributes partially in a nuclear fraction of neurones in culture. J Neurochem. 2001;78:1168–1178. doi: 10.1046/j.1471-4159.2001.00516.x. [DOI] [PubMed] [Google Scholar]
- 53.Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 54.Shariati SA, De Strooper B. Redundancy and divergence in the amyloid precursor protein family. FEBS Lett. 2013;587:2036–2045. doi: 10.1016/j.febslet.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 55.Reinhard C, et al. The amyloid-beta precursor protein: integrating structure with biological function. Embo J. 2005;24:3996–4006. doi: 10.1038/sj.emboj.7600860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small DH, et al. A heparin-binding domain in the amyloid protein precursor of Alzheimer's disease is involved in the regulation of neurite outgrowth. J Neurosci. 1994;14:2117–2127. doi: 10.1523/JNEUROSCI.14-04-02117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rossjohn J, et al. Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein. Nature structural biology. 1999;6:327–331. doi: 10.1038/7562. [DOI] [PubMed] [Google Scholar]
- 58.Stahl R, et al. Shedding of APP limits its synaptogenic activity and cell adhesion properties. Frontiers in cellular neuroscience. 2014;8:410. doi: 10.3389/fncel.2014.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaden D, et al. Subcellular localization and dimerization of APLP1 are strikingly different from APP and APLP2. J Cell Sci. 2009;122:368–377. doi: 10.1242/jcs.034058. [DOI] [PubMed] [Google Scholar]
- 60.Reinhard C, et al. Soluble amyloid-beta precursor protein binds its cell surface receptor in a cooperative fashion with glypican and syndecan proteoglycans. J Cell Sci. 2013;126:4856–4861. doi: 10.1242/jcs.137919. [DOI] [PubMed] [Google Scholar]
- 61.Sarrazin S, et al. Heparan sulfate proteoglycans. Cold Spring Harbor perspectives in biology. 2011:3. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsuo I, Kimura-Yoshida C. Extracellular distribution of diffusible growth factors controlled by heparan sulfate proteoglycans during mammalian embryogenesis. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2014:369. doi: 10.1098/rstb.2013.0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gralle M, et al. Neuroprotective secreted amyloid precursor protein acts by disrupting amyloid precursor protein dimers. J Biol Chem. 2009;284:15016–15025. doi: 10.1074/jbc.M808755200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young-Pearse TL, et al. Secreted APP regulates the function of full-length APP in neurite outgrowth through interaction with integrin beta1. Neural Dev. 2008;3:15. doi: 10.1186/1749-8104-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chasseigneaux S, Allinquant B. Functions of Abeta, sAPPalpha and sAPPbeta : similarities and differences. J Neurochem. 2011;120(Suppl 1):99–108. doi: 10.1111/j.1471-4159.2011.07584.x. [DOI] [PubMed] [Google Scholar]
- 66.Wade A, et al. Proteoglycans and their roles in brain cancer. FEBS J. 2013;280:2399–2417. doi: 10.1111/febs.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hacker U, et al. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 68.Jin LW, et al. Peptides containing the RERMS sequence of amyloid beta/A4 protein precursor bind cell surface and promote neurite extension. J Neurosci. 1994;14:5461–5470. doi: 10.1523/JNEUROSCI.14-09-05461.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maruyama IN. Mechanisms of activation of receptor tyrosine kinases: monomers or dimers. Cells. 2014;3:304–330. doi: 10.3390/cells3020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Endres NF, et al. Emerging concepts in the regulation of the EGF receptor and other receptor tyrosine kinases. Trends Biochem Sci. 2014;39:437–446. doi: 10.1016/j.tibs.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 71.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoefgen S, et al. Heparin induced dimerization of APP is primarily mediated by E1 and regulated by its acidic domain. Journal of structural biology. 2014;187:30–37. doi: 10.1016/j.jsb.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Ha Y. The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Mol Cell. 2004;15:343–353. doi: 10.1016/j.molcel.2004.06.037. [DOI] [PubMed] [Google Scholar]
- 74.Sosa LJ, et al. Amyloid precursor protein is an autonomous growth cone adhesion molecule engaged in contact guidance. PLoS One. 2013;8:e64521. doi: 10.1371/journal.pone.0064521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sosa LJ, et al. Dosage of amyloid precursor protein affects axonal contact guidance in Down syndrome. FASEB J. 2014;28:195–205. doi: 10.1096/fj.13-232686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baumkotter F, et al. Amyloid precursor protein dimerization and synaptogenic function depend on copper binding to the growth factor-like domain. J Neurosci. 2014;34:11159–11172. doi: 10.1523/JNEUROSCI.0180-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Z, et al. Presynaptic and postsynaptic interaction of the amyloid precursor protein promotes peripheral and central synaptogenesis. J Neurosci. 2009;29:10788–10801. doi: 10.1523/JNEUROSCI.2132-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jepson S, et al. LINGO-1, a transmembrane signaling protein, inhibits oligodendrocyte differentiation and myelination through intercellular self-interactions. J Biol Chem. 2012;287:22184–22195. doi: 10.1074/jbc.M112.366179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cobret L, et al. Targeting the cis-dimerization of LINGO-1 with low MW compounds affects its downstream signalling. Br J Pharmacol. 2015;172:841–856. doi: 10.1111/bph.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaden D, et al. The amyloid precursor protein and its homologues: structural and functional aspects of native and pathogenic oligomerization. European journal of cell biology. 2012;91:234–239. doi: 10.1016/j.ejcb.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 81.Mayer MC, et al. Novel zinc-binding site in the E2 domain regulates amyloid precursor-like protein 1 (APLP1) oligomerization. J Biol Chem. 2014;289:19019–19030. doi: 10.1074/jbc.M114.570382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scheinfeld MH, et al. Processing of beta-amyloid precursor-like protein-1 and −2 by gamma-secretase regulates transcription. J Biol Chem. 2002;277:44195–44201. doi: 10.1074/jbc.M208110200. [DOI] [PubMed] [Google Scholar]
- 83.Kuhn PH, et al. Secretome protein enrichment identifies physiological BACE1 protease substrates in neurons. EMBO J. 2012;31:3157–3168. doi: 10.1038/emboj.2012.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Munter LM, et al. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fluhrer R, et al. The alpha-helical content of the transmembrane domain of the British dementia protein-2 (Bri2) determines its processing by signal peptide peptidase-like 2b (SPPL2b) J Biol Chem. 2012;287:5156–5163. doi: 10.1074/jbc.M111.328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsuda S, et al. The familial dementia BRI2 gene binds the Alzheimer gene amyloid-beta precursor protein and inhibits amyloid-beta production. J Biol Chem. 2005;280:28912–28916. doi: 10.1074/jbc.C500217200. [DOI] [PubMed] [Google Scholar]
- 87.Perrin J, et al. TM9 family proteins control surface targeting of glycine-rich transmembrane domains. J Cell Sci. 2015;128:2269–2277. doi: 10.1242/jcs.164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsuda S, et al. Maturation of BRI2 generates a specific inhibitor that reduces APP processing at the plasma membrane and in endocytic vesicles. Neurobiol Aging. 2011;32:1400–1408. doi: 10.1016/j.neurobiolaging.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fotinopoulou A, et al. BRI2 interacts with amyloid precursor protein (APP) and regulates amyloid beta (Abeta) production. J Biol Chem. 2005;280:30768–30772. doi: 10.1074/jbc.C500231200. [DOI] [PubMed] [Google Scholar]
- 90.Matsuda S, et al. BRI2 inhibits amyloid beta-peptide precursor protein processing by interfering with the docking of secretases to the substrate. J Neurosci. 2008;28:8668–8676. doi: 10.1523/JNEUROSCI.2094-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamayev R, et al. APP heterozygosity averts memory deficit in knockin mice expressing the Danish dementia BRI2 mutant. EMBO J. 2011;30:2501–2509. doi: 10.1038/emboj.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Endres NF, et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bocharov EV, et al. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- 94.Caille I, et al. Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult subventricular zone. Development. 2004;131:2173–2181. doi: 10.1242/dev.01103. [DOI] [PubMed] [Google Scholar]
- 95.Hoefgen S, et al. The Amyloid Precursor Protein Shows a pH-Dependent Conformational Switch in Its E1 Domain. Journal of molecular biology. 2015;427:433–442. doi: 10.1016/j.jmb.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 96.Jiang S, et al. Trafficking regulation of proteins in Alzheimer's disease. Mol Neurodegener. 2014;9:6. doi: 10.1186/1750-1326-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gersbacher MT, et al. Turnover of amyloid precursor protein family members determines their nuclear signaling capability. PLoS One. 2013;8:e69363. doi: 10.1371/journal.pone.0069363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park JH, et al. Alzheimer precursor protein interaction with the Nogo-66 receptor reduces amyloid-beta plaque deposition. J Neurosci. 2006;26:1386–1395. doi: 10.1523/JNEUROSCI.3291-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou X, et al. Interaction between amyloid precursor protein and Nogo receptors regulates amyloid deposition. FASEB J. 2011;25:3146–3156. doi: 10.1096/fj.11-184325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leem JY, et al. A role for presenilin 1 in regulating the delivery of amyloid precursor protein to the cell surface. Neurobiol Dis. 2002;11:64–82. doi: 10.1006/nbdi.2002.0546. [DOI] [PubMed] [Google Scholar]
- 101.Hoe HS, et al. DAB1 and Reelin effects on amyloid precursor protein and ApoE receptor 2 trafficking and processing. J Biol Chem. 2006;281:35176–35185. doi: 10.1074/jbc.M602162200. [DOI] [PubMed] [Google Scholar]
- 102.Trommsdorff M, et al. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 103.Chaufty J, et al. Intracellular amyloid precursor protein sorting and amyloid-beta secretion are regulated by Src-mediated phosphorylation of Mint2. J Neurosci. 2012;32:9613–9625. doi: 10.1523/JNEUROSCI.0602-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuan YH, et al. PAT1a modulates intracellular transport and processing of amyloid precursor protein (APP), APLP1, and APLP2. J Biol Chem. 2006;281:40114–40123. doi: 10.1074/jbc.M605407200. [DOI] [PubMed] [Google Scholar]
- 105.Zheng P, et al. PAT1, a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein. Proc Natl Acad Sci U S A. 1998;95:14745–14750. doi: 10.1073/pnas.95.25.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dilsizoglu Senol A, et al. PAT1 inversely regulates the surface Amyloid Precursor Protein level in mouse primary neurons. BMC neuroscience. 2015;16:10. doi: 10.1186/s12868-015-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ullrich S, et al. The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. J Biol Chem. 2010;285:20664–20674. doi: 10.1074/jbc.M109.055608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vetrivel KS, et al. Dual roles of the transmembrane protein p23/TMP21 in the modulation of amyloid precursor protein metabolism. Mol Neurodegener. 2007;2:4. doi: 10.1186/1750-1326-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kumar P, et al. CHIP and HSPs interact with beta-APP in a proteasome-dependent manner and influence Abeta metabolism. Hum Mol Genet. 2007;16:848–864. doi: 10.1093/hmg/ddm030. [DOI] [PubMed] [Google Scholar]
- 110.Watanabe T, et al. FBL2 regulates amyloid precursor protein (APP) metabolism by promoting ubiquitination-dependent APP degradation and inhibition of APP endocytosis. J Neurosci. 2012;32:3352–3365. doi: 10.1523/JNEUROSCI.5659-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 112.Higashiyama S, et al. Membrane-anchored growth factors, the epidermal growth factor family: beyond receptor ligands. Cancer Sci. 2008;99:214–220. doi: 10.1111/j.1349-7006.2007.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vetrivel KS, et al. Spatial segregation of gamma-secretase and substrates in distinct membrane domains. J Biol Chem. 2005;280:25892–25900. doi: 10.1074/jbc.M503570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vetrivel KS, Thinakaran G. Membrane rafts in Alzheimer's disease beta-amyloid production. Biochim Biophys Acta. 2010;1801:860–867. doi: 10.1016/j.bbalip.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 116.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 117.Okamoto T, et al. A simple structure encodes G protein-activating function of the IGF-II/mannose 6-phosphate receptor. Cell. 1990;62:709–717. doi: 10.1016/0092-8674(90)90116-v. [DOI] [PubMed] [Google Scholar]
- 118.Timossi C, et al. Functional significance of the BBXXB motif reversed present in the cytoplasmic domains of the human follicle-stimulating hormone receptor. Mol Cell Endocrinol. 2004;223:17–26. doi: 10.1016/j.mce.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 119.Tamayev R, et al. The interactome of the amyloid beta precursor protein family members is shaped by phosphorylation of their intracellular domains. Mol Neurodegener. 2009;4:28. doi: 10.1186/1750-1326-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Muller T, et al. The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer's disease. Prog Neurobiol. 2008;85:393–406. doi: 10.1016/j.pneurobio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 121.Schettini G, et al. Phosphorylation of APP-CTF-AICD domains and interaction with adaptor proteins: signal transduction and/or transcriptional role - relevance for Alzheimer pathology. J Neurochem. 2010;115:1299–1308. doi: 10.1111/j.1471-4159.2010.07044.x. [DOI] [PubMed] [Google Scholar]
- 122.Gandy S, et al. Phosphorylation of Alzheimer disease amyloid precursor peptide by protein kinase C and Ca2+/calmodulin-dependent protein kinase II. Proc Natl Acad Sci U S A. 1988;85:6218–6221. doi: 10.1073/pnas.85.16.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Slomnicki LP, Lesniak W. A putative role of the Amyloid Precursor Protein Intracellular Domain (AICD) in transcription. Acta neurobiologiae experimentalis. 2008;68:219–228. doi: 10.55782/ane-2008-1691. [DOI] [PubMed] [Google Scholar]
- 124.Sumioka A, et al. Role of 14-3-3gamma in FE65-dependent gene transactivation mediated by the amyloid beta-protein precursor cytoplasmic fragment. J Biol Chem. 2005;280:42364–42374. doi: 10.1074/jbc.M504278200. [DOI] [PubMed] [Google Scholar]
- 125.Scheinfeld MH, et al. Amyloid beta protein precursor is phosphorylated by JNK-1 independent of, yet facilitated by, JNK-interacting protein (JIP)-1. J Biol Chem. 2003;278:42058–42063. doi: 10.1074/jbc.M304853200. [DOI] [PubMed] [Google Scholar]
- 126.Muresan Z, Muresan V. Coordinated transport of phosphorylated amyloid-beta precursor protein and c-Jun NH2-terminal kinase-interacting protein-1. J Cell Biol. 2005;171:615–625. doi: 10.1083/jcb.200502043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chiba K, et al. Quantitative analysis of APP axonal transport in neurons: role of JIP1 in enhanced APP anterograde transport. Mol Biol Cell. 2014;25:3569–3580. doi: 10.1091/mbc.E14-06-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fu MM, Holzbaur EL. JIP1 regulates the directionality of APP axonal transport by coordinating kinesin and dynein motors. J Cell Biol. 2013;202:495–508. doi: 10.1083/jcb.201302078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vagnoni A, et al. Loss of c-Jun N-terminal kinase-interacting protein-1 does not affect axonal transport of the amyloid precursor protein or Abeta production. Hum Mol Genet. 2013;22:4646–4652. doi: 10.1093/hmg/ddt313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lai A, et al. Characterization of sorting signals in the beta-amyloid precursor protein cytoplasmic domain. J Biol Chem. 1995;270:3565–3573. [PubMed] [Google Scholar]
- 131.Russo C, et al. Signal transduction through tyrosine-phosphorylated carboxy-terminal fragments of APP via an enhanced interaction with Shc/Grb2 adaptor proteins in reactive astrocytes of Alzheimer's disease brain. Annals of the New York Academy of Sciences. 2002;973:323–333. doi: 10.1111/j.1749-6632.2002.tb04660.x. [DOI] [PubMed] [Google Scholar]
- 132.Tarr PE, et al. Tyrosine phosphorylation of the beta-amyloid precursor protein cytoplasmic tail promotes interaction with Shc. J Biol Chem. 2002;277:16798–16804. doi: 10.1074/jbc.M110286200. [DOI] [PubMed] [Google Scholar]
- 133.Zhou D, et al. Growth factor receptor-bound protein 2 interaction with the tyrosine-phosphorylated tail of amyloid beta precursor protein is mediated by its Src homology 2 domain. J Biol Chem. 2004;279:25374–25380. doi: 10.1074/jbc.M400488200. [DOI] [PubMed] [Google Scholar]
- 134.Leissring MA, et al. A physiologic signaling role for the gamma -secretase-derived intracellular fragment of APP. Proc Natl Acad Sci U S A. 2002;99:4697–4702. doi: 10.1073/pnas.072033799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matrone C. A new molecular explanation for age-related neurodegeneration: the Tyr682 residue of amyloid precursor protein. BioEssays : news and reviews in molecular, cellular and developmental biology. 2013;35:847–852. doi: 10.1002/bies.201300041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tarr PE, et al. Evidence for a role of the nerve growth factor receptor TrkA in tyrosine phosphorylation and processing of beta-APP. Biochemical and biophysical research communications. 2002;295:324–329. doi: 10.1016/s0006-291x(02)00678-2. [DOI] [PubMed] [Google Scholar]
- 137.Araki Y, et al. Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid beta-protein precursor metabolism. J Biol Chem. 2003;278:49448–49458. doi: 10.1074/jbc.M306024200. [DOI] [PubMed] [Google Scholar]
- 138.Takei N, et al. Cytoplasmic Fragment of Alcadein alpha Generated by Regulated Intramembrane Proteolysis Enhances Amyloid beta-Protein Precursor (APP) Transport into the Late Secretory Pathway and Facilitates APP Cleavage. J Biol Chem. 2015;290:987–995. doi: 10.1074/jbc.M114.599852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Young-Pearse TL, et al. Biochemical and functional interaction of disrupted-in-schizophrenia 1 and amyloid precursor protein regulates neuronal migration during mammalian cortical development. J Neurosci. 2010;30:10431–10440. doi: 10.1523/JNEUROSCI.1445-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nakaya T, Suzuki T. Role of APP phosphorylation in FE65-dependent gene transactivation mediated by AICD. Genes to cells : devoted to molecular & cellular mechanisms. 2006;11:633–645. doi: 10.1111/j.1365-2443.2006.00968.x. [DOI] [PubMed] [Google Scholar]
- 141.Scheinfeld MH, et al. JNK-interacting protein-1 promotes transcription of A beta protein precursor but not A beta precursor-like proteins, mechanistically different than Fe65. Proc Natl Acad Sci U S A. 2003;100:1729–1734. doi: 10.1073/pnas.0437908100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gao Y, Pimplikar SW. The gamma -secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc Natl Acad Sci U S A. 2001;98:14979–14984. doi: 10.1073/pnas.261463298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Biederer T, et al. Regulation of APP-dependent transcription complexes by Mint/X11s: differential functions of Mint isoforms. J Neurosci. 2002;22:7340–7351. doi: 10.1523/JNEUROSCI.22-17-07340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ryan KA, Pimplikar SW. Activation of GSK-3 and phosphorylation of CRMP2 in transgenic mice expressing APP intracellular domain. J Cell Biol. 2005;171:327–335. doi: 10.1083/jcb.200505078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou F, et al. The APP intracellular domain (AICD) inhibits Wnt signalling and promotes neurite outgrowth. Biochim Biophys Acta. 2012;1823:1233–1241. doi: 10.1016/j.bbamcr.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 146.Goodger ZV, et al. Nuclear signaling by the APP intracellular domain occurs predominantly through the amyloidogenic processing pathway. J Cell Sci. 2009;122:3703–3714. doi: 10.1242/jcs.048090. [DOI] [PubMed] [Google Scholar]
- 147.Norstrom EM, et al. Identification of NEEP21 as a ss-amyloid precursor protein-interacting protein in vivo that modulates amyloidogenic processing in vitro. J Neurosci. 2010;30:15677–15685. doi: 10.1523/JNEUROSCI.4464-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kohli BM, et al. Interactome of the amyloid precursor protein APP in brain reveals a protein network involved in synaptic vesicle turnover and a close association with Synaptotagmin-1. Journal of proteome research. 2012;11:4075–4090. doi: 10.1021/pr300123g. [DOI] [PubMed] [Google Scholar]
- 149.Del Prete D, et al. APP is cleaved by Bace1 in pre-synaptic vesicles and establishes a pre-synaptic interactome, via its intracellular domain, with molecular complexes that regulate pre-synaptic vesicles functions. PLoS One. 2014;9:e108576. doi: 10.1371/journal.pone.0108576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fanutza T, et al. APP and APLP2 interact with the synaptic release machinery and facilitate transmitter release at hippocampal synapses. eLife. 2015:4. doi: 10.7554/eLife.09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bai Y, et al. The in vivo brain interactome of the amyloid precursor protein. Mol Cell Proteomics. 2008;7:15–34. doi: 10.1074/mcp.M700077-MCP200. [DOI] [PubMed] [Google Scholar]
- 152.Milosch N, et al. Holo-APP and G-protein-mediated signaling are required for sAPPalpha-induced activation of the Akt survival pathway. Cell death & disease. 2014;5:e1391. doi: 10.1038/cddis.2014.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fogel H, et al. APP homodimers transduce an amyloid-beta-mediated increase in release probability at excitatory synapses. Cell reports. 2014;7:1560–1576. doi: 10.1016/j.celrep.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 154.Cheng G, et al. Phosphatidylinositol-3-kinase-Akt kinase and p42/p44 mitogen-activated protein kinases mediate neurotrophic and excitoprotective actions of a secreted form of amyloid precursor protein. Exp Neurol. 2002;175:407–414. doi: 10.1006/exnr.2002.7920. [DOI] [PubMed] [Google Scholar]
- 155.Eckert GP, et al. Liposome-incorporated DHA increases neuronal survival by enhancing non-amyloidogenic APP processing. Biochim Biophys Acta. 2011;1808:236–243. doi: 10.1016/j.bbamem.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 156.Jimenez S, et al. Age-dependent accumulation of soluble amyloid beta (Abeta) oligomers reverses the neuroprotective effect of soluble amyloid precursor protein-alpha (sAPP(alpha)) by modulating phosphatidylinositol 3-kinase (PI3K)/Akt-GSK-3beta pathway in Alzheimer mouse model. J Biol Chem. 2011;286:18414–18425. doi: 10.1074/jbc.M110.209718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guo Q, et al. Secreted beta-amyloid precursor protein counteracts the proapoptotic action of mutant presenilin-1 by activation of NF-kappaB and stabilization of calcium homeostasis. J Biol Chem. 1998;273:12341–12351. doi: 10.1074/jbc.273.20.12341. [DOI] [PubMed] [Google Scholar]
- 158.Greenberg SM, et al. Amino-terminal region of the beta-amyloid precursor protein activates mitogen-activated protein kinase. Neurosci Lett. 1995;198:52–56. doi: 10.1016/0304-3940(95)11944-r. [DOI] [PubMed] [Google Scholar]
- 159.Venezia V, et al. Amyloid precursor protein modulates ERK-1 and −2 signaling. Annals of the New York Academy of Sciences. 2006;1090:455–465. doi: 10.1196/annals.1378.048. [DOI] [PubMed] [Google Scholar]
- 160.Nizzari M, et al. Amyloid precursor protein and Presenilin1 interact with the adaptor GRB2 and modulate ERK 1,2 signaling. J Biol Chem. 2007;282:13833–13844. doi: 10.1074/jbc.M610146200. [DOI] [PubMed] [Google Scholar]
- 161.Chasseigneaux S, et al. Secreted amyloid precursor protein beta and secreted amyloid precursor protein alpha induce axon outgrowth in vitro through Egr1 signaling pathway. PLoS One. 2011;6:e16301. doi: 10.1371/journal.pone.0016301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Townsend M, et al. Soluble Abeta inhibits specific signal transduction cascades common to the insulin receptor pathway. J Biol Chem. 2007;282:33305–33312. doi: 10.1074/jbc.M610390200. [DOI] [PubMed] [Google Scholar]
- 163.Magdesian MH, et al. Amyloid-beta binds to the extracellular cysteine-rich domain of Frizzled and inhibits Wnt/beta-catenin signaling. J Biol Chem. 2008;283:9359–9368. doi: 10.1074/jbc.M707108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Llorens-Martin M, et al. GSK-3beta, a pivotal kinase in Alzheimer disease. Frontiers in molecular neuroscience. 2014;7:46. doi: 10.3389/fnmol.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pietrzik CU, et al. From differentiation to proliferation: the secretory amyloid precursor protein as a local mediator of growth in thyroid epithelial cells. Proc Natl Acad Sci U S A. 1998;95:1770–1775. doi: 10.1073/pnas.95.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Krause K, et al. Evidence for a role of the amyloid precursor protein in thyroid carcinogenesis. The Journal of endocrinology. 2008;198:291–299. doi: 10.1677/JOE-08-0005. [DOI] [PubMed] [Google Scholar]
- 167.Beher D, et al. Regulation of amyloid protein precursor (APP) binding to collagen and mapping of the binding sites on APP and collagen type I. J Biol Chem. 1996;271:1613–1620. doi: 10.1074/jbc.271.3.1613. [DOI] [PubMed] [Google Scholar]
- 168.Kibbey MC, et al. beta-Amyloid precursor protein binds to the neurite-promoting IKVAV site of laminin. Proc Natl Acad Sci U S A. 1993;90:10150–10153. doi: 10.1073/pnas.90.21.10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Williamson TG, et al. Secreted glypican binds to the amyloid precursor protein of Alzheimer's disease (APP) and inhibits APP-induced neurite outgrowth. J Biol Chem. 1996;271:31215–31221. doi: 10.1074/jbc.271.49.31215. [DOI] [PubMed] [Google Scholar]
- 170.Ho A, Sudhof TC. Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proc Natl Acad Sci U S A. 2004;101:2548–2553. doi: 10.1073/pnas.0308655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lorenzo A, et al. Amyloid beta interacts with the amyloid precursor protein: a potential toxic mechanism in Alzheimer's disease. Nat Neurosci. 2000;3:460–464. doi: 10.1038/74833. [DOI] [PubMed] [Google Scholar]
- 172.Van Nostrand WE, et al. Localization of a fibrillar amyloid beta-protein binding domain on its precursor. J Biol Chem. 2002;277:36392–36398. doi: 10.1074/jbc.M204676200. [DOI] [PubMed] [Google Scholar]
- 173.Hoe HS, et al. Interaction of reelin with amyloid precursor protein promotes neurite outgrowth. J Neurosci. 2009;29:7459–7473. doi: 10.1523/JNEUROSCI.4872-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Osterfield M, et al. Interaction of amyloid precursor protein with contactins and NgCAM in the retinotectal system. Development. 2008;135:1189–1199. doi: 10.1242/dev.007401. [DOI] [PubMed] [Google Scholar]
- 175.Rice HC, et al. Systematic evaluation of candidate ligands regulating ectodomain shedding of amyloid precursor protein. Biochemistry. 2013;52:3264–3277. doi: 10.1021/bi400165f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rice HC, et al. Pancortins interact with amyloid precursor protein and modulate cortical cell migration. Development. 2012;139:3986–3996. doi: 10.1242/dev.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bhattacharyya R, et al. Palmitoylation of amyloid precursor protein regulates amyloidogenic processing in lipid rafts. J Neurosci. 2013;33:11169–11183. doi: 10.1523/JNEUROSCI.4704-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nusse R. Wnts and Hedgehogs: lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development. 2003;130:5297–5305. doi: 10.1242/dev.00821. [DOI] [PubMed] [Google Scholar]
- 179.Steinhauer J, Treisman JE. Lipid-modified morphogens: functions of fats. Current opinion in genetics & development. 2009;19:308–314. doi: 10.1016/j.gde.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cisse M, et al. Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature. 2011;469:47–52. doi: 10.1038/nature09635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cisse M, Checler F. Eph receptors: new players in Alzheimer's disease pathogenesis. Neurobiol Dis. 2015;73:137–149. doi: 10.1016/j.nbd.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 182.Lourenco FC, et al. Netrin-1 interacts with amyloid precursor protein and regulates amyloid-beta production. Cell death and differentiation. 2009;16:655–663. doi: 10.1038/cdd.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rama N, et al. Amyloid precursor protein regulates netrin-1-mediated commissural axon outgrowth. J Biol Chem. 2012;287:30014–30023. doi: 10.1074/jbc.M111.324780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fischer DF, et al. Activation of the Notch pathway in Down syndrome: cross-talk of Notch and APP. FASEB J. 2005;19:1451–1458. doi: 10.1096/fj.04-3395.com. [DOI] [PubMed] [Google Scholar]
- 185.Oh SY, et al. Amyloid precursor protein interacts with notch receptors. J Neurosci Res. 2005;82:32–42. doi: 10.1002/jnr.20625. [DOI] [PubMed] [Google Scholar]
- 186.Asada-Utsugi M, et al. N-cadherin enhances APP dimerization at the extracellular domain and modulates Abeta production. J Neurochem. 2011;119:354–363. doi: 10.1111/j.1471-4159.2011.07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kam TI, et al. FcgammaRIIb mediates amyloid-beta neurotoxicity and memory impairment in Alzheimer's disease. J Clin Invest. 2013;123:2791–2802. doi: 10.1172/JCI66827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kim T, et al. Human LilrB2 is a beta-amyloid receptor and its murine homolog PirB regulates synaptic plasticity in an Alzheimer's model. Science. 2013;341:1399–1404. doi: 10.1126/science.1242077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hasebe N, et al. Soluble beta-amyloid Precursor Protein Alpha binds to p75 neurotrophin receptor to promote neurite outgrowth. PLoS One. 2013;8:e82321. doi: 10.1371/journal.pone.0082321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nikolaev A, et al. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 191.Matrone C, et al. APP is phosphorylated by TrkA and regulates NGF/TrkA signaling. J Neurosci. 2011;31:11756–11761. doi: 10.1523/JNEUROSCI.1960-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Abel T, Nguyen PV. Regulation of hippocampus-dependent memory by cyclic AMP-dependent protein kinase. Prog Brain Res. 2008;169:97–115. doi: 10.1016/S0079-6123(07)00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhao WQ, et al. Amyloid beta oligomers induce impairment of neuronal insulin receptors. FASEB J. 2008;22:246–260. doi: 10.1096/fj.06-7703com. [DOI] [PubMed] [Google Scholar]
- 194.Hooper C, et al. The GSK3 hypothesis of Alzheimer's disease. J Neurochem. 2008;104:1433–1439. doi: 10.1111/j.1471-4159.2007.05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Obregon D, et al. Soluble amyloid precursor protein-alpha modulates beta-secretase activity and amyloid-beta generation. Nature communications. 2012;3:777. doi: 10.1038/ncomms1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hemming ML, et al. Identification of beta-secretase (BACE1) substrates using quantitative proteomics. PLoS One. 2009;4:e8477. doi: 10.1371/journal.pone.0008477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Olsen O, et al. Genetic analysis reveals that amyloid precursor protein and death receptor 6 function in the same pathway to control axonal pruning independent of beta-secretase. J Neurosci. 2014;34:6438–6447. doi: 10.1523/JNEUROSCI.3522-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ma QH, et al. A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat Cell Biol. 2008;10:283–294. doi: 10.1038/ncb1690. [DOI] [PubMed] [Google Scholar]
- 199.Um JW, et al. Alzheimer amyloid-beta oligomer bound to postsynaptic prion protein activates Fyn to impair neurons. Nat Neurosci. 2012;15:1227–1235. doi: 10.1038/nn.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 201.Westaway D, Jhamandas JH. The P's and Q's of cellular PrP-Abeta interactions. Prion. 2012;6:359–363. doi: 10.4161/pri.20675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lauren J, et al. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gimbel DA, et al. Memory impairment in transgenic Alzheimer mice requires cellular prion protein. J Neurosci. 2010;30:6367–6374. doi: 10.1523/JNEUROSCI.0395-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Um JW, et al. Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer abeta oligomer bound to cellular prion protein. Neuron. 2013;79:887–902. doi: 10.1016/j.neuron.2013.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Renner M, et al. Deleterious effects of amyloid beta oligomers acting as an extracellular scaffold for mGluR5. Neuron. 2010;66:739–754. doi: 10.1016/j.neuron.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hsieh H, et al. AMPAR removal underlies Abeta-induced synaptic depression and dendritic spine loss. Neuron. 2006;52:831–843. doi: 10.1016/j.neuron.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Innocent N, et al. NMDA receptor/amyloid precursor protein interactions: a comparison between wild-type and amyloid precursor protein mutations associated with familial Alzheimer's disease. Neurosci Lett. 2012;515:131–136. doi: 10.1016/j.neulet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 208.Snyder EM, et al. Regulation of NMDA receptor trafficking by amyloid-beta. Nat Neurosci. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 209.Wang D, et al. Binding of amyloid beta peptide to beta2 adrenergic receptor induces PKA-dependent AMPA receptor hyperactivity. FASEB J. 2010;24:3511–3521. doi: 10.1096/fj.10-156661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Fu W, et al. Amyloid beta (Abeta) peptide directly activates amylin-3 receptor subtype by triggering multiple intracellular signaling pathways. J Biol Chem. 2012;287:18820–18830. doi: 10.1074/jbc.M111.331181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rezaei-Ghaleh N, et al. Interaction between amyloid beta peptide and an aggregation blocker peptide mimicking islet amyloid polypeptide. PLoS One. 2011;6:e20289. doi: 10.1371/journal.pone.0020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nelson CD, Sheng M. Gpr3 stimulates Abeta production via interactions with APP and beta-arrestin2. PLoS One. 2013;8:e74680. doi: 10.1371/journal.pone.0074680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang D, et al. Amyloid beta peptide-(1-42) induces internalization and degradation of beta2 adrenergic receptors in prefrontal cortical neurons. J Biol Chem. 2011;286:31852–31863. doi: 10.1074/jbc.M111.244335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kang DS, et al. Role of beta-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr Opin Cell Biol. 2014;27:63–71. doi: 10.1016/j.ceb.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 216.Girnita L, et al. Something old, something new and something borrowed: emerging paradigm of insulin-like growth factor type 1 receptor (IGF-1R) signaling regulation. Cell Mol Life Sci. 2014;71:2403–2427. doi: 10.1007/s00018-013-1514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Pandey KN. Endocytosis and Trafficking of Natriuretic Peptide Receptor-A: Potential Role of Short Sequence Motifs. Membranes. 2015;5:253–287. doi: 10.3390/membranes5030253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Morcavallo A, et al. Ligand-mediated endocytosis and trafficking of the insulinlike growth factor receptor I and insulin receptor modulate receptor function. Frontiers in endocrinology. 2014;5:220. doi: 10.3389/fendo.2014.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.DeWire SM, et al. Beta-arrestins and cell signaling. Annual review of physiology. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 220.Small SA, Gandy S. Sorting through the cell biology of Alzheimer's disease: intracellular pathways to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Burgos PV, et al. Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex. Developmental cell. 2010;18:425–436. doi: 10.1016/j.devcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tian Y, et al. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci U S A. 2013;110:17071–17076. doi: 10.1073/pnas.1315110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dumanis SB, et al. FE65 as a link between VLDLR and APP to regulate their trafficking and processing. Mol Neurodegener. 2012;7:9. doi: 10.1186/1750-1326-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pietrzik CU, et al. The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. EMBO J. 2002;21:5691–5700. doi: 10.1093/emboj/cdf568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kanekiyo T, et al. Neuronal clearance of amyloid-beta by endocytic receptor LRP1. J Neurosci. 2013;33:19276–19283. doi: 10.1523/JNEUROSCI.3487-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Alvira-Botero X, et al. Megalin interacts with APP and the intracellular adapter protein FE65 in neurons. Mol Cell Neurosci. 2010;45:306–315. doi: 10.1016/j.mcn.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 227.Hoe HS, et al. F-spondin interaction with the apolipoprotein E receptor ApoEr2 affects processing of amyloid precursor protein. Mol Cell Biol. 2005;25:9259–9268. doi: 10.1128/MCB.25.21.9259-9268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Fuentealba RA, et al. ApoER2 expression increases Abeta production while decreasing Amyloid Precursor Protein (APP) endocytosis: Possible role in the partitioning of APP into lipid rafts and in the regulation of gamma-secretase activity. Mol Neurodegener. 2007;2:14. doi: 10.1186/1750-1326-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Willnow TE, Andersen OM. Sorting receptor SORLA--a trafficking path to avoid Alzheimer disease. J Cell Sci. 2013;126:2751–2760. doi: 10.1242/jcs.125393. [DOI] [PubMed] [Google Scholar]
- 230.Mehmedbasic A, et al. SorLA CR-Domains Protect the Amyloid Precursor Protein against Processing. J Biol Chem. 2014 doi: 10.1074/jbc.M114.619940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Caglayan S, et al. Lysosomal sorting of amyloid-beta by the SORLA receptor is impaired by a familial Alzheimer's disease mutation. Science translational medicine. 2014;6:223ra220. doi: 10.1126/scitranslmed.3007747. [DOI] [PubMed] [Google Scholar]
- 232.Schmidt V, et al. Quantitative modelling of amyloidogenic processing and its influence by SORLA in Alzheimer's disease. EMBO J. 2012;31:187–200. doi: 10.1038/emboj.2011.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kanekiyo T, Bu G. The low-density lipoprotein receptor-related protein 1 and amyloid-beta clearance in Alzheimer's disease. Frontiers in aging neuroscience. 2014;6:93. doi: 10.3389/fnagi.2014.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pietrzik CU, et al. FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J Neurosci. 2004;24:4259–4265. doi: 10.1523/JNEUROSCI.5451-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Liu CC, et al. Deficiency in LRP6-mediated Wnt signaling contributes to synaptic abnormalities and amyloid pathology in Alzheimer's disease. Neuron. 2014;84:63–77. doi: 10.1016/j.neuron.2014.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 237.Christianson HC, Belting M. Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix biology : journal of the International Society for Matrix Biology. 2014;35:51–55. doi: 10.1016/j.matbio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 238.Zhang GL, et al. Towards understanding the roles of heparan sulfate proteoglycans in Alzheimer's disease. BioMed research international. 2014;2014:516028. doi: 10.1155/2014/516028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kanekiyo T, et al. Heparan sulphate proteoglycan and the low-density lipoprotein receptor-related protein 1 constitute major pathways for neuronal amyloid-beta uptake. J Neurosci. 2011;31:1644–1651. doi: 10.1523/JNEUROSCI.5491-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Suresh B, et al. Diverse roles of the scaffolding protein RanBPM. Drug discovery today. 2012;17:379–387. doi: 10.1016/j.drudis.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 241.Lakshmana MK, et al. Novel role of RanBP9 in BACE1 processing of amyloid precursor protein and amyloid beta peptide generation. J Biol Chem. 2009;284:11863–11872. doi: 10.1074/jbc.M807345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Woo JA, et al. Pivotal role of RanBP9 in integrin-dependent focal adhesion signaling and assembly. FASEB J. 2012;26:1672–1681. doi: 10.1096/fj.11-194423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.de Laat R, et al. LINGO-1 promotes lysosomal degradation of amyloid-beta protein precursor. Pathobiology of aging & age related diseases. 2015;5:25796. doi: 10.3402/pba.v5.25796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Mi S, et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7:221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 245.Meabon JS, et al. LINGO-1 Protein Interacts with the p75 Neurotrophin Receptor in Intracellular Membrane Compartments. J Biol Chem. 2015;290:9511–9520. doi: 10.1074/jbc.M114.608018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Mandai K, et al. LIG family receptor tyrosine kinase-associated proteins modulate growth factor signals during neural development. Neuron. 2009;63:614–627. doi: 10.1016/j.neuron.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Marchese A, Trejo J. Ubiquitin-dependent regulation of G protein-coupled receptor trafficking and signaling. Cell Signal. 2013;25:707–716. doi: 10.1016/j.cellsig.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Morel E, et al. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nature communications. 2013;4:2250. doi: 10.1038/ncomms3250. [DOI] [PMC free article] [PubMed] [Google Scholar]