Abstract
Long-chain fatty acid amides are signaling lipids found in mammals and other organisms; however, details of the metabolic pathways for the N-acylglycines and primary fatty acid amides (PFAMs) have remained elusive. Heavy-labeled precursor and subtraction lipidomic experiments in mouse neuroblastoma N18TG2 cells, a model cell line for the study of fatty acid amide metabolism, establish the biosynthetic pathways for the N-acylglycines and the PFAMs. We provide evidence that the N-acylglycines are formed by a long-chain specific glycine-conjugating enzyme, glycine N-acyltransferase-like 3 (GLYATL3). siRNA knockdown of GLYATL3 in the N18TG2 cells resulted in a decrease in the levels of the N-acylglycines and the PFAMs. This is the first report of an enzyme responsible for long-chain N-acylglycine production in cellula. The production of the PFAMs in N18TG2 cells was reported to occur by the oxidative cleavage of the N-acylglycines, as catalyzed by peptidylglycine α-amidating monooxygenase (PAM). siRNA knockdown of PAM resulted in an accumulation of [13C18]N-oleoylglycine and decreased levels of [13C18]oleamide when the N18TG2 cells were grown in the presence of [13C18]oleic acid. The addition of [1-13C]palmitate to the N18TG2 cell growth media led to the production of a family of [1-13C]palmitoylated fatty acid amides, consistent with the biosynthetic pathways detailed herein.
Keywords: siRNA knockdown, neuroblastoma cells, N-acylamide, arachidonic acid, brain lipids, lipids, eicosanoids, mass spectrometry, palmitoylation, oleamide
The long-chain fatty acid amides are an emerging family of bioactive lipids with members that include N-acyl amino acids, primary fatty acid amides (PFAMs), N-acylarylalkylamides, N-acylethanolamines, and N-monoacylpolyamines. Fatty acid amides were first identified from biological sources over 50 years ago with the isolation and identification of N-palmitoylethanolamine from egg yolk in 1957 (1) and, in 1965, N-palmitoylethanolamine and N-stearoylethanolamine in several tissues from rat and guinea pig (2). The discovery of N-arachidonoylethanolamine (anandamide) as the endogenous ligand for the mammalian brain cannabinoid receptor CB1 sparked a dramatic interest in the fatty acid amides (3). Since these early discoveries, a diversity of long-chain fatty acid amides have been identified in mammals and, more recently, in invertebrates as well (4–8).
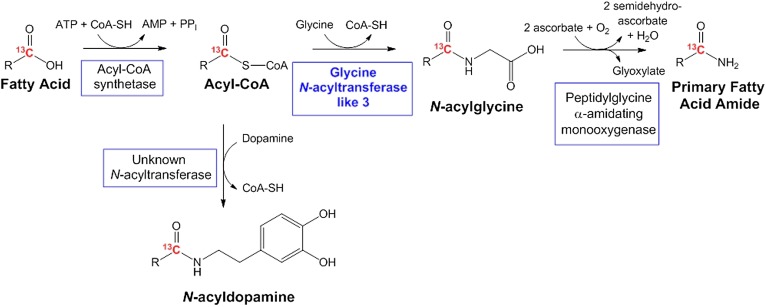
The N-fatty acylglycines, a subclass of the N-fatty acyl amino acids, are an important class of cell signaling lipids that are distributed throughout the central nervous system and the rest of the body (6, 7, 9). Identification of glycine conjugates dates back to the 1820s with the identification of N-benzoylglycine (hippurate) as a mammalian metabolite (10). N-arachidonoylglycine was the first long-chain N-acylglycine identified from a mammalian source and was determined to have anti-nociceptive and anti-inflammatory effects in rat models of pain (11). Since 2001, other N-acylglycines with varying long-chain N-acyl groups have been identified in mammals and invertebrates, and have been shown to be important signaling molecules (4, 5, 7, 12–15). However, the details of long-chain N-acylglyine metabolism have remained elusive. There are two proposed pathways for N-acylgycine biosynthesis: a) sequential oxidation of an N-acylethanolamine by alcohol dehydrogenase and aldehyde dehydrogenase (16–19); and b) activation of a fatty acid by an acyl-CoA synthetase followed by the conjugation with glycine via a long-chain specific glycine N-acyltransferase (GLYAT) (Fig. 1) (5, 9, 12, 19, 20).
Fig. 1.
Proposed pathways for the biosynthesis of the N-acylglycines, PFAMs, and N-acyldopamines in mammalian systems. The novel enzyme identified in this work, mGLYATL3, is shown in blue. The red carbons represent the metabolic flow of the 13C atom based on data reported herein.
GLYAT catalyzes the conjugation of short-chain acyl-CoA thioesters and glycine to produce short-chain N-acylglycines (C2-C10) (21–23). We proposed that a GLYAT-like (GLYATL) enzyme would catalyze the formation of long-chain N-acylglycines in a similar reaction in vivo (12, 19). In 2010, Waluk, Schultz, and Hunt (24) described four human GLYATs named hGLYAT, hGLYATL1, hGLYATL2, and hGLYATL3. Interestingly, hGLYATL2 prefers oleoyl-CoA as a substrate and catalyzes the synthesis of N-oleoylglycine from oleoyl-CoA and glycine in vitro. A Basic Local Alignment Search Tool (BLAST) search of the hGLYATL orthologs against the mouse genome yielded two GLYATs, mouse (m)GLYAT and mGLYATL3. Recently, Dempsey et al. (23) characterized recombinant mGLYAT with glycine serving as the best amino donor substrate and benzoyl-CoA serving as the best amino acceptor substrate. The formation of N-acylglycines possessing acyl chains longer than six carbon atoms was not observed. Therefore, we proposed that the remaining GLYATL ortholog, mGLYATL3, might be the enzyme responsible for long-chain N-acylglycine formation in vivo. To test this hypothesis, we analyzed fatty acid amide levels before and after knocking down mGLYATL3 expression with siRNA, a technique termed subtraction lipidomics. We applied this subtraction lipidomic technique to mouse N18TG2 cells because these cells produce fatty acid amides, including the long-chain N-acylglycines and PFAMs (12), and express many of the enzymes proposed to function in fatty acid amide metabolism (12, 25, 26). Knockdown of mGLYATL3 in the N18TG2 cells resulted in decreases in the cellular levels of both the long-chain N-acylglycines and the PFAMs.
Merkler et al. (12) proposed that the long-chain N-acylglycines are intermediates in the PFAM biosynthetic pathway: fatty acid → fatty acyl-CoA → N-fatty acylglycine → PFAM (Fig. 1). In this pathway, a long-chain N-acylglycine is oxidatively cleaved to the corresponding PFAM and glyoxylate in a reaction catalyzed by peptidylglycine α-amidating monooxygenase (PAM). PAM knockdown in the N18TG2 cells resulted in a decrease in the cellular levels of the PFAMs and an increase in cellular levels of the long-chain N-acylglycines; results consistent with the proposed pathway in Fig. 1. Our research provides key new information regarding the enzyme-catalyzed reactions that occur in cellula to convert fatty acids into the long-chain N-acylglycines and the PFAMs.
MATERIALS AND METHODS
Materials
N-oleoylglycine, N-palmitoylglycine, N-arachidonoylglycine-d8, N-oleoylethanolamine, N-palmitoyldopamine, N-oleoyldopamine, and N-arachidonoyldopamine were from Cayman Chemical Co. siPORTTM amine transfection reagent and Silencer® Select Predesigned siRNA against mouse (m)PAM and mGLYATL2 were from Life Technologies. PAM (S-16) primary antibody was from Santa Cruz Biotechnologies, Inc. Donkey anti-goat secondary antibody conjugated with horseradish peroxidase was from ICN Laboratories. mGLYATL3 primary antibody was custom made by Genscript. Goat anti-rabbit secondary antibody conjugated with horseradish peroxidase and β-actin loading control antibody were from Genscript. N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) and silica were from Supelco. Heptadecanoic acid-d33 was from C/D/N Isotopes. Fatty acid-free BSA (BSA), [13C18]oleic acid, [1-13C]palmitic acid, protease inhibitor cocktail (P8340), and 6-thioguanine were from Sigma-Aldrich. DMEM and penicillin/streptomycin were from Mediatech Cellgro. Mouse neuroblastoma N18TG2 cells were from DSMZ (Deutsche Sammlung von Mikrooganism und Zellkuturen GmBH). Polyvinylidene fluoride membranes were from Millipore. SuperSignal chemiluminescent horseradish peroxidase substrate was from Pierce. MagicMark XP molecular weight ladder was from Invitrogen. All other reagents and cell culture supplies were of the highest quality available from commercial suppliers.
Methods
N18TG2 cell culture.
Cells were grown in 225 cm2 culture flasks in an incubator with a temperature of 37°C and 5% CO2. The medium was DMEM supplemented with 100 μM 6-thioguanine, 10% FBS, 100 IU/ml penicillin, and 1.0 mg/ml streptomycin. Aliquots of cells were taken for cell counting using a hemocytometer and trypan blue dye.
Identification and quantification of endogenous long-chain fatty acid amides in N18TG2 cells by LC/QTOF-MS.
Lipids, including the long-chain fatty acid amides, were extracted from the N18TG2 cells using a previously published extraction method (27, 28). A solid-phase extraction method described earlier (5, 27) was subsequently used to further purify the long-chain fatty acid amides from the dried lipid extracts. The fatty acid amide-containing fractions were combined, dried under N2 at 40°C, and then stored at −20°C until analyzed in more detail. An Agilent 1260 LC system with a Kinetex™ 2.6 μm C18 100 Å (50 × 2.1 mm) reverse phase column was connected to an Agilent 6540 quadrupole TOF (QTOF) mass spectrometer equipped with a Dual Agilent Jetstream ESI source in positive ion mode. The LC/QTOF-MS methods that we used are described in Jeffries et al. (5). Extracted ion chromatograms were obtained from the total ion chromatograms for each of the long-chain fatty acid amides using Agilent MassHunter Qualitative Analysis B.04.00. The fatty acid amides in each extraction were identified by comparison to synthetic standards by molecular ion m/z and retention time. Synthetic standards of the N-acylglycines, N-acyldopamines, and N-acylethanolamines were commercially available. The PFAMs were synthesized according to Jeffries et al. (5). Run-to-run variation for all metabolites was less than ±0.1 min.
Quantification of each identified fatty acid amide was performed by integrating the area under the chromatographic peak and comparing that value against a standard curve constructed using the same fatty acid amide. Standard curves were in the linear range of 0.1–10 pmoles on the column (r2 > 0.99). N-arachidonoylglycine-d8, 1 pmole per 10 μl injection, was spiked into each extraction to measure instrument performance and for data normalization. Solvent and slip additive blanks were run to evaluate background oleamide and palmitamide levels (29). In addition, blank samples were run between standards and extracts to eliminate memory effects from the HPLC column. Background levels of these PFAMs were subtracted from analyte levels and were <10% of the endogenous levels in the N18TG2 cells. The levels of the N-acyldopamine reported herein represent the sum total of the amount of the N-acyldopamine and the amount of the N-acyldopamine quinone. The amounts of endogenous fatty acid amides in the N18TG2 cells were reported as the average of the determinations along with the standard deviation. For statistical analyses, the Holm-Šidák method was used to correct for multiple comparisons (30, 31).
RT-PCR of GLYATL3 transcripts.
Ambion MicroPoly(A) Purist kit was used to purify mRNA from N18TG2 cells grown as described above. An Ambion Retroscript kit was used to generate a cDNA library for RT-PCR experiments. Identification of mGLYATL3 transcripts was completed by nested RT-PCR. The outer PCR product was generated with a denaturation step at 95°C for 5 min followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. A final extension step was performed at 72°C for 10 min. The nested product was subsequently generated with the same PCR conditions. The outer primers (sense, CAAATACAAGGGTTGCAAAGTG; antisense, TAACCTTTCCTGCGATGGTC) were designed and synthesized by Eurofins MWG Operon to amplify a 603 bp product. The amplified product from the RT-PCR reaction was purified from the outer primers using ultrafiltration with a 100 kDa cut-off filter. The nested primers (sense, GCGAGAGAGAGAGGCTGAGA; antisense, ATGGAAGCCAGGTTGTCATC) were designed and synthesized by Eurofins MWG Operon to amplify a 380 bp product. This product was analyzed by a 1.3% agarose gel and the band was visualized using 0.5 μg/ml ethidium bromide under UV light. The positive bands at 380 bp were cut out of the gel, purified by the Promega Wizard SV Gel and PCR Clean-up system, and sequenced by Eurofins MWG Operon.
siRNA knockdown of GLYATL3 in N18TG2 cells.
The N18TG2 cells were transfected with anti-mGLYATL3 siRNA using siPORT amine transfection reagent in order to achieve knockdown of mGLYATL3. N18TG2 cells were grown to 80% confluency and then collected by scraping into fresh medium. In each well of a 6-well plate, 2 × 105 cells were transfected with 10 μl transfection reagent and 20 nM siRNA (sense, GGUUGCAAAGUGAAUUAUATT; antisense, UAUAAUUCACUUUGCAACCCT) in growth medium containing 0% FBS via a “reverse transfection method.” Cultures were incubated for 5, 8, 12, or 24 h before the cells were collected by scraping and centrifugation. After washing with PBS, the cell pellets were stored at −80°C until the long-chain fatty acid amides were extracted, purified, and analyzed by LC/QTOF-MS as described above. Negative controls include: a) incubation with transfection reagent only or b) transfection reagent with Silencer® Select Negative Control No. 1 (scrambled siRNA). Three separate independent experiments were performed for each set of conditions.
siRNA knockdown of PAM and purification of N-oleoylglycine in N18TG2 cells.
The N18TG2 cells were transfected with anti-PAM siRNA to achieve knockdown of mouse PAM. N18TG2 cells were grown to 80% confluency in 6-well plates. The medium was removed and replaced with 0.5% FBS medium containing 2.5 mM [13C18]oleate and 2.5 mM BSA. After 8 h, anti-mPAM siRNA (sense, GCGGGAAGUUCAUCACUCATT; antisense, UGAGUGAUGAACUUCCCGCTT) was added to each well in medium with 0% FBS. After a total incubation time of 56 h, the cells were collected by scraping and centrifugation. After washing with PBS, cell pellets were stored at −80°C until the labeled fatty acid amides were extracted. Labeled and unlabeled oleamide was extracted from the cell pellet and subsequently purified using the methods of Farrell et al. (28). Labeled and unlabeled N-oleoylglycine was extracted and purified using preparative TLC. Dried lipid extracts were applied to a prewashed (chloroform:acetone 1:1) 10 cm EMD analytical normal phase TLC plate. Chloroform:methanol:acetic acid (95:5:1) was used until the mobile phase reached 5.5 cm from the bottom of the plate. The plate was dried and placed in the second running solvent, hexane:diethyl ether:acetone (60:40:5), and run until the mobile phase reached 8 cm. After drying, the final solvent, hexane:diethyl ether (97:3), was run until the mobile phase reached 9.5 cm. An N-oleoylglycine standard was run alongside the cell extract and visualized using KMnO4 to test for position of the N-acylglycines. The appropriate area was scraped from the TLC plate into anhydrous isopropanol and sonicated in a water bath sonicator for 5 min. After initial sonication, the sample was centrifuged to pellet the silica. The supernatant liquid was syringe filtered with a 0.22 μm filter and dried under nitrogen at 40–50°C. The silica was subjected to another round of sonication in isopropanol, centrifugation, and syringe filtering before adding to the previous supernatant. Dried samples were trimethylsilylated with 100 μl BSTFA. Samples were flushed with dry N2, BSTFA added, flushed briefly again, and allowed to react at 55–60°C for 1 h before running on GC-MS. Samples were spiked with an internal standard, heptadecanoic acid-d33, to monitor instrument performance and then injected onto a Shimadzu QP-5000 GC-MS using the conditions described in Farrell et al. (28). Quantification of analytes was achieved by integration of the area under the chromatographic peak compared with a standard curve. The major product of oleamide derivitization was oleonitrile, and oleamide-TMS was also observed (28). The observed sum of these products represents the level of oleamide in the N18TG2 cell extract, as determined by standards run with chemically pure oleamide. The level of the N-oleoylglycine reported herein represents the sum total of the amount of the observed mono- and di-TMS derivatives of N-oleoylglycine. Methods were validated by running spiked controls, and extraction efficiency was determined to be approximately 90% from picomole levels of metabolites in solution.
Western blotting for RNAi confirmation.
Knockdown resulting from the RNAi experiments was monitored via Western blotting. Ten percent SDS-PAGE was run and, subsequently, electroblotted to a polyvinylidene fluoride membrane. The membranes were blocked with 5% nonfat dry milk, incubated with the respective primary antibody, and treated with a secondary antibody conjugated to horseradish peroxidase. Visualization of bands was achieved with the SuperSignal chemiluminescent system on radiographic film. β-Actin antibody was used as a loading control.
[1-13C]palmitate feeding study in N18TG2 cells.
N18TG2 cells were grown to 80% confluency and fresh medium containing 0.5% FBS, 2.5 mM [1-13C]palmitate, and 2.5 mM BSA was added to the culture flask. BSA, prepared as described in Farrell et al. (28), was used as a carrier for [1-13C]palmitate into the N18TG2 cells. After a 48 h incubation, the medium was removed and the cells were washed with PBS. Cells were collected by scraping, followed by centrifugation and washing with PBS. Cell pellets were stored at −80°C until analyzed by LC/QTOF-MS, as described above. Similar to the endogenous analysis of the N-acyldopamines, the levels of [13C]N-acyldopamine and [13C]N-acyldopamine quinone were combined and represented as a total amount for [13C]acyldopamine.
RESULTS AND DISCUSSION
Identification and quantification of endogenous long-chain fatty acid amides in N18TG2 cells by LC/QTOF-MS
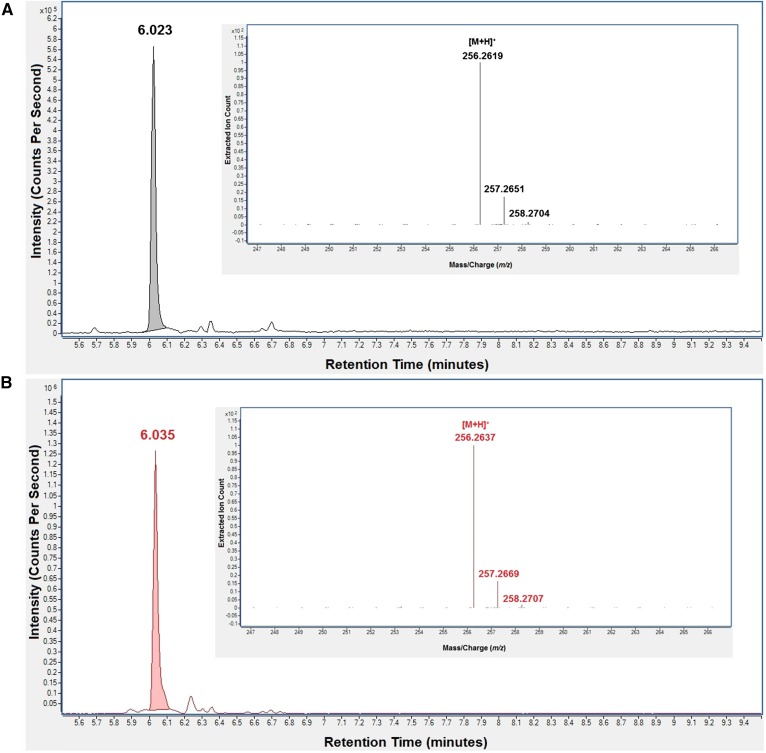
A panel of long-chain fatty acid amides was identified in N18TG2 cell extracts by comparison of molecular ion m/z values and chromatographic retention times against synthetic standards via LC/QTOF-MS analysis (Table 1). The chromatographic retention time and mass spectrum of endogenous palmitamide identified in an N18TG2 cell extract matched those of a palmitamide synthetic standard (Fig. 2). These data are representative of the data collected for each endogenous fatty acid amide identified in N18TG2 cells. Tandem MS analyses further validated the identification of these metabolites (supplementary Table 1). The identification of the PFAMs and N-acylglycines is consistent with the previous identification of these metabolites in N18TG2 cell extracts, as well as their proposed role as brain signaling lipids (12, 28, 32, 33). This is the first report of the identification of N-palmitoyldopamine, N-oleoyldopamine, and N-arachidonoyldopamine in N18TG2 cells. A previous report identified N-arachidonoylethanolamine in the N18TG2 cells at levels below our limit of detection (32). Endogenous amounts of the identified long-chain fatty acid amides were obtained by integrating the area under the chromatographic peak and comparing the integrated value against a standard curve prepared using the respective synthetic standard (supplementary Table 2). Known to be potent cell-signaling lipids, these long-chain fatty acid amides are found at cellular levels on the order of picomoles per 107 cells. Palmitamide was the most abundant fatty acid amide with an endogenous amount of 1,300 ± 98 pmoles per 107 cells, while palmitoleamide was the least abundant.
TABLE 1.
Identification of endogenous long-chain fatty acid amides in N18TG2 cell extracts by LC/QTOF-MS
| Fatty Acid Amide | Standard | N18TG2 cells | ||
| [M+H]+ (m/z) | Retention Time (minutes) | [M+H]+ (m/z) | Retention Time (minutes) | |
| N-acylglycines | ||||
| N-palmitoylglycine | 314.2966 | 5.892 | 314.2487 | 5.838 |
| N-oleoylglycine | 340.2929 | 5.995 | 340.2895 | 5.963 |
| N-acylethanolamines | ||||
| N-oleoylethanolamine | 326.3079 | 5.982 | 326.2736 | 5.996 |
| PFAMs | ||||
| Palmitamide | 256.2619 | 6.023 | 256.2637 | 6.035 |
| Palmitoleamide | 254.2487 | 5.768 | 254.2282 | 5.755 |
| Oleamide | 282.2869 | 6.177 | 282.2578 | 6.129 |
| Linoleamide | 280.2646 | 5.902 | 280.2378 | 5.838 |
| N-acyldopamines | ||||
| N-palmitoyldopamine | 392.3163 | 6.053 | 392.2726 | 6.054 |
| N-palmitoyldopamine quinone | 390.2958 | 6.148 | 390.2714 | 6.118 |
| N-oleoyldopamine | 418.3367 | 6.132 | 418.2215 | 6.121 |
| N-oleoyldopamine quinone | 416.3412 | 6.256 | 416.2446 | 6.245 |
| N-arachidonoyldopamine | 440.3198 | 5.885 | 440.2375 | 5.896 |
| N-arachidonoyldopamine quinone | 438.3832 | 5.948 | 438.3310 | 5.946 |
Fig. 2.
Identification of palmitamide in an N18TG2 cell extract by LC/QTOF-MS. The extracted ion chromatograms peak and mass spectrum of a synthetic standard, palmitamide (A), matched those of endogenous palmitamide identified in the N18TG2 extract (B). These data are representative of the data collected for each long-chain fatty acid amide identified in N18TG2 extract herein.
N18TG2 cells express GLYATL3 transcripts
Nested PCR primers were designed to amplify a 380 bp mGLYATL3 product from a cDNA library constructed from the mRNA extracted from an N18TG2 cell pellet. After the first round of PCR, the product was filtered and then used as the template in a second reaction with nested primers. Nested primers were necessary due to the low abundance of mGLYATL3 transcripts in N18TG2 cells. The 380 bp product isolated from the agarose electrophoresis gel was positively identified to be mGLYATL3 by DNA sequencing (supplementary Fig. 1). These data indicate that N18TG2 cells express mGLYATL3 transcripts.
siRNA knockdown of GLYATL3 results in decreased levels of N-acylglycines and PFAMs
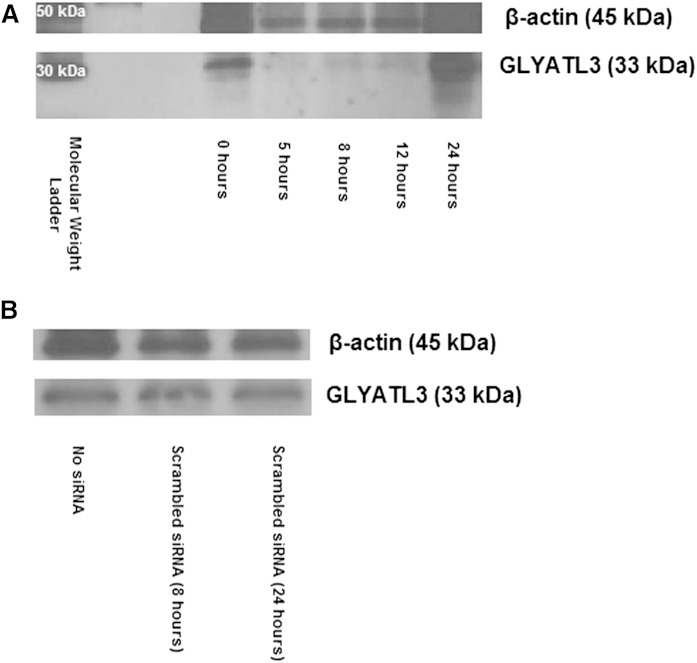
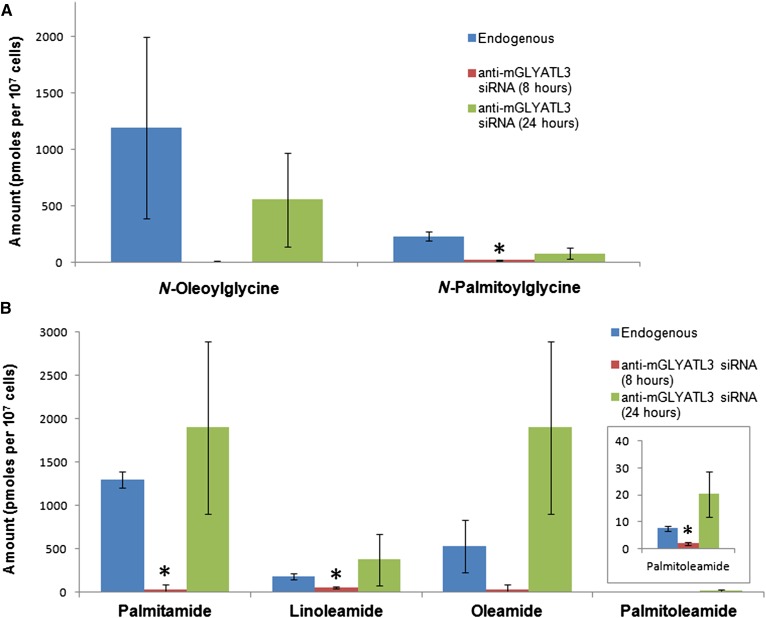
Transfection of siRNA against mGLYATL3 into N18TG2 cells resulted in the knockdown of mGLYATL3 expression for 12 h (Fig. 3). While mGLYATL3 expression levels decreased 103 ± 13% during knockdown, β-actin levels only changed 12 ± 7% (Fig. 3). After 24 h, the expression of mGLYATL3 returned to normal levels, which limited our ability to do a heavy-labeled precursor study during mGLYATL3 knockdown. The mGLYATL3 primary antibody was specific for recombinant maltose binding protein-tagged mGLYATL3, but not recombinant mGLYAT (23) (supplementary Fig. 2). After 8 and 24 h, the transfected cells were collected and the long-chain fatty acid amides extracted, purified, and analyzed by LC/QTOF-MS. A panel of identified fatty acid amides were quantified and compared with endogenous levels (supplementary Table 2). At the 8 h time point, mGLYATL3 expression was silenced and there were significant changes in N-acylglycine and PFAM levels. N-oleoylglycine levels decreased 99.4% relative to the untransfected control, while oleamide levels decreased 93.6%. Similarly, N-palmitoylglycine levels decreased 90.4% relative to the untransfected control, while palmitamide levels decreased 86.9%. These data (Fig. 4) indicate that: a) mGLYATL3 has a major role in N-acylglycine formation in these cells; and b) that the N-acylglycines are precursors for PFAMs. Both are consistent with the biosynthetic pathways for the N-acylglycines and PFAMs that we have proposed (Fig. 1). The differences in the percentage changes between oleoylated or palmitoylated amides are unclear and there are many factors contributing to these differences. One possibility could be differences between the rates of hydrolysis of N-acylglycines relative to PFAMs by fatty acid amide hydrolase (FAAH) (34). Other than changing the cellular levels of the N-acylglycines and PFAMs, the knockdown of mGLYATL3 expression resulted in no change in N-oleoylethanolamine levels and an increase in N-acyldopamine levels. The increases in N-acyldopamine levels were unexpected, but could be due to an increase in acyl-CoAs available for conjugation to dopamine by an unidentified arylalkylamine N-acyltransferase-like (AANATL) enzyme. In Drosophila, AANATL2 catalyzes the formation of long-chain N-acyldopamines in vitro (35, 36) and we propose that a mammalian AANATL2 ortholog could be responsible for N-acyldopamine formation in the N18TG2 cells.
Fig. 3.
Western blot analysis of mGLYATL3 RNAi in N18TG2 cells. A: All lanes contain 22 μg of N18TG2 cell lysate and were incubated with anti-mGLYATL3 siRNA for the respective number of hours. B: RNAi controls in N18TG2 cells. All lanes contain 20 μg of N18TG2 cell lysate and were incubated with no siRNA or scrambled siRNA for the respective number of hours. β-Actin was used as a loading control.
Fig. 4.
Subtraction lipidomics: quantification of endogenous long-chain fatty acid amides in N18TG2 cell extracts by LC/QTOF-MS with mGLYATL3 RNAi. Endogenous amounts of the N-acylglycines along with the amounts with mGLYATL3 RNAi are in (A). Endogenous amounts of the long-chain PFAMs along with the amounts with mGLYATL3 RNAi are in (B). Error bars represent ±standard deviation, n = 3. Statistically significant changes (P < 0.05) are marked with an asterisk (*).
Controls for this siRNA experiment indicate that overall protein expression was unaffected by the knockdown of mGLYATL3, as β-actin expression was unchanged (Fig. 3). Average cell viability during knockdown was 80% relative to the experiment with cells transfected with scrambled siRNA. At the 24 h time point, average cell viability was 88% when the cells were returning to normal metabolism. Negative controls included incubation with transfection reagent only or transfection reagent with scrambled siRNA. Long-chain fatty acid amide levels were unchanged in these control experiments (Table 2). When the transient knockdown of mGLYATL3 is no longer present in the cell culture (at the 24 h time point), N-acylglycine and PFAM levels increase. This further validates the role of mGLYATL3 in the production of the N-acylglycines as intermediates in the biosynthesis of the PFAMs.
TABLE 2.
RNAi controls: quantification of long-chain fatty acid amides in N18TG2 cell extracts by LC/QTOF-MS
| Fatty Acid Amide | Amounta (picomoles per 107 cells) | |||
| (+) Scrambled siRNA (+) Transfection Reagentb | (+) Scrambled siRNA (+) Transfection Reagentc | (−) Scrambled siRNA (+) Transfection Reagentc | (−) Scrambled siRNA (−) Transfection Reagentc | |
| N-acylglycines | ||||
| N-palmitoylglycine | 90 ± 18 | 69 ± 18 | 92 ± 6.0 | 120 ± 72 |
| N-oleoylglycine | 530 ± 250 | 870 ± 320 | 2000 ± 480 | 450 ± 240 |
| N-acylethanolamines | ||||
| N-oleoylethanolamine | 26 ± 16 | 43 ± 34 | 16 ± 15 | 2.8 ± 2.0 |
| PFAMs | ||||
| Palmitamide | 510 ± 100 | 640 ± 400 | 620 ± 39 | 320 ± 170 |
| Palmitoleamide | 12 ± 3.4 | 19 ± 14 | 24 ± 1.8 | 10 ± 5.6 |
| Oleamide | 1300 ± 150 | 1,200 ± 310 | 1,200 ± 1,100 | 1,200 ± 320 |
| Linoleamide | 280 ± 32 | 230 ± 35 | 410 ± 220 | 320 ± 61 |
| N-acyldopamines | ||||
| N-palmitoyldopamine | 500 ± 190 | 290 ± 310 | 930 ± 360 | 570 ± 62 |
| N-oleoyldopamine | 29 ± 6.6 | 24 ± 8.2 | 83 ± 33 | 65 ± 48 |
| N-arachidonoyldopamine | 20 ± 7.9 | 17 ± 3.9 | 31 ± 12 | 8.8 ± 0.5 |
No statistically significant changes between any two groups were reported (P > 0.05).
Eight hour incubation.
Twenty-four hour incubation.
siRNA knockdown of PAM results in an accumulation of N-oleoylglycine and a decrease in oleamide levels
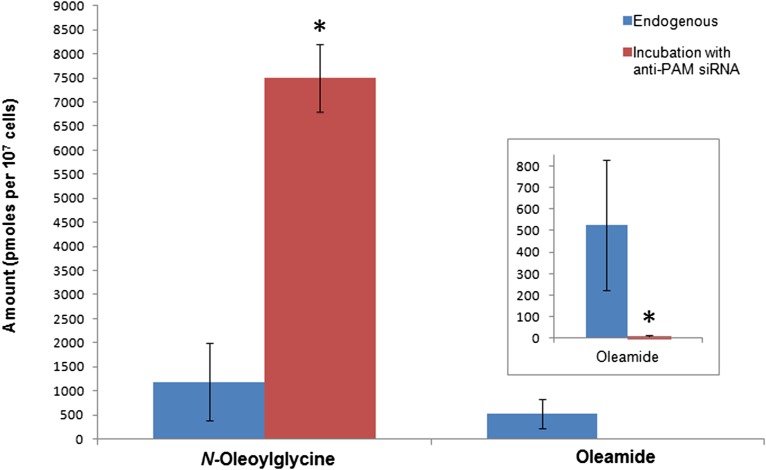
Transfection of siRNA against PAM into N18TG2 cells resulted in the knockdown of PAM expression for 48 h (Fig. 5). The average viability after 48 h was 56% relative to untransfected cells. Because a large number of peptide hormones are α-amidated for biological activation (37), a decrease in cell viability was expected upon the knockdown of PAM. After a total of 56 h of incubation with anti-PAM siRNA and 48 h with [13C18]oleate, the cells were collected and the amounts of labeled and unlabeled N-oleoylglycine and oleamide were analyzed and quantified. In a previous study, the incubation of N18TG2 cells with a PAM inhibitor, trans-4-phenyl-3-butenoic acid, resulted in the accumulation of N-oleoylglycine (12). Consistent with these earlier results, decreased expression of PAM resulted in the accumulation of N-oleoylglycine and a decrease in oleamide levels relative to endogenous amounts (Fig. 6). These data provide additional evidence that PAM is involved in the conversion of N-acylglycines to PFAMs in cellula.
Fig. 5.

Western blot analysis of PAM RNAi in N18TG2 cells. N18TG2 cells were exposed to anti-PAM siRNA or a scrambled siRNA for the respective number of hours.
Fig. 6.
PAM RNAi. Quantification of total (labeled and unlabeled) N-oleoylglycine and oleamide in N18TG2 cell extracts after incubation with [13C18]oleate and anti-PAM siRNA. Error bars represent ±standard deviation, n = 3. Statistically significant changes (P < 0.05) are marked with an asterisk (*).
N18TG2 cells incubated with [1-13C]palmitate lead to a family of [1-13C]palmitoylated fatty acid amides
The N18TG2 cells were incubated with [1-13C]palmitate in order to investigate metabolic flux after being exposed to a heavy-labeled precursor. After a 48 h incubation with [1-13C]palmitate using BSA as a carrier, the lipids were extracted from the cells to analyze for fatty acid amides that contained the 13C-N-palmitoyl chain as well as the other unlabeled metabolites. [13C]N-palmitoylglycine, [13C]N-palmitamide, and [13C]N-palmitoyldopamine were identified in cell extracts, showing that these metabolites are synthesized from the exogenous [1-13C]palmitate (Table 3). The total palmitamide (13C-labeled and unlabeled) in the N18TG2 cells that were incubated with [1-13C]palmitate was 990 ± 480 pmoles per 107 cells, consistent with the endogenous amount of palmitamide reported herein, 1,300 ± 98 pmoles per 107 cells. A similar trend was also observed for N-palmitoyldopamine. A low percentage, ∼0.1%, of the incubated [1-13C]palmitate was converted to labeled N-palmitoylglycine, palmitamide, or N-palmitoyldopamine by N18TG2 cells.
TABLE 3.
Amounts of long-chain fatty acid amides in N18TG2 cells after [1-13C]palmitic acid incubation
| Fatty Acid Amide | Amount (picomoles per 107 cells) | |
| Endogenous | After [1-13C]palmitic Acid Incubation | |
| N-acylglycines | ||
| N-palmitoylglycine | 230 ± 40 | 76 ± 46 |
| 13C-N-palmitoylglycine | N.D. | 32 ± 24 |
| Total N-palmitoylglycine | N/A | 110 ± 52 |
| N-oleoylglycine | 1200 ± 800 | 1000 ± 800 |
| N-acylethanolamines | ||
| N-oleoylethanolamine | 13 ± 1.6 | 2.1 ± 0.9 |
| PFAMs | ||
| Palmitamide | 1,300 ± 98 | 840 ± 470 |
| 13C-palmitamide | N.D. | 150 ± 71 |
| Total palmitamide | N/A | 990 ± 480 |
| Palmitoleamide | 7.6 ± 1.7 | 14 ± 0.70 |
| Oleamide | 530 ± 300 | 420 ± 140 |
| Linoleamide | 180 ± 32 | 42 ± 3.4 |
| N-acyldopamines | ||
| N-palmitoyldopamine | 350 ± 200 | 140 ± 87 |
| 13C-N-palmitoyldopamine | N.D. | 140 ± 120 |
| Total N-palmitoyldopamine | N/A | 280 ± 150 |
| N-oleoyldopamine | 510 ± 410 | 120 ± 40 |
| N-arachidonoyldopamine | 1,800 ± 1,100 | 330 ± 62 |
N.D., not detected; N/A, not applicable.
CONCLUSIONS
mGLYATL3 catalyzes the formation of long-chain N-acylglycines in cellula. Subtraction lipidomic data observed a decrease in N-acylglycine and PFAM levels in N18TG2 cells after the siRNA-mediated knockdown of mGLYATL3. These data support the proposed biosynthetic pathway of activation of a fatty acid by acyl-CoA synthetase followed by the conjugation of the resulting acyl-CoA with glycine. It is known that long-chain fatty acid amides can be produced by more than one route; there is evidence for their biosynthesis by the sequential oxidation of N-acylethanolamines (16–18, 20). There is evidence that this alternative pathway does take place in the N18TG2 cells (28). The subtraction lipidomic data herein suggests that the glycine-dependent route is the predominant route for long-chain N-acylglycine synthesis in these cells because N-acylgycine levels were not rescued by an alternative biosynthetic pathway during mGLYATL3 knockdown. This is the first report of an enzyme responsible for the biosynthesis of long-chain N-acylglycines in cellula. In vitro data obtained using purified recombinant GST-tagged mGLYATL3 shows the production of N-oleoylglycine from oleoyl-CoA and glycine providing support for our in cellula results (supplementary Table 3). Furthermore, Waluk, Schultz, and Hunt (24) previously described a human enzyme, hGLYATL2, that catalyzes the conjugation of long-chain acyl-CoA thioesters to glycine in vitro. Interestingly, our BLAST searches did not identify orthologs of hGLYATL2 in organisms known to produce long-chain N-acylglycines, such as the mouse and the rabbit. However, these species did have putative orthologs of mGLYATL3. A similar strategy employed by Saghatelian and Cravatt [discovery metabolic profiling, see (38)] led to the identification of N-acyltaurines as substrates for FAAH, despite the relatively low kcat/KM values obtained in vitro for the FAAH-catalyzed hydrolysis for the N-acyltaurines. As these authors point out (39), in vitro substrate specificity data obtained using a purified enzyme may not perfectly predict in vivo substrates for the same enzyme because proteins often function as partners in multi-protein complexes and networks and may be regulated in ways not fully captured in in vitro experiments. A subsequent decrease in PFAM levels with the knockdown of mGLYATL3 gives support to the proposed role for the N-acylglycines as intermediates in the biosynthesis of the long-chain PFAMs. These data are further validated by the siRNA-mediated knockdown of the N-acylglycine amidating enzyme, PAM. Knockdown of PAM resulted in a decrease in oleamide levels and an increase in N-oleoylglycine levels. Lastly, the results of our heavy-labeled precursor feeding study were consistent with the following biosynthetic pathway: fatty acid → fatty acyl-CoA → N-fatty acylglycine → PFAM (Fig. 1).
Supplementary Material
Footnotes
Abbreviations:
- AANATL
- arylalkylamine N-acyltransferase-like
- BSTFA
- N,O-bis(trimethylsilyl)trifluoroacetamide
- FAAH
- fatty acid amide hydrolase
- GLYAT
- glycine N-acyltransferase
- GLYATL3
- glycine N-acyltransferase-like 3
- hGLYAT
- human glycine N-acyltransferase
- mGLYAT
- mouse glycine N-acyltransferase
- PAM
- peptidylglycine α-amidating monooxygenase
- PFAM
- primary fatty acid amide
- QTOF
- quadrupole TOF
This work was supported, in part, by Office of Extramural Research, National Institutes of Health Grant R03-DA034323, National Institutes of Health Grant R15-GM107864, and the Florida Center for Excellence for Biomolecular Identification and Targeted Therapeutics (FCoE-BITT Grant GALS020). This work also received support from the Mass Spectrometry and Peptide Facility, Department of Chemistry, University of South Florida. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Keuhl F. A. Jr., Jacob T. A., Ganley H. O., Ormond R. E., and Meisinger M. A. P.. 1957. The identification of N-(-2-hydroxyethyl)-palmitamide as a naturally occurring anti-inflammatory agent. J. Am. Chem. Soc. 79: 5577–5578. [Google Scholar]
- 2.Bachur N. R., Masek K., Melmon K. L., and Udenfriend S.. 1965. Fatty acid amides of ethanolamine in mammalian tissues. J. Biol. Chem. 240: 1019–1024. [PubMed] [Google Scholar]
- 3.Devane W. A., Hanuš L., Breuer A., Pertwee R. G., Stevenson L. A., Griffin G., Gibson D., Mandelbaum A., Etinger A., and Mechoulam R.. 1992. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 258: 1946–1949. [DOI] [PubMed] [Google Scholar]
- 4.Tortoriello G., Rhodes B. P., Takacs S. M., Stuart J. M., Basnet A., Raboune S., Widlanski T. S., Doherty P., Harkany T., and Bradshaw H. B.. 2013. Targeted lipidomics in Drosophila melanogaster identifies novel 2-monoacylglycerols and N-acyl amides. PLoS One. 8: e67865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeffries K. A., Dempsey D. R., Behari A. L., Anderson R. L., and Merkler D. J.. 2014. Drosophila melanogaster as a model system to study long-chain fatty acid amide metabolism. FEBS Lett. 588: 1596–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell E. K., and Merkler D. J.. 2008. Biosynthesis, degradation and pharmacological importance of the fatty acid amides. Drug Discov. Today. 13: 558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradshaw H. B., Rimmerman N., Hu S. S-J., Burstein S., and Walker J. M.. 2009. Novel endogenous N-acyl glycines: identification and characterization. Vitam. Horm. 81: 191–205. [DOI] [PubMed] [Google Scholar]
- 8.Hanuš L., Shohami E., Bab I., and Mechoulam R.. 2014. N-Acyl amino acids and their impact on biological processes. Biofactors. 40: 381–388. [DOI] [PubMed] [Google Scholar]
- 9.Waluk D. P., Battistini M. R., Dempsey D. R., Farrell E. K., Jeffries K. A., Mitchell P. Jr., Hernandez L. W., McBride J. C., Merkler D. J., and Hunt M. C.. 2014. Mammalian fatty acid amides of the brain and CNS. In Omega-3 Fatty Acids in Neurological Health. R. R. Watson and F. De Meester, editors. Elsevier, London. 87–107. [Google Scholar]
- 10.Mulder G. J. 2003. Conjugation reactions. In Drug Metabolism: An Integrated Approach. Taylor & Francis Inc., Bristol, PA. 274–275. [Google Scholar]
- 11.Huang S. M., Bisogno T., Petros T. J., Chang S. Y., Zavitsanos P. A., Zipkin R. E., Sivakumar R., Coop A., Maeda D. Y., De Petrocellis L., et al. 2001. Identification of a new class of molecules, the arachidonyl amino acids, and characterization of one member that inhibits pain. J. Biol. Chem. 276: 42639–42644. [DOI] [PubMed] [Google Scholar]
- 12.Merkler D. J., Chew G. H., Gee A. J., Merkler K. A., Sorondo J. P. O., and Johnson M. E.. 2004. Oleic acid derived metabolites in mouse neuroblastoma N18TG2 cells. Biochemistry. 43: 12667–12674. [DOI] [PubMed] [Google Scholar]
- 13.Liu P., Duan J., Wang P., Qian D., Guo J., Shang E., Su S., Tang Y., and Tang Z.. 2013. Biomarkers of primary dysmenorrhea and herbal formula intervention: an exploratory metabonomics study of blood plasma and urine. Mol. Biosyst. 9: 77–87. [DOI] [PubMed] [Google Scholar]
- 14.Rimmerman N., Bradshaw H. B., Hughes H. V., Chen J. S-C., Hu S. S-J., McHugh D., Vefring E., Jahnsen J. A., Thompson E. L., Masuda K., et al. 2008. N-Palmitoyl glycine, a novel endogenous lipid that acts as a modulator of calcium influx and nitric oxide production in sensory neurons. Mol. Pharmacol. 74: 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan B., Yu Y. W., Monn M. F., Hughes H. V., O’Dell D. K., and Walker J. M.. 2009. Targeted lipidomics approach for endogenous N-acyl amino acids in rat brain tissue. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2890–2894. [DOI] [PubMed] [Google Scholar]
- 16.Burstein S. H., Rossetti R. G., Yagen B., and Zurier R. B.. 2000. Oxidative metabolism of anandamide. Prostaglandins Other Lipid Mediat. 61: 29–41. [DOI] [PubMed] [Google Scholar]
- 17.Aneetha H., O’Dell D. K., Tan B., Walker J. M., and Hurley T. D.. 2009. Alcohol dehydrogenase-catalyzed in vitro oxidation of anandamide to N-arachidonoyl glycine, a lipid mediator: synthesis of N-acyl glycinals. Bioorg. Med. Chem. Lett. 19: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivkovic M., Dempsey D. R., Handa S., Hilton J. H., Lowe E. W., and Merkler D. J.. 2011. N-Acylethanolamines as novel alcohol dehydrogenase 3 substrates. Arch. Biochem. Biophys. 506: 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merkler D. J., Merkler K. A., Stern W., and Fleming F. F.. 1996. Fatty acid amide biosynthesis: a new role for peptidylglycine α-amidating enzyme and acyl-coenzyme A:glycine N-acyltransferase. Arch. Biochem. Biophys. 330: 430–434. [DOI] [PubMed] [Google Scholar]
- 20.Bradshaw H. B., Rimmerman N., Hu S. S-J., Benton V. M., Stuart J. M., Masuda K., Cravatt B. F., O’Dell D. K., and Walker J. M.. 2009. The endocannabinoid anandamide is a precursor for the signaling lipid N-arachidonoyl glycine by two distinct pathways. BMC Biochem. 10: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregersen N., Kølvraa S., and Mortensen P. B.. 1986. Acyl-CoA-glycine N-acyltransferase: in vitro studies on the glycine conjugation of straight-chained and branched-chained acyl-CoA esters in human liver. Biochem. Med. Metab. Biol. 35: 210–218. [DOI] [PubMed] [Google Scholar]
- 22.Kelley M., and Vessey D. A.. 1993. Isolation and characterization of mitochondrial acyl-CoA:glycine N-acyltransferases from kidney. J. Biochem. Toxicol. 8: 63–69. [DOI] [PubMed] [Google Scholar]
- 23.Dempsey D. R., Bond J. D., Carpenter A-M., Rodriguez Ospina S., and Merkler D. J.. 2014. Expression, purification, and characterization of mouse glycine N-acyltransferase in Escherichia coli. Protein Expr. Purif. 97: 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waluk D. P., Schultz N., and Hunt M. C.. 2010. Identification of glycine N-acyltransferase-like 2 (GLYATL2) as a transferase that produces N-acyl glycines in humans. FASEB J. 24: 2795–2803. [DOI] [PubMed] [Google Scholar]
- 25.Ritenour-Rodgers K. J., Driscoll W. J., Merkler K. A., Merkler D. J., and Mueller G. P.. 2000. Induction of peptidylglycine α-amidating monooxygenase in N18TG2 cells: A model for studying oleamide biosynthesis. Biochem. Biophys. Res. Commun. 267: 521–526. [DOI] [PubMed] [Google Scholar]
- 26.Farrell E. K. 2010. Biosynthesis of Fatty Acid Amides. PhD Dissertation. University of South Florida, Tampa, FL.
- 27.Sultana T., and Johnson M. E.. 2006. Sample preparation and gas chromatography of primary fatty acid amides. J. Chromatogr. A. 1101: 278–285. [DOI] [PubMed] [Google Scholar]
- 28.Farrell E. K., Chen Y. D., Barazanji M., Jeffries K. A., Cameroamortegui F., and Merkler D. J.. 2012. Primary fatty acid amide metabolism: conversion of fatty acids and an ethanolamine in N18TG2 and SCP cells. J. Lipid Res. 53: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald G. R., Hudson A. L., Dunn S. M., You H., Baker G. B., Whittal R. M., Martin J. W., Jha A., Edmondson D. E., and Holt A.. 2008. Bioactive contaminants leach from disposable laboratory plasticware. Science. 322: 917. [DOI] [PubMed] [Google Scholar]
- 30.Šidák Z. 1967. Rectangular confidence regions for the means of multivariate normal distributions. J. Am. Stat. Assoc. 62: 626–633. [Google Scholar]
- 31.Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6: 65–70. [Google Scholar]
- 32.Di Marzo V., De Petrocellis L., Sepe N., and Buono A.. 1996. Biosynthesis of anandamide and related acylethanolamides in mouse J774 macrophages and N18 neuroblastoma cells. Biochem. J. 316: 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bisogno T., Sepe N., DePetrocellis L., Mechoulam R., and DiMarzo V.. 1997. The sleep inducing factor oleamide is produced by mouse neuroblastoma cells. Biochem. Biophys. Res. Commun. 239: 473–479. [DOI] [PubMed] [Google Scholar]
- 34.Maurelli S., Bisogno T., DePetrocellis L., DiLuccia A., Marino G., and DiMarzo V.. 1995. Two novel classes of neuroactive fatty acid amides are substrates for mouse neuroblastoma ‘anandamide amidohydrolase’. FEBS Lett. 377: 82–86. [DOI] [PubMed] [Google Scholar]
- 35.Dempsey D. R., Jeffries K. A., Anderson R. L., Carpenter A. M., Opsina S. R., and Merkler D. J.. 2014. Identification of an arylalkylamine N-acyltransferase from Drosophila melanogaster that catalyzes the formation of long-chain N-acylserotonins. FEBS Lett. 588: 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dempsey D. R., Carpenter A-M., Ospina S. R., and Merkler D. J.. 2015. Probing the chemical mechanism and critical regulatory amino acid residues of Drosophila melanogaster arylalkylamine N-acyltransferase like 2. Insect Biochem. Mol. Biol. 66: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eipper B. A., and Mains R. E.. 1988. Peptide α-amidation. Annu. Rev. Physiol. 50: 333–344. [DOI] [PubMed] [Google Scholar]
- 38.Saghatelian A., and Cravatt B. F.. 2005. Discovery metabolie profiling – forging functional connections between the proteome and the metabolome. Life Sci. 77: 1759–1766. [DOI] [PubMed] [Google Scholar]
- 39.Saghatelian A., and Cravatt B. F.. 2005. Global strategies to integrate the proteome and metabolome. Curr. Opin. Chem. Biol. 9: 62–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.