Abstract
Chaperones assist the folding of many proteins in the cell. While the most well studied chaperones use cycles of ATP binding and hydrolysis to assist protein folding, a number of chaperones have been identified that promote protein folding in the absence of high-energy cofactors. Precisely how ATP-independent chaperones accomplish this feat is unclear. Here we have characterized the kinetic mechanism of substrate folding by the small, ATP-independent chaperone, Spy. Spy rapidly associates with its substrate, Immunity protein 7 (Im7), eliminating its potential for aggregation. Remarkably, Spy then allows Im7 to fully fold into its native state while remaining bound to the surface of the chaperone. These results establish a potentially widespread mechanism whereby ATP-independent chaperones can assist in protein refolding. They also provide compelling evidence that substrate proteins can fold while continuously bound to a chaperone.
Chaperones are essential for maintaining protein-folding homeostasis and play important roles in the cellular stress response. By binding to aggregation-sensitive folding intermediates, chaperones inhibit aberrant interactions between proteins. The most intensively studied folding chaperones, such as the GroEL–ES and DnaK systems, facilitate substrate protein folding through ATP- and cofactor-driven conformational changes1–5. These conformational changes convert ATP-dependent folding chaperones from a state with a high affinity for substrates to a state with a low affinity for substrates, which allows substrate proteins to be released. It is widely thought that this change in affinity is needed to release bound substrates into bulk solution, or in the case of GroEL–ES, into the unbound interior of the chaperonin cavity, where folding of the substrate protein occurs1,3–6.
In contrast to ATP dependent chaperones, a number of chaperones have been identified that can assist protein folding in the absence of ATP7–11. The mechanism by which ATP-independent chaperones promote substrate protein folding without a means of regulating substrate binding and release is not clear. One recently discovered ATP-independent chaperone is Spy, which was identified through a genetic selection to enhance in vivo protein stability in Escherichia coli10. Spy is a cradle-shaped, dimeric, α-helical protein that is massively and rapidly upregulated upon exposure to protein folding stresses such as butanol or tannins10,12,13. Spy levels change from near zero under non-stress conditions to comprising 20-50% of the periplasmic proteome during stress10,12,13. This major investment of cellular resources toward the production of Spy under stressful conditions implies that there is a pressing need for Spy in vivo during stress. In vitro experiments revealed that Spy both inhibits the aggregation of proteins and enhances the refolding yield of proteins in an ATP-independent manner10,14.
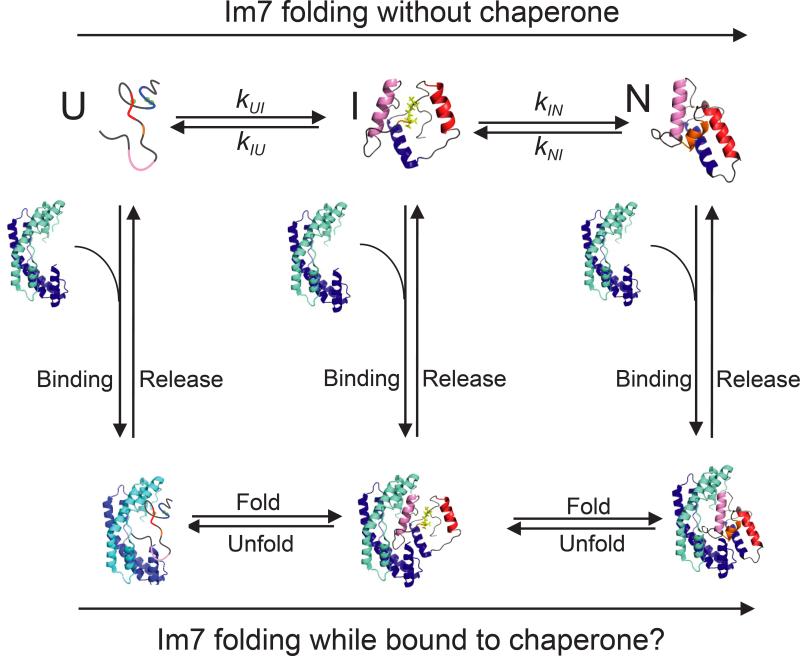
The mechanism by which Spy and the growing class of ATP-independent chaperones function to refold proteins in the absence of energy cofactors is an unresolved question. We addressed this by determining how Spy affects the folding pathway of the protein that was used to discover Spy—immunity protein 7 (Im7). Im7 has a number of attributes that facilitate its study. It is small, monomeric, and folds via a well-characterized mechanism that involves the population of a partially-folded on pathway intermediate prior to reaching the native state15–20. The fluorescence of the lone tryptophan residue in Im7 is a sensitive reporter of the folding status of the substrate: the unfolded state of Im7 (Im7U) is moderately fluorescent, the partially-folded intermediate state (Im7I) is hyperfluorescent, and the native state (Im7N) is weakly fluorescent16,20. In addition, variants of Im7 have been developed that mimic Im7U or Im7I. Under physiological pH and salt conditions, the Im7U mimic, Im7-L18A L19A L37A, is unfolded, and the Im7I mimic, Im7-L53A I54A, closely resembles the hyperfluorescent intermediate state21–24. The small size and biochemical accessibility of the Spy-Im7 system, as diagramed in Figure 1, provide a powerful system to address the open question of exactly how ATP-independent chaperones affect the folding of proteins. In particular, we decided to address whether proteins can fold while bound to a chaperone or are instead arrested at a defined state in the folding energy landscape, relying upon release for continued folding. We set out to analyze the ability of Spy to bind to three low energy states that comprise the Im7 folding pathway, as well as the effect of Spy on the kinetic mechanism of Im7 folding (Fig. 1). We show here that Spy binds the unfolded, intermediate, and native states of Im7 with comparable (μM) affinities, allowing Im7 to fold completely into the native state while bound to the chaperone. By doing so, Spy minimizes the concentration of unbound unfolded or partially-folded proteins, suppressing their tendency to aggregate. Once the folding stress is removed, Spy is presumably diluted by degradation or cell growth, releasing bound substrate without the need for energy provided by ATP binding and/or hydrolysis.
Figure 1.
Folding of the protein Im7 in the presence and absence of the chaperone Spy. Im7 is shown as a multicolored protein that is helical in both the folding intermediate (I) and in the native folded state (N) and lacks any persistent secondary structure in the unfolded state (U). Spy is shown as a blue cradle-shaped homodimer. Spy binding and release are shown as vertical arrows.
RESULTS
Spy binds the three states on the Im7 folding energy landscape
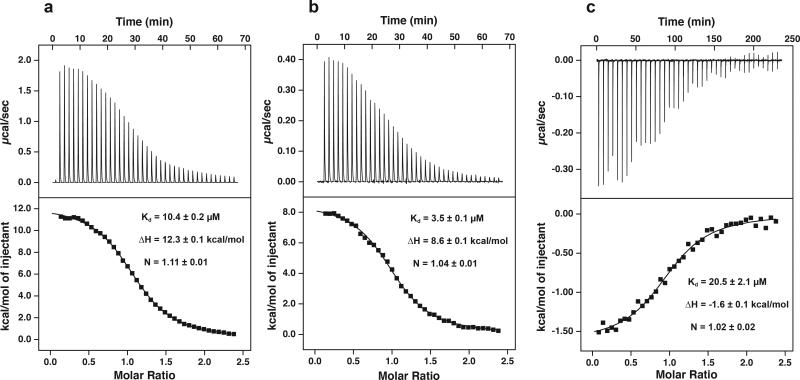
We first investigated whether Spy is able to interact with three different low energy states that comprise the Im7 folding pathway (Fig. 1). We did this by using isothermal titration calorimetry (ITC) to monitor binding of Spy to Im7-L18A L19A L37A (Im7U), Im7-L53A I54A (Im7I), and native wild-type Im7 (Im7N). We found that Spy binds to all three variants of Im7, with affinities of 10.4 μM, 3.5 μM, and 20.5 μM for Im7-L18A L19A L37A (Im7U), Im7-L53A I54A (Im7I), and Im7-WT (Im7N), respectively (Fig. 2). In all cases, we observed a stoichiometry of 1 Spy dimer per Im7. Therefore, Spy binds all three populated states on the Im7 folding energy landscape with comparable affinities.
Figure 2.
ITC titrations for Spy binding to wild-type Im7 and variants. (a) Titration of 150 μM Im7-L18A L19A L37A (Im7U) in the cell with 1.65 mM Spy dimer in the syringe. (b) Titration of 50 μM Im7-L53A I54A (Im7I) in the cell with 550 μM Spy dimer in the syringe. (c) Titration of 230 μM Im7-WT (Im7N) in the cell with 2.53 mM Spy dimer in the syringe. All titrations were performed in 40 mM HEPES-KOH pH 7.5, 100 mM NaCl at 10 °C. The black lines in the bottom panels are the best fit of the data to a one-site model. Values reported are the mean ± s.e. of the fit.
Spy rapidly binds unfolded Im7
We decided to further examine Spy-Im7 binding, and more importantly, to look at how the binding reaction might alter the distribution of Im7 between the different states on the folding energy landscape (Fig. 1). To do this, we first measured the kinetics of the interaction between Spy and Im7-L18A L19A L37A (Im7U), Im7-L53A I54A (Im7I), and Im7-WT (Im7N) by stopped-flow fluorescence using the tryptophan of Im7 as a reporter. Since Spy lacks tryptophan residues, the signal change reflects the environment of the lone tryptophan (W75) in Im7, which in turn reports on the folding state of the protein substrate.
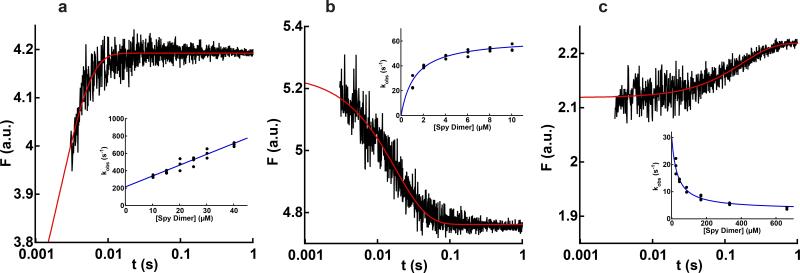
When Spy binds the constitutively unfolded variant Im7-L18A L19A L37A, the tryptophan fluorescence intensity rapidly increases (Fig. 3a). The observed rate constant (kobs) of the association reaction increased linearly with Spy concentration (Fig. 3a, inset), consistent with a bimolecular reaction in which Spy binds directly to Im7U. Fitting kobs against Spy concentration to a straight line gives a bimolecular association rate constant (kon) of 1.3 ± 0.2 × 107 M−1s−1 and a dissociation rate constant (koff) of 210 ± 20 s−1. Under butanol or tannin stress, the concentration of Spy is ~2 mM in vivo10,12,13. At this concentration, association between Spy and Im7U would be very rapid (occurring with a half-time of 26 μs). An important in vivo consequence of the rapid association between Spy and Im7U is that under stress conditions, Im7 (and presumably other unfolded, aggregation-prone proteins) would have very little time to aggregate prior to being bound by Spy.
Figure 3.
Kinetics of Spy binding to Im7U, Im7I, and Im7N. Graphs show the change in tryptophan fluorescence (F) associated with Spy binding to (a) 1 μM Im7-L18A L19A L37A (Im7U), (b) 0.5 μM Im7-L53A I54A (Im7I), and (c) 4.8 μM Im7-WT (Im7N). Note the logarithmic timescale. The red lines are the best fit of each data set to a single exponential. The insets show the change in observed rate constant with Spy concentration for each variant of Im7. Each data point in the insets represents the average of 10-15 traces. For Spy binding to Im7-L18A L19A L37A, kobs increased linearly with Spy concentration, indicating that Spy can bind Im7U, and giving kon of 1.3 ± 0.1 × 107 M−1s−1 and koff of 210 ± 20 s−1. For Spy binding to Im7-L53A I54A, kobs increased hyperbolically with Spy concentration, reaching a limiting value of 62 ± 2 s−1, which is consistent with Spy binding Im7I followed by partial folding or unfolding of Im7 within the complex. For Spy binding to Im7-WT, kobs decreased with Spy concentration, reaching a limiting value of 3.3 ± 0.9 s−1. Values reported are the mean ± s.e. of the fit. .In combination with the burst phase when Spy binds Im7-WT (Fig. 4a), this decreasing kobs suggests that Spy can bind Im7N followed by partial unfolding to Im7I while bound.
Binding to Spy alters the conformation of the Im7 intermediate
In contrast to the fluorescence increase that occurs upon Spy binding to the Im7U mimic, Im7-L18A L19A L37A, the addition of Spy to the Im7I mimic, Im7-L53A I54A, caused a fluorescence decrease (Fig. 3b). This change could reflect quenching of the fluorescence of W75 in the Spy–Im7-L53A I54A complex or a conformational change in Im7-L53A I54A that occurs following binding to Spy.
These two possibilities can be distinguished by measuring the dependence of kobs of the fluorescence change on Spy concentration. If the change in fluorescence occurs upon Spy binding, kobs should increase linearly with Spy concentration, consistent with a bimolecular reaction. Instead, we observed that kobs increased hyperbolically with Spy concentration (Fig. 3b, inset), consistent with a two-step mechanism wherein Spy first binds the Im7I mimic Im7-L53A I54A, followed by a conformational change within the complex25,26. Extrapolation of the traces back to time zero showed that the initial fluorescence of the binding reaction is identical to the fluorescence of Im7-L53A I54A alone (Supplementary Fig. 1); i.e., the fluorescence of Im7-L53A I54A alone is equal to the fluorescence of the Spy–Im7-L53A I54A complex prior to the conformational change. Thus, it appears that the initial bimolecular step in which Spy binds Im7-L53A I54A is not detectable by fluorescence. Instead, it is the subsequent conformational change within the complex that can be observed using fluorescence. Since Im7I is hyperfluorescent relative to both Im7U and Im7N, the fluorescence decrease associated with this conformational change would be consistent with either unfolding of the intermediate or folding to the native state while bound to Spy. Thus, Im7I can either fold or unfold while bound to Spy.
Spy binds to native Im7 and unfolds it while bound
The conformational change that occurs in the Im7I mimic while it is bound to Spy led us to explore the possibility that a conformational change may also occur within native Im7 while it is bound to Spy. Again using stopped-flow fluorescence, we found that the fluorescence intensity of Im7-WT increases upon binding Spy (Fig. 3c). The initial fluorescence intensities observed in the dead time (3.5 ms) of Spy binding to Im7-WT were higher than the fluorescence of Im7-WT alone (Fluorescence of 1.78 a.u. in Fig. 4a), indicating that at least one step occurred within the dead time of the instrument (Fig. 4a). This burst in fluorescence reached a saturating value at high Spy concentrations, indicating that it likely reflects the rapid binding of Spy to Im7N, which causes an increase in the tryptophan fluorescence of Im7N (Supplementary Fig. 2a). This rapid binding is followed by a small, slow exponential increase in fluorescence. The kobs for the slower, post-burst phase step decreased hyperbolically with Spy concentration, asymptotically reaching a limiting value (~3 s−1) (Fig. 3c, inset). The hyperbolically decreasing kobs is consistent with the post-burst phase step monitoring a change in the equilibrium constant between Im7N and another conformation of Im7 that is more fluorescent than Im7N and caused by Spy binding.25,26 This is likely Im7I, since Im7I is hyperfluorescent relative to Im7N. The magnitude of the fluorescence increase that occurs after Spy binds Im7-WT is much smaller than the difference in fluorescence between Im7I and Im7N, indicating that only a minor proportion of Im7-WT is unfolded to Im7I upon binding Spy (Supplementary Fig. 2b). Since the binding affinity of Spy for Im7-L53AI54A (Im7I) is ~6-fold higher than the affinity of Spy for Im7-WT (Fig. 2b,c), Spy should shift the equilibrium of Im7N towards Im7I. Consistent with the known differences in Kd, the amplitude of the post-burst phase exponential when Spy binds Im7-WT comprises only 3% of the total fluorescence intensity difference between Im7N and Im7I (Supplementary Fig. 2b). Most notably, the kobs for the post-burst phase step does not decrease to 0 s−1 at high Spy concentrations (Fig 3c, inset). This observation suggests that Im7 actually folds and unfolds while bound to Spy (Supplementary Fig. 3). In summary, our kinetic data suggest that Spy is capable of binding to Im7U, Im7I, and Im7N—three substantially different conformations21–24—and is able to promote folding and/or unfolding of the intermediate and native conformers in the bound state.
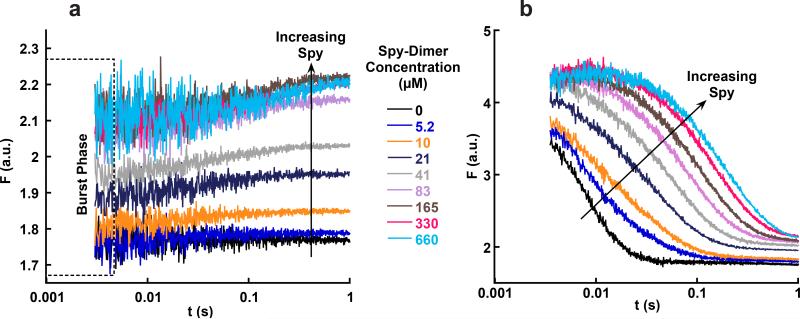
Figure 4.
Stopped-flow fluorescence traces of Spy binding Im7-WT and Im7 folding in the presence of Spy. (a) Traces for Spy binding to Im7-WT under native conditions. The fluorescence of the first data point for each trace is higher than the fluorescence of Im7-WT alone (black trace) and reaches a saturating fluorescence at high Spy concentrations. This burst phase is consistent with Spy binding Im7N within the dead time of the instrument. (b) Traces for Im7-WT folding in the presence of different Spy concentrations. Im7 folding slows as the concentration of Spy is increased. An increase in fluorescence in the first 20 ms occurs in the traces at the highest Spy concentrations; this phase represents the conversion of Im7U to Im7I during Im7 folding that is slowed by Spy. The Spy dimer concentrations used in a and b are identical for each color. Note that the y axes are different in a and b, but that the final fluorescence at 1 s is the same in both experiments, suggesting that the same equilibrium is reached.
Im7 folds while bound to Spy
To further examine whether Im7 can fold or unfold while bound to Spy, we next directly investigated how Spy affects Im7 folding (Fig. 1). Im7-WT was denatured in 8 M urea, and folding was monitored in a stopped-flow fluorimeter by diluting the unfolded Im7-WT into buffer containing various concentrations of Spy (Fig. 4b). Folding of Im7 slowed as the concentration of Spy increased. While the fluorescence decrease for Im7 folding in the absence of Spy (reflecting the I to N transition) could be fitted to a single exponential, the fluorescence decrease for Im7 folding in the presence of an equimolar concentration (5μM) of Spy required two exponentials to achieve a reasonable fit, and at ≥160 μM Spy, an additional third phase, which increased in fluorescence, appeared at the beginning of the traces (Supplementary Fig. 4a–f). The kobs values for all of the phases decreased with increasing concentrations of Spy, but did not go to zero (Supplementary Fig. 4g–i). This combination of results indicates that Im7 is able to fold in the presence of Spy, even at concentrations of the chaperone well above the Kd for the interaction; i.e., where essentially all of the Im7 is chaperone-bound. The final fluorescence intensity obtained in these refolding experiments at various concentrations of Spy was indistinguishable from those when Spy is added to native Im7, suggesting that both experiments reached the same thermodynamic end point.
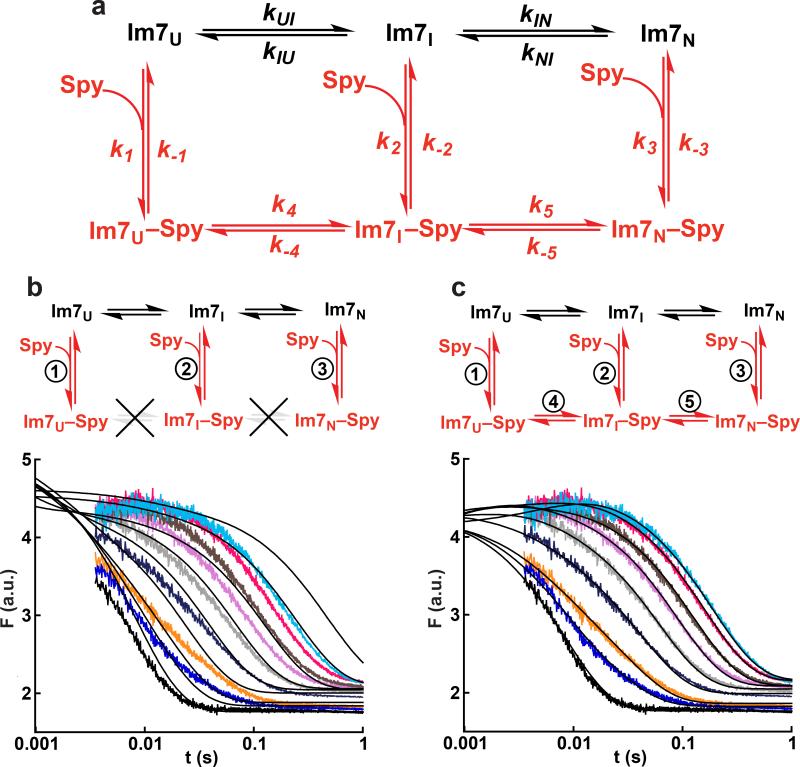
Using all of the kinetic measurements, we next determined the complete protein folding mechanism of Im7 in the presence of Spy by globally fitting the fluorescence transients for the two experiments (the folding of Im7-WT in the presence of Spy and the binding of Spy to Im7-WT) to various possible models in KinTek Explorer27. The same kinetic mechanism should govern both experiments, as the only difference between the two was the initial folding status of Im7. Convergence when globally fitting the raw fluorescence traces is rigorous in that it requires that the model adequately explain both the changes in kobs and the kinetic amplitudes as a function of Spy concentration for all transients. The kinetic mechanisms tested were built upon the well-characterized three-state on-pathway mechanism for Im7 folding in the absence of Spy15–20; we then increased the complexity of the tested models in a step-wise fashion until a mechanism was identified that could adequately describe all of the data (Fig. 5a). To help constrain the global fitting, we fixed the equilibrium constant for the conversion of unbound Im7U to unbound Im7I (KUI) and the forward and reverse rate constants (kIN and kNI, respectively) for the conversion of unbound Im7I to unbound Im7N to the known values obtained from the urea dependence for Im7 folding and unfolding (Supplementary Fig. 5).
Figure 5.
Global fitting of Im7+Spy kinetic data. (a) The global fitting was built upon the well-characterized mechanism15–20 for Im7 folding in the absence of Spy (black path). The experimental fluorescence traces for Spy binding to Im7-WT and Im7-WT folding in the presence of Spy (Fig. 4) were globally fit to different mechanisms containing various combinations of the steps in red. In the global fitting, the equilibrium constant for the Im7U to Im7I step (KUI = kUI/kIU) and the forward and reverse rate constants for the Im7I to Im7N step were fixed to the values determined from the urea dependence of Im7 folding in the absence of Spy (Supplementary Fig. 5). (b, c) Attempted global fitting to the mechanism that omits and allows, respectively, the folding of Im7 while bound to Spy. For clarity, only the traces for Im7 folding in the presence of Spy are shown. The black lines in the plots are the best fit to the data. The mechanism that completely omits folding of Im7 while bound to Spy (b) fails to fit the data, whereas the mechanism that allows folding of Im7 while bound (c) can successfully fit the data. Global fitting to additional mechanisms and the best fit for the Spy binding Im7-WT data can be found in Supplementary Fig. 6.
Strikingly, the mechanism shown in Figure 5b that omits folding of Im7 while bound to Spy (steps 4 and 5) could not fit the experimental traces. This is because, in this mechanism, Im7 must first be released from Spy before it can continue folding, making Spy a competitive inhibitor to Im7 folding; as a result, the Figure 5b mechanism predicts that kobs for all phases should reach 0 s−1 at high Spy concentrations, in marked contrast with our experimental observations. This result shows that chaperones do not necessarily need to release bound substrates from their surface in order for substrate folding to occur, as has been posited to be the case for several ATP-dependent chaperones1.
A good fit was only achieved when we globally fit the data to the kinetic mechanism that allows both folding steps 4 and 5, i.e., complete folding of Im7 while bound to Spy, making this the simplest kinetic mechanism that can explain all of the experimental data (Fig. 5c). None of the 11 other mechanisms we tested resulted in a good fit (Supplementary Fig. 6). As a final validation of the mechanism in Figure 5c, we attempted to fit the three datasets obtained simultaneously to a common kinetic mechanism; ie the raw fluorescence transients for the urea dependence of Im7 folding and unfolding, the transients for Im7 folding in the presence of Spy and kinetic traces obtained during the binding of Spy to Im7-WT were all fitted globally to the mechanism shown in Figure 5c (Supplementary Fig. 7). Doing so further constrains the model by simultaneously enforcing the same fluorescence scaling factors and rate constants upon all of the raw data for all of the experiments used in the analysis. If such a fit can be achieved, this represents a rigorous test of the kinetic model27–30. Gratifyingly, an excellent fit was achieved (Table 1 and Supplementary Fig. 7). The Kd values for Spy binding to Im7U, Im7I, and Im7N obtained from the global fit are similar to the experimental values obtained by ITC for Spy binding to the Im7 variants Im7-L18A L19A L37A, Im7-L53A I54A, and Im7-WT (Supplementary Table 1). In addition, the relative fluorescence intensities generated from the fit for the different species in the kinetic mechanism were similar to the relative fluorescence intensities of Im7-L18A L19A L37A (Im7U), Im7-L53A I54A (Im7I), and Im7-WT, and their complexes with Spy, further validating the mechanism (Supplementary Table 1). The global analysis also satisfies the thermodynamic cycles of the mechanism in Figure 5c by maintaining a zero net free energy change around each cycle.
Table 1.
Rate constants obtained from global fit of kinetic data to mechanism in Fig. 5c that allows complete folding of Im7 while bound to Spy
| Rate constanta (s−1 or M−1s−1) | |
|---|---|
| kUI | 1610 ± 210 |
| kIU | 1940 ± 180 |
| kIN | 286 ± 23 |
| kNI | 0.99 ± 0.01 |
| k1 | 1.4 ± 0.2 × 107 |
| k−1 | 65 ± 14 |
| k2 | 1.7 ± 0.2 × 107 |
| k−2 | 79 ± 17 |
| k3 | 3.3 ± 0.4 × 107 |
| k−3 | 1000b |
| k4 | 51 ± 12 |
| k−4 | 62 ± 11 |
| k5 | 10.1 ± 2.1 |
| k−5 | 0.23 ± 0.01 |
Values in 0.7 M urea. Determined by global analysis of all data (Supplementary Fig. 7). Values reported are the mean ± s.e. of the fit.
Fixed at 1000 s−1.
DISCUSSION
Our data indicate that Spy can bind to multiple conformers of Im7 and that Spy actually allows Im7 to fold while bound to the chaperone surface. The forward rate constants for Im7 folding bound to Spy obtained by global analysis are 30–40 times lower than the corresponding rate constants for Im7 folding in solution (Table 1), indicating that there is a kinetic penalty associated with Im7 folding while bound to Spy. However, the rate of Im7 folding while bound to Spy still occurs on a sub-second timescale, much faster than transcription or translation; thus, fast enough to be physiologically relevant. This ability of Spy to allow substrates to fold while bound is likely an essential component of its function in vivo—if substrate folding while bound to Spy were not possible, folding of proteins would be inhibited at the ~2 mM concentration of Spy that is present in vivo during stress. Previously, barnase was shown to fold while bound to the cavity of the GroEL ring in the absence of ATP and GroES, and at a slower rate than barnase folds when free in solution31,32. More recently, a mutant of the model protein substrate, Fyn SH3 domain (SH3Mut), was shown by NMR to fold and unfold into a partially folded intermediate state while bound within the double ring structure of GroEL, in the absence of ATP and GroES at rates substantially faster than SH3Mut folds and unfolds when free in solution33. GroEL can therefore accelerate or decelerate substrate folding, depending on the protein, whereas Spy, thus far, has only been shown to decelerate the folding of Im7. The folding rate constant of SH3Mut was ~3 times higher while bound to GroEL whereas the unfolding rate constant was ~500 times higher while bound, leading to a dramatic stabilization of the intermediate state over the native state of SH3Mut and contributing to GroEL's intrinsic unfoldase activity33. In contrast, Spy binds Im7I and Im7U with only a ~6-fold higher affinity than Im7N and therefore does not substantially destabilize the native state of Im7 when bound. By contrast with Spy, which has to function in vivo in the absence of an energy source and the assistance of cochaperones, GroEL and in many other chaperone systems, utilize ATP and cochaperones (like GroES) to control substrate binding and release1,6,34–36. However, the fact that both Spy and GroEL allow protein folding while bound raises the intriguing possibility that folding while flexibly bound may represent the underlying basis for the action of multiple chaperones. The evolutionary addition of ATP and co-chaperone dependencies would then serve as enhancements to help control chaperone action. It has previously been thought that only ATP dependent “foldase” chaperones can actively facilitate protein folding. The ATP independent “holdase” chaperones were thought to play a more passive role, holding onto aggregation-sensitive folding intermediates and in so doing inhibiting aggregation until they can transfer these intermediates to foldases which then actively fold the substrate proteins. Our finding that proteins can fold while loosely bound to the surface of Spy suggests that ATP independent chaperones may play a more active role in protein folding than was previously surmised.
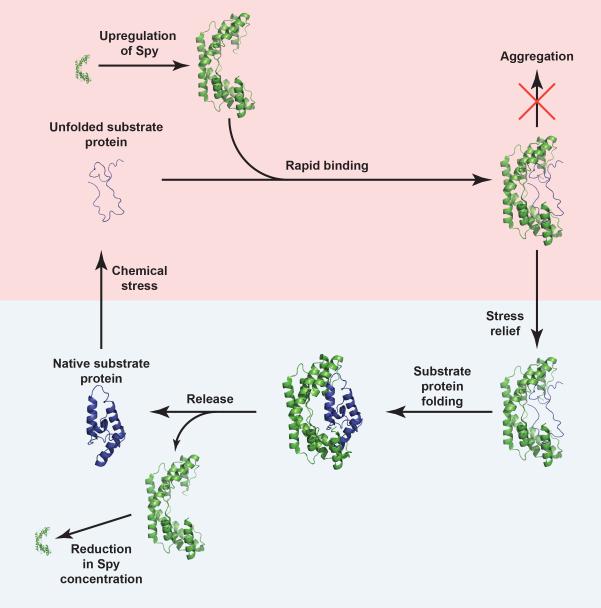
In light of our findings, we propose the following model for how Spy operates in vivo (Fig. 6). Upon chemical stress such as butanol or tannins, Spy production is upregulated, rapidly transforming the periplasmic concentration from near zero under non-stress conditions to ~2 mM in the presence of stress10,12,13. At these concentrations, Spy rapidly (on a sub-ms timescale) associates most tightly with proteins that are at least partially or transiently unfolded by the stressors. At high Spy concentrations, the vast majority of these unfolded proteins are bound to Spy, preventing their aggregation. Once the stress is removed, refolding of the substrate protein becomes energetically more favorable. We have shown that Spy allows Im7 to fold to its native state while still remaining bound to the chaperone. This mechanism minimizes the concentration of unbound, aggregation-sensitive protein folding intermediates, while still allowing the timely folding of substrates to their aggregation-insensitive native state prior to release. The dilution of Spy through degradation or cell growth after return to non-stress conditions is expected to gradually lower the Spy concentration, leading to the release of substrate proteins in their native state by mass action (Supplementary Movie). Spy thus provides proteins with a sanctuary from stress that allows refolding while avoiding aggregation.
Figure 6.
Proposed mechanism for how Spy inhibits aggregation and assists protein folding in vivo. Clockwise from the bottom left (native substrate protein), exposure to chemical stress (denoted by the pink box) causes proteins in the periplasm of Escherichia coli to unfold. The stress induces the production of Spy, which rapidly associates with its substrate proteins, thereby preventing their aggregation. Once the stress is removed (denoted by the light blue box), dilution of Spy through cell growth or degradation lowers the Spy concentration, allowing the native substrate protein to be released.
In summary, we have provided evidence that the ATP-independent chaperone Spy rapidly associates with many different conformers (U, I, and N) of its substrate proteins, at least as exemplified by Im7, the protein for which Spy was first discovered as a chaperone10. Spy allows folding while bound, thereby preventing the irreversible aggregation of proteins that may otherwise detrimentally affect the cell. Spy allows Im7 to fold while bound without substantially altering its folding energy landscape by binding to all three populated states with similar affinities. The surface of Spy therefore is tuned to accommodate not only multiple substrate proteins10,14, but also drastically different conformations of the same protein. Spy binds substrates with a relatively modest (μM) affinity, which is likely necessary for the chaperone to promiscuously bind multiple substrates and multiple folding conformations of the same protein. This finding is consistent with a theoretical model for how chaperones work in which loose binding allows substrate folding37. We speculate that the principle of binding multiple conformations and allowing protein folding while loosely bound may form the primordial basis of chaperone action.
ONLINE METHODS
Protein expression and purification
WT Spy was purified as described previously10, with the exception that Ni-HisTrap columns (GE Healthcare) were used in place of the Ni-NTA beads and mini-chromatography column. Following the Ni-HisTrap column, WT Spy was treated with ULP1 to cleave the SUMO-tag while dialyzing into 40 mM Tris HCl, pH 8, 300 mM NaCl. After dialysis, Spy was passed over the HisTrap column to remove the cleaved SUMO-tag. The flow-through was exchanged into 20 mM Tris HCl, pH 8 by five cycles of concentration and dilution in a 10 kDa cutoff Amicon centrifugal filter unit. Spy was then passed over a HiTrap Q column. Spy has an isoelectric point of 9.5 and therefore was collected in the flow-through. The flow-through containing Spy was buffer-exchanged into 50 mM sodium phosphate buffer, pH 6.5, loaded onto a HiTrap SP column, and eluted using a 0–1 M NaCl gradient in 50 mM sodium phosphate buffer, pH 6.5. Spy was then further purified using a HiLoad 75 gel-filtration column in 40 mM HEPES-KOH, pH 7.5, 100 mM NaCl. Fractions containing Spy were concentrated, frozen in liquid nitrogen, and stored at −80 °C. Im7-L18A L19A L37A, Im7-L53A I54A, and Im7-WT were purified using the same protocol, except that the Im7 was retained by the Q column (the isoelectric point of Im7 is 4.5). Im7 was eluted from the Q column using a 0–1 M NaCl gradient. The SP column step was not used in purifying Im7-WT or the Im7 variants.
Isothermal titration calorimetry (ITC)
ITC was performed using a MicroCal iTC200 in 40 mM HEPES-KOH, pH 7.5, 100 mM NaCl at 10 °C with Im7 in the cell and Spy in the titration syringe. 150 μM Im7-L18A L19A L37A, 50 μM Im7-L53A I54A, and 230 μM Im7-WT were titrated with 1.65 mM, 550 μM, and 2.53 mM Spy dimer, respectively. 1.0 μL injections were used for the titrations with Im7-L18A L19A L37A and Im7-L53A I54A. 1.5 μL injections were used for the Im7-WT titration. ITC thermograms were fit to a one-site model using the Origin software provided with the instrument.
Stopped-flow fluorescence experiments
Stopped-flow experiments were performed at 10 °C in 40 mM HEPES-KOH, pH 7.5, 100 mM NaCl using a KinTek SF-2004 or an Applied Photophysics SX18.MV stopped-flow instrument. Fluorescence of the single tryptophan of Im7 was excited at 296 nm with a 4 nm bandpass and the fluorescence emission signal was collected through a 340 ± 10 nm band-pass filter. A 1:1 mix was used for the experiments monitoring the binding of Spy to the different Im7 variants. Final Im7 concentrations were 5 μM, 0.5 μM, and 4.8 μM for Im7-L18A L19A L37A, Im7-L53A I54A, and Im7-WT, respectively. The urea dependence of Im7 folding and unfolding in the absence of Spy was performed as described previously15,16,20. To measure folding of Im7 in the presence of different Spy concentrations, a 1:10.5 mix was used to mix 55 μM of Im7-WT in 8 M urea against buffer containing various concentrations of Spy (0–660 μM dimer after mixing). The final Im7-WT concentration after mixing was 4.8 μM, and the urea concentration after mixing was 0.7 M. For the Spy binding to Im7-WT experiment, 0.7 M urea was also included in the buffer so that the buffer conditions were identical to the final buffer conditions in the Im7 folding experiments in the presence of Spy experiment. 10-15 traces were averaged for each data point in the Spy binding to the Im7 variants experiments, and 7-10 traces were averaged for each data point in the Im7 folding experiments. The tyrosine of Spy contributed some background fluorescence to the stopped-flow traces, especially at higher Spy concentrations. To remove this background fluorescence, shots of Spy diluted against buffer were collected at each Spy concentration, which was then subtracted from the traces for experiments containing Im7. Observed rate constants were obtained by fitting individual traces to sums of exponentials using KaleidaGraph (Synergy Software).
Simulations of Spy binding Im7-WT kinetics (Supplementary Fig. 3)
Simulations of the change in kobs with Spy concentration for the two mechanisms in Supplementary Fig. 3 were run in Berkeley Madonna (University of California at Berkeley), which can numerically solve the differential equations describing the two kinetic schemes depicted in this figure. Simulations of experimental traces were generated by defining the kinetic scheme and initial conditions at several different Spy concentrations. The forward and reverse rate constants for the Im7I to Im7N step used in the simulations were the values determined at 0.7 M urea from the denaturant dependence of Im7 folding in the absence of Spy (Supplementary Fig. 5). kon and koff for Spy binding Im7I and Im7N were chosen so that the affinity of Spy for Im7I and Im7N would equal the values determined from the ITC titrations (Fig. 2) and so that binding of Spy to Im7 would be rapid relative to the conformational change step (koff and the apparent kon rate constants are much greater than the rate constants for the conformational change). The folding and unfolding rate constants for Im7 bound to Spy in Supplementary Fig. 3b were selected to maintain a zero net free energy around the thermodynamic cycle and so that the sum of the forward and reverse rate constants for the Im7I to Im7N step while bound would equal 3.5 s−1. 4.785 μM Im7N and 0.015 μM Im7I were used as the initial conditions (consistent with the equilibrium constant of 319 in 0.7 M urea for the Im7I to Im7N step, as determined by the urea dependence of Im7 folding and unfolding in the absence of Spy, Supplementary Fig. 5). Traces of the slow step in the simulation were subsequently fit in KaleidaGraph (Synergy Software) to a single exponential to obtain the observed rate constant at different Spy concentrations. The limiting rate constant of the slow step at infinite Spy concentrations was determined by fitting plots of kobs against Spy concentration from the simulated traces to an inverse square hyperbola.
Global fitting of kinetic data
The stopped-flow fluorescence traces for Spy binding to Im7-WT under non-denaturing conditions and the traces for Im7 folding in the presence of different Spy concentrations were globally fit to various kinetic mechanisms in KinTek Explorer (KinTek Corporation)27, version 5.1.15078, which can constrain thermodynamic cycles by maintaining a zero net free energy around the cycle. This program fits kinetic data by direct numerical integration of the rate equations that define the kinetic model; fitting in KinTek Explorer involves minimizing the global χ2 to obtain the microscopic rate constants and the fluorescence signals of the individual species of the kinetic mechanism. The mechanisms tested were built upon the known mechanism for Im7 folding15–20, which passes through an intermediate state prior to reaching the native state. When initially globally fitting the Im7 folding in the presence of Spy and Spy binding to native Im7 data to different mechanisms, the kIN and kNI rate constants were fixed at 345 s−1 and 1.08 s−1, respectively, and the KUI equilibrium constant (kUI/kIU) was fixed at 0.57; these were the values in 0.7 M urea obtained from the Im7 folding/unfolding chevron plots (Supplementary Fig. 5). When fitting the Spy binding to Im7-WT traces, the Im7 was allowed to equilibrate for 10 s prior to the addition of Spy so the 4.8 μM Im7 could distribute between Im7U, Im7I, and Im7N to the concentrations dictated by the equilibrium constants generated from the fitting before the addition of Spy. 4.8 μM Im7U was used as the initial conditions in globally fitting the Im7 folding in the presence of Spy traces. The fluorescence of Im7I and the Im7I-Spy complex were set equal to one another; this constraint is consistent with the fact that there is no change in fluorescence prior to the conformational change when Spy binds Im7-L53A 54A (i.e., there is no burst phase, Supplementary Fig. 1). Initial global fitting indicated that the association (kon) and dissociation (koff) rate constants for Spy binding Im7N were poorly constrained by the data since Spy binding Im7N occurred within the dead time in the Spy binding Im7-WT experiment. Accordingly, koff for the Spy binding Im7N step was fixed at 1000 s−1 (a reasonable lower limit given the dead time) to allow the program to simply fit the Kd for this step. During the process of global fitting, individual traces were scaled using a correction factor (<4%) to correct for slight fluctuations in lamp intensity between traces. The mechanism depicted in Figure 5c, which includes steps for Spy binding to Im7U, Im7I, and Im7N and allows for the complete folding of Im7 while bound to Spy, was the simplest model able to satisfactorily describe the data based on the goodness-of-fit (χ2) and visual inspection of the fit. The fluorescence traces for Im7 folding and unfolding at different urea concentrations were fitted assuming that a rate constant varies with urea concentration based on the expression kxy = kxy H2Oe(mxy/RT)[Urea] where kxy is the rate constant for the formation of y from x, kxy H2O is the rate constant in 0 M urea, and mxy defines the urea dependence for this step15. It was also assumed that the fluorescence of Im7U, Im7I, and Im7N changed linearly with urea concentration. From the simultaneous fitting of all of the fluorescence traces, the fluorescence of Im7U, Im7I, and Im7N was 0.725 + 0.055[urea], 1.070 + 0.067[urea], and 0.335 + 0.041[urea], respectively. The rate constants reported in Table 1 are rounded to 2-3 significant figures.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Horowitz for critical reading of the manuscript. This work was funded by a Boehringer Ingelheim Fonds fellowship (P.K.) and US National Institutes of Health Grant GM102829 (J.C.A.B. and S.E.R.), which also funded J.R.H. J.C.A.B. is supported as a Howard Hughes Investigator.
Footnotes
AUTHOR CONTRIBUTIONS
F.S., P.K., and J.R.H. performed the experiments. All authors analyzed the data. F.S., S.E.R., and J.C.A.B. designed the study. F.S. wrote the manuscript, with contributions from all other authors.
REFERENCES
- 1.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 2.Buchner J. Hsp90 & Co.– a holding for folding. Trends Biochem. Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 3.Zuiderweg ERP, et al. Allostery in the Hsp70 chaperone proteins. Top. Curr. Chem. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Priya S, Sharma SK, Goloubinoff P. Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides. FEBS Lett. 2013;587:1981–1987. doi: 10.1016/j.febslet.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013;14:630–42. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwich AL, Fenton WA. Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 7.Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J. Biol. Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- 8.Tapley TL, Franzmann TM, Chakraborty S, Jakob U, Bardwell JCA. Protein refolding by pH-triggered chaperone binding and release. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1071–1076. doi: 10.1073/pnas.0911610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann A, Bukau B, Kramer G. Structure and function of the molecular chaperone Trigger Factor. Biochim. Biophys. Acta. 2010;1803:650–661. doi: 10.1016/j.bbamcr.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Quan S, et al. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat. Struct. Mol. Biol. 2011;18:262–269. doi: 10.1038/nsmb.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahl J-U, et al. HdeB functions as an acid-protective chaperone in bacteria. J. Biol. Chem. 2015;290:65–75. doi: 10.1074/jbc.M114.612986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brynildsen MP, Liao JC. An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol. Syst. Biol. 2009;5:277. doi: 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutherford BJ, et al. Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl. Environ. Microbiol. 2010;76:1935–1945. doi: 10.1128/AEM.02323-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quan S, et al. Super Spy variants implicate flexibility in chaperone action. Elife. 2014;3:e01584. doi: 10.7554/eLife.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson N, Capaldi AP, James R, Kleanthous C, Radford SE. Rapid folding with and without populated intermediates in the homologous four-helix proteins Im7 and Im9. J. Mol. Biol. 1999;286:1597–1608. doi: 10.1006/jmbi.1998.2548. [DOI] [PubMed] [Google Scholar]
- 16.Capaldi AP, Shastry MCR, Kleanthous C, Roder H, Radford SE. Ultrarapid mixing experiments reveal that Im7 folds via an on-pathway intermediate. Nat. Struct. Biol. 2001;8:68–72. doi: 10.1038/83074. [DOI] [PubMed] [Google Scholar]
- 17.Capaldi AP, Kleanthous C, Radford SE. Im7 folding mechanism: misfolding on a path to the native state. Nat. Struct. Biol. 2002;9:209–16. doi: 10.1038/nsb757. [DOI] [PubMed] [Google Scholar]
- 18.Friel CT, Capaldi AP, Radford SE. Structural analysis of the rate-limiting transition states in the folding of Im7 and Im9: Similarities and differences in the folding of homologous proteins. J. Mol. Biol. 2003;326:293–305. doi: 10.1016/s0022-2836(02)01249-4. [DOI] [PubMed] [Google Scholar]
- 19.Knowling SE, Figueiredo AM, Whittaker SBM, Moore GR, Radford SE. Amino acid insertion reveals a necessary three-helical intermediate in the folding pathway of the colicin E7 immunity protein Im7. J. Mol. Biol. 2009;392:1074–1086. doi: 10.1016/j.jmb.2009.07.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friel CT, Smith DA, Vendruscolo M, Gsponer J, Radford SE. The mechanism of folding of Im7 reveals competition between functional and kinetic evolutionary constraints. Nat. Struct. Mol. Biol. 2009;16:318–324. doi: 10.1038/nsmb.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spence GR, Capaldi AP, Radford SE. Trapping the on-pathway folding intermediate of Im7 at equilibrium. J. Mol. Biol. 2004;341:215–226. doi: 10.1016/j.jmb.2004.05.049. [DOI] [PubMed] [Google Scholar]
- 22.Gsponer J, et al. Determination of an ensemble of structures representing the intermediate state of the bacterial immunity protein Im7. Proc. Natl. Acad. Sci. 2006;103:99–104. doi: 10.1073/pnas.0508667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whittaker SBM, Spence GR, Günter Grossmann J, Radford SE, Moore GR. NMR analysis of the conformational properties of the trapped on- pathway folding intermediate of the bacterial immunity protein Im7. J. Mol. Biol. 2007;366:1001–1015. doi: 10.1016/j.jmb.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pashley CL, et al. Conformational properties of the unfolded state of Im7 in nondenaturing conditions. J. Mol. Biol. 2012;416:300–318. doi: 10.1016/j.jmb.2011.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogt AD, Di Cera E. Conformational selection or induced fit? A critical appraisal of the kinetic mechanism. Biochemistry. 2012;51:5894–5902. doi: 10.1021/bi3006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gianni S, Dogan J, Jemth P. Distinguishing induced fit from conformational selection. Biophys. Chem. 2014;189:33–39. doi: 10.1016/j.bpc.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Johnson KA, Simpson ZB, Blom T. Global Kinetic Explorer: A new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 28.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 29.Huddleston JP, Schroeder GK, Johnson K. a., Whitman CP. A pre- steady state kinetic analysis of the αY60W mutant of trans-3-chloroacrylic acid dehalogenase: Implications for the mechanism of the wild-type enzyme. Biochemistry. 2012;51:9420–9435. doi: 10.1021/bi3010686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroeder GK, et al. Reaction of cis -3-chloroacrylic acid dehalogenase with an allene substrate, 2,3-butadienoate: Hydration via an enamine. J. Am. Chem. Soc. 2012;134:293–304. doi: 10.1021/ja206873f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray TE, Fersht AR. Refolding of barnase in the presence of GroE. J. Mol. Biol. 1993;232:1197–1207. doi: 10.1006/jmbi.1993.1471. [DOI] [PubMed] [Google Scholar]
- 32.Corrales FJ, Fersht AR. The folding of GroEL-bound barnase as a model for chaperonin-mediated protein folding. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5326–5330. doi: 10.1073/pnas.92.12.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Libich DS, Tugarinov V, Clore GM. Intrinsic unfoldase/foldase activity of the chaperonin GroEL directly demonstrated using multinuclear relaxation-based NMR. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8817–8823. doi: 10.1073/pnas.1510083112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fink AL. Chaperone-mediated protein folding. Physiol. Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 35.Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jewett AI, Shea JE. Folding on the chaperone: Yield enhancement through loose binding. J. Mol. Biol. 2006;363:945–957. doi: 10.1016/j.jmb.2006.08.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.