Cdc42 is believed to play an important role in controlling the polarity of migrating cells, but it has not been possible to directly determine the effects of localized Cdc42 activity. Optogenetic activation of Cdc42 at one side of the cell was used to identify local and distal signaling responses that contribute to directed cell migration.
Abstract
Migratory immune cells use intracellular signaling networks to generate and orient spatially polarized responses to extracellular cues. The monomeric G protein Cdc42 is believed to play an important role in controlling the polarized responses, but it has been difficult to determine directly the consequences of localized Cdc42 activation within an immune cell. Here we used subcellular optogenetics to determine how Cdc42 activation at one side of a cell affects both cell behavior and dynamic molecular responses throughout the cell. We found that localized Cdc42 activation is sufficient to generate polarized signaling and directional cell migration. The optically activated region becomes the leading edge of the cell, with Cdc42 activating Rac and generating membrane protrusions driven by the actin cytoskeleton. Cdc42 also exerts long-range effects that cause myosin accumulation at the opposite side of the cell and actomyosin-mediated retraction of the cell rear. This process requires the RhoA-activated kinase ROCK, suggesting that Cdc42 activation at one side of a cell triggers increased RhoA signaling at the opposite side. Our results demonstrate how dynamic, subcellular perturbation of an individual signaling protein can help to determine its role in controlling polarized cellular responses.
INTRODUCTION
Migrating cells exhibit spatially polarized intracellular signaling, with distinct biochemical events confined to the front or back of a cell (Artemenko et al., 2014). Signaling proteins and lipids have been identified that localize to either the front or back (Cai and Devreotes, 2011), but how this spatial polarization is generated, maintained, and reversed remains poorly understood. These are inherently dynamic cellular processes, but they have mostly been studied using genetic perturbations that lack temporal or subcellular spatial control. In general, it is not known how changing the activity of a given signaling protein affects the rest of the signaling network both locally and distally within a cell. Here we use an optogenetic approach to dynamically control endogenous Cdc42 activity and determine its ability to control signaling events at the front and back of migratory immune cells.
The Rho-family monomeric G protein Cdc42 is considered a master regulator of cell polarity (Etienne-Manneville, 2004). It has been implicated in both G protein–coupled receptor (GPCR)– and receptor tyrosine kinase (RTK)–stimulated chemotaxis in cell types ranging from leukocytes to fibroblasts (Chou et al., 2003; Li et al., 2003; Cau and Hall, 2005). Cdc42 activity is found at the leading edge of migrating cells (Li et al., 2003), where it contributes to the generation of cytoskeleton-driven cellular protrusions by regulating Wiskott–Aldrich syndrome protein (WASP) and subsequently Arp2/3 (Leung and Rosen, 2005). This would suggest a role in generating cell locomotion. However, studies using gene knockdown or a dominant-negative Cdc42 mutant suggest that Cdc42 is not required for cell motility but instead plays a critical role in controlling the directionality of cell migration (Allen et al., 1998; Li et al., 2003; Srinivasan et al., 2003; Hind et al., 2014). These observations indicate that Cdc42 may play an important role in controlling the formation of front–back polarity in migrating cells. Indeed, it has been suggested that Cdc42 activation at the leading edge not only stimulates local “frontness” responses but can also produce long-range effects that modulate signaling events at the cell rear (Van Keymeulen et al., 2006; Szczur et al., 2009; Kumar et al., 2012). However, it has not been possible to test this hypothesis directly due to a lack of methods for dynamic subcellular control over Cdc42 activity.
Subcellular optogenetics can overcome this limitation (Karunarathne et al., 2015). We use subcellular optogenetic control to activate Cdc42 at one side of a migratory immune cell to address several questions regarding its role in establishing front–back polarity and directional cell migration. Is direct activation of Cdc42, independent of parallel signaling pathways that may be activated by chemoattractant receptors, sufficient to generate cell motility and directional migration? Is Cdc42 activation sufficient to generate localized “frontness” responses such as actin polymerization–driven membrane protrusions? Can Cdc42 activation at one side of a cell trigger events at the opposite side that define its rear, such as the formation of actomyosin bundles that generate retraction? We address these questions by dynamically controlling Cdc42 activity optically while simultaneously measuring molecular and cellular responses by live-cell imaging.
RESULTS
Chemoattractant-sensing GPCRs trigger Cdc42 activation in RAW 264.7 macrophage cells
We previously demonstrated that the macrophage-like cell line RAW 264.7 provides a useful model system for optogenetic studies of immune cell migration (Karunarathne et al., 2013b; O’Neill and Gautam, 2014). We showed that migration of these cells can be controlled by optically stimulating G protein–coupled blue opsin (Karunarathne et al., 2013b) or directly generating intracellular signaling gradients through the use of novel constructs for subcellular optogenetic inhibition of heterotrimeric G protein subunits (O’Neill and Gautam, 2014). Farther downstream in the chemoattractant-stimulated signaling cascade, Rho-family monomeric G proteins play important roles in controlling cell morphology through regulation of cytoskeletal dynamics. Here we sought to identify the contributions of the Rho-family protein Cdc42 to cell migration by applying optogenetics in RAW cells. First, we performed experiments to verify that chemoattractant receptors trigger Cdc42 activation in these cells.
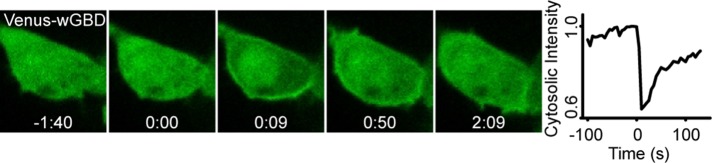
Cdc42 activation downstream of chemoattractant-sensing GPCRs has been reported in a variety of cell types (Benard et al., 1999; Li et al., 2003; Ueda et al., 2008; Runne and Chen, 2013). In RAW 264.7 cells, a dominant-negative Cdc42 mutant inhibited cytoskeletal responses to GPCR stimulation (Cox et al., 1997), but GPCR-mediated activation of Cdc42 was not measured directly. Therefore, we used a live-cell imaging approach to detect GPCR stimulation of Cdc42 in RAW cells. Cells were transfected with the chemoattractant receptor CXCR4 and the Cdc42 biosensor Venus-wGBD. The latter contains the G protein–binding domain from WASP (wGBD), which selectively binds to activated Cdc42, resulting in translocation from the cytosol to the plasma membrane upon Cdc42 activation (Kim et al., 2000; Benink and Bement, 2005). Stimulation of cells with 50 ng/ml SDF-1α resulted in subtle but discernible translocation of Venus-wGBD to the plasma membrane, and the translocation was more pronounced in cells overexpressing wild-type Cdc42 (Figure 1). The translocation was transient, suggesting that the Cdc42 response to a global stimulus exhibits adaptation. This is consistent with several signaling responses that have been reported to exhibit adaptation to spatially uniform activation of chemoattractant receptors (Devreotes and Horwitz, 2015). Thus chemoattractant receptors activate Cdc42 in RAW 264.7 cells, consistent with findings in other types of migratory cells.
FIGURE 1:
Chemoattractant receptors activate Cdc42 in RAW 264.7 cells. The image sequence shows a RAW cell transfected with CXCR4, Cdc42, and Venus-wGBD (Cdc42 sensor). SDF-1α was added at t = 0. Venus-wGBD selectively binds to activated Cdc42, resulting in translocation from the cytosol to the plasma membrane. The plot shows the transient decrease in cytosolic fluorescence after CXCR4 activation. Time is shown in minutes:seconds.
Subcellular optogenetic activation of Cdc42 generates directional migration
Next we sought to determine directly the effects of localized Cdc42 activation on RAW cell migration. To optically activate Cdc42 independent of upstream signaling events, we used light- inducible dimerization to optically recruit a Cdc42-selective guanine nucleotide exchange factor (GEF) to the plasma membrane (Figure 2; Guntas et al., 2015). The approach is based on optical control of the interaction between the bacterial protein SspB and the peptide SsrA (Guntas et al., 2015). The SsrA peptide is fused to the C-terminus of the LOV-Jα helix, and the fusion is referred to as iLID. In the dark, iLID adopts a conformation that prevents SsrA from interacting with SspB. Photoactivation with blue light changes the iLID conformation to make SsrA accessible, resulting in light-inducible dimerization with SspB. We used a fusion of SspB with the DHPH domain of ITSN (Guntas et al., 2015), which is a Cdc42-selective GEF (Jaiswal et al., 2013). In cells coexpressing the ITSN construct with a membrane-targeted iLID (iLID-CaaX), optical activation with blue light confined to a subcellular region triggers local recruitment of the Cdc42 GEF to the plasma membrane, resulting in increased levels of Cdc42 activity.
FIGURE 2:
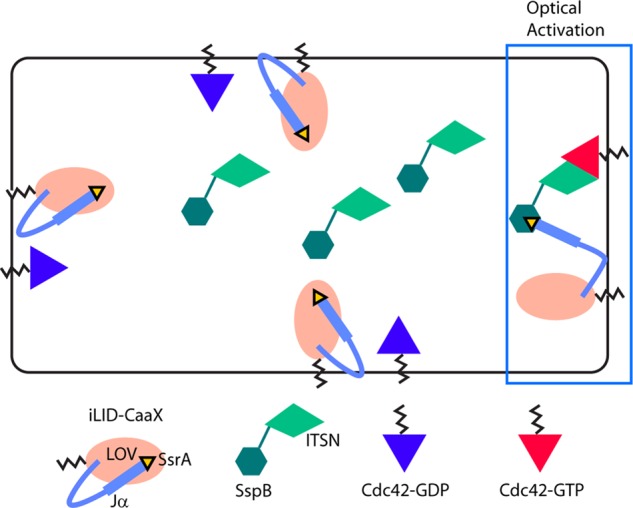
Optical stimulation of Cdc42 using a light-inducible dimerization pair. A light-inducible dimerization pair is used to achieve rapid temporal and subcellular spatial control over the membrane localization of a Cdc42 GEF (Guntas et al., 2015). The SsrA peptide is fused to the C-terminus of the Jα helix of the LOV domain. This fusion also contains a membrane-targeting domain and is referred to as iLID-CaaX. In the dark, iLID-CaaX adopts a conformation that prevents the interaction between SsrA and SspB. On optical activation, it changes conformation to make SsrA accessible for SspB binding. Optical activation is applied at one side of the cell, resulting in local recruitment of SspB to the plasma membrane. A Cdc42-selective GEF domain from ITSN is fused to SspB, so that local membrane recruitment results in local activation of Cdc42.
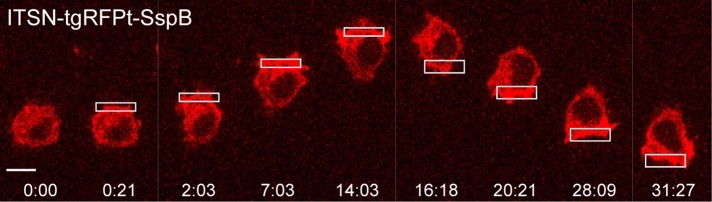
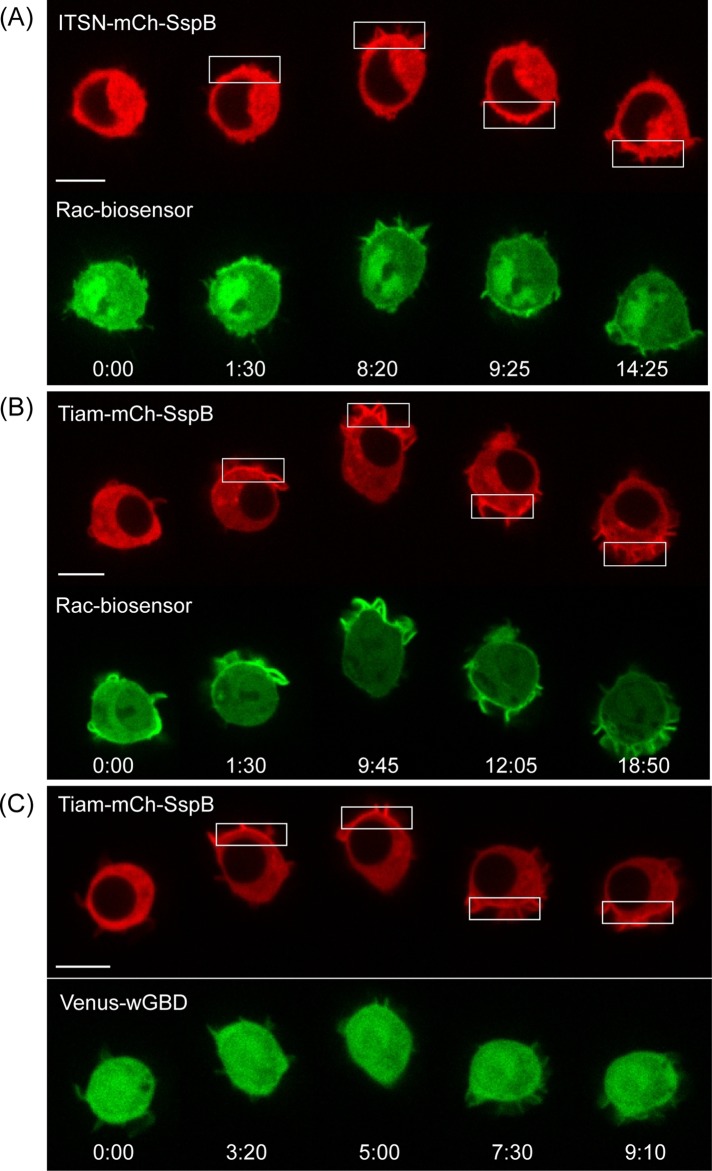
RAW cells were transfected with ITSN-tgRFPt-SspB and iLID-CaaX. We used the R73Q mutant SspB, which has reduced interaction with iLID in the dark relative to wild-type SspB (Guntas et al., 2015). The effect of localized Cdc42 activation was then determined by live-cell imaging (Figure 3 and Supplemental Movie S1). Localized photoactivation with 445-nm light triggered translocation of the ITSN construct from the cytosol to the plasma membrane in the photoactivated region within seconds. Activation of endogenous Cdc42 by the membrane-recruited ITSN domain generated all of the stereotypical features of cell migration. It initiated cells to adopt a polarized morphology, with lamellipodia restricted to the photoactivated side of the cell. The membrane protrusions exhibited dynamic cycles of extension and retraction, as well as lateral movement at the leading edge of the cell. Cells migrated toward the photoactivated side, and the directional response could be maintained by continued photoactivation at the leading edge. The migration could also be reversed rapidly by changing the side of photoactivation. Retraction of the cell rear was especially pronounced upon reversal of the direction of migration, making it clear that the Cdc42 at the leading edge does not simply drag the cell forward but also actively controls signaling at the cell rear required for retraction.
FIGURE 3:
Subcellular optical activation of endogenous Cdc42 generates directional, reversible cell migration. The image sequence shows a RAW cell transfected with ITSN-tgRFPt-SspB and iLID-CaaX. The white box marks the region photoactivated with 445-nm light. Scale bar, 10 μm. Time is given in minutes:seconds. See also Supplemental Movie S1.
Cdc42-driven cell migration broadly reflects GPCR-driven migration
Immune cell migration is driven natively by activation of GPCRs and RTKs that sense chemoattractants. We examined migration driven by GPCR activation relative to direct activation of Cdc42. Such a comparison can identify whether Cdc42 activation recapitulates GPCR activated migration or is distinctly different as a result of the activation of the downstream element alone. We compared migration driven by optical activation of Cdc42 to that driven by optical activation of blue opsin. Blue opsin is a light-activated GPCR from cone photoreceptor cells that we showed is capable of activating Gi/o heterotrimeric G proteins in heterologous cell types and directing immune cell migration (Karunarathne et al., 2013a, b).
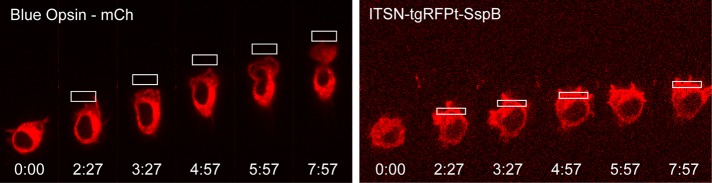
Asymmetric activation of blue opsin resulted in a directionally sensitive migration of RAW cells that was broadly similar to the migration elicited by direct activation of Cdc42 (Figure 4 and Supplemental Movie S2). The ability of direct Cdc42 activation to coordinate several responses, including generation of lamellipodia and retraction of the cell rear to control migration similar to GPCR-driven migration, reveals that Cdc42 generates “frontness” responses in the region where it is activated, as well as “backness” signals at the opposite side of the cell that lead to retraction of the cell rear. We explored both of these aspects of Cdc42 signaling in more detail in the experiments to be described.
FIGURE 4:
Comparison of GPCR-driven vs. Cdc42-driven migration. Optically stimulated migration of RAW cells through either localized photoactivation of GPCR signaling using blue opsin-mCherry or direct activation of Cdc42 using ITSN-tgRFPt-SspB together with iLID-CaaX. Both are capable of generating directional migration, but cells are more elongated along the direction of migration upon GPCR stimulation relative to Cdc42 activation. We found previously that asymmetric optical activation is best achieved when the optical input is positioned slightly outside of the cell using the laser power reported here (Karunarathne et al., 2013b). Here we found that asymmetric activation of the iLID construct, on the other hand, was optimized by positioning the optical input at the edge of the cell. This difference likely reflects different sensitivities of the chromophores present in opsins and LOV domains. See also Supplemental Movie S2.
Verification that optically triggered membrane recruitment of ITSN’s GEF domain locally activates Cdc42
The established selectivity of ITSN GEF activity for Cdc42 over other monomeric G proteins (Jaiswal et al., 2013), together with a lack of responses in negative control cells expressing an mCh-SspB construct lacking the ITSN domain (Supplemental Figure S1), suggests that the migration described earlier occurred due to activation of Cdc42 as intended. Optically triggered membrane recruitment of the ITSN-DHPH domain has been demonstrated to activate Cdc42 in other cell types, but it has never been tested in migratory immune cells (Levskaya et al., 2009; Guntas et al., 2015; Valon et al., 2015). We therefore directly tested for Cdc42 activation by live-cell imaging of the Cdc42 biosensor Venus-wGBD.
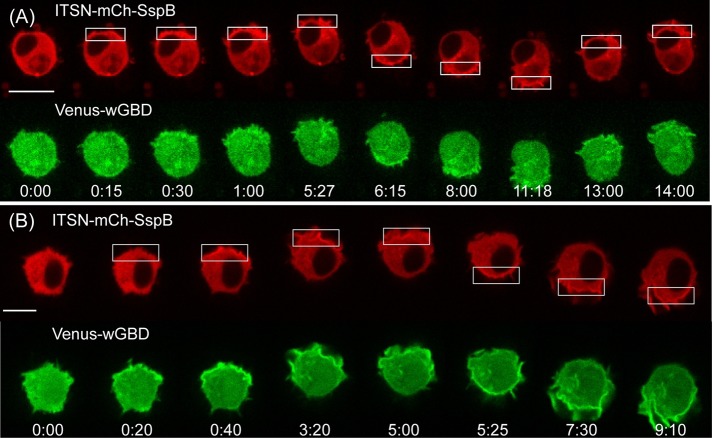
RAW cells were transfected with ITSN-mCh-SspB, iLID-CaaX, and Venus-wGBD. As expected, localized optical activation resulted in translocation of both the ITSN construct and Venus-wGBD to the plasma membrane in the optically activated region (Figure 5A and Supplemental Movie S3). However, light-induced Venus-wGBD translocation was difficult to detect (one of seven cells), even when they exhibited clear migratory responses. We suspected that Cdc42 was activated in these cells but at levels below the sensitivity range of the Venus-wGBD sensor. Indeed, cotransfection with wild-type Cdc42 resulted in detectable wGBD translocation in most cells (seven of nine cells; Figure 5B and Supplemental Figure S4). Of note, optical control of endogenous Cdc42 was sufficient to generate cell migration, as well as the molecular responses described in the remaining sections.
FIGURE 5:
Optically triggered membrane recruitment of ITSN generates localized Cdc42 activation. (A) Image sequence showing a RAW cell transfected with ITSN-mCh-SspB, iLID-CaaX, and Venus-wGBD. Membrane recruitment of the ITSN construct results in translocation of Venus-wGBD to the site of optical activation, demonstrating local activation of endogenous Cdc42. (B) Overexpression of wild-type Cdc42 enhances the effect. Whereas wGBD translocation in response to optical activation was only clearly detected in one of seven cells containing endogenous Cdc42 (A), it was detected in seven of nine cells when wild-type Cdc42 was cotransfected (B). Time is given in minutes:seconds. Scale bars, 10 μm. See also Supplemental Figure S4 and Supplemental Movie S3.
The localization of Venus-wGBD on the membrane closely resembled that of the ITSN construct, indicating that Cdc42 activity was spatially confined to the site of optical activation. This suggests that Cdc42 deactivation by GTP hydrolysis is fast enough to inhibit the diffusive spread of activated Cdc42 to other regions of the cell.
Cdc42 triggers actin polymerization at the leading edge
Given the ability of Cdc42 to regulate actin dynamics through WASP and Arp2/3 (Leung and Rosen, 2005), we anticipated that protrusions generated by optical activation of Cdc42 resulted from a local increase in actin polymerization. We tested this hypothesis by combining optical control of Cdc42 with live-cell imaging of mTopaz-Lifeact. Lifeact contains a 17-residue peptide sequence that selectively binds to filamentous actin (Riedl et al., 2008).
RAW cells were transfected with ITSN-mCh-SspB, iLID-CaaX, and mTopaz-Lifeact. Lifeact was initially distributed uniformly around the cell periphery, marking the cortical actin cytoskeleton. Localized optical activation resulted in increased Lifeact at the leading edge (six of six cells), consistent with a role for Cdc42 control of actin dynamics in driving cell protrusions (Figure 6, Supplemental Figure S5, and Supplemental Movie S4). Reversing the side of optical activation resulted in rapid reversal of the side of actin polymerization, consistent with the formation of a new leading edge and the ability of the cell to reverse the direction of migration.
FIGURE 6:
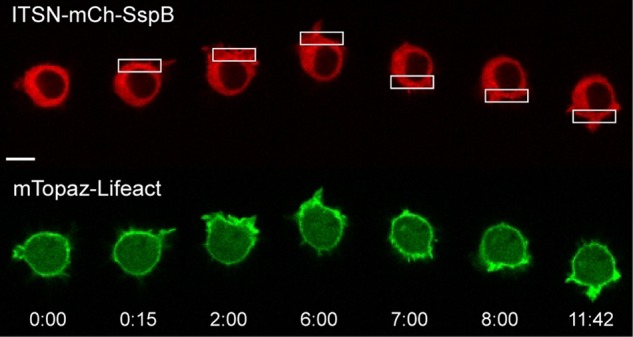
Local Cdc42 activation triggers actin polymerization at the leading edge. RAW cell transfected with ITSN-mCh-SspB, iLID-CaaX, and mTopaz-Lifeact. Optical activation of one side of the cell generates actin polymerization at the leading edge (six of six cells). Time is given in minutes:seconds. Scale bar, 10 μm. See also Supplemental Figure S5 and Supplemental Movie S4.
Cdc42 locally activates Rac but not vice versa
In several cell types, Cdc42 has been reported to trigger Rac activation (Nobes and Hall, 1995; Nishimura et al., 2005). However, how Cdc42 spatially and temporally controls Rac activity is unknown because previous experiments used methods that did not provide dynamic control over Cdc42 activity in living cells. Is Rac activity spatially confined to the region of the cell where Cdc42 is activated, or does local Cdc42-activation result in increased Rac activity throughout the entire cell? How rapidly does Cdc42 activity generate Rac activity, and how quickly can it be reversed?
We used a Rac biosensor to examine the effect of Cdc42 on Rac activation. The Rac biosensor was based on a previously reported sensor consisting of a monomeric cerulean, two tandem p21-binding domains of Pak1, monomeric Venus, and full-length Rac1 (Moshfegh et al., 2014). Although the biosensor was designed to exhibit fluorescence resonance energy transfer (FRET) changes upon Rac1 activation, we found that it also translocated from the cytosol to the plasma membrane upon Rac activation. Translocation was observed for CXCR4-mediated activation of Rac (Supplemental Figure S2) and direct optogenetic activation of Rac using optically triggered membrane recruitment of the Rac-selective GEF Tiam1 (eight cells; Figure 7B and Supplemental Figure S6).
FIGURE 7:
Cdc42 activation triggers Rac activation but not vice versa. RAW cells were transfected with the constructs specified in the image sequences, along with iLID-CaaX. (A) Optically triggered membrane recruitment of ITSN to activate endogenous Cdc42 resulted in local translocation of a Rac biosensor, suggesting that Rac can be activated downstream of Cdc42 in RAW cells. (B) The Tiam construct also activates Rac, consistent with Tiam’s known activity as a Rac-selective GEF. (C) The Tiam construct does not generate a detectable Cdc42 response, suggesting that Rac does not activate Cdc42 in these cells. Scale bars, 10 μm. Time is given in minutes:seconds.
To achieve spectral selectivity that is required for subcellular optical activation of Cdc42, we used translocation of the sensor rather than FRET changes to detect Rac activation. FRET imaging required blue excitation light, which photoactivated iLID throughout the entire cell and prevented spatial and temporal control over Cdc42 activation. In contrast, translocation of the Rac sensor could be measured using 515-nm excitation of Venus, which did not detectably photoactivate iLID at the intensity used here. This allowed us to combine selective optical activation of Cdc42 with dynamic measurement of Rac activation.
RAW cells were transfected with ITSN-mCh-SspB, iLID-CaaX, wild-type Rac1, and the Rac1 biosensor. Localized optical activation at one side of a cell resulted in an increase in the level of Rac1 sensor intensity at the plasma membrane within the same region within 5 s (six of eight cells; Figure 7A and Supplemental Figure S6). On continued photoactivation, the increased Rac1 sensor intensity remained spatially confined to the photoactivated region. On reversing the side of optical activation, the Rac1 sensor rapidly switched sides within 10 s. These results suggest that Cdc42 activation triggers Rac1 activity that is spatially confined to the same side of the cell, consistent with reports that activation of both Rac and Cdc42 is enriched at the leading edge of migrating cells (Itoh et al., 2002).
Whereas optical activation of Cdc42 generated an increase in Rac activity, the converse was not detected. In RAW cells transfected with Tiam-mCh-SspB, iLID-CaaX, wild-type Cdc42, and Venus-wGBD, localized optical activation resulted in membrane recruitment of the Tiam construct, but translocation of Venus-wGBD was not detected (six cells; Figure 7C and Supplemental Figure S4). Thus the Rac-selective GEF Tiam failed to generate detectable Cdc42 activation under conditions in which the Cdc42-selective GEF ITSN generated readily detectable Cdc42 activation. The ability of the Tiam construct to clearly activate Rac1 without activating Cdc42 suggests that Rac does not trigger Cdc42 activation in these cells.
Localized activation of Cdc42 or Rac alone does not generate increased phosphatidylinositol (3,4,5)-trisphosphate
Positive and negative feedback loops play important roles in controlling dynamic responses within signaling networks. Identifying such feedback loops requires the ability to perturb signaling on time scales that are faster than the kinetics of the feedback loop. Whereas classical gene-knockdown approaches lead to changes on the time scale of hours or days, rapid chemical or optogenetic perturbations can lead to changes within seconds. The optogenetic approach additionally provides the ability to spatially confine the perturbation to one side of cell, which is important for studying chemotaxis signaling, because cells are known to generate distinct responses depending on whether a stimulus is applied uniformly or asymmetrically across a cell.
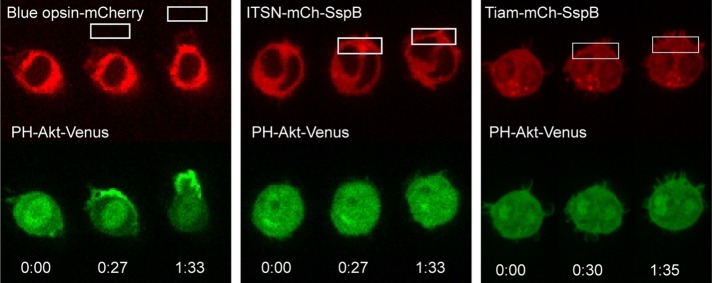
A potential feedback loop that has received interest in studies of chemotaxis involves Rac and phosphatidylinositol (3,4,5)-trisphosphate (PIP3). PIP3 is a signaling lipid that is enriched at the leading edge of many types of migratory cells. Originally, PIP3 was believed to act upstream of Cdc42/Rac, but experiments using expression of constitutively active mutants suggested that Rac can generate increased PIP3, indicating the presence of a positive feedback loop in neutrophils (Srinivasan et al., 2003). However, spatially uniform activation of Rac using chemically driven translocation of the Tiam GEF domain to the plasma membrane failed to generate increased PIP3 in neutrophils (Inoue and Meyer, 2008). It is unknown whether spatially localized activation of Rac or Cdc42 would generate a different result and whether the findings in neutrophils hold for macrophages. We therefore tested whether optical activation of Cdc42 or Rac at one side of a cell induced PIP3 changes in RAW cells.
We found that the PIP3 response to Cdc42/Rac activation differs markedly from the response to GPCR activation (Figure 8). Consistent with our previous findings (Karunarathne et al., 2013b), activation of the entire chemotaxis signaling network using blue opsin, a light- activated GPCR, generated a strong localized PIP3 response in 13 of 14 cells (Figure 8, left panel, and Supplemental Figure S7A). In contrast, direct optical activation of Cdc42 failed to generate any detectable increase in PIP3 in 11 cells tested (Figure 8, middle panel, and Supplemental Figure S7B). Similarly, optical activation of Rac failed to generate a PIP3 increase in any of 12 cells tested (Figure 8, right panel, and Supplemental Figure S7C). This difference shows that upstream signaling is required to generate increased PIP3 at the leading edge and that Cdc42/Rac and PIP3 alone do not comprise a positive feedback loop.
FIGURE 8:
Localized activation of Cdc42 or Rac alone does not generate increased PIP3. RAW cells transfected with the PIP3 sensor PH-Akt-Venus and blue opsin-mCherry, ITSN-mCh-SspB and iLID-CaaX, or Tiam-mCh-SspB and iLID-CaaX. Localized optical activation of the entire GPCR stimulated signaling network using blue opsin resulted in a strong PIP3 response at the front of the cell (13 of 14 cells). In contrast, localized activation of Cdc42 or Rac using the ITSN and Tiam constructs failed to generate a detectable PIP3 response (n = 11 and 12, respectively). See also Supplemental Figure S7.
Cdc42 activity at the leading edge causes myosin II–driven retraction of the cell rear
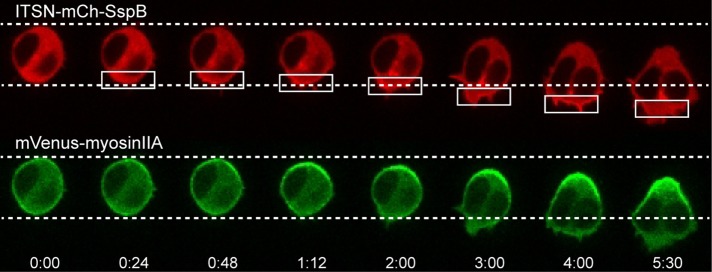
Because localized Cdc42 activation generated not only membrane protrusion at the leading edge, but also retraction of the cell rear, we anticipated that it was capable of directing the formation of actomyosin bundles at the far end of the cell. We tested this possibility by combining optical control over Cdc42 with live-cell imaging of tagged myosin. RAW cells were transfected with ITSN-mCh-SspB-R73Q, iLID-CaaX, and Venus-myosin IIA. Optical activation of Cdc42 at one side of the cell resulted in myosin accumulation at the opposite side (Figure 9, Supplemental Figure S3, and Supplemental Movie S5). In many cells, we observed that after the initiation of the localized optical input, myosin first accumulated in a crescent at the cell rear, and its spatial distribution became more compact over the time course of a few minutes. This process often resulted in the formation of a focal spot enriched with myosin and localized directly opposite from the side of optical activation (Supplemental Figure S9). This process correlated with retraction of the cell rear, consistent with the formation of force-generating actomyosin bundles. Reversing the side of optical activation caused the myosin to relocalize to the opposite side of the cell and correlated with retraction of the new cell rear. Myosin IIB was similarly found to accumulate at the opposite side of the cell relative to optical activation (Supplemental Figure S10).
FIGURE 9:
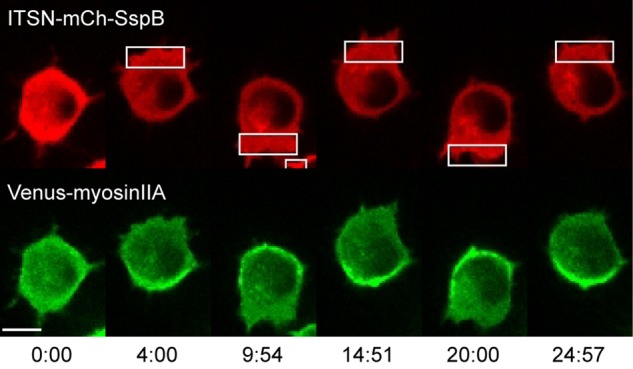
Cdc42 activity at the leading edge induces myosin accumulation at the cell rear. RAW cell transfected with ITSN-mCh-SspB, iLID-CaaX, and Venus-myosinIIA. Optically triggered activation of Cdc42 at one side of the cell generates myosin accumulation at the opposite side. Changing the side of optical activation causes myosin to redistribute to the new cell rear. Myosin accumulation opposite the side of Cdc42 activation was observed in 56 of 63 cells. Time is given in minutes:seconds. Scale bar, 10 μm. See also Supplemental Movie S5 and Supplemental Figure S8.
A benefit of the subcellular optogenetic approach is that it can help to determine the temporal order of events involved in generating cell polarity. On optical activation of Cdc42 at one side of a cell, we observed that the accumulation of myosin at the cell rear occurs even before the generation of protrusions at the front (Figure 10). This suggests that the ability of Cdc42 activity at the front to trigger actomyosin bundle formation at the rear does not depend on the formation of membrane protrusions at the leading edge.
FIGURE 10:
Myosin kinetics. Cdc42-triggered changes at the cell rear occur before the generation of visible protrusions at the leading edge. A RAW cell was transfected with the constructs specified in the image sequence, together with iLID-CaaX. On localized optical activation, myosin accumulation at the cell rear can be observed before any significant protrusions occur at the cell front. In addition, retraction of the cell rear occurs before the cell front moves forward. Times are minutes:seconds.
Although myosin accumulation at the cell rear was observed in the majority of cells (56 of 63 cells; Supplemental Figure S8), a subset of cells exhibited increased myosin IIA levels at the leading edge rather than the cell rear (seven of 63 cells; Supplemental Figure S11). The cause for this variation among cells remains to be determined but may be related to reports that macrophages can exhibit both amoebic and mesenchymal modes of migration (Van Goethem et al., 2010). Whereas myosin II has been reported to localize to the back of migrating neutrophils (Wong et al., 2007), which exhibit amoebic migration, myosin IIA has been reported to localize to the leading edge in fibroblasts undergoing mesenchymal migration (Vicente-Manzanares et al., 2008).
Rho-activated kinase (Rho-associated, coiled coil–containing kinase) is required downstream of Cdc42 for retracting the cell rear, as well as for regulating protrusions at the leading edge
How does increased Cdc42 activity at one side of a cell trigger myosin accumulation at the opposite side? One possibility is that it controls the canonical RhoA–Rho-associated, coiled coil–containing kinase (ROCK)–myosin pathway (Artemenko et al., 2014). Cdc42 and RhoA are both members of the Rho family of monomeric G proteins, and there are many examples of cross-regulation within this family (Burridge and Wennerberg, 2004; Guilluy et al., 2011), and long-range activation of RhoA by Cdc42 has been suggested but never tested directly (Van Keymeulen et al., 2006). RhoA activates ROCK, which in turn regulates myosin through phosphorylation of myosin light chain (Vicente-Manzanares et al., 2009). We therefore sought to test whether a RhoA/ROCK pathway is required for the ability of Cdc42 activation at the front to generate myosin II accumulation at the back.
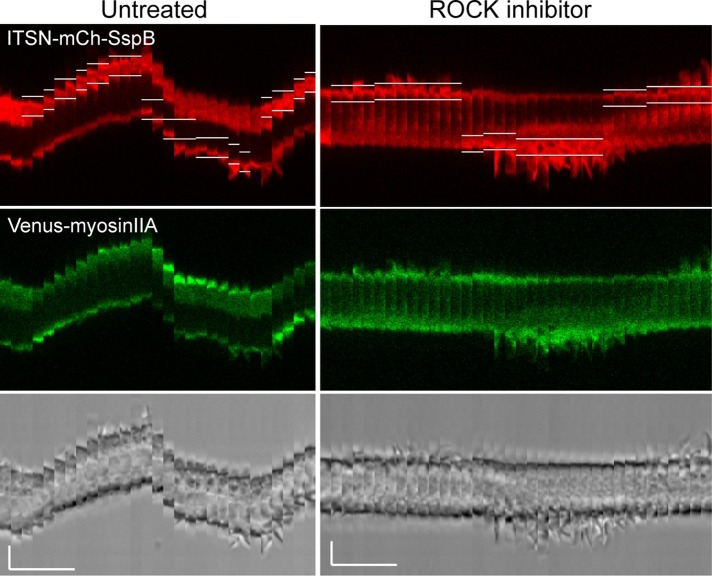
RAW cells were transfected with ITSN-mCh-SspB-R73Q, iLID-CaaX, and mVenus-myosin IIA. First, localized optical activation was performed to verify the cell’s ability to generate directional movement and myosin accumulation at the cell rear. Then 100 μM ROCK inhibitor Y27632 was added to the dish. After 10 min, localized optical activation was applied again to the same cell. After treatment with the ROCK inhibitor, optical activation of Cdc42 still generated localized cell protrusions at the leading edge, but myosin no longer accumulated at the back, and the cell failed to retract its rear (seven of seven cells; Figure 11 and Supplemental Movie S6). This suggests that Cdc42 activation at the leading edge is capable of controlling the canonical RhoA/ROCK/myosin signaling pathway at the cell rear.
FIGURE 11:
Cdc42-mediated effects at both the front and back of a migrating cell involve ROCK. Kymographs showing a single RAW cell transfected with ITSN-mCh-SspB, iLID-CaaX, and Venus-myosinIIA. Localized photoactivation was performed before and after addition of 100 μM Y-27632 to the bath for 10 min to inhibit endogenous ROCK. The kymographs show that ROCK inhibition blocks myosin accumulation at the back and retraction of the cell rear. ROCK-inhibited cells also display more-pronounced membrane protrusions at the leading edge, but the cell fails to move forward. Similar results were observed for all cells tested (n = 7). White lines in the red images show the boundaries of the photoactivated regions. The brightness level has been increased for the red and green images on the right relative to those on the left in order to correct for some fluorescence decrease observed after addition of Y-27632. Scale bars, 5 μm (vertical), 5 min (horizontal). See also Supplemental Movie S6.
ROCK inhibition also resulted in much longer membrane protrusions at the leading edge (Figure 11 and Supplemental Movie S6). This shows that Cdc42 activates ROCK-independent signaling that drives extension of the leading edge, as well as ROCK-dependent signaling that controls the length and dynamics of the extensions. Whereas cells treated with the ROCK inhibitor generated longer lamellipodia, they failed to generate any forward movement of the cell body itself. This suggests that ROCK is required for processes that allow the cell to make use of its lamellipodia to pull the front of the cell body forward. These results suggest that Cdc42 activity at the leading edge acts through ROCK to differentially regulate myosin dynamics at both the front and back of the cell to coordinate forward migration.
DISCUSSION
Cdc42 has been implicated in polarized cell behaviors ranging from yeast budding to the directed migration of mammalian immune cells (Etienne-Manneville, 2004). Genetic and biochemical studies have identified important molecular interactions between Cdc42 and “polarity proteins” (Cau and Hall, 2005; Nishimura et al., 2005; Welchman et al., 2007), and an understanding of how Cdc42 activity becomes polarized is beginning to emerge in some simple model systems (Goryachev and Pokhilko, 2008; Kozubowski et al., 2008). However, little is known about how localized Cdc42 activity exerts spatial and temporal control over a network of signaling molecules to dynamically orient polarized cellular responses. Here we used optogenetic control over endogenous Cdc42 to examine its role in generating and reversing polarity and directional migration in macrophages.
Molecular mechanisms by which chemoattractant receptors activate Cdc42 have been reported (Li et al., 2003), but it has not been possible to isolate the Cdc42-regulated portion of the chemotaxis signaling network to study it independently of receptor activation. The optogenetic approach overcomes this limitation and provides subcellular spatial control over Cdc42 activity that can be reversed in <1 min. Our results show that localized activation of Cdc42 generates directed cell migration through a combination of signaling responses generated at the front and back of the cell.
At the front of the cell, Cdc42 generates increased Rac activation, increased actin polymerization, and cellular protrusions. These “frontness” responses are confined spatially to the region of increased Cdc42 activation, indicating that the cell regulates these processes to inhibit their diffusive spread to other regions of the cell. Of note, we find that such regulation does not require parallel signaling controlled by chemoattractant receptors. This is consistent with migration induced by direct activation of Cdc42 broadly reflecting the migration characteristics induced by local activation of a GPCR.
Cdc42 also exerts long-range effects on the back of the cell, directing the formation of actomyosin bundles that generate retraction of cell rear. This process requires the RhoA-activated kinase ROCK, suggesting that Cdc42 signaling at the front of the cell causes increased RhoA signaling at the back. There have been numerous reports of mutual antagonism between Cdc42/Rac and RhoA (Burridge and Wennerberg, 2004; Guilluy et al., 2011), but our results provide the first direct evidence that Cdc42 signaling at one side of a cell can cause an increase in RhoA signaling at the far side. This raises the question of how an increase in Cdc42 activation at one side of a cell generates increased RhoA activation at the opposite side. One possible mechanism involves signaling through integrins: extensive cross-talk between integrins and Rho signaling is well established (Huveneers and Danen, 2009; Shen et al., 2012), and Cdc42 activation of WASP at the leading edge of migrating neutrophils reportedly triggers WASP translocation to the cell rear, where it regulates integrins (Szczur et al., 2009; Kumar et al., 2012). An alternative mechanism could involve Cdc42 activation of GEFs and GTPase- activating proteins (GAPs) that act on RhoA. If Cdc42 activates a RhoGEF that diffuses throughout the cell and a RhoGAP that remains at the leading edge, then the net effect would be increased RhoA activity at the cell rear. In the future, efforts to differentiate between these and other potential mechanisms for front–back communication will benefit from the expanding collection of optogenetic constructs available for activating or inhibiting select signaling proteins.
Localized Cdc42 activation in cells treated with the ROCK inhibitor also generated much longer membrane protrusions at the leading edge compared with untreated cells. We suspect that Cdc42 acts through ROCK to regulate myosin at the front of the cell, allowing it to act as a brake on the membrane extensions at the leading edge. ROCK is also known to act through Lim kinase to regulate coffilin (Arber et al., 1998; Maekawa et al., 1999). Thus it is also possible that Cdc42 acts through RhoA/ROCK to regulate actin depolymerization at the leading edge. These results show that Cdc42 activity at the front of the cell acts on RhoA/ROCK at both the front and back of the cell to regulate completely different functions in these spatially distinct regions.
There has been a growing appreciation that the simple view in which each Rho-family protein has one specific function is inadequate. Instead, they can exhibit distinct functions defined by spatiotemporal signaling modules that include various GEFs, GAPs, and effectors (Pertz, 2010). Our results demonstrate how optical control of signaling can help to identify distinct functions of these different modules. Combining optogenetics with pharmacological and genetic perturbations and live-cell imaging will help to better define the molecular compositions of these different signaling modules. More generally, our results illustrate how subcellular optogenetics can provide new insights into the dynamic interactions within signaling networks that control polarity and directional cellular responses like migration.
MATERIALS AND METHODS
DNA constructs
The Rac biosensor was kindly provided by Louis Hodgson (Albert Einstein College of Medicine of Yeshiva University, Bronx, NY) and is a modified version of a published Rac biosensor (Moshfegh et al., 2014). The following constructs were obtained through Addgene (Cambridge, MA): ISTN(64-473)-tgRFPt-SspB-R73Q (plasmid 60418), Tiam1(1159-1509)-tgRFPt-SspB-R73Q (plasmid 60420), Venus-iLID-CaaX (plasmid 60411), GFP-wGBD (plasmid 26734), mTopaz-Lifeact (plasmid 54661), mVenus-myosinIIA (plasmid 56389), and GFP-myosinIIB (35691). PH-Akt-Venus was previously described (O’Neill and Gautam, 2014). For both the ITSN-mCherry-SspB and Tiam-mCherry-SspB constructs, tgRFPt was replaced with mCherry using the EcoR1 and BspE1 sites in the corresponding GEF-tgRFPt-SspB construct. Venus-wGBD was made by ligating a HindIII-KpnI PCR product of Venus with a KpnI/EcoR1 wGBD into pcDNA3.1. A PCR product of iLID-CaaX was cloned into the KpnI/EcoR1 sites of pcDNA3.1.
Cell culture
RAW 264.7 cells were obtained from the Washington University Tissue Culture Support Center and cultured in DMEM (D6429; Sigma-Aldrich, St. Louis, MO) with 10% dialyzed fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA) and 1% penicillin–streptomycin at 37°C and 5% CO2. RAW cells ranging from passage 3 to passage 12 were used for experiments.
Transfections
RAW cells were transfected by electroporation in Amaxa Nucleofector Solution V using the D-032 or T-020 setting on an Amaxa Nucleofector 2b device (Lonza, Basel, Switzerland). Each electroporation was performed on 2–4 million cells in 100 μl of Nucleofector solution, followed immediately by addition of 500 μl of warm culture medium. The cells were then plated in 5–10 glass bottom dishes, placed in an incubator at 37°C and 5% CO2, and imaged 3–10 h after electroporation.
Imaging
Imaging and optical activation were performed using a spinning-disk confocal imaging system consisting of a Leica DMI6000B microscope with adaptive focus control, a Yokogawa CSU-X1 spinning-disk unit, an Andor iXon electron-multiplying charge-coupled device camera, a laser combiner with 445-, 488-, 515-, and 594-nm solid-state lasers, and an Andor FRAPPA unit for photoactivation of manually selected regions of the sample in real time, all controlled using Andor iQ2 software (Andor Technologies, Belfast, United Kingdom). For optical activation of iLID, the 445-nm laser was used at 5 μW and scanned across the selected region at a rate of 0.9 ms/μm2. This was performed once every 3–5 s. Laser wavelengths of 515 and 594 nm were used for excitation of Venus and mCherry, respectively. Emission filters were Venus 528/20 and mCherry 628/20 (Semrock). All images were acquired using a 63× oil immersion objective. A single confocal plane was imaged at a rate of 1 frame/3 s or 1 frame/5 s. All imaging was performed inside a temperature-controlled chamber held at 37°C. The chamber was also maintained at 5% CO2 during longer-duration experiments, that is, for samples kept on the microscope before and after treatment with Y-27632.
Supplementary Material
Acknowledgments
We thank Louis Hodgson for kindly providing cDNA for the Rac biosensor. This work was funded by the National Institutes of Health through National Institute of General Medical Sciences Grants GM069027 and GM107370.
Abbreviations used:
- Cdc42
cell division control protein 42 homologue
- GEF
guanine nucleotide exchange factor
- GPCR
G protein–coupled receptor
- ROCK
Rho-associated coiled coil–containing kinase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-12-0832) on March 3, 2016.
REFERENCES
- Allen WE, Zicha D, Ridley AJ, Jones GE. A role for Cdc42 in macrophage chemotaxis. J Cell Biol. 1998;141:1147–1157. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Artemenko Y, Lampert TJ, Devreotes PN. Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell Mol Life Sci. 2014;71:3711–3747. doi: 10.1007/s00018-014-1638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J Biol Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- Benink HA, Bement WM. Concentric zones of active RhoA and Cdc42 around single cell wounds. J Cell Biol. 2005;168:429–439. doi: 10.1083/jcb.200411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- Cai H, Devreotes PN. Moving in the right direction: how eukaryotic cells migrate along chemical gradients. Semin Cell Dev Biol. 2011;22:834–841. doi: 10.1016/j.semcdb.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau J, Hall A. Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J Cell Sci. 2005;118:2579–2587. doi: 10.1242/jcs.02385. [DOI] [PubMed] [Google Scholar]
- Chou J, Burke NA, Iwabu A, Watkins SC, Wells A. Directional motility induced by epidermal growth factor requires Cdc42. Exp Cell Res. 2003;287:47–56. doi: 10.1016/s0014-4827(03)00119-8. [DOI] [PubMed] [Google Scholar]
- Cox D, Chang P, Zhang Q, Reddy PG, Bokoch GM, Greenberg S. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J Exp Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P, Horwitz AR. Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol. 2015;7:a005959. doi: 10.1101/cshperspect.a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42—the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]
- Guilluy C, Garcia-Mata R, Burridge K. Rho protein crosstalk: another social network. Trends Cell Biol. 2011;21:718–726. doi: 10.1016/j.tcb.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci USA. 2015;112:112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind LE, Mackay JL, Cox D, Hammer DA. Two-dimensional motility of a macrophage cell line on microcontact-printed fibronectin. Cytoskeleton. 2014;71:542–554. doi: 10.1002/cm.21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S, Danen EH. Adhesion signaling—crosstalk between integrins, Src and Rho. J Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- Inoue T, Meyer T. Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PLoS One. 2008;3:e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal M, Dvorsky R, Ahmadian MR. Deciphering the molecular and functional basis of Dbl family proteins: a novel systematic approach toward classification of selective activation of the Rho family proteins. J Biol Chem. 2013;288:4486–4500. doi: 10.1074/jbc.M112.429746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne WK, Giri L, Kalyanaraman V, Gautam N. Optically triggering spatiotemporally confined GPCR activity in a cell and programming neurite initiation and extension. Proc Natl Acad Sci USA. 2013a;110:E1565–E1574. doi: 10.1073/pnas.1220697110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne WK, Giri L, Patel AK, Venkatesh KV, Gautam N. Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc Natl Acad Sci USA. 2013b;110:E1575–E1583. doi: 10.1073/pnas.1220755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne WK, O’Neill PR, Gautam N. Subcellular optogenetics - controlling signaling and single-cell behavior. J Cell Sci. 2015;128:15–25. doi: 10.1242/jcs.154435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Li Z, Sacks DB. E-cadherin-mediated cell-cell attachment activates Cdc42. J Biol Chem. 2000;275:36999–37005. doi: 10.1074/jbc.M003430200. [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol. 2008;18:1719–1726. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Xu J, Perkins C, Guo F, Snapper S, Finkelman FD, Zheng Y, Filippi MD. Cdc42 regulates neutrophil migration via crosstalk between WASp, CD11b, and microtubules. Blood. 2012;120:3563–3574. doi: 10.1182/blood-2012-04-426981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Rosen MK. The nucleotide switch in Cdc42 modulates coupling between the GTPase-binding and allosteric equilibria of Wiskott-Aldrich syndrome protein. Proc Natl Acad Sci USA. 2005;102:5685–5690. doi: 10.1073/pnas.0406472102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hannigan M, Mo ZC, Liu B, Lu W, Wu Y, Smrcka AV, Wu GQ, Li L, Liu MY, et al. Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of cdc42. Cell. 2003;114:215–227. doi: 10.1016/s0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol. 2014;16:574–586. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, Hoshino M, Kaibuchi K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol. 2005;7:270–277. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- O’Neill PR, Gautam N. Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol Biol Cell. 2014;25:2305–2314. doi: 10.1091/mbc.E14-04-0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O. Spatio-temporal Rho GTPase signaling—where are we now. J Cell Sci. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runne C, Chen S. PLEKHG2 promotes heterotrimeric G protein betagamma-stimulated lymphocyte migration via Rac and Cdc42 activation and actin polymerization. Mol Cell Biol. 2013;33:4294–4307. doi: 10.1128/MCB.00879-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Delaney MK, Du X. Inside-out, outside-in, and inside-outside-in: G protein signaling in integrin-mediated cell adhesion, spreading, and retraction. Curr Opin Cell Biol. 2012;24:600–606. doi: 10.1016/j.ceb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Wang F, Glavas S, Ott A, Hofmann F, Aktories K, Kalman D, Bourne HR. Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J Cell Biol. 2003;160:375–385. doi: 10.1083/jcb.200208179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczur K, Zheng Y, Filippi MD. The small Rho GTPase Cdc42 regulates neutrophil polarity via CD11b integrin signaling. Blood. 2009;114:4527–4537. doi: 10.1182/blood-2008-12-195164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Nagae R, Kozawa M, Morishita R, Kimura S, Nagase T, Ohara O, Yoshida S, Asano T. Heterotrimeric G protein beta gamma subunits stimulate FLJ00018, a guanine nucleotide exchange factor for Rac1 and Cdc42. J Biol Chem. 2008;283:1946–1953. doi: 10.1074/jbc.M707037200. [DOI] [PubMed] [Google Scholar]
- Valon L, Etoc F, Remorino A, di Pietro F, Morin X, Dahan M, Coppey M. Predictive spatiotemporal manipulation of signaling perturbations using optogenetics. Biophys J. 2015;109:1785–1797. doi: 10.1016/j.bpj.2015.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem E, Poincloux R, Gauffre F, Maridonneau-Parini I, Le Cabec V. Matrix architecture dictates three-dimensional migration modes of human macrophages: differential involvement of proteases and podosome-like structures. J Immunol. 2010;184:1049–1061. doi: 10.4049/jimmunol.0902223. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Wong K, Knight ZA, Govaerts C, Hahn KM, Shokat KM, Bourne HR. To stabilize neutrophil polarity, PIP3 and Cdc42 augment RhoA activity at the back as well as signals at the front. J Cell Biol. 2006;174:437–445. doi: 10.1083/jcb.200604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Koach MA, Whitmore L, Lamers ML, Horwitz AF. Segregation and activation of myosin IIB creates a rear in migrating cells. J Cell Biol. 2008;183:543–554. doi: 10.1083/jcb.200806030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev. Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman DP, Mathies LD, Ahringer J. Similar requirements for CDC-42 and the PAR-3/PAR-6/PKC-3 complex in diverse cell types. Dev Biol. 2007;305:347–357. doi: 10.1016/j.ydbio.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, Van Keymeulen A, Bourne HR. PDZRhoGEF and myosin II localize RhoA activity to the back of polarizing neutrophil-like cells. J Cell Biol. 2007;179:1141–1148. doi: 10.1083/jcb.200706167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.