Temporal control of dendritogenesis is poorly understood. Mutual feedback between NFIA temporal occupancy and ETV1 drives the timing of gene expression associated with dendrite formation in maturing neurons. A sequential timing model is proposed in which ETV1 autoregulation precedes activation of downstream NFIA/ETV1 coregulated genes.
Abstract
Nuclear Factor One (NFI) transcription factors regulate temporal gene expression required for dendritogenesis and synaptogenesis via delayed occupancy of target promoters in developing cerebellar granule neurons (CGNs). Mechanisms that promote NFI temporal occupancy have not been previously defined. We show here that the transcription factor ETV1 directly binds to and is required for expression and NFI occupancy of a cohort of NFI-dependent genes in CGNs maturing in vivo. Expression of ETV1 is low in early postnatal cerebellum and increases with maturation, mirroring NFI temporal occupancy of coregulated target genes. Precocious expression of ETV1 in mouse CGNs accelerated onset of expression and NFI temporal occupancy of late target genes and enhanced Map2(+) neurite outgrowth. ETV1 also activated expression and NFI occupancy of the Etv1 gene itself, and this autoregulatory loop preceded ETV1 binding and activation of other coregulated target genes in vivo. These findings suggest a potential model in which ETV1 activates NFI temporal binding to a subset of late-expressed genes in a stepwise manner by initial positive feedback regulation of the Etv1 gene itself followed by activation of downstream coregulated targets as ETV1 expression increases. Sequential transcription factor autoregulation and subsequent binding to downstream promoters may provide an intrinsic developmental timer for dendrite/synapse gene expression.
INTRODUCTION
Timing mechanisms are now recognized as fundamentally important requirements for neuronal development (Martynoga et al., 2012; Kumamoto et al., 2013). In particular, neurons elaborate dendrites and form numerous synaptic connections, and the timing of these events is critical for establishing appropriate neural circuitry (Deguchi et al., 2011; Tripodi and Arber, 2012). This requires precise temporal regulation of numerous genes (Balamotis et al., 2012), and disruption of these timing mechanisms can dramatically alter neural circuitry (Hippenmeyer et al., 2005). Further, dysregulated gene expression during critical temporal windows of synaptic maturation has been implicated in neurodevelopmental disorders (NDs; Meredith et al., 2012). The mechanisms that temporally coordinate dendrite and synapse formation and related gene expression remain poorly understood.
Cerebellar granule neurons (CGNs) undergo a well-defined postnatal program of development in which subsets of genes are expressed in distinct temporal patterns linked to specific stages of maturation—for example, progenitor proliferation within the external germinal layer (EGL), axon formation in the premigratory zone, radial migration, and finally dendrite/synapse formation by postmigratory CGNs within the internal granule cell layer (IGL) (Goldowitz and Hamre, 1998; Furuichi et al., 2011). Further, much of this developmentally regulated gene expression is reproduced in primary CGN cultures (Ding et al., 2013), providing an excellent system for elucidating mechanisms underlying temporal programming of dendrite and synapse formation.
Transcription factor and chromatin interactions have a central role in dendrite and synapse formation during neuronal development (Santiago and Bashaw, 2014; Vogel-Ciernia and Wood, 2014), and evidence for their involvement in the timing of these events has recently come to light. Nuclear Factor One (NFI) proteins, and in particular NFIA, regulate CGN dendritogenesis both in vivo and in purified cultures (Wang et al., 2007, 2010). Recent studies revealed that this is mediated by an NFI-dependent developmental switch that controls the timing of dendritogenesis in postmigratory CGNs maturing within the internal granule cell layer during the second and third postnatal weeks (Ding et al., 2013). Although NFI proteins are constitutively expressed in the nucleus throughout CGN differentiation and in vitro NFI DNA binding activity is similar in early- and later-maturing cerebellum (Wang et al., 2011), late gene activation is specifically linked to temporally up-regulated occupancy of NFI sites within late gene promoters. Onset of NFI temporal occupancy is orchestrated in part by voltage-sensitive and maturation-dependent dismissal of NFATc4 repressor bound to late gene promoters, permitting subsequent NFI occupancy of late-expressed genes (Ding et al., 2013).
The aforementioned studies left open the question of whether additional factors control NFI temporal binding and switch programming in CGNs. For example, up-regulation of specific subsets of late-expressed genes likely occurs with distinct time frames, requiring the operation of additional mechanisms to fine-tune late temporal gene expression. This may include trans-activators that promote these events subsequent to NFATc4 dismissal from NFI-late genes. ETV1 is a member of the ETS family of trans-factors that is temporally up-regulated in postmigratory CGNs maturing within the IGL (Sato et al., 2005; Schuller et al., 2006). Knockdown recently implicated ETV1 in the activity-dependent up-regulation of gene expression during CGN maturation (Abe et al., 2011). Here we used gene-knockout mice, chromatin immunoprecipitation (ChIP), and lentiviral transduction of neuronal cultures to investigate the role of ETV1 in promoting the NFI switch program and NFI temporal binding in maturing CGNs.
RESULTS
Etv1 regulates the NFI switch in CGNs maturing in vivo
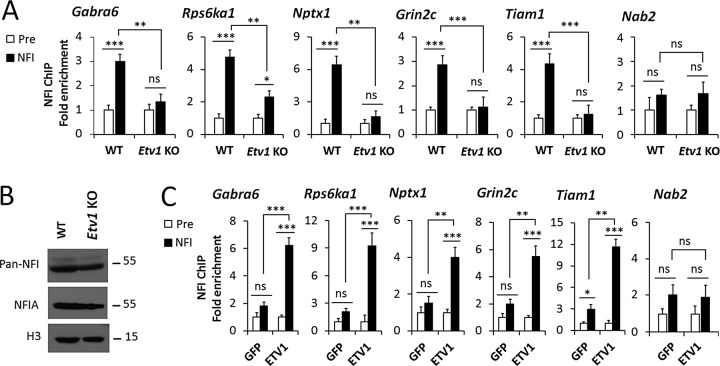
Etv1-knockout mice survive into the second postnatal week (Cave et al., 2010), during which the NFI switch program and dendrite formation are underway in CGNs within the IGL (Altman and Bayer, 1997). Because Etv1 gene expression is detected only in CGNs and not other cerebellar cell types in the mouse (Sato et al., 2005; Schuller et al., 2006), cerebella from these mice should reflect largely CGN-intrinsic deficits in ETV1 function. Transcripts for the NFI-switch late genes Gabra6, Rps6ka1, Nptx1, Wnt7a, and Ets2 were markedly down-regulated in Etv1-null mouse cerebellum at postnatal day 10 (P10) and P11 (Figure 1A). Note that expression of each of these genes is either unique or highly predominant in CGNs within the IGL of the developing mouse cerebellum (Sato et al., 2005; Cerebellar Development Transcriptome Database [www.cdtdb.neuroinf.jp/CDT/Top.jsp]; MGI:3511933 [www.informatics.jax.org/assay/MGI:3511933]). In contrast, the non-NFI switch gene Rbfox3/NeuN, which is a temporally up-regulated specific marker for postmitotic mouse CGNs (Weyer and Schilling, 2003), was unaffected by Etv1 deficiency (Figure 1A). Thus these results did not reflect generalized changes in cerebellar or CGN gene expression. Consistent with this, no significant differences were observed in the thickness and cell densities of the EGL, molecular layer, and IGL of P10 Etv1-deficient and wild-type mice (Supplemental Figure S1). Further, the early-expressed NFI-switch genes Dcx, Neurod6, and Tgfb2 were up-regulated in Etv1-knockout cerebellum (Figure 1A), indicating that ETV1 also regulates a switch program that overlaps with the NFI-switch regulon (Ding et al., 2013).
FIGURE 1:
ETV1 and NFI coregulate a subset of late temporal genes. (A) RT-qPCR results for selected NFI switch-program and control genes in P10 and P11 Etv1-knockout cerebellum. (B) RT-qPCR assays of Grin2c and Tiam1 transcripts in wild-type (WT) and Nfia-knockout (Nfia KO) cerebellum at P15 and of 6-DIV CGN cultures transduced on 0 DIV with lentiviruses expressing NFI dominant repressor (NFI/EnR) or its negative control (EnR). (C) ChIP-qPCR analysis of NFI binding to the Tiam1 and Grin2c promoters in the developing cerebellum at P7 and P21, expressed as the enrichment relative to control values (set as 1). Foxd1 genomic sequences were assayed as a negative control. Pre, preimmune serum. ***p < 0.001, **p < 0.01, *p < 0.05; ns, no significant difference.
The effects of in vivo Etv1 deficiency on the NFI-late genes Gabra6, Nptx1, Ets2, and Wnt7a mirror those found in CGN cultures using Etv1 siRNAs and are consistent with ETV1 activation of several NFI-late gene promoters in transfection studies (Abe et al., 2011). Of interest, the NFI-late gene Nab2 was not significantly affected in Etv1-knockout mouse cerebellum (Figure 1A), revealing selectivity in ETV1 regulation of NFI-late genes in vivo.
Grin2c and Tiam1 are Etv1-dependent NFI-switch late genes
Promoter cotransfection experiments and small interfering RNA (siRNA) studies previously identified Grin2c and Tiam1 as ETV1-regulated late target genes in maturing CGNs (Abe et al., 2011). We extend these results here by showing that both genes are strongly down-regulated in Etv1-null mouse cerebella (Figure 1A). We also find that Grin2c and Tiam1 are regulated as part of the NFI switch program, using an NFI dominant repressor lentivirus, which represses genes activated by all NFI family members, in CGN cultures and by analysis of P15 Nfia-knockout mouse cerebellum (Figure 1B). Further, both genes undergo NFI temporal occupancy (Figure 1C). Thus Grin2c and Tiam1 are part of the ETV1/NFI temporal coregulon.
Effects of precocious ETV1 expression in immature CGNs
ETV1 protein and Etv1 mRNA are low in immature CGNs and increase with maturation (Sato et al., 2005; Abe et al., 2012). We therefore examined the effect of elevated early expression of ETV1 on endogenous late genes in immature CGNs. Premature expression of ETV1 increased late gene expression at 2 and 3 d in vitro (DIV), when endogenous Etv1 mRNA is normally low (Abe et al., 2012; Figure 2A). This included both Grin2c and Tiam1 but not Rbfox3 (Figure 2A). Collectively these gain- and loss-of-function results indicate both a requirement and an activating role for ETV1 in NFI-late gene regulation in immature CGNs.
FIGURE 2:
Overexpression of ETV1 up-regulates NFI-late genes and neurite formation in maturing CGNs. (A) Transcript levels for the indicated genes for wild-type 6-DIV CGN cultures transduced on 0 DIV with lentiviruses expressing GFP or ETV1 protein and assayed on 2 and 3 DIV. (B) Left, photomicrographs of Map2 immunofluorescence in wild-type 2-DIV CGNs transduced on 0 DIV with GFP- or ETV1-expressing lentiviruses. Bar, 100 μm. Right, quantification of the effects of ETV1 overexpression on Map2(+) neurite length in wild-type CGN cultures expressed relative to control (GFP). Data are from three biological replicates (averages of 32 [GFP] and 26 [ETV1] transduced cells were analyzed in each experiment) and expressed relative to GFP(+) cells. ***p < 0.001, **p < 0.01; ns, no significant difference.
The NFI switch program enhances dendritogenesis in developing CGNs (Wang et al., 2007; Ding et al., 2013). We therefore examined whether ETV1 itself regulates the maturation of neurites expressing Map2, a dendritic marker. Transduction of 0-DIV CGN cultures with ETV1 produced a modest but significant increase in Map2(+) neurite length at 2 DIV (Figure 2B). Thus elevation of ETV1 expression in immature CGNs accelerates both neurite formation and NFI-late gene expression linked to this process. These in cellulo gain-of-function results are consistent with reduced maturation of CGN primary dendrites observed in vivo using Etv1 siRNAs (Abe et al., 2011).
ETV1 promotes NFI temporal occupancy
Because NFI temporal binding is a hallmark of the NFI late gene program, we next asked whether ETV1 acts by enhancing NFI binding to target promoters. ChIP assays revealed that NFI occupancy was dramatically reduced in P10 Etv1-knockout cerebellum for Gabra6, Rps6ka1, Nptx1, Grin2c, and Tiam1, whereas no specific NFI binding was observed for the Nab2 gene at this early age in either wild-type or Etv1-null mice (Figure 3A). Reduced NFI occupancy of late genes was not related to decreased amounts of total NFI or NFIA proteins in Etv1-knockout cerebella (Figure 3B). Conversely, early expression of ETV1 in immature CGNs stimulated NFI binding to these late genes at 2 DIV (Figure 3C).
FIGURE 3:
ETV1 facilitates NFI occupancy of NFI-late gene promoters. (A) ChIP assays of NFI binding to several target genes in P10 Etv1(–/–) (Etv1 KO) cerebellum. The Nab2 gene showed no specific occupancy at P10 or at P11 (unpublished data). Pre, preimmune serum. (B) Western blot analysis of NFIA and total NFI proteins (pan-NFI) in P10 cerebella of Etv1-knockout mice (Etv1 KO) relative to wild type (WT). Histone H3 (H3) served as a loading control. Identical results were obtained in two biological replicate experiments. (C) ChIP analysis of NFI binding to late genes in 2-DIV wild-type CGN cultures transduced on 0 DIV with lentiviral vectors expressing ETV1 protein relative to GFP. ***p < 0.001, **p < 0.01, *p < 0.05; ns, no significant difference.
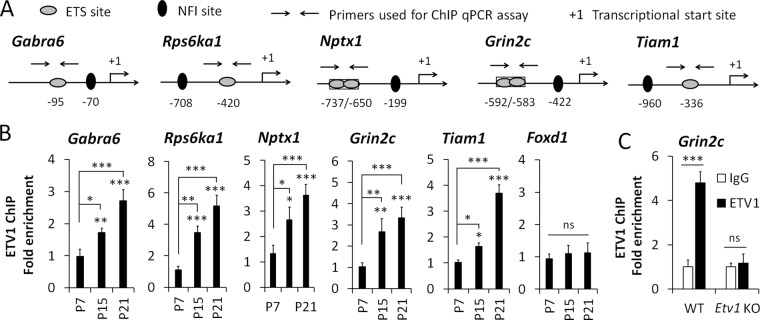
ETV1 was shown to activate promoter constructs for numerous late genes (Grin2c, Gabra6, Tiam1, Nptx1) in a heterologous cell line, and it binds to the Gabra6 and Grin2c genes in P21 mouse cerebellum (Abe et al., 2011). We confirmed that ETV1 occupies sites within these two gene promoters in mouse cerebellum, as well as those for the late genes Rps6ka1, Nptx1, and Tiam1 (Figure 4, A and B). For each gene, ETV1 binding was temporally up-regulated in parallel with its increased expression (P7–P21), whereas no specific binding was observed for nonexpressed Foxd1 genomic sequences. Further, specific ETV1 occupancy was substantially reduced in cerebella of Etv1-knockout mice (Figure 4C). Together with promoter transfection studies (Abe et al., 2011), these findings support a direct activating role for ETV1 in regulating a substantial segment of the NFI late gene program and, critically, in promoting NFI temporal occupancy of numerous late genes as CGNs mature.
FIGURE 4:
ETV1 binding to coregulated NFI-late genes increases as CGNs mature. (A) Schematic of ETV1- and NFI-binding sites in the proximal promoter regions of several NFI-switch late genes. (B) ChIP analysis of ETV1 binding to the indicated genes during postnatal development of the mouse cerebellum. (C) ChIP assays showing depletion of ETV1 binding to the Grin2c gene in P11 Etv1-null mouse cerebella. ***p < 0.001, **p < 0.01, *p < 0.05; ns, no significant difference.
Etv1 is an NFI-switch late gene
We previously identified Etv1 as a potential NFI-late gene in microarray studies (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42018). We first confirmed that Etv1 gene expression is up-regulated as CGNs mature in vivo (P7 vs. P21) and in culture (0 DIV, consisting of CGN progenitors and immature premigratory CGNs; 6 DIV, more-mature CGNs; Figure 5A). Western analyses further confirmed that nuclear levels of ETV1 protein (∼50–55 kDa) increased with cerebellar maturation between P7 and P21 (Figure 5B). Two isoforms of ETV1 were detected, one with a faster migration predominating at P7, and the other a slower and more abundant form present at P15 and P21. This more abundant species may reflect in vivo–phosphorylated ETV1, which exhibits increased transcriptional activity and has been detected in CGN cultures after brain-derived neurotrophic factor (BDNF) stimulation (Abe et al., 2012). This is not further addressed here. In contrast, nuclear levels of NFIA were relatively unchanged (or slightly declined), and nuclear NFATc4 strongly decreased between P7 and P21 (Figure 5B). The reduction in nuclear NFATc4 is consistent with decreasing NFATc4 occupancy of late genes and declining calcineurin phosphatase activity with maturation of the mouse cerebellum (Ding et al., 2013).
FIGURE 5:

Etv1 is an NFI-regulated late gene. (A) Etv1 transcripts increase as CGNs mature in developing mouse cerebellum (P7/P21) and culture (0/6 DIV). (B) Comparison of developmental changes in nuclear levels of ETV1, NFIA, and NFATc4 proteins in the developing cerebellum (P7, P15, and P21). A transition from a faster- to a slower-migrating form of ETV1 is apparent with maturation (shown by bars). TBP served as a soluble nuclear protein loading control. Data are reflective of two biological replicate experiments. (C) Etv1 gene expression in wild-type 6-DIV CGN cultures transduced on 0 DIV with lentiviruses expressing NFI dominant repressor (NFI/EnR) relative to EnR control (EnR), as well as in the cerebellum of P15 Nfia-knockout (Nfia KO) mice relative to wild type (WT). (D) Top, schematic showing an NFI-binding site within the proximal Etv1 5′-flanking region relative to the transcription start site (+1) and the position of primers (arrows) used for ChIP qPCR assays. Bottom, ChIP analysis of NFIA binding to the Etv1 promoter in the developing mouse cerebellum at P7 and P21. ***p < 0.001; ns, no significant difference.
Etv1 gene expression was strongly reduced in 6-DIV CGNs transduced on 0 DIV with an NFI-dominant repressor lentivirus and also in the cerebella of P15 Nfia-knockout mice, when expression of late genes is normally elevated (Figure 5C). We also identified an NFI consensus site within the proximal Etv1 promoter. Because the NFI ChIP antibody used here recognizes both NFIA and NFIB (Ding et al., 2013), we performed ChIP analysis of this site using an NFIA-specific antibody. As observed for other NFI-late genes (Ding et al., 2013), NFIA exhibited delayed occupancy of the Etv1 promoter region in vivo (Figure 5D).
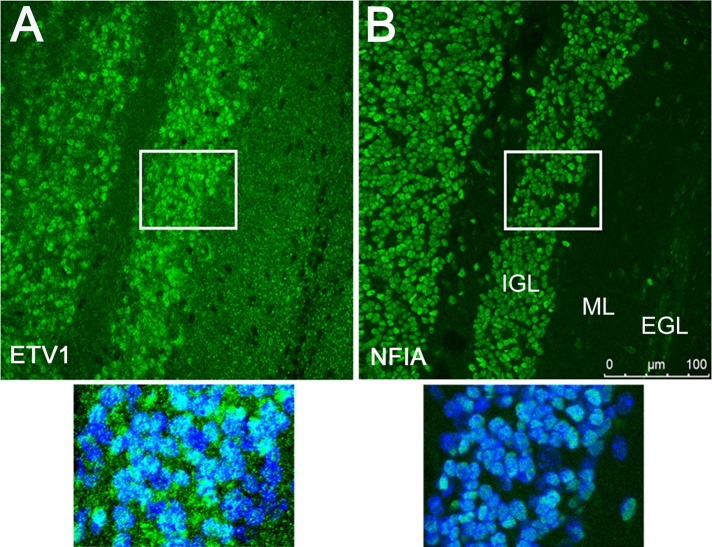
Consistent with this regulatory interaction, both ETV1 and NFIA proteins are widely expressed in CGN nuclei within the IGL of the P15 mouse cerebellum (Figure 6). This agrees with previous in situ hybridization results showing broad but selective detection of Etv1 mRNA within the mouse IGL (Sato et al., 2005; Schuller et al., 2006), as well as widespread immunostaining of NFIA within the IGL of P6 mouse cerebellum (Wang et al., 2007). Because only rabbit antibodies were found suitable for immunohistochemistry for either protein, it was not possible to show NFIA and ETV1 coexpression directly in the same cells. However, immunostaining for the two proteins was detected in nearly all CGN nuclei throughout the IGL (Figure 6), consistent with the presence of NFIA and ETV1 together in nuclei of most postmigratory CGNs in the IGL.
FIGURE 6:
ETV1 and NFIA are both widely expressed within the IGL of the developing mouse cerebellum. Immunostaining for ETV1 (A) and NFIA (B) proteins was performed on adjacent coronal sections of P15 mouse cerebella using rabbit antibodies in both cases (shown in green). Top, widespread staining of both proteins within the IGL, which consists mainly of maturing postmigratory CGNs. Bottom, higher-magnification images of IGL immunostaining (regions indicated by white rectangles at top) and costaining with the nuclear marker DAPI (blue). Background signal was higher for ETV1 immunostaining due to reduced signal-to-noise relative to the NFIA antibody. No–primary antibody controls yielded no detectable signal.
To study the functional effect of NFIA and ETV1 coexpression on late-gene transcription, we performed transient transfections using a 6-kb mouse Gabra6 promoter-luciferase reporter plasmid (Wang et al., 2004). Transfections were performed in the JEG3 cell line, since these cells have low endogenous NFI protein levels (Chaudhry et al., 1998). Both ETV1 and NFIA stimulated Gabra6 promoter activity when transfected individually (Figure 7A). To study the effects of coexpression, we expressed NFIA with and without increasing amounts of ETV1 vector to mirror developmental changes occurring in CGNs. Under these conditions, Gabra6 promoter activity increased relative to its activation with either factor alone, and this effect was more than additive at lower levels of ETV1 expression vector (Figure 7B). Thus ETV1/NFIA exhibited a transcriptional interaction under these conditions.
FIGURE 7:

ETV1 and NFIA coactivate late-gene transcription. (A) Expression plasmids for ETV1, NFIA, or GFP were transfected individually into JEG-3 cells along with a 6-kb mouse Gabra6 promoter construct (left) or promoterless pGL3 plasmid (right). (B) The effect of ETV1 and NFIA coexpression on Gabra6 promoter activity was determined using a constant amount of NFIA plasmid and increasing amounts of ETV1 expression plasmid. The dotted line highlights promoter activity above that for the GFP control plasmid for each condition. Luciferase activity is expressed as the fold increase of GFP control activity throughout. ***p < 0.001, **p < 0.01, *p < 0.05; ns, no significant difference.
We also attempted to determine whether NFIA and ETV1 proteins directly interacted in the developing mouse cerebellum. However, we were unable to detect NFIA and ETV1 interactions on chromatin using sequential ChIP and native coimmunoprecipitations in P21 mouse cerebella. These results may reflect in part the limited sensitivity in detecting ETV1 protein using available antibodies. Transient or indirect functional interactions between NFIA and ETV1 in vivo cannot be ruled out.
ETV1 regulates NFI occupancy of its own promoter in maturing CGNs
Similar to other NFI-regulated late genes, lentiviral expression of human ETV1 in early- developing CGNs (2–3 DIV) increased endogenous mouse Etv1 transcripts (Figure 8A). ChIP assays also revealed ETV1 binding to a region of the proximal Etv1 promoter containing multiple ETS binding sites during mouse cerebellar development that increased in parallel with temporal expression of ETV1 (Figure 8B). These findings were consistent with the notion that temporal increases in Etv1 expression in maturing CGNs are driven at least partly by direct ETV1 binding and autoregulation. This is further supported by ETV1 activation of the transfected Etv1 promoter (Abe et al., 2011).
FIGURE 8:
The Etv1 gene undergoes autoregulated NFI temporal occupancy in maturing CGNs. (A) Wild-type CGN cultures were transduced with lentiviruses expressing GFP or human ETV1 protein on 0 DIV and then assayed by RT-qPCR using primers to noncoding mouse Etv1 sequences not present in the lentiviral expression vector. (B) Top, locations of multiple ETS- binding sites and the NFI-binding site within the Etv1 proximal promoter. Arrows show the positions of primers used for ChIP qPCR. Bottom, ChIP assays of ETV1 occupancy of the Etv1 promoter in the maturing mouse cerebellum. (C) ChIP qPCR assay of NFI binding to its site within the Etv1 promoter in wild-type 3-DIV CGN cultures transduced on 0 DIV with GFP- or ETV1-expressing lentivirus. (D) NFI binding to the Etv1 promoter in P10 Etv1-knockout (Etv1 KO) mouse cerebellum. ***p < 0.001, **p < 0.01, *p < 0.05; ns, no significant difference.
Next we examined the effect of ETV1 on NFI binding to the Etv1 promoter. Early ETV1 expression increased NFI occupancy of the Etv1 promoter in immature CGNs (Figure 8C), and NFI binding to this site decreased in P10 Etv1-knockout mouse cerebellum (Figure 8D). Collectively these findings argued that ETV1 temporally up-regulates its own gene expression in maturing CGNs via positive feedback involving in part ETV1 enhancement of NFI binding to the Etv1 promoter.
Variable timing patterns for ETV1 occupancy in vivo
Etv1 mRNA is already detectable throughout the IGL of P7 mouse cerebellum (Sato et al., 2005; Schuller et al., 2006), suggesting that Etv1 expression precedes activation of downstream NFI-late genes. Temporal onset of Etv1 transcripts was in fact accelerated during early IGL maturation (P8–P13) compared with several NFI-late genes (Figure 9A) and strongly so relative to Gabra6 and Grin2c. Further, ETV1 occupancy of its own promoter preceded and was temporally distinguishable from that of other NFI-late genes based on ChIP (Figure 9B). Of note, there was ∼1-d latency between increases in Etv1 mRNA and ETV1 occupancy of its own promoter that preceded ETV1 binding of other late-gene promoters (compare fractional binding levels between P9 and P10; Figure 9B). Thus ETV1 preferentially binds to its own promoter relative to sites in the other NFI-late genes during earlier stages of cerebellar development, likely contributing to more-accelerated Etv1 gene expression.
FIGURE 9:
Temporal expression and ETV1 occupancy of the Etv1 gene are accelerated relative to other NFI-late genes. (A) Transcripts for Etv1 and several other NFI-late genes were assayed at different postnatal ages by RT-qPCR. (B) ChIP assays of ETV1 temporal binding to the Etv1 and other NFI-late genes. Data are expressed relative to values at P21, when IGL formation and CGN late gene expression are largely complete (Altman and Bayer, 1997; Furuichi et al., 2011; Ding et al., 2013). Data were derived from three biological replicates, and significance is shown for the difference between Etv1 values and that for the gene showing the least significant difference at each time point. ***p < 0.001, **p < 0.01, *p < 0.05; ns, no significant difference.
These differential rates of ETV1 binding are consistent with an autoregulatory mechanism that contributes to accelerated Etv1 expression relative to other NFI-late genes targeted by ETV1. Further, they suggest that ETV1 autoregulation is an important determinant of the timing of NFI occupancy and expression of ETV1-dependent NFI-late genes. Of interest, gene expression was considerably more delayed for Nab2 relative to other NFI-late genes (Figure 9A). The more protracted temporal pattern for Nab2 may account for its weak NFI occupancy at P11 (Figure 1A) and be related to its insensitivity to Etv1 deficiency (Figure 3A).
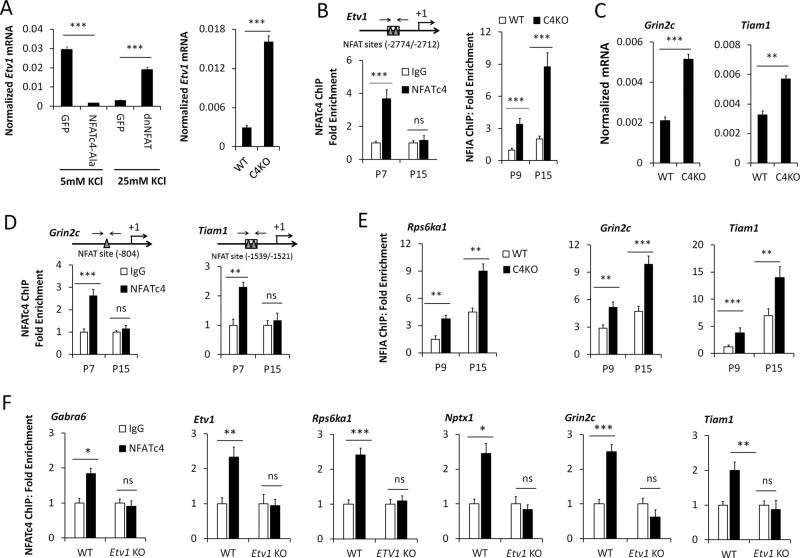
NFATc4 controls Etv1 expression in maturing CGNs
NFATc4 represses NFI-switch late genes in immature CGNs by binding to and preventing NFI occupancy of their promoters (Ding et al., 2013). It is subsequently dismissed from these promoters as CGNs differentiate. We therefore examined whether Etv1 gene expression was similarly regulated by NFATc4. In CGN cultures, Etv1 expression was strongly suppressed by constitutively active NFATc4 (Figure 10A), as previously found for other NFI-late genes (Ding et al., 2013). Etv1 mRNA was also inhibited by culturing CGNs in membrane- depolarizing concentrations of KCl (25 mM; Figure 10A), which enhances NFATc4 nuclear localization via activation of calcineurin (Ding et al., 2013). Further, expression of a dominant-interfering form of NFATc4 substantially reversed this depolarization-induced inhibition (Figure 10A). Conversely, Etv1 mRNA was strongly up-regulated in Nfatc4-knockout cerebellum during the postnatal week 2 (Figure 10A), when NFATc4 normally binds to and represses late genes in CGNs (Ding et al., 2013).
FIGURE 10:
NFATc4 regulates expression and NFI occupancy of the Etv1, Grin2c, and Tiam1 genes in maturing CGNs. (A) RT-qPCR assays of the regulation of Etv1 mRNA by NFAT proteins. Left, CGNs were transduced on 0 DIV with lentiviruses expressing GFP, constitutively active NFATc4 (NFATc4-Ala), or dominant-negative NFAT (dnNFAT) and then cultured to 6 DIV in medium containing either normal (5 mM) or depolarizing (25 mM) KCl concentrations. Right, assay of Nfatc4-knockout (C4KO) mouse cerebellum at P10 relative to wild type (WT). (B) Left, NFATc4 binding to the Etv1 gene in developing mouse cerebellum between P7 and P15. Shown above is the location of ChIP PCR primers relative to NFAT-binding sites examined in the Etv1 promoter. Right, NFIA binding to the Etv1 promoter in Nfatc4-knockout (C4KO) mouse cerebellum relative to WT at P9 and P15. (C) RT-qPCR assays of the Tiam1 and Grin2c genes in cerebella of C4KO compared with WT at P10. (D) ChIP assays of NFIA binding to the indicated genes in C4KO cerebellum relative to WT at P9 and P15. The locations of PCR primers relative to NFAT-binding sites are shown above. (E) NFATc4 temporal ChIP in the developing mouse cerebellum for the Rps6ka1, Grin2c, and Tiam1 genes. (F) ChIP assays of NFATc4 binding to the Etv1 and several other NFI-late genes in Etv1-knockout mouse cerebellum at P10. ***p < 0.001, **p < 0.01, *p < 0.05; ns, no significant difference.
NFAT consensus sites were identified upstream of the Etv1 promoter, and NFATc4 occupied this region in the P7 but not the P15 mouse cerebellum (Figure 10B). Of significance, NFIA occupancy of the Etv1 promoter was enhanced in Nfatc4-null cerebella (Figure 10B). Thus NFATc4 binds to the Etv1 gene and represses its expression and NFI occupancy in vivo, and NFATc4 departure coincides with up-regulation of these events during mouse cerebellar development. We also found that the NFI/ETV1 late genes Grin2c and Tiam1 were up-regulated in P10 mouse cerebella of Nfatc4-null mice (Figure 10C) and that NFATc4 temporally occupies their promoters in P7 but not P15 mouse cerebellum (Figure 10D). Further, loss of NFATc4 enhanced NFIA occupancy of the Grin2c and Tiam1 genes in the developing cerebellum (Figure 10E). NFIA occupancy was also increased for the Rps6ka1 gene in Nfatc4-null mouse cerebellum (Figure 10E), which was not previously documented.
These findings raised the possibility that ETV1 promoted NFI occupancy of the Etv1 and other late genes indirectly by displacing bound NFATc4. However, ChIP analysis revealed that NFATc4 binding to late-gene promoters decreased in Etv1-knockout cerebellum (Figure 10F), indicating that the effects of Etv1 deficiency on NFI occupancy were not mediated via enhanced NFATc4 occupancy. This result may reflect compensatory mechanisms activated in response to the loss of late gene expression in maturing Etv1-deficient CGNs. NFATc4 mRNA levels were not significantly altered in P10 and P11 Etv1-null mouse cerebella (Supplemental Figure S2), indicating that the effects of Etv1 deficiency on NFATc4 binding lie downstream of Nfatc4 gene regulation.
DISCUSSION
Postmigratory CGNs elaborate dendrites and form synapses with excitatory mossy fibers and inhibitory Golgi type II cells postnatally (Zheng et al., 1993; Mellor et al., 1998). These events are important for cerebellar circuitry and are temporally linked to the onset of eyeblink conditioning and increased motor activity (Tia et al., 1996). NFI proteins, and in particular NFIA, control the temporal expression of genes required for dendritogenesis and synapse formation in postmigratory CGNs, and the calcineurin/NFATc4 pathway regulates the timing of this expression by blocking NFI occupancy and late-gene expression in immature CGNs (Ding et al., 2013). Other NFI family members, including NFIB, also may contribute to NFI temporal occupancy.
The present findings provide deeper insight into regulation of the NFI temporal occupancy program during postmigratory CGN development. We identify ETV1 as a positive regulator of NFI temporal binding to several dendrite/synapse-related genes, showing that ETV1 regulates, directly binds to, and facilitates NFI occupancy of a subset of NFI-late gene targets. Together with promoter transfection studies done here and by others (Abe et al., 2011), these findings support a role for ETV1 in temporal activation of NFI binding and expression of coregulated genes in CGNs. Of importance, Etv1 is itself an NFI-late gene, and ETV1 autoregulates its expression in maturing CGNs and promotes NFI binding to its own promoter. Also of importance, Etv1 gene autoregulation and its occupancy by NFI precede the activation of downstream ETV1-dependent NFI-late genes. Further, our data also revealed that loss of NFATc4 occupancy, which normally leads to up-regulated NFI-late gene expression and NFI binding (Ding et al., 2013), is not sufficient by itself to stimulate these events in the absence of ETV1. This is consistent with an activating function of ETV1 in promoting NFI occupancy of late genes.
ETV1 and NFIA are present in most nuclei of postmigratory CGNs, and both proteins bind to and regulate the Etv1 and several other coregulated late genes and promoters in the postnatal cerebellum (see also Abe et al., 2011). These findings argue that ETV1 enhances NFI binding to coregulated late genes within individual CGN nuclei. Our data do not allow us to distinguish whether these events involve direct physical interactions between NFIA and ETV1 or occur via intermediary factors such as chromatin-remodeling complexes or other trans-factors.
Our findings suggest a possible stepwise model for temporal activation of NFI-late genes involving reciprocal, mutually dependent interactions between NFI occupancy and ETV1 (Figure 11). In premigratory and immature postmigratory CGNs (P7), NFATc4 occupies the Etv1 and other NFI-late genes, repressing NFI binding and gene expression. ETV1 protein and its occupancy of target genes are low at this stage. As maturation proceeds in the IGL (P8–P11), NFATc4 is dismissed from the Etv1 and other late-gene promoters due to declining calcineurin activity, which is permissive for NFI binding (Ding et al., 2013). Initial up-regulation of ETV1 and its binding enhances NFI occupancy of the Etv1 promoter, which in turn further increases Etv1 gene expression via a positive feedback loop (Figure 11, step 1). In an overlapping second phase (P11–P21), elevated ETV1 expression and binding activates other NFI-late genes in part by promoting NFI occupancy (Figure 11, step 2).
FIGURE 11:
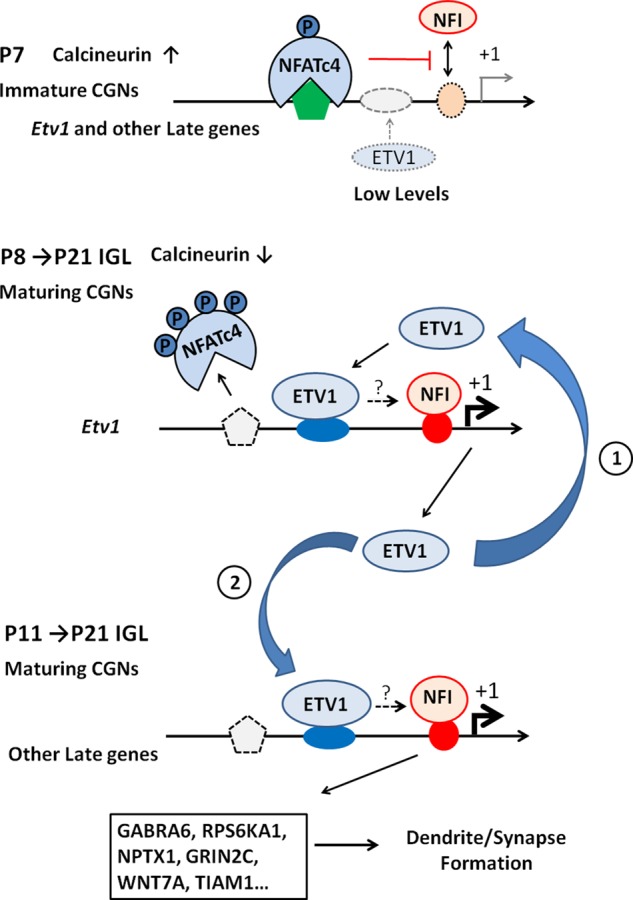
Possible sequential model for developmental regulation of NFI switch programming by ETV1. (A) In the early postnatal cerebellum (P7), elevated calcineurin (CaN) activity promotes NFATc4 translocation to the nucleus and its occupancy of late genes, preventing NFI binding and gene activation. ETV1 protein is low at this stage. (B) As CGNs mature (P8–P21), decreasing calcineurin activity reduces NFATc4 occupancy. This is permissive for NFI binding, resulting in increased NFI occupancy of the Etv1 promoter and enhanced gene expression. Initially, increasing ETV1 protein preferentially targets its own gene promoter and further facilitates NFI occupancy and Etv1 gene expression in CGNs in a positive feedback manner (step 1). In step 2 (P11–P21), elevated ETV1 protein subsequently binds to and promotes NFI occupancy of a subset of other NFI-late genes, resulting in enhanced mature gene expression and dendrite/synapse formation. ETV1 may promote NFI occupancy (dotted arrows) in CGNs via direct interactions with NFI and/or indirectly via other transcriptional mechanisms (see Discussion). This uncertainty is indicated by question marks above the arrows.
A significant implication of this model is that initiation of Etv1 autoregulation is a prerequisite for expression of dendrite/synapse-related genes coregulated by ETV1 and NFI. Diminished calcineurin signaling and NFATc4 departure is necessary but not sufficient to activate this subset of NFI-late genes (Figure 10F). We show here that both expression and NFI occupancy of this gene subset are modulated by ETV1. Furthermore, this model implies that NFI occupancy of this late-gene subset is itself autoregulatory. That is, the NFI occupancy program simultaneously is dependent on and promotes Etv1 autoregulation and vice versa, reflecting an intrinsic interdependence of these processes.
Based on our model, ETV1 and its autoregulation may function as an integral timing mechanism for NFI occupancy and temporal expression of dendrite/synapse-related genes. Preferential binding of ETV1 to its own promoter would increase its occupancy by NFI and drive accelerated up-regulation of Etv1 expression in early-maturing CGNs, thereby regulating the timing of its own expression. Increasing ETV1 levels in turn would promote temporally coordinated NFI occupancy and expression of a subset of dendrite/synapse-related genes that function in postmigratory CGNs maturing within the IGL. Some NFI-regulated late genes are not dependent on ETV1 in vivo and likely have different regulatory timing requirements and functions. ETV1 may thus provide both a selective and delayed timing mechanism for activating a subset of NFI-late genes in addition to the earlier and more widespread temporal effects on late-gene expression mediated by calcineurin/NFATc4 derepression. Of interest, precocious expression of ETS transcription factors perturbs synaptic connectivity of mouse sensory neurons (Hippenmeyer et al., 2005). Thus ETS-factor temporal expression affects synaptogenesis in both developing central and peripheral neurons. Whether ETV1 and NFIA also regulate the magnitude of late-gene expression in the mature cerebellum and not just its timing remains an open question, since neither Etv1- nor Nfia-null mice survive beyond the second to third postnatal week.
Multiple mechanisms may contribute to preferential early binding of ETV1 to its own promoter, including unique synergism between ETV1-binding sites (Figure 8B) and their interactions with coregulator sites as well as with chromatin-modifying complexes. ETV1 autoregulation also may be modulated by posttranslational modifications in maturing CGNs. BDNF, a regulator of CGN differentiation, transiently phosphorylates ETV1 in CGNs and increases ETV1 transcriptional activity in heterologous cells (Abe et al., 2012). BDNF/TRKB signaling may therefore augment ETV1 autoregulation in immature CGNs by stimulating ETV1 activity when its expression levels are low.
CGN differentiation and its timing, including the NFI switch, are reproduced in purified CGN cultures (Trenkner et al., 1984; Gao and Fritschy, 1995; Mellor et al., 1998; Yacubova and Komuro, 2002; Ding et al., 2013), implicating cell-intrinsic processes in these events. Thus CGNs provide a valuable system for elucidating how distinct temporal stages are controlled in developing neurons (Kilpatrick et al., 2012). Here we used this system to provide evidence that a transcription factor autoregulatory loop coupled with differential occupancy rates may provide a timing mechanism for developmental stage–specific gene expression in late-maturing neurons. Transcriptional positive feedback loops play important roles in diverse cellular events, including circadian oscillators (Shearman et al., 2000), commitment to cell division (Eser et al., 2011), and specification of peripheral nervous system cell types (Grocott et al., 2012). Of interest, the ETV1 autoactivation loop preceding activation of downstream genes observed here in CGNs mirrors the “positive feedback first” events seen in autoregulatory control of commitment to cell cycle progression (Eser et al., 2011).
Altered timing of dendrite and synapse maturation and circuitry formation occurs in several NDs (Geschwind and Levitt, 2007; Leonardo and Hen, 2008; Harlow et al., 2010; LeBlanc and Fagiolini, 2011; Penzes et al., 2011; Addington and Rapoport, 2012; Till et al., 2012; Uhlhaas and Singer, 2012), and disrupted synaptogenic gene expression has been implicated in these disorders (Brown, 2011; Addington and Rapoport, 2012; Meredith et al., 2012; Tsigelny et al., 2013). In addition to movement control, the cerebellum participates in language, cognition, mood, and attention, likely via cerebellar-thalamo-cortical connections (O’Halloran et al., 2012). Alterations in cerebellar structure and its connectivity also occur in several NDs (Fatemi et al., 2012; O’Halloran et al., 2012; Rogers et al., 2013a, b). Further, NFIA gene variants and deletions have been linked to delayed acquisition of language/motor skills and NDs (Mikhail et al., 2011; Iossifov et al., 2012; Prasad et al., 2012; Lee et al., 2013). An intriguing question is whether disruption of NFI-dependent synaptogenic timing mechanisms in CGNs contributes to altered cerebellar circuitry and associated ND phenotypes.
MATERIALS AND METHODS
Animals and primary cultures
Cerebellar tissues were obtained from postnatal knockout mice and wild-type littermates of varying genetic backgrounds: Etv1(–/–) were 129/Sv and C57B/6J hybrids (Arber et al., 2000), Nfatc4(–/–) (Ding et al., 2013) were C57Bl/6, and Nfia(–/–) were C57Bl/6NTac (Shu et al., 2003). Mouse CGNs were prepared from P6 CD1 mouse pups of either sex and cultured as previously described (Ding et al., 2013; Selvakumar and Kilpatrick, 2013). Individual culture experiments were performed using cells prepared from the same litter. Cells were plated at a density of 5 × 104 cells/cm2 onto chamber slides or cell culture dishes coated with poly-d-lysine/laminin (Invitrogen, Carlsbad, CA) in Neurobasal medium (Invitrogen) containing B-27 serum-free supplement (50×; Invitrogen).
Plasmids and cell lines
Plasmid FU-ETV1-CRW containing full-length human ETV1 coding sequences was generously provided by Owen N. Witte (University of California, Los Angeles, Los Angeles, CA). Self-inactivating lentiviruses expressing green fluorescent protein (GFP), hemagglutinin (HA)-tagged NFI dominant repressor (NFI/EnR), or Drosophila engrailed repressor domain alone (EnR), FLAG-tagged NFATc4 proteins (constitutively active [NFATc4-Ala] and dominant-negative [dnNFAT]) were described previously (Wang et al., 2004; Ding et al., 2013). Human embryonic kidney 293T cells were grown in DMEM (Invitrogen) containing 10% heat-inactivated fetal bovine serum (Invitrogen).
Lentivirus preparation and transduction of primary CGNs
Lentiviruses were generated by cotransfection of 293T cells with lentiviral expression plasmid, psPAX2, and pCMV-VSVG and then concentrated and titered as previously described (Wang et al., 2005; Ding and Kilpatrick, 2013b). CGN cultures were transduced on day 0 in vitro using multiplicity of infection of ∼3, yielding 80–90% cell transduction.
RNA isolation and real-time quantitative PCR
RNA was extracted from tissues or cultured cells, and cDNAs were prepared as described previously (Ding et al., 2013). Real-time quantitative PCR (RT-qPCR) was performed in triplicate using primers, SYBR Green PCR master mix (Qiagen, Valencia, CA), and a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA). Transcript data were analyzed using the 2−ΔCt method and then normalized to 18S rRNA as previously described (Ding et al., 2013). PCR primer sequences are available upon request.
ChIP assays
ChIP was performed as previously described (Ding et al., 2013). For tissues, nuclei were first purified by Percoll gradient centrifugation (Wang et al., 2011; Ding and Kilpatrick, 2013a) or differential centrifugation (1400 × g for 3 min, 4°C) in 0.25 M sucrose. Samples were assayed by real-time PCR, and data from triplicate biological replicates were expressed as the fold enrichment relative to antibody controls. ChIP antibodies were to ETV1 (sc-28681; Santa Cruz Biotechnology, Dallas, TX), Xenopus NFIB1, which recognizes mammalian NFIA and NFIB (Ding et al., 2013), NFIA (39036; Active Motif, Carlsbad, CA), and NFATc4 (sc-1153; Santa Cruz Biotechnology). The term NFI is used to distinguish ChIP assays employing the Xenopus NFIB1 antibody from those specifically assaying NFIA occupancy. Note that the NFI and NFIA antibodies both detect binding to the same genomic regions in ChIP assays (Ding et al., 2013). Preimmune serum or normal rabbit immunoglobulin G (IgG; PP64; Millipore, Billerica, MA) was used as negative control. PCR primer sequences for ChIP assays are available upon request.
Immunostaining
Immunofluorescence of neuronal cultures was performed as in previous studies (Ding et al., 2013). Briefly, primary CGNs were cultured on chamber slides coated with poly-d-lysine/laminin (Invitrogen) and then fixed on 2 DIV with 4% paraformaldehyde in phosphate-buffered saline (PBS). Cells were permeabilized with 1% Triton X-100 solution and incubated with 5% normal goat serum followed by primary antibodies at 4°C overnight and then with Cy3-conjugated goat anti-rabbit IgG (NG1807775; Millipore). Anti-Map2 antibody (AB5622; Millipore) was used to stain neurites, and bisbenzimide (1 μg/ml; Sigma-Aldrich, St. Louis, MO) was used to stain nuclei. Pictures were captured with a fluorescence and phase contrast microscope (Leica DM IRE2; Leica Microsystems, Buffalo Grove, IL). Neurite length was measured with Image Pro Plus 6.0 software. Lentiviral titrations used antibodies for ETV1 (sc-28681; Santa Cruz Biotechnology), HA (NFI/EnR and EnR; C29F4, Cell Signaling Technology, Boston, MA), or FLAG (NFATc4 proteins; M2 antibody; Sigma-Aldrich).
For tissue immunostaining, cerebella were dissected from P15 mouse pups and fixed in 4% paraformaldehyde and 0.1 M phosphate buffer, pH 7.2, washed with PBS, and equilibrated in 30% sucrose and 0.1 M sodium phosphate buffer, pH 7.2, overnight at 4°C. Cerebella were frozen in OCT, and cryostat sections (10 μm) were mounted onto slides and blocked using 5% normal goat serum, 0.5% Triton X-100, and 0.02% sodium azide in PBS. Separate slides containing adjacent sections were incubated with rabbit primary antibodies against ER81/ETV1 (1:15,000; PRB-362C; Covance, Dedham, MA) or NFIA (1:500; 39036; Active Motif) or with no primary antibody overnight at 4°C in 2% normal goat serum, 0.1% Triton X-100, 0.02% sodium azide, and PBS. Only rabbit antibodies were found to be suitable for immunostaining of ETV1 and NFIA proteins. After being washed in PBS, slides were incubated with goat Alexa Fluor 488–conjugated anti-rabbit IgG F(ab′)2 (1:500; A11070; Invitrogen), and 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml). Confocal fluorescence microscopy was performed using a Nikon A1 confocal microscope.
Western blotting
For analyses of knockout mice, proteins were prepared from frozen cerebellar tissues of postnatal Etv1(–/–) mice and wild-type littermates by extraction with RIPA buffer (10 mM Tris-HCl, pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS) containing protease inhibitors (11 873 580 001; 454 Life Sciences, Branford, CT). Extracts were centrifuged at 1000 × g for 10 min at 4°C, and the supernatant was collected. Protein samples were separated on 8% SDS–polyacrylamide gels and transferred onto Pure Nitrocellulose membranes (GE Water & Process Technologies, Boulder, CO). After blocking for 1 h, blots were incubated with primary antibodies at 4°C overnight and then with horseradish peroxidase–conjugated secondary antibody at room temperature for 1 h. Bound antibodies were detected with a chemiluminescent substrate (Thermo Scientific, Waltham, MA). For temporal analyses, nuclear extracts were prepared from P7, P15, and P21 mouse cerebella as previously described (Wang et al., 2011). Primary antibodies used were as follows: pan-NFI antibody (1:1000; sc-5567; Santa Cruz Biotechnology), anti–histone H3 (1:1000; PAB0653; Abnova, Taipei City, Taiwan), NFIA (39036; Active Motif), NFATc4 (ab99431; Abcam, Cambridge, MA), ETV1 (sc-28681; Santa Cruz Biotechnology), and TBP (8515; Cell Signaling).
Promoter cotransfections
JEG-3 cells were transfected with a luciferase reporter plasmid containing 6 kb of the mouse Gabra6 gene inserted into pGL3 (Wang et al., 2004) along with various combinations of expression plasmids for NFIA (Wang et al., 2004) and ETV1 or GFP expression vectors using TransIT-LT1 reagent (Mirus Bio, Madison, WI). pBluescript plasmid was used to maintain a constant amount of transfected DNA in all samples. This DNA did not affect promoter activity or its activation by expression vectors. Promoterless pGL3 plasmid was used as a negative control. Luciferase activity was measured ∼72 h after initiating transfection using a firefly luciferase assay kit (Mirus Bio) according to the manufacturer’s instructions.
Statistics
Unpaired two-tailed Student’s t tests were run for comparisons of one experimental sample with its control. One-way analysis of variance was performed when comparing multiple samples, with either a Tukey (when comparing samples to each other) or Dunnett (when comparing samples to a control) post hoc tests. Results were expressed as mean ± SD, and p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank George Gagnon and Jason Osinski for essential technical assistance in this work’s completion. This work was supported by Public Health Service Grant NS063047 to D.L.K., Public Health Service Grant DCD008955 and the Burke Medical Research Institute to J.W.C., and HL08624 (National Heart, Lung and Blood Institute) and C026429 (New York Stem Cell Science) to R.M.G.
Abbreviations used:
- CGN
cerebellar granule neuron
- DIV
days in vitro
- IGL
internal granule cell layer
- ND
neurodevelopmental disorder
- NFI
Nuclear Factor One.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0476) on March 3, 2016.
REFERENCES
- Abe H, Okazawa M, Nakanishi S. The Etv1/Er81 transcription factor orchestrates activity-dependent gene regulation in the terminal maturation program of cerebellar granule cells. Proc Natl Acad Sci USA. 2011;108:12497–12502. doi: 10.1073/pnas.1109940108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Okazawa M, Nakanishi S. Gene regulation via excitation and BDNF is mediated by induction and phosphorylation of the Etv1 transcription factor in cerebellar granule cells. Proc Natl Acad Sci USA. 2012;109:8734–8739. doi: 10.1073/pnas.1206418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addington AM, Rapoport JL. Annual research review: impact of advances in genetics in understanding developmental psychopathology. J Child Psychol Psychiatry. 2012;53:510–518. doi: 10.1111/j.1469-7610.2011.02478.x. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the Cerebellar System in Relation to Its Evolution, Structure and Function. Boca Raton, FL: CRC Press; 1997. [Google Scholar]
- Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- Balamotis MA, Tamberg N, Woo YJ, Li J, Davy B, Kohwi-Shigematsu T, Kohwi Y. Satb1 ablation alters temporal expression of immediate early genes and reduces dendritic spine density during postnatal brain development. Mol Cell Biol. 2012;32:333–347. doi: 10.1128/MCB.05917-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS. The environment and susceptibility to schizophrenia. Prog Neurobiol. 2011;93:23–58. doi: 10.1016/j.pneurobio.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave JW, Akiba Y, Banerjee K, Bhosle S, Berlin R, Baker H. Differential regulation of dopaminergic gene expression by Er81. J Neurosci. 2010;30:4717–4724. doi: 10.1523/JNEUROSCI.0419-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry AZ, Vitullo AD, Gronostajski RM. Nuclear factor I (NFI) isoforms differentially activate simple versus complex NFI-responsive promoters. J Biol Chem. 1998;273:18538–18546. doi: 10.1074/jbc.273.29.18538. [DOI] [PubMed] [Google Scholar]
- Deguchi Y, Donato F, Galimberti I, Cabuy E, Caroni P. Temporally matched subpopulations of selectively interconnected principal neurons in the hippocampus. Nat Neurosci. 2011;14:495–504. doi: 10.1038/nn.2768. [DOI] [PubMed] [Google Scholar]
- Ding B, Kilpatrick DL. Chromatin immunoprecipitation assay of brain tissues using Percoll gradient-purified nuclei. Methods Mol Biol. 2013a;1018:199–209. doi: 10.1007/978-1-62703-444-9_19. [DOI] [PubMed] [Google Scholar]
- Ding B, Kilpatrick DL. Lentiviral vector production, titration, and transduction of primary neurons. Methods Mol Biol. 2013b;1018:119–131. doi: 10.1007/978-1-62703-444-9_12. [DOI] [PubMed] [Google Scholar]
- Ding B, Wang W, Selvakumar T, Xi HS, Zhu H, Chow CW, Horton JD, Gronostajski RM, Kilpatrick DL. Temporal regulation of nuclear factor one occupancy by calcineurin/NFAT governs a voltage-sensitive developmental switch in late maturing neurons. J Neurosci. 2013;33:2860–2872. doi: 10.1523/JNEUROSCI.3533-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol Cell. 2011;43:515–527. doi: 10.1016/j.molcel.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Aldinger KA, Ashwood P, Bauman ML, Blaha CD, Blatt GJ, Chauhan A, Chauhan V, Dager SR, Dickson PE, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11:777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi T, Shiraishi-Yamaguchi Y, Sato A, Sadakata T, Huang J, Shinoda Y, Hayashi K, Mishima Y, Tomomura M, Nishibe H, et al. Systematizing and cloning of genes involved in the cerebellar cortex circuit development. Neurochem Res. 2011;36:1241–1252. doi: 10.1007/s11064-011-0398-1. [DOI] [PubMed] [Google Scholar]
- Gao B, Fritschy JM. Cerebellar granule cells in vitro recapitulate the in vivo pattern of GABAA-receptor subunit expression. Brain Res Dev Brain Res. 1995;88:1–16. doi: 10.1016/0165-3806(95)00062-i. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- Grocott T, Tambalo M, Streit A. The peripheral sensory nervous system in the vertebrate head: a gene regulatory perspective. Dev Biol. 2012;370:3–23. doi: 10.1016/j.ydbio.2012.06.028. [DOI] [PubMed] [Google Scholar]
- Harlow EG, Till SM, Russell TA, Wijetunge LS, Kind P, Contractor A. Critical period plasticity is disrupted in the barrel cortex of FMR1 knockout mice. Neuron. 2010;65:385–398. doi: 10.1016/j.neuron.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick DL, Wang W, Gronostajski R, Litwack ED. Nuclear factor I and cerebellar granule neuron development: an intrinsic-extrinsic interplay. Cerebellum. 2012;11:41–49. doi: 10.1007/s12311-010-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto T, Toma K, Gunadi, McKenna WL, Kasukawa T, Katzman S, Chen B, Hanashima C. Foxg1 coordinates the switch from nonradially to radially migrating glutamatergic subtypes in the neocortex through spatiotemporal repression. Cell Rep. 2013;3:931–945. doi: 10.1016/j.celrep.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc JJ, Fagiolini M. Autism: a “critical period” disorder. Neural Plast. 2011;2011:921680. doi: 10.1155/2011/921680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Woo HG, Greenwood TA, Kripke DF, Kelsoe JR. A genome-wide association study of seasonal pattern mania identifies NF1A as a possible susceptibility gene for bipolar disorder. J Affect Disord. 2013;145:200–207. doi: 10.1016/j.jad.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo ED, Hen R. Anxiety as a developmental disorder. Neuropsychopharmacology. 2008;33:134–140. doi: 10.1038/sj.npp.1301569. [DOI] [PubMed] [Google Scholar]
- Martynoga B, Drechsel D, Guillemot F. Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harb Perspect Biol. 2012;4:a0083. doi: 10.1101/cshperspect.a008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor JR, Merlo D, Jones A, Wisden W, Randall AD. Mouse cerebellar granule cell differentiation: electrical activity regulates the GABAA receptor alpha 6 subunit gene. J Neurosci. 1998;18:2822–2833. doi: 10.1523/JNEUROSCI.18-08-02822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RM, Dawitz J, Kramvis I. Sensitive time-windows for susceptibility in neurodevelopmental disorders. Trends Neurosci. 2012;35:335–344. doi: 10.1016/j.tins.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, Rutledge SL, Korf BR, Carroll AJ. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A. 2011;155A:2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- O’Halloran CJ, Kinsella GJ, Storey E. The cerebellum and neuropsychological functioning: a critical review. J Clin Exp Neuropsychol. 2012;34:35–56. doi: 10.1080/13803395.2011.614599. [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Merico D, Thiruvahindrapuram B, Wei J, Lionel AC, Sato D, Rickaby J, Lu C, Szatmari P, Roberts W, et al. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda) 2012;2:1665–1685. doi: 10.1534/g3.112.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers TD, Dickson PE, McKimm E, Heck DH, Goldowitz D, Blaha CD, Mittleman G. Reorganization of circuits underlying cerebellar modulation of prefrontal cortical dopamine in mouse models of autism spectrum disorder. Cerebellum. 2013a:547–556. doi: 10.1007/s12311-013-0462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers TD, McKimm E, Dickson PE, Goldowitz D, Blaha CD, Mittleman G. Is autism a disease of the cerebellum? An integration of clinical and pre-clinical research. Front Syst Neurosci. 2013b;7:15. doi: 10.3389/fnsys.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago C, Bashaw GJ. Transcription factors and effectors that regulate neuronal morphology. Development. 2014;141:4667–4680. doi: 10.1242/dev.110817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suzuki K, Yamazaki H, Nakanishi S. A pivotal role of calcineurin signaling in development and maturation of postnatal cerebellar granule cells. Proc Natl Acad Sci USA. 2005;102:5874–5879. doi: 10.1073/pnas.0501972102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller U, Kho AT, Zhao Q, Ma Q, Rowitch DH. Cerebellar “transcriptome” reveals cell-type and stage-specific expression during postnatal development and tumorigenesis. Mol Cell Neurosci. 2006;33:247–259. doi: 10.1016/j.mcn.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Selvakumar T, Kilpatrick DL. Culturing mouse cerebellar granule neurons. Methods Mol Biol. 2013;1018:49–59. doi: 10.1007/978-1-62703-444-9_5. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tia S, Wang JF, Kotchabhakdi N, Vicini S. Developmental changes of inhibitory synaptic currents in cerebellar granule neurons: role of GABA(A) receptor alpha 6 subunit. J Neurosci. 1996;16:3630–3640. doi: 10.1523/JNEUROSCI.16-11-03630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till SM, Wijetunge LS, Seidel VG, Harlow E, Wright AK, Bagni C, Contractor A, Gillingwater TH, Kind PC. Altered maturation of the primary somatosensory cortex in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21:2143–2156. doi: 10.1093/hmg/dds030. [DOI] [PubMed] [Google Scholar]
- Trenkner E, Smith D, Segil N. Is cerebellar granule cell migration regulated by an internal clock. J Neurosci. 1984;4:2850–2855. doi: 10.1523/JNEUROSCI.04-11-02850.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripodi M, Arber S. Regulation of motor circuit assembly by spatial and temporal mechanisms. Curr Opin Neurobiol. 2012;22:615–623. doi: 10.1016/j.conb.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Tsigelny IF, Kouznetsova VL, Baitaluk M, Changeux JP. A hierarchical coherent-gene-group model for brain development. Genes Brain Behav. 2013;12:147–165. doi: 10.1111/gbb.12005. [DOI] [PubMed] [Google Scholar]
- Uhlhaas PJ, Singer W. Neuronal dynamics and neuropsychiatric disorders: toward a translational paradigm for dysfunctional large-scale networks. Neuron. 2012;75:963–980. doi: 10.1016/j.neuron.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Vogel-Ciernia A, Wood MA. Neuron-specific chromatin remodeling: a missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology. 2014;80:18–27. doi: 10.1016/j.neuropharm.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Crandall JE, Litwack ED, Gronostajski RM, Kilpatrick DL. Targets of the nuclear factor I regulon involved in early and late development of postmitotic cerebellar granule neurons. J Neurosci Res. 2010;88:258–265. doi: 10.1002/jnr.22199. [DOI] [PubMed] [Google Scholar]
- Wang W, Mullikin-Kilpatrick D, Crandall JE, Gronostajski RM, Litwack ED, Kilpatrick DL. Nuclear factor I coordinates multiple phases of cerebellar granule cell development via regulation of cell adhesion molecules. J Neurosci. 2007;27:6115–6127. doi: 10.1523/JNEUROSCI.0180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Qu Q, Smith FI, Kilpatrick DL. Self-inactivating lentiviruses: versatile vectors for quantitative transduction of cerebellar granule neurons and their progenitors. J Neurosci Methods. 2005;149:144–153. doi: 10.1016/j.jneumeth.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Wang W, Shin Y, Shi M, Kilpatrick DL. Temporal control of a dendritogenesis-linked gene via REST-dependent regulation of nuclear factor I occupancy. Mol Biol Cell. 2011;22:868–879. doi: 10.1091/mbc.E10-10-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Stock RE, Gronostajski RM, Wong YW, Schachner M, Kilpatrick DL. A role for nuclear factor I in the intrinsic control of cerebellar granule neuron gene expression. J Biol Chem. 2004;279:53491–53497. doi: 10.1074/jbc.M410370200. [DOI] [PubMed] [Google Scholar]
- Weyer A, Schilling K. Developmental and cell type-specific expression of the neuronal marker NeuN in the murine cerebellum. J Neurosci Res. 2003;73:400–409. doi: 10.1002/jnr.10655. [DOI] [PubMed] [Google Scholar]
- Yacubova E, Komuro H. Intrinsic program for migration of cerebellar granule cells in vitro. J Neurosci. 2002;22:5966–5981. doi: 10.1523/JNEUROSCI.22-14-05966.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Santi MR, Bovolin P, Marlier LN, Grayson DR. Developmental expression of the alpha 6 GABAA receptor subunit mRNA occurs only after cerebellar granule cell migration. Brain Res Dev Brain Res. 1993;75:91–103. doi: 10.1016/0165-3806(93)90068-l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.