Abstract
Protein synthesis in eukaryotes is regulated by diverse reprogramming mechanisms that expand the coding capacity of individual genes. Here, we exploit one such mechanism termed −1 programmed ribosomal frameshifting (−1 PRF) to engineer ligand-responsive RNA switches that regulate protein expression. First, efficient −1 PRF stimulatory RNA elements were discovered by in vitro selection; then, ligand-responsive switches were constructed by coupling −1 PRF stimulatory elements to RNA aptamers using rational design and in vivo directed evolution. We demonstrate that −1 PRF switches tightly control the relative stoichiometry of two distinct protein outputs from a single mRNA, exhibiting consistent ligand response across whole populations of cells. Furthermore, −1 PRF switches were applied to build single-mRNA logic gates and an apoptosis module in yeast. Together, these results showcase the potential for harnessing translation-reprogramming mechanisms for synthetic biology, and establish −1 PRF switches as powerful RNA tools for controlling protein synthesis in eukaryotes.
INTRODUCTION
The ribosome coordinates the biosynthesis of proteins from mRNA templates according to a standard translational program. While the ribosome typically executes translation uniformly and with high fidelity1, in some cases, the program is temporarily altered in order to change the protein output of a given gene2,3. This ‘reprogramming’ endows the translation apparatus with expanded synthetic capabilities, enabling the expression of proteins containing non-canonical amino acids (such as selenocysteine or pyrrolysine) or the regulated expression of multiple distinct protein products from a single mRNA transcript4. Some forms of translation reprogramming have been directly adopted for biotechnology, including internal ribosome entry sites (IRES)5 and co-translational cleaving 2A peptides6. Moreover, substantial effort has been directed toward the redefinition of codons to specify unnatural amino acids7. While significant progress has been made in these areas, other modes of translation reprogramming remain largely unexplored despite their potential applications to synthetic biology.
Notably, many reprogramming mechanisms utilize cis-acting RNA elements embedded within mRNAs. Recently, other RNA-based gene-expression frameworks have emerged as powerful tools for engineering biological systems8. Over two decades of SELEX and related in vitro selection experiements9–11 have yielded synthetic RNA molecules, termed aptamers, that bind to diverse ligands12,13. These aptamers have been coupled to RNA-based expression platforms to construct ligand-controlled gene-regulatory tools such as allosteric ribozymes14–18. These RNA devices have been applied for cellular computation19, regulation of gene expression20, and phenotypic control21,22. The apparent modularity of device construction suggests that other RNA gene-expression frameworks could be exploited to engineer new classes of RNA devices with distinct regulatory opportunities.
We identified −1 programmed ribosomal frameshifting (−1 PRF) as a potentially powerful gene-regulatory mechanism for RNA device engineering (Fig. 1). Eukaryotic −1 PRF signals contained within mRNA transcripts are composed of two principal features: (i) a heptanucleotide slippery site where the frameshift event occurs, with the general sequence X-XXY-YYZ (dashes indicate original frame; X denotes any nucleotide; Y denotes A or U; Z denotes A, C or U); and (ii) a downstream stimulatory RNA structure, typically a hairpin or pseudoknot23. When encountering a −1 PRF signal in an mRNA, a fraction of translating ribosomes slip back by a single nucleotide, placing the translation apparatus in the −1 reading frame. This, in turn, alters the amino acid composition of the polypeptide that is synthesized downstream of the frameshift site.
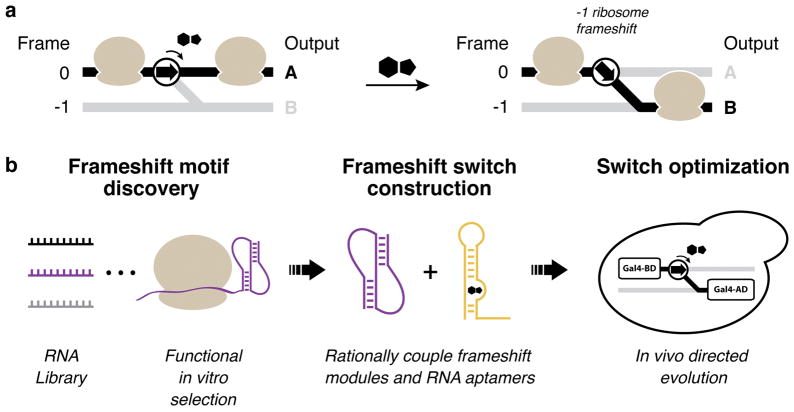
Figure 1.
Design of ligand-responsive −1 PRF switches. (a) Translation control scheme. The protein output of an mRNA is dictated by the translation reading frame. −1 PRF switches direct the ribosome’s translation reading frame depending on the presence or absence of a ligand. (b) Methodological approach to build −1 PRF switches. Active frameshift stimulatory elements are discovered from large RNA libraries using a functional in vitro selection. Frameshift stimulatory elements (purple) are then coupled to RNA aptamer modules (gold) by rational design to create frameshift switches. Lastly, frameshift switch devices are optimized by in vivo directed evolution using a frameshift-dependent growth selection.
−1 PRF has been well studied in retroviruses, including HIV, where it serves to establish a precise ratio of Gag to Gag-Pol proteins24. Regardless of variation in mRNA transcript levels or translational activity, the stoichiometry of frameshift to non-frameshift protein products remains constant for a given −1 PRF signal. While viral −1 PRF signals have fixed frameshift activities, it may be possible to engineer frameshift signals to respond to environmental ligands25 (Fig. 1a). Previous studies26,27 in vitro and in mammalian cell culture demonstrated the feasibility of small molecule regulated −1 PRF using metabolite sensing transcriptional riboswitches that adopt frameshift stimulatory pseudoknot conformations in the presence of their cognate ligands. However, the ligand binding domains of these bacterial riboswitches are integral components that cannot be exchanged with other ligand binding RNA aptamer domains and are not easily modified to recognize entirely new ligands. As a result, no general design strategy currently exists for assembling synthetic −1 PRF devices that respond exclusively to an orthogonal small molecule of choice.
Here, we establish a modular platform for engineering ligand-responsive −1 PRF switches and demonstrate the applicability of such devices for gene regulation in vivo. We describe a functional in vitro selection for −1 PRF stimulatory element discovery, bringing together the combinatorial complexity of a classical in vitro RNA selection and the biochemical complexity of a cell lysate. We further leverage rational design and in vivo directed evolution to construct ligand-responsive −1 PRF switches (Fig. 1b). Our synthetic −1 PRF switches display robust performance as a result of balanced protein output stoichiometry, enabling the assembly of logic gates and a phenotypic control module that responds with precision at the individual cell level.
RESULTS
In vitro selection for −1 PRF stimulatory elements
To establish a −1 PRF toolkit for synthetic biology, we set out to compile a collection of active RNA frameshift stimulatory elements that is suitable for downstream applications. Ideally, these elements are short in length, have well-defined structures, trigger high efficiency −1 PRF, and are readily exchanged with other frameshift elements of similar size and composition. However, natural −1 PRF stimulatory elements are diverse in both size and sequence4, and stimulate frameshifting at levels that are tuned for optimal viral replication (typically 5–10%), not maximal efficiency3. The above limitations place constraints on the scope of available parts, the modularity for engineering, and the achievable dynamic range of ligand-responsive devices. Therefore, with the intent to generate −1 PRF stimulators optimized for engineering, we developed a selection strategy to discover novel −1 PRF stimulatory elements from large libraries of sequence variants derived from a uniformly compact scaffold.
Our directed evolution discovery approach utilizes mRNA display (Fig. 2a), a well-established in vitro selection technology28. During in vitro translation of mRNA-display templates, the amplifiable genotype of the mRNA is linked to the phenotype of the polypeptide through a critical puromycin ligation reaction. While mRNA-display is conventionally employed to select for functional proteins and peptides, we repurposed it to select for the translation reprogramming activity of an mRNA29 (Fig. 2b). Due to the strong distance dependence of the puromycin reaction30, mRNA-peptide fusion occurs only if the ribosome translates the entire mRNA transcript. As a result, upstream stop codons that terminate translation preclude mRNA-peptide fusion formation31. We exploited this specificity of puromycin reactivity to differentiate frameshift stimulating sequences from inactive sequences.
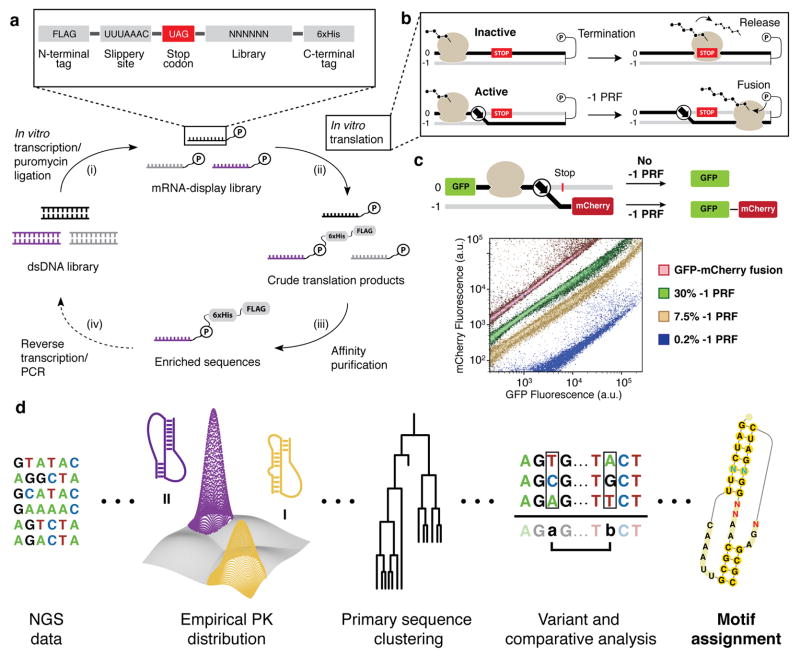
Figure 2.
In vitro selection for −1 PRF stimulatory elements. (a) The mRNA display selection. The display construct encodes an N-terminal FLAG tag, a heptanucleotide slippery site, an in-frame stop codon, the stimulatory element library, and a C-terminal His6-tag encoded in the −1 frame. The selection cycle comprises four stages: (i) RNA is in vitro transcribed from DNA library templates, then ligated to the puromycin adaptor; (ii) mRNA display templates are translated in vitro with rabbit reticulocyte lysate; (iii) mRNA-peptide fusions are isolated from non-fused RNA by affinity purification of the peptide tags; (iv) enriched sequences are reverse transcribed and PCR amplified for subsequent rounds of selection. (b) Translation reprogramming selection principle. Ribosomes that terminate translation upstream of the designated fusion point will fail to produce mRNA-peptide fusions (upper). Frameshifting enables bypass of encoded stop codons (lower). (c) Dual-FP reporter assay in S. cerevisiae. The frameshift variant is cloned between a green fluorescent protein (GFP) and the red fluorescent protein variant mCherry. The ratio of FP signals reflects bulk −1 PRF efficiency. Flow cytometry of individual clones harboring -1 PRF stimulatory elements of varying efficiencies is shown. (d) NGS analysis workflow for library selection products. Selected sequences are grouped into pseudoknot (PK) families, analyzed for post-selection enrichment, and clustered based on primary sequence identity. Motifs can be analyzed by comparative analysis, or individual sequences can be analyzed for single and pair-wise nucleotide changes (see Supplementary Fig. 5).
An mRNA library was designed such that only active −1 PRF signals form mRNA-peptide fusions and become enriched. Starting from a prokaryotic riboswitch scaffold26, 14 of 35 nucleotides were randomized to generate 2.68×108 sequence variants (Supplementary Fig. 1). Notably, a library of this size is easily accommodated by the mRNA-display technology, which allows for upwards of 1014 input sequences30. To enrich active −1 PRF stimulatory elements, the library was encoded downstream of the heptanucleotide slippery site U-UUA-AAC and an in-frame UAG termination codon. mRNA display templates were translated in vitro in rabbit reticulocyte lysate and purified based solely on the presence of the peptide epitope tags. Unlike traditional mRNA display, which uses the purified mRNA–peptide fusions for subsequent selections, the functional selection for −1 PRF activity is complete at this stage of the cycle.
After three rounds of in vitro selection, assaying of selection products in a dual-fluorescent protein (dual-FP) reporter in Saccharomyces cerevisiae revealed enrichment for active −1 PRF stimulatory elements with in vivo efficiencies of up to 30% (Fig. 2c and Supplementary Fig. 2). Moreover, flow cytometry of individual clones revealed that the ratio of fluorescent proteins remained constant for a given population of cells harboring the same −1 PRF signal, irrespective of total protein synthesis. As a result, two populations of yeast with frameshifting efficiencies that differ by only 3- to 4-fold are highly resolvable, despite overall expression levels that span several orders of magnitude.
The in vitro selection products were submitted for next-generation sequencing (NGS) to characterize the landscape of frameshift stimulatory elements, with a particular focus on pseudoknot structures (Supplementary Note 1). A computational pipeline was implemented to identify promising sequence motifs for downstream engineering applications (Fig. 2d). Sequences were grouped on the basis of compatibility with different hairpin-type (H-type) pseudoknot geometries (Supplementary Fig. 3), assessed for structure subtype enrichment (Supplementary Fig. 4), and lastly clustered into motifs based on primary sequence identity. The differential abundances of sequence variants within a motif were used to identify nucleotide preferences at variable sites and mutation intolerant positions. This analysis provides strong support for further engineering of −1 PRF motifs, and forms the basis for rational switch engineering.
Rational design and directed evolution of −1 PRF switches
To engineer ligand-responsive devices (Fig. 3), we pursued a modular strategy of rationally coupling −1 PRF signals to small molecule binding RNA aptamers (Fig. 3a). For the stimulatory element, we chose sequence FS-2 from the in vitro selection, which was the second most abundant sequence overall in the NGS, displayed high −1 PRF efficiency in yeast (30%), and has a confidently predicted pseudoknot fold (Supplementary Fig. 5). As aptamer domains, we chose the theophylline12 and neomycin32 binding aptamers based on their previous successes for in vivo applications21.
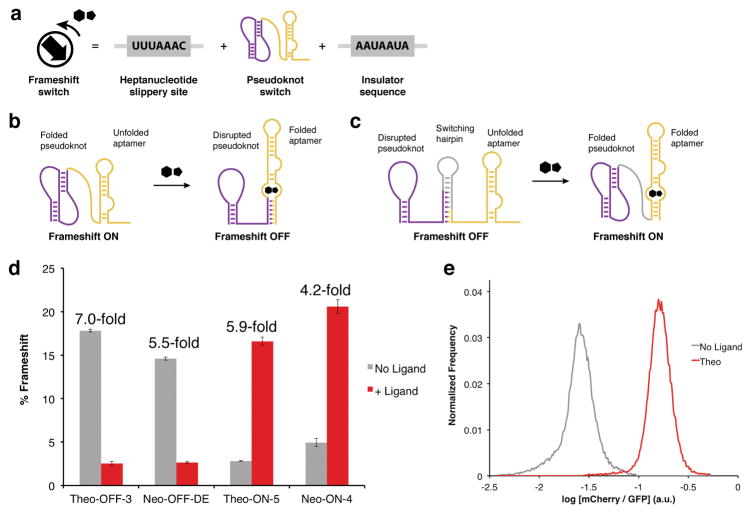
Figure 3.
Rational design of frameshift switches. (a) Architecture of frameshift switch devices. Each device contains a 5′ heptanucleotide slippery site, the pseudoknot switch, and a 3′ insulator sequence. (b) OFF-switch design. In the absence of ligand, the stimulatory pseudoknot (purple) is energetically dominant, producing high frameshift levels. Ligand binding induces aptamer (gold) folding, which disrupts the pseudoknot structure leading to lowered frameshift levels. (c) ON-switch design. A switching hairpin (gray) is installed to disrupt the pseudoknot and lower basal frameshifting. In the presence of ligand, the aptamer folds and destabilizes the switching hairpin, allowing the pseudoknot to re-fold and restore frameshift activity. (d) The ligand responsiveness of four −1 PRF switches assayed in the dual-FP reporter. Error bars represent the s.e.m. for n = 3 biological replicates derived from individual yeast transformants. Theophylline was used at a concentration of 40 mM; neomycin was used at a concentration of 550 μM. (e) Flow cytometry of yeast harboring the Theo-ON-5 switch in the absence and presence of theophylline (40 mM).
In the OFF-switch design (Fig. 3b), the RNA aptamer and FS-2 pseudoknot sequences overlap and thus compete for folding. In the absence of ligand, the active pseudoknot predominates and stimulates high −1 PRF activity. However, in the presence of ligand, the folded aptamer is stabilized by ligand binding energy and disrupts the FS-2 pseudoknot, resulting in lowered −1 PRF. We designed several constructs by varying the length and composition of the aptamer stems. As a general trend, we found that increasing the thermodynamic stability of the aptamer lowered frameshift activity (Supplementary Fig. 6). This simple approach led to the discovery of a high-performing theophylline OFF-switch that displays a 7-fold reduction in −1 PRF in the presence of ligand (Fig. 3d, Theo-OFF-3).
Rational optimization of aptamer stability did not result in adequate neomycin OFF-switch responsiveness (Supplementary Fig. 6). Therefore, we devised an in vivo directed evolution platform for optimizing switch performance that exploits the unique output of −1 PRF (Supplementary Fig. 7). Briefly, −1 PRF devices were inserted between the open reading frames of a Gal4 minimal DNA binding domain (BD) and minimal activation domain (AD)33. Overall, expression of Gal4-responsive genes in a yeast two-hybrid strain34 was a function of the Gal4(BD) to Gal4(BD-AD) ratio, which in turn was dictated by −1 PRF efficiency (Supplementary Fig. 8). We designed a library of neomycin OFF-switches and performed selection within the Gal4 yeast two-hybrid system, yielding several switches with improved performance (Supplementary Fig. 9). The best switch responded to neomycin with a 5.5-fold reduction in −1 PRF (Fig. 3d, Neo-OFF-DE). In addition to directed evolution applications, this Gal4 system could be applicable for small molecule regulated transcriptional activation.
To engineer ON-switches, a “switching hairpin” was introduced to compete with FS-2 pseudoknot folding and reduce basal −1 PRF levels. By design, structural rearrangements stimulated by ligand-aptamer recognition serve to destabilize the switching hairpin, leading to coincident refolding of the pseudoknot and restoration of −1 PRF activity (Fig. 3c). Several constructs were created to tune the relative stabilities of the ON and OFF states by varying the lengths and compositions of the aptamer stems and switching hairpins. These constructs were assessed using RNA secondary structure prediction (NUPACK35) and tested experimentally in the dual-FP assay to correlate hairpin and aptamer stability to switch activity (Supplementary Note 2 and Supplementary Figs. 10–11). With minimal optimization, this approach led to efficient ON-switches that respond to theophylline (5.9-fold) or neomycin (4.2-fold) (Fig. 3d, Theo-ON-5 and Neo-ON-4). Notably, for the Theo-ON-5 switch, the population of cells exposed to theophylline is >99% resolved from the population of untreated cells (Fig. 3e) due to stoichiometrically controlled outputs.
−1 PRF logic gates and phenotypic controllers
We envisioned exploiting translation reading frames by layering −1 PRF devices within individual mRNAs to create logic gates and phenotypic controllers. Logic gates are important genetic devices for executing cellular computation and programming biological systems. While diverse logic gate architectures have been reported19,36,37, most require the expression of multiple components to transduce small-molecule inputs into a gene expression output. We recognized the opportunity to construct highly condensed logic gates from a single mRNA transcript using −1 PRF switches (Fig. 4).
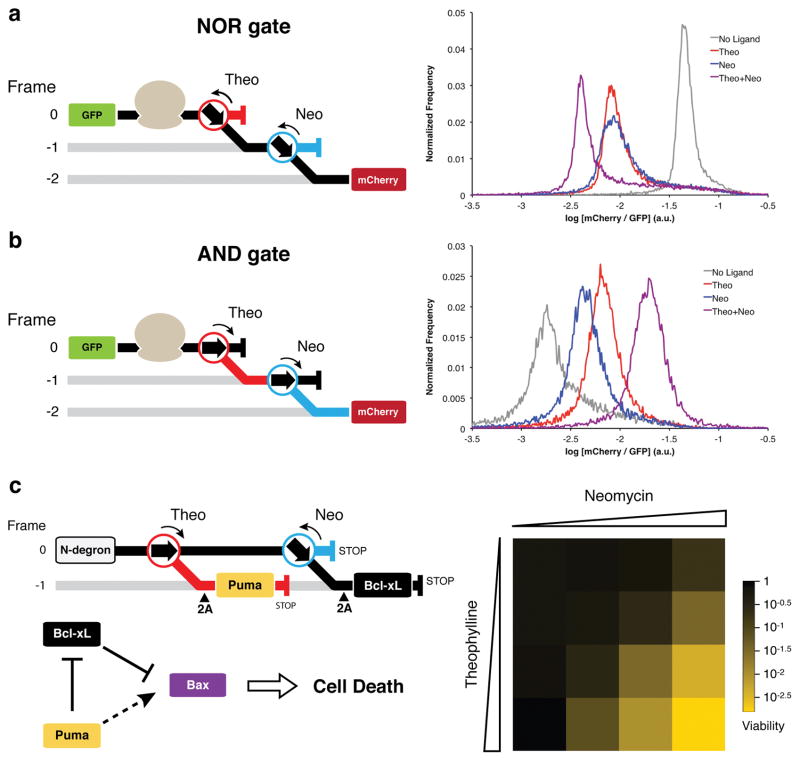
Figure 4.
Construction of logic gates and an apoptosis module with layered −1 PRF switches. For all constructs, translation in the absence of a ligand follows the black path; theophylline directs translation down the red path; neomycin directs translation down the blue path. (a) The NOR gate is composed of Theo-OFF-3 and Neo-OFF-DE switches. (b) The AND gate is composed Theo-ON-5 and Neo-ON-4 switches. For both gates, mCherry is encoded in the -2 frame with respect to GFP. Gate function was assessed within the dual-FP reporter in yeast by flow cytometry. Theophylline was used at a concentration of 40 mM; neomycin was used at a concentration of 550 μM. (c) The apoptosis module construct encodes Puma under the control of a theophylline-responsive ON-switch and Bcl-xL under the control of the Neo-OFF-DE switch. 2A peptides encoded 5′ to the Puma and Bcl-xL open reading frames cleave the functional proteins from the nonsense translation products of the switch devices and alternative reading frames. The latter products are targeted for degradation by an N-degron signal at the N-terminus of the polypeptide. Relative production of Puma and Bcl-xL controls the ability of Bax to induce cell death. Cells expressing Bax and the apoptosis module were grown in various concentrations of neomycin (0 μM, 44 μM, 165 μM, 715 μM) and theophylline (0 mM, 1 mM, 5 mM, 20 mM) and assessed for viability by plating efficiency, reported as the mean of three technical replicates.
First, a NOR gate (Fig. 4a) and an AND gate (Fig. 4b) were constructed using fluorescent protein reporters. Predictions of logic gate outputs based on individual switch activities were found to be in good agreement with the experimentally obtained results, demonstrating that the different frameshift switch devices function independent of one another and their context. Moreover, ON/OFF states are well distinguished by flow cytometry, particularly for the NOR gate. Though not demonstrated here, other more complex logic gates could be constructed by layering −1 PRF switches in alternative configurations within mRNAs (Supplementary Fig. 12).
To extend this concept to logic gates with phenotypically meaningful outputs, we designed an apoptosis module in yeast using mammalian Bcl-2 family proteins38. Though naturally absent from S. cerevisiae, heterologously expressed Bcl-2 family members retain their basic functions in this host39. Bax expression in S. cerevisiae has been shown to induce cell death through mitochondrial membrane permeabilization40. Moreover, co-expression of the pro-survival protein Bcl-xL protects yeast from Bax-mediated lethality39, consistent with its function in mammalian systems. A third subset of factors, termed the BH3-only proteins, is proposed to inhibit the pro-survival family members by direct interaction and perhaps activate Bax. Therefore, it has been proposed that cell fate is dictated by the relative expression of BH3-only and pro-survival proteins in a background of basal Bax expression38.
While previous studies in yeast have demonstrated that the BH3-only protein Puma enhances Bax-mediated lethality, co-expression of pro-survival Bcl-xL mitigated all lethal effects41. Thus, it is unclear to what extent Bcl-xL expression is required to rescue cells from Bax-induced cell death and how the relative production of Puma and Bcl-xL influence cell viability. Our −1 PRF expression system could offer insight into this process by precise ligand-dependent alteration of protein stoichiometry. We designed an expression construct that allows for the simultaneous control of the BH3-only protein Puma by a theophylline-responsive ON-switch and the pro-survival protein Bcl-xL by a neomycin-responsive OFF-switch (Fig. 4c). Using the small molecule ligands, we titrated the relative levels of Puma and Bcl-xL in a background of Bax expression and observed the effects on cell viability.
In the absence of ligands, cells were protected from Bax-mediated killing through expression of Bcl-xL. Decreasing Bcl-xL synthesis with neomycin had only a minor effect on cell viability at maximal concentrations (4-fold decrease). Treatment with theophylline alone to increase Puma production while maintaining high Bcl-xL had no influence on Bax-mediated killing, consistent with previous reports41. However, when cells were treated with both ligands, we observed a cooperative effect building up to a >300-fold decrease in cell viability at maximal concentrations of theophylline and neomycin (Fig. 4c). These results support the hypothesis that apoptosis is regulated by the relative balance between BH3-only proteins and pro-survival proteins42, and establish this system as an efficient AND gate controlled kill switch in S. cerevisiae.
DISCUSSION
Our results demonstrate that translation reprogramming can be co-opted and engineered for the fabrication of gene-regulatory devices in eukaryotes. Reprogramming mechanisms that utilize RNA are amenable to various directed evolution strategies and rational engineering with modular RNA components. Notably, our method applies a combination of these approaches to construct −1 PRF switches that control protein synthesis in yeast. While −1 PRF switches should find utility in higher eukaryotes, it is likely that new switches will need to be fashioned based on the particular properties of the biological setting of interest. Nonetheless, our general methodological approach should be applicable to designing −1 PRF switches that are optimized for performance in mammalian cells and other eukaryotic systems where frameshifting naturally occurs.
We employed an in vitro directed evolution strategy that selects for a complex functional phenotype within a cell lysate. Interestingly, some previous attempts to improve allosteric ribozymes by in vitro directed evolution resulted in devices that were non-functional in vivo18. This was attributed to differences between the in vitro and cellular environments. However, our in vitro selections were performed within a cell lysate, enriching for a function that requires direct interaction with the cell’s complex biochemical machinery. It is possible that this improved the likelihood that selection products would retain activity in vivo. Analogous to previously reported allosteric selections17, this mRNA-display directed evolution platform might also be adapted for the selection of ligand-responsive frameshift devices or other modes of translation reprogramming.
Ligand-responsive −1 PRF switches provide distinct advantages over other gene-control strategies. Since −1 PRF switches are based on modular RNA parts, it is feasible to scale the number of devices using alternative aptamer and frameshift components. The discovery of new RNA aptamers through SELEX and related approaches will be critical to this scaling effort. In comparison to other RNA devices, such as recently optimized allosteric ribozymes16, our first-generation −1 PRF switches demonstrate comparable fold-changes in gene expression but have the benefit of regulating multi-protein output stoichiometry. The utility of the latter feature is best highlighted by our apoptosis module, which displays a >300-fold change in cell viability using −1 PRF switches that respond to ligand with moderate 5- to 6-fold changes in activity. Interestingly, natural biological mechanisms may utilize a similar principle of stoichiometric control that provides uniform phenotypic response across heterogeneous cell populations despite relatively small fold changes in molecular response43. −1 PRF switches also perform robustly in various contexts, whether independently in the setting of a fluorescent protein reporter or a transcription factor, or in conjunction with other −1 PRF switches in the context of logic gates or an apoptosis module.
While these first-generation −1 PRF switches should find broad applicability, switch performance could be improved by increasing ligand sensitivity and fold activation. The ligand detection threshold of a thermodynamically controlled RNA switch will be fundamentally limited by the binding affinity of its aptamer. However, as has occurred for many natural riboswitches, it should be possible to optimize the conversion of ligand binding energy into productive conformational rearrangements so that −1 PRF switches respond to ligand concentrations closer to the Kd of their aptamers. Additionally, minimizing alternative conformations in the ON and OFF states, either through context optimization or exploration of other −1 PRF stimulatory elements, would likely improve the sensitivity and the dynamic range of these devices. The approaches outlined in this work, particularly the in vivo directed evolution strategies, establish a framework for improving −1 PRF switch performance. Overall, we believe that the tools and methodology developed here will bolster synthetic biology efforts to develop customized cellular programs and advance our capabilities to study and manipulate biological systems.
ONLINE METHODS
Nucleotide sequences used in this study are detailed in Supplementary Tables 2–5.
Frameshift stimulator library design
The starting library was synthesized (IDT) as oligonucleotide AVA107 encoding a modified PreQ1-class I riboswitch26 with 14 positions randomized in the region surrounding the ligand cavity44 (see Supplementary Fig. 1). This library has a theoretical diversity of 2.68 × 108 unique sequences. The selection construct for in vitro transcription was assembled from four oligonucleotides (AVA105, AVA106, AVA107 and AVA108) by PCR with Vent Polymerase (NEB).
In vitro selection for −1 PRF
The mRNA display protocol was carried out similar to previous reports30,45. Briefly, the DNA library was in vitro transcribed with T7 RNA polymerase and purified by denaturing polyacrylamide gel electrophoresis (8.5% Urea-PAGE). The resulting RNA was ligated to phospho-dA27dCdC-puromycin (TriLink Biotechnologies) by splint ligation30 with oligonucleotide AVA95 and T4 DNA ligase. The puromycin conjugated mRNA templates were purified from non-ligated RNA by 8.5% Urea-PAGE gel. 20 pmoles (1.2 × 1013 molecules) of mRNA display templates were translated at 30° C for 1 hour in 100 μL of 40% rabbit reticulocyte lysate (nuclease treated, Promega) supplemented with amino acids, Mg(OAc)2 (0.5 mM final), and KCl (100 mM final). After incubation at 30° C, the translation reaction was treated with 38 μL of puromycin salt mix (31.2 μL of 2.5M KCl + 6.8 μL 1M MgCl2) and incubated at room temperature for 15 minutes, then placed on ice. mRNA-peptide fusions were FLAG purified using anti-FLAG M2 Affinity Gel (Sigma) as described45, then purified with Ni-NTA resin (Qiagen) as described30. Following purification, mRNA-peptide fusions were buffer exchanged into reverse transcription buffer using 30,000-MW cutoff spin concentrators (Amicon). cDNA was reverse transcribed from purified mRNA with Superscript II (Life Technologies) for 50 minutes at 42° C (300 μL reaction containing: purified mRNA; 1 μM RT primer AVA95; 0.5 mM dNTP mix; 1X First-strand buffer; 10 mM DTT; 1 μL RNasin; 1 μL Superscript II Reverse Transcriptase). The reverse transcription reaction was spin-concentrated, then amplified with GoTaq polymerase (Promega) using primer pair AVA109·AVA108 in a 0.5 mL reaction to generate dsDNA for subsequent rounds of selection. Of note, primer AVA109 includes the entire 5′ segment of the selection construct leading up to the first in frame TAG stop codon in order to correct frameshift insertion mutations enriched in the previous round of selection.
Yeast dual-fluorescent protein reporter
The reporter plasmid was constructed by inserting the yEGFP open reading frame (amplified with primer pair AVA111·AVA112) between the GPD promoter and the open reading frame of a yeast-optimized mCherry46 (amplified from yEmRFP) flanked by a GPD terminator in a pRS425 backbone (high copy, LEU2 marker). NheI and AatII restriction sites were encoded between the GFP and mCherry open reading frames for replacement of the intervening sequence with a −1 PRF insert. To assay library −1 PRF activity, dsDNA products from the in vitro selection were PCR amplified in two steps (primer pairs AVA117·AVA118 and AVA119·AVA118) to add homology to the NheI/AatII digested dual-FP reporter plasmid. The library was cloned by in vivo gap repair into yeast strain Fy251 using high efficiency transformation as previously described47 and plated on selective medium (synthetic complete agar with leucine dropout containing 2% dextrose; SC-Agar-(Gluc) L-). Individual colonies were isolated and grown in liquid SC-(Gluc) L- media to mid-log phase and the −1 PRF efficiency was determined by comparing quantified GFP and mCherry fluorescence signals to a fusion protein control (calibrated to 100% frameshift). For analysis of the pooled selection products, the rescued transformation was grown in liquid SC-(Gluc) L- selection media for 3 days to enrich for transformants, then analyzed directly by flow cytometry (LSRII).
NGS of in vitro selection products
NGS was performed using the Illumina HiSeq platform (Columbia Genome Center). DNA was prepared for sequencing by PCR in two separate batches containing either five or nine variable positions at the start site of the HiSeq read to avoid identical base calls during the initialization phase of sequencing. The in vitro selection products were PCR amplified with primer pair AVA317·AVA322 or AVA318·AVA322. Both PCR products were then mixed and amplified with primer pair AVA319·AVA321 to add full HiSeq adaptor sequences. After sequencing, the raw fastq file was trimmed to the scaffold of interest (see Supplementary Fig. 3), and the copy number of each unique sequence was computed (step 1 of sequence analysis pipeline, see Supplementary Note 1).
Analysis of NGS for motif classification
The scaffold and its 14 variable positions define a restricted region of sequence space (2.68 × 108). We constructed a pseudoknot (PK) feature space based on a combination of the nucleotide constraints and our user-defined segment constraints, generating a set of 2,068 individual PK features (Supplementary Fig. 3). We enumerated the full set of unique sequences theoretically present in the initial library and calculated their compatibilities with the 2,068 PK features. The feature with the greatest amount of base pairing (including G·U) was associated to each sequence. PK compatibilities were similarly calculated for the sequences observed in the in vitro selection products. Highly enriched PK features could then be measured by comparing the two distributions. Of the top 10% of PK features by enrichment, those with the highest absolute representation in the selection library defined the broad motif categories. Within a set of PK-compatible sequences, a greedy clustering algorithm was used to divide the set of sequences into the final motifs. The modes of the different motif sets were further characterized in terms of the abundances and entropies of their immediate neighborhood in sequence space. In particular, the modes from the highest abundance motifs were assessed for single- and pairwise- nucleotide variant sensitivity, to reveal potential base pair position and tertiary interactions within a given PK geometry. See Supplementary Note 1 for further details, and Supplementary Data 1 for the motif table.
Rational design and in vivo characterization of −1 PRF switches
Switch constructs were evaluated using RNA secondary structure predictions and thermodynamic calculations (NUPACK and pKiss) as described in Supplementary Note 2, Supplementary Table 1, and Supplementary Data 2. Constructs were assembled from synthesized oligonucleotides (IDT). Switches were cloned into the dual-FP reporter by in vivo gap repair of NheI/SalI digested dual-FP reporter plasmid using the −1 PRF switch fragment and an mCherry PCR product containing homology to the switch. Colonies were grown overnight in SC-(Gluc) L- media, then used to seed SC-(Gluc) L- cultures (starting OD600 ~ 0.02) with or without the small molecule ligand. Cultures were grown to mid-log phase and measured for −1 PRF activity by quantification of GPF and mCherry fluorescence signals.
Gal4 selection system for in vivo directed evolution
The Gal4 selection plasmid was constructed by inserting a minimal Gal4 transcriptional activator between the ADH1 promoter and ADH1 terminator in a pRS425 vector backbone. Gal4 was PCR amplified from yeast genomic DNA in two pieces with primer pairs MS049·AVA155 (Gal4 minimal DNA biding domain, amino acids 1-147) and AVA158·MS050 (Gal4 minimal activation domain, amino acids 768-881). Between the Gal4 binding and activation domains, BamHI and HindIII restriction sites were encoded for replacement of the intervening sequence with a −1 PRF insert. The yeast two-hybrid background strain MaV203 (Invitrogen) was used for all growth assays and growth selections. Control −1 PRF sequences were cloned into the Gal4 selection vector by in vivo gap repair of the BamHI/HindIII digested plasmid −1 PRF inserts (two step PCR, first with AVA156·AVA162, then AVA161·AVA162). The Neo-OFF libraries were constructed by PCR assembly of oligonucleotides (Library 1: AVA345·AVA347, then AVA156·AVA346, then AVA161·AVA276; Library 2: AVA345·AVA348, then AVA156·AVA346, then AVA161·AVA276). This generated 2,048 combined theoretical variants. The library was cloned by in vivo gap repair into the Gal4 selection plasmid, and the rescued transformation culture was grown in SC-(Gluc) L- at 30 °C to enrich for transformants. Positive selection media (SC-(Gluc) HL-, 80 mM 3-AT, 10 mL) was inoculated with the library to a starting OD600 = 0.01 and grown for 60 hours at 30 °C to OD600 = 1.2. Then, SC-(Gluc) L- containing 500 μg/mg neomycin was inoculated with positive selection products and grown from OD600 = 0.1 to OD600 = 1.5 to allow for protein turnover. This culture was then used to inoculate counterselection media (SC-(Gluc) L-, 500 μg/mg neomycin, 0.05% 5-fluoroorotic acid) to a starting OD600 = 0.02 and grown for 84 hours at 30 °C to a final OD600 ~ 1.3. DNA was isolated from the counterselection culture and used as PCR template for amplification of the −1 PRF switches and cloning into the dual-FP reporter for assaying frameshift activity (AVA117·AVA357, then AVA119·AVA367, gap repaired with mCherry AVA358·AVA208 PCR product).
Logic gates
The NOR gate was assembled by fusion PCR of the Theo-OFF-3 switch fragment (AVA119·AVA406) and the Neo-OFF-DE switch fragment (AVA246·AVA247). The AND gate was assembled by fusion PCR of the Theo-ON-5 switch fragment (AVA119·AVA245) and the Neo-ON-4 switch fragment (AVA246·AVA247). Gate inserts were cloned by gap repair of the NheI/SalI digested dual-FP reporter plasmid using the gate PCR fragment and an mCherry PCR product (AVA248·AVA208). Colonies were grown overnight in SC-(Gluc) L- media, then used to seed SC-(Gluc) L- cultures (starting OD600 ~ 0.02) with or without the small molecule ligand. Cultures were grown to mid-log phase and analyzed by flow cytometry.
Apoptosis module and cell viability assay
pBM272-339648 encoding wild-type mouse Bax under the control of the Gal10 promoter (CEN plasmid, Ura) was obtained from Addgene. The human Puma open reading frame (BBC3, Gene ID: 27113) nucleotide sequence lacking the ATG start codon was codon optimized for yeast expression, manually recoded to remove stop codons from its +1 frame, and ordered synthesized (IDT). The nucleotide sequence for human Bcl-xL (BCL2L1, Gene ID: 598) was codon optimized for yeast expression and ordered synthesized (IDT). The N-degron signal (Ubiquitin-Arg-LacI(1–37)) was amplified from an existing plasmid with primer pair AVA462·AVA364. The full construct (Supplementary Table 4) was assembled by conventional molecular biology techniques and chromosomally integrated at the LEU2 locus in an Fy251 background strain giving AA01 (leu2Δ::LEU2-pTDH3-[Apoptosis Module]). This strain was then transformed with pBM272-3396, giving strain AA02. For control viability assays, cultures of AA02 were grown for 72 hours at 30 °C without Bax induction in synthetic complete media with uracil dropout containing 2% raffinose (SC-(Raf) U-) and either no ligand, theophylline (20 mM), neomycin (650 μg/mL), or both theophylline (20 mM) and neomycin (650 μg/mL). For assessing viability with Bax expression, cultures of AA02 were grown in synthetic complete media with uracil dropout containing 2% raffinose and 2% galactose (SC-(Gal) U-) with varying concentrations of theophylline (0 mM, 1 mM, 5 mM, or 20 mM) and neomycin (0 μg/mL, 40 μg/mL, 150 μg/mL, or 650 μg/mL) for 72 hours at 30 °C. After this time, the OD of each culture was measured, diluted to a standard cell density, and then plated at the appropriate dilution in triplicate on SC-Agar-(Gluc) U- plates. Colonies from each plate for the same condition were counted and averaged (Supplementary Table 6).
Code Availability
The software and resources for the analysis pipeline can be accessed at: https://github.com/szairis/frameshift.
Supplementary Material
Acknowledgments
We thank N. Dean (Stony Brook University, USA) for providing the yEmRFP plasmid encoding yeast-optimized mCherry. We thank R.L. Gonzalez for valuable discussions and feedback, and N. Ostrov for critical comments on the manuscript. This work was supported by funds from the US National Institutes of Health (NIH) (grants R01 GM090126 and R01 AI110794 to V.W.C.). S.Z. and R.R. were supported by the Center for Topology of Cancer Evolution and Heterogeneity (NIH U54 CA193313). A.V.A. was supported by a US National Institutes of Health F30 fellowship (F30CA174357). S.Z. was supported by a TL1 Precision Medicine fellowship (5TL1TR000082). Research reported in this publication was performed in the CCTI Flow Cytometry Core, supported in part by US National Institutes of Health award S10RR027050.
Footnotes
AUTHOR CONTRIBUTIONS
A.V.A. conceived the project, designed and performed experiments, analyzed data, and wrote the manuscript. A.J.L. performed experiments and analyzed data. S.Z. and R.R. developed custom software to analyze the NGS data. V.W.C. supervised the research and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gesteland RF, Atkins JF. Recoding: Dynamic Reprogramming of Translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 3.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkins JF, Gesteland RF. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer; New York: 2010. [Google Scholar]
- 5.Mountford PS, Smith AG. Internal ribosome entry sites and dicistronic RNAs in mammalian transgenesis. Trends Genet. 1995;11:179–184. doi: 10.1016/S0168-9525(00)89040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Felipe P, Hughes LE, Ryan MD, Brown JD. Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. J Biol Chem. 2003;278:11441–11448. doi: 10.1074/jbc.M211644200. [DOI] [PubMed] [Google Scholar]
- 7.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 8.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 9.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 10.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 11.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 12.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 13.Berens C, Thain A, Schroeder R. A tetracycline-binding RNA aptamer. Bioorg Med Chem. 2001;9:2549–2556. doi: 10.1016/s0968-0896(01)00063-3. [DOI] [PubMed] [Google Scholar]
- 14.Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 15.Klauser B, Atanasov J, Siewert LK, Hartig JS. Ribozyme-based aminoglycoside switches of gene expression engineered by genetic selection in S. cerevisiae. ACS Synth Biol. 2015;4:516–525. doi: 10.1021/sb500062p. [DOI] [PubMed] [Google Scholar]
- 16.Townshend B, Kennedy AB, Xiang JS, Smolke CD. High-throughput cellular RNA device engineering. Nat Methods. 2015;12:989–994. doi: 10.1038/nmeth.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koizumi M, Soukup GA, Kerr JN, Breaker RR. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat Struct Biol. 1999;6:1062–1071. doi: 10.1038/14947. [DOI] [PubMed] [Google Scholar]
- 18.Link KH, et al. Engineering high-speed allosteric hammerhead ribozymes. Biol Chem. 2007;388:779–786. doi: 10.1515/BC.2007.105. [DOI] [PubMed] [Google Scholar]
- 19.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ausländer S, et al. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat Methods. 2014;11:1154–1160. doi: 10.1038/nmeth.3136. [DOI] [PubMed] [Google Scholar]
- 21.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci USA. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galloway KE, Franco E, Smolke CD. Dynamically reshaping signaling networks to program cell fate via genetic controllers. Science. 2013;341:1235005. doi: 10.1126/science.1235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brierley I. Ribosomal frameshifting on viral RNAs. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 24.Jacks T, Madhani HD, Masiarz FR, Varmus HE. Signals for ribosomal frameshifting in the rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov IP, Gesteland RF, Atkins JF. Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 2000;28:3185–3196. doi: 10.1093/nar/28.17.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu CH, Luo J, Iwata-Reuyl D, Olsthoorn RCL. Exploiting preQ1 riboswitches to regulate ribosomal frameshifting. ACS Chem Biol. 2013;8:733–740. doi: 10.1021/cb300629b. [DOI] [PubMed] [Google Scholar]
- 27.Hsu HT, Lin YH, Chang KY. Synergetic regulation of translational reading-frame switch by ligand-responsive RNAs in mammalian cells. Nucleic Acids Res. 2014;42:14070–14082. doi: 10.1093/nar/gku1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frankel A, Roberts RW. In vitro selection for sense codon suppression. RNA. 2003;9:780–786. doi: 10.1261/rna.5350303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu R, Barrick JE, Szostak JW, Roberts RW. Optimized synthesis of RNA-protein fusions for in vitro protein selection. Methods Enzymol. 2000;318:268–293. doi: 10.1016/s0076-6879(00)18058-9. [DOI] [PubMed] [Google Scholar]
- 31.Cho G, Keefe AD, Liu R, Wilson DS, Szostak JW. Constructing high complexity synthetic libraries of long ORFs using in vitro selection. J Mol Biol. 2000;297:309–319. doi: 10.1006/jmbi.2000.3571. [DOI] [PubMed] [Google Scholar]
- 32.Weigand JE, et al. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 34.Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zadeh JN, et al. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 36.Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying genetic logic gates. Science. 2013;340:599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 37.Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–253. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 39.Sato T, et al. Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc Natl Acad Sci USA. 1994;91:9238–9242. doi: 10.1073/pnas.91.20.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Priault M, Camougrand N, Kinnally KW, Vallette FM, Manon S. Yeast as a tool to study Bax/mitochondrial interactions in cell death. FEMS Yeast Res. 2003;4:15–27. doi: 10.1016/S1567-1356(03)00143-0. [DOI] [PubMed] [Google Scholar]
- 41.Gallenne T, et al. Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J Cell Biol. 2009;185:279–290. doi: 10.1083/jcb.200809153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 43.Lee REC, Walker SR, Savery K, Frank DA, Gaudet S. Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol Cell. 2014;53:867–879. doi: 10.1016/j.molcel.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang M, Peterson R, Feigon J. Structural insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 45.Seelig B. mRNA display for the selection and evolution of enzymes from in vitro-translated protein libraries. Nat Protoc. 2011;6:540–552. doi: 10.1038/nprot.2011.312. [DOI] [PubMed] [Google Scholar]
- 46.Keppler-Ross S, Noffz C, Dean N. A new purple fluorescent color marker for genetic studies in saccharomyces cerevisiae and candida albicans. Genetics. 2008;179:705–710. doi: 10.1534/genetics.108.087080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirakitikulr N, Ostrov N, Peralta-Yahya P, Cornish VW. PCRless library mutagenesis via oligonucleotide recombination in yeast. Protein Sci Publ Protein Soc. 2010;19:2336–2346. doi: 10.1002/pro.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gross A, et al. Biochemical and genetic analysis of the mitochondrial response of yeast to BAX and BCL-X(L) Mol Cell Biol. 2000;20:3125–3136. doi: 10.1128/mcb.20.9.3125-3136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.