Abstract
Colorectal cancer (CRC) is the second most common cancer in women and the third most common in men globally. CRC arises from one or a combination of chromosomal instability, CpG island methylator phenotype, and microsatellite instability. Genetic instability is usually caused by aneuploidy and loss of heterozygosity. Mutations in the tumor suppressor or cell cycle genes may also lead to cellular transformation. Similarly, epigenetic and/or genetic alterations resulting in impaired cellular pathways, such as DNA repair mechanism, may lead to microsatellite instability and mutator phenotype. Non-coding RNAs, more importantly microRNAs and long non-coding RNAs have also been implicated at various CRC stages. Understanding the specific mechanisms of tumorigenesis and the underlying genetic and epigenetic traits is critical in comprehending the disease phenotype. This paper reviews these mechanisms along with the roles of various non-coding RNAs in CRCs.
Keywords: Colorectal cancer, chromosomal instability, microsatellite instability, non-coding RNA mismatch repair
Introduction
Globally, colorectal cancer (CRC) is the second most common cancer in women (614, 000 cases per year) and the third most common in men (746, 000 cases per year). The incidence rates are higher in more developed countries (737, 000 cases per year) than in less developed ones (624, 000 cases per year). However, mortality is higher in the latter (52% of total deaths), which indicates poor survival. In 2015, the GLOBOCAN online analysis tool has predicted 61, 228 new CRC cases for Asia. Accordingly, 25, 816 of these cases are associated with people who are less than 65 years old1.
Mechanisms of carcinogenesis
CRCs can arise from one or a combination of three different mechanisms, namely chromosomal instability (CIN), CpG island methylator phenotype (CIMP), and microsatellite instability (MSI). According to Fearon2, the classical CIN pathway begins with the acquisition of mutations in the adenomatous polyposis coli (APC), followed by the mutational activation of oncogene KRAS and the inactivation of the tumor suppressor gene, TP53. Aneuploidy and loss of heterozygosity (LOH) are the major players in CIN tumors, which not only constitute most of the sporadic tumors (85%) but also involve familial adenomatous polyposis cases associated with germline mutations in the APC gene3. The CIMP pathway is characterized by promoter hypermethylation of various tumor suppressor genes, most importantly MGMT and MLH1. This hypermethylation is often associated with BRAF mutation and microsatellite instability4. The MSI pathway involves the inactivation of genetic alterations in short repeated sequences. This activation occurs in CRCs in DNA mismatch repair (MMR) genes, and is a hallmark condition in familial Lynch syndrome (LS), which also appears in ~15% of the sporadic CRC cases. In addition, the hypermethylation of the MMR genes may lead to MSI. This mechanism is often associated with the CIMP pathway5. MSI tumors are often associated with proximal colon and poor differentiation but better prognosis6. Moreover, the three mechanisms often overlap in CRC. Figure 1 illustrates important molecular, genetic, and epigenetic changes with respect to disease progression. A detailed account of which is provided in the sections that follow2,7-9.
Figure1.
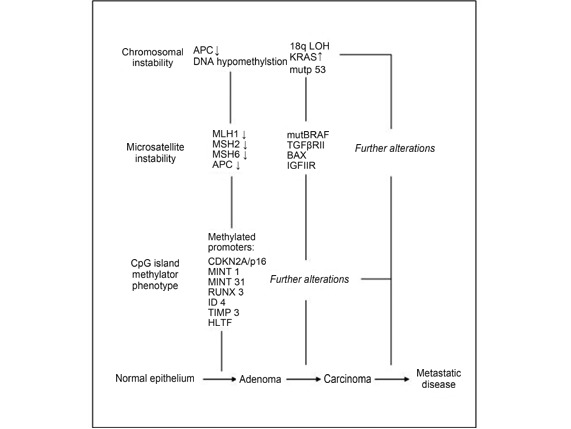
Important molecular, genetic and epigenetic changes with respect to disease progression
Chromosomal instability
Chromosomal instability is associated with 65%-70% of sporadic CRCs. This pathway comprises aneuploidy, which is an imbalance in the chromosome number, and LOH10. Defects in chromosomal segregation, DNA damage repair, and telomere function along with specific mutations in certain oncogenes and tumor suppressor genes may be responsible for such instability. The mechanisms underlying genetic instability and other causative mutations in certain genes are reviewed elsewhere10. This review discusses updated findings relevant to these mechanisms.
Aneuploidy arises because of defects in the mitotic checkpoint, which cause chromosome mis-segregation. Mutational upregulation or downregulation of various mitotic checkpoint players, such as hRod, hZwilch, hZw10, Ding, budding uninhibited by benzimidazoles (Bub) R1, centromere-associated protein E (CENP-E), and mitotic arrest deficient (MAD) 1, can result in CIN11-15. A deleterious increase in aneuploidy may also lead to cell death. Other centromere protein genes, such as CENP-A and CENP-H, may also be overexpressed in CRCs, which leads to mis-localizations on non-centromeric chromatin positions16,17.
The centrosome-associated kinases, Aurora (AURK) and Polo-like (Plk), are also implicated in CRCs. AURKA overexpression causes arrested mitosis and multi-nucleation. This kinase also has a positive association with the degree of instability in CRCs18and activates Plk1 overexpressed in 73% of primary CRCs and associated with poor prognosis19,20. Similarly, Aurora-B regulates histone modification, chromosome segregation, and cytokinesis. Aurora-B is also overexpressed and associated with advanced stage tumors21. The overexpression of Aurora-C prevents the activation of the mitotic checkpoint leading to tumorigenesis22.
Disruptions in the DNA damage pathway and excess telomere breakage can lead to chromosomal instability. The damaged DNA is repaired by four mechanisms, namely base excision repair (BER), double-stranded break repair (DSBR), mismatch repair (MMR), and nucleotide excision repair (NER). The polymorphisms in BER-associated genes XRCC1, OGG1, and MUTYH have been correlated with reduced oxidative DNA damage repair efficiency in CRC23. Similarly, the MRE11 gene involved with the DSBR contributes to genomic instability11. DNA damage can activate two different signaling pathways, namely ataxia telangiectasia mutated-checkpoint kinase 2 (ATM-Chk2) and ataxia telangiectasia and Rad3-related-checkpoint kinase 1 (ATR-Chk1)24. Inactivating mutations in Chk2 along with AURKA overexpression results in increased microtubule assembly rates as noted in 73% of CRCs25. Excess telomere breakage is responsible for the transition from adenoma to carcinoma in CRCs26. A comparison with adjacent normal tissues has also shown that shorter chromosomes are found in both adenomatous polyps and carcinomas27. These results imply that telomere shortening initially causes instability. However, mutations in the APC gene and an increase in the telomerase activity drive metastasis.
Accordingly, LOH also leads to CIN. LOH is the loss of one of the parental alleles, which is caused by mitotic non-disjunction, recombination between two homologous or non-homologous chromosomes, or a deletion or gene conversion event. In CRCs, LOH is frequently observed in at least five different loci (i.e. 1, 5, 8, 17, and 1828). Previous studies have linked various CRC forms with losses at 1p, 4q, 5q, 8p, 9q, 11q, 14q, 15q, 17p, 18p, and 18q. These findings are reviewed elsewhere29. Mutations are also observed in these locations. Inactivating the mutations in the tumor suppressor APC gene (5q21) is considered to be a carcinogenesis-initiating event2. This process is usually followed by the activation of KRAS mutations (12p12). This step only promotes tumor progression in combination with the APC mutations2,30. Mutations in other genes, such as TP53 (17q13), PIK3CA (3q26), and TGF-Î (3p22), are also acquired31,2,32-34. These carcinogenesis-promoting mutations are further described in the sections that follow.
Mutational landscape in chromosomal instability
Mutations in the APC activate the Wnt signaling pathway by increasing Î-catenin levels. Î-catenin is translocated to the nucleus and enhances the transcription of various oncogenes with T-cell factor (TCF) transcription factors35. High Î-catenin levels are noted in gastrointestinal tumors36.
APC inactivation may occur through germline and somatic mutations or the hypermethylation of its promoter. Germline mutations often occur in FAP. Somatic mutations occur in 72% of sporadic tumors, 30%-70% of sporadic adenomas, and 5% of dysplastic aberrant crypt foci (ACF). Hypermethylation-led inactivation occurs in about 12% of the primary carcinomas and adenomas37-42.
Around 75% of CRCs have mutations or LOH in the APC gene. Most of these mutations are clustered in the mutation cluster region between codons 1282 and 1581. However, a complete inactivation is not required. Mutations sufficient for tumorigenesis differ between the proximal and distal CRCs43. The effects of APC restoration in mice are demonstrated on tumor regression by the conversion of cancer cells back to normal, which indicates a similar possibility in humans44. Mutations in other genes of this pathway, particularly in Î-catenin, may also lead to CIN. These mutations are found in 48% of CRCs without APC mutations45. Î-catenin activates a set of 162 Wnt pathway target genes in a colon cancer cell line. However, no conclusions could be drawn for their effect on disease prognosis46.
Point mutations in codons 12, 13, and 61 of exons 2 and 3 of KRAS activate the enzyme increasing RAS signaling. This enzyme is mutated in 30%-40% of CRCs and 60%-90% of hyperplastic or non-dysplastic ACF47,48. Mutations in KRAS are not precursory unlike APC mutations48. Mutations in codon 12 have higher chances for lymph node metastasis associated with the advanced form of the disease49. The activated RAS further activates the Raf-MEK-ERK and PI3K/AKT/PKB pathways or Ral small GTPases10. PI3K is first activated by Ras and then by the receptor tyrosine kinases as apparent in the incomplete inactivation of PI3K by KRAS knockdown50. However, most CRC cases with PI3K mutations also carry mutations in KRAS51. The activated PI3K further activates AKT1 and AKT2, which then enhances tumor growth by promoting epithelial to mesenchymal transition (EMT)52,53. Loss-of-function mutations in PTEN, which is a tumor suppressor and an antagonist of the PI3K/AKT pathway, induce AKT-regulated metastasis in CRCs54. The MEK/ERK and PI3K/AKT pathways often converge to activate a cap-dependent translation through survivin knockdown, which can inhibit metastasis55. Therefore, targeting both pathways together is more clinically effective56.
The tumor suppressor transcription factor 53 (TP53) is located on chromosome 17, which is activated under stress. TP53 targets cell cycle inhibitors such as GADD45 and 14-3-3 and pro-apoptotic factors including BAX, KILLER (DR5), FAS (APO1), and PIG357,58. However, p53 is dysfunctional in the majority of human tumors. Certain tumors show a gain-of-function mutation in p53, which results in mutated proteins, notably the mutp53 isoform. This isoform causes chronic transcription factor NF-ÎB activation in mice models, which enhances inflammation and accelerates tumorigenesis that finally result in invasive carcinoma59. This activation is independent of the loss of wild-type p53. Mutp53 may also inactivate the tumor suppressor RasGAP disabled 2 interacting protein, which makes the cancer cells more responsive toward inflammatory cytokines. Paradoxically, this increases the cancer cells' invasive behavior but reduces their aggressiveness60. The transactivation domain of p53 under normal conditions is bound and ubiquitinated by MDM2, E3-ubiquitin ligase, and MDM4. MDM2 can also bind and inactivate the mutp53 isoform. A comparison with the adjacent normal tissues shows that mRNA and protein overexpression of the spliced isoform MDM2-B is observed in CRCs. This overexpression is mainly correlated with the mutp53 protein as MDM2-B binds to MDM2, thereby allowing mutp53 accumulation in CRC61. The loss-of-function of p53 is reported in 44.9% of colorectal adenoma and 42.22% of single and 43.75% of multiple primary CRCs62. The downregulation of p53 via IL-6-dependent rRNA transcription enhancement leads to the development of EMT specific changes that are also seen in ulcerative colitis patients63.
Cyclooxygenase-2 (COX-2) is overexpressed in 43% of adenomas and 86% of carcinomas64. The deletion of a single allele causes 34% reduction, whereas a complete knockdown causes 86% reduction in the number of intestinal polyps in APC716 knockout mice65. Increased levels of both COX-2 and prostaglandin E2 (PGE2), which is the enzymatic product of COX-2, are found in CRCs66-67. Another study found 51 SNPs in HPGD, SLCO2A, and ABCC4 aside from Cox-2, which code for 15-PGDH (Cox-2 antagonist), MRP4 (Cox-2 transporter), and PGT (Cox-2 transporter), respectively. Seven of these SNPs are important. Four are validated for gene-gene interaction68.
The loss of 18q chromosomal region has been correlated with poorer survival rate in stage II CRCs69. The region carrying the deleted in colorectal cancer gene (a dependence factor) is more frequently lost in advanced cancers70. The mutations in similar to mothers against decapentaplegic (SMAD) homologproteins, namely SMAD2, SMAD3, SMAD4 and SMAD7, which are regulators of TGF Î signaling, have been associated with CRC71-73. SMAD 4 loss predicts worse prognosis for fluorouracil-based therapies74.
Microsatellite instability
Microsatellite instability occurs because of inactivating mutations in the DNA mismatch repair genes that are responsible for correcting DNA replication errors. The important components of the DNA mismatch repair system are ATPases hMSH2, hMSH6, hMSH3, hMLH1, hPMS2, hPMS1, and hMLH375-80. The germline mutations that may render these proteins dysfunctional can predispose to cancer as in the case of LS81. MSI is found in 15% of colorectal cancers, with only 3% associated with LS. The rest are sporadic cases caused by the hypermethylation of the MLH1 gene promoter82.
In 1997, the National Cancer Institute held a workshop where a five-marker MSI panel was validated. This panel included two mononucleotide markers, namely BAT25 and BAT26, and three dinucleotide markers, namely D5S346, D2S123, and D17S250. Tumors with instability in ≥30% of markers are called MSI-high (MSI-H). Those with instability in <30% are called MSI-low (MSI-L), while those without microsatellite instability are called MSI stable (MSS)83. Mutations in MLH1, MSH2, MSH6, and PMS2 have been associated with a risk of developing LS. By the age of 70, an estimated risk for developing LS of 38% and 31% is observed in males and females, respectively. An additional cumulative risk of 41%, 48%, and 12% for mutated hMLH1, MSH2, and MSH684, respectively, have been found.
More than 1, 500 germline variants have been found in the MMR genes along with promoter methylation, somatic deletions, or point mutations85-87. The germline hypermethylations in MLH1 and MSH2 may also increase susceptibility88-90. Furthermore, 3' deletions in EPCAM, which is a gene upstream of MSH2, encompass the termination signal, thereby reading through MSH2 and silencing it in colonic tissues. Moreover, a correlation between MSH2 promoter methylation and EPCAM deletion is observed in both CRC tissues and adjacent normal ones91,92. Large variations are also observed in MLH1 and MSH2 genes although they are not detected by conventional exon-specific methods such as screening. Deletions have been confirmed in 27% of patients; 12% of these deletions belong to those lacking MLH1 and 56% to those lacking the MSH2 expression93. A 10Mb inversion has been identified in MSH2 in samples lacking the MSH2 protein94. Other related genes must be tested in a similar manner.
The somatic mutations in the MMR genes and in EPCAM, POLE, and POLD1 have also been associated with CRCs. A recent study has found somatic mutations in at least two of these genes in almost 70% of patient tumors95. In another study, two acquired mutations in MLH1 and MSH2 have been observed in 52% (13/25) of the patients studied96.
The hypermethylation of the MLH1 promoter in MSI-H sporadic CRCs is found in 83%-100% of tumors97-100. The same finding is obtained in 15% of LS cases, which makes differentiation from sporadic CRCs difficult101. In such cases, the V600E mutation in v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) gene eliminates the possibility of LS102.
CpG island methylator phenotype
Global DNA hypomethylation and localized promoter hypermethylation are common epigenetic events that occur in cancer. Hypomethylation refers to a marked global decrease in methylation on cytosine bases103-106 that is observed in hyperplastic and adenomatous polyps and carcinomas107,108. Hypomethylation in the repetitive DNA sequences, such as in satellite regions, can lead to genomic instability. Furthermore, loss of imprinting or promoter demethylation could reactivate the retrotransposons. The demethylation of the long interspersed nuclear element- promoter has been suggested as an early event. However, demethylation has also been observed in the normal colonic mucosa of the same patients109-111. The loss of imprinting of insulin-like growth factor 2 (IGF2) is seen in almost 40% of CRC tumors, which leads to microsatellite instability in younger patients112,113.
The CpG Island (CGI) hypermethylation in the promoter region results in the transcriptional inactivation of genes that have tumor suppressive roles or are involved in the cell cycle114. Mutations in the BRAF gene appear to be an early event in the CIMP tumors. The BRAF V600E mutation is strongly correlated with MLH1 hypermethylation and has a frequency of 20.3% in unselected and 18.7% in sporadic cases103,116. Another study has confirmed MLH1 hypermethylation in 80% of MSI-H sporadic CRC, with loss of expression and without a known germline mutation in the MMR genes116.
The hypermethylation of other gene promoters (e.g. CDKN2A) has led to the development of a commonly used five-marker gene panel. This panel includes MLH1, CDKN2A, methylated in tumors 1 (MINT1), MINT2, and MINT31 and has been extensively studied102,114-117. This classic panel is used to term tumors as CIMP-high (CIMP-H) if two or more of these promoters are hypermethylated. A new panel has also been designed from 195 different CIMP markers comprising CACNA1G, IGF2, SOCS1, RUNX3, and NEUROG1. Using which, CIMP positive tumors are defined as those with three or more of these panel gene promoters methylated4. Studies comparing the two panels show various results. The new panel accurately predicts CIMP1 (positive) but not CIMP2 (CIMP low) tumors118. Another study has concluded that the new panel underperformed in determining the clinicopathological features of the tumors. Accordingly, the classic panel could better predict the clinical outcomes119. However, another study has tested eight markers. Five of these markers belong in the old panel. MLH1, CDKN2A, and CRABP1 have been added. Tumors with six or more methylated promoters are identified as CIMP-H. MLH1, RUNX3, IGF2, and CACNA1G have been considered to be the most specific and sensitive out of these eight markers. This set creates yet another panel120. A recent study has also combined markers with different functions and analyzed their methylator phenotype including DNA repair gene MGMT, tumor suppressors (i.e. CDKN2A, HLTF, GATA5, ID4, and TSLC1), metastasis suppressors (i.e. CDH4, CDH13, and TIMP3), apoptosis-related genes (i.e. CACNA1G, HRK, and RSASF1A), and angiogenesis inhibitors (i.e. TSP1). This study classifies the carcinomas as CIMP-H, CIMP-low (CIMP-L), and CIMP-normal (CIMP-N) if more than six, less than six, or none of the genes are methylated, respectively121.
Hypermethylated promoters are associated with BRAF mutations. A total of 759 hypermethylated regions are found. Accordingly, 96% of these regions occur in BRAF mutant tumors. Out of these, 229 regions are localized in the promoter regions enhancing five different pathways, namely the Wnt signaling, hedgehog signaling, bZip transcription regulation, PI3 kinase, and IGF-protein kinase B signaling pathways122. Early Wnt signaling activation has been attributed to APC mutations in CIN tumors. However, promoter hypermethylation of the Wnt antagonists suggests a role in the Wnt activation at later stages. Hypermethylation is noted in seven gene promoters in the normal to adenoma transition, and in four of these seven genes from adenoma to carcinoma123.
Similarly, ten eleven translocation 1 (TET1) methylation is an early event linked to BRAF mutations in CIMP+ tumors and polyps. The TET family proteins regulate demethylation by catalyzing the conversion of 5-methylcytosine to 5-hydroxymethylcytosine. Consequently, the inactivation results in hypermethylation leading toward CIMP124.
The aberrations in chromatin-remodeling genes, such as ATP-dependent chromatin remodelers, chromodomain helicase 7 (CHD7) and CHD8, may also be associated with CIMP tumors. These mutations may lead to chromatin structure modification and deregulation, which contributes to CIMP125.
Clinical implications of the disease molecular mechanisms in CRC
CIN, MSI, and CIMP often overlap in molecular tumor subtypes, which have considerable prognostic implications126. In one study, tumors were initially classified as MSI or MSS tumors based on 509 CRC cases. The latter group was further classified as CIN-only, CIMP-only, CIN+CIMP, and triple negatives. As expected, MSI tumors had the lowest frequency for APC and KRAS mutations, the second lowest for p53 mutations, and the highest for BRAF V600E mutations. By contrast, CIN-only tumors showed the highest frequency for p53 mutations and the lowest for BRAF V600E mutations.
The patient survival outcome was recently associated with specific molecular subtypes classified based on the MSI, CIMP, BRAF-mutation, and KRAS-mutation status. The MSI-H tumors (types 1 and 5) had the highest five-year disease specific survival (89.5% and 93.1%), followed by the MSI-L/MSS tumors (type 4; 82.5%) without CIMP or BRAF and KRAS mutations, and the tumors with only KRAS mutations (type 3; 72.4%). The tumors with CIMP and BRAF mutations had the worst survival (type 2; 49.2%)127. CIMP was associated in another study with poor disease free survival and overall survival rate irrespective of the MSI status128. Another six-subtype classification (C1-C6) was proposed based on the mutational landscape. The C1, C5, and C6 tumors frequently had chromosomal instability, TP53 mutations, and were distally located lacking the mutator phenotype. The other three subtypes had proximally located tumors often associated with CIMP. Furthermore, C2 had BRAF mutations and deficient MMR, while C3 had KRAS mutations129.
Classifying tumors according to the particular mutations present is necessary in determining the treatment regimen to be offered. A combination of either 5-fluorouracil (5-FU) or capecitabine is usually offered with irinotecan or oxaliplatin as the main line of treatment. Targeted therapies such as vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR) inhibitors have recently been found effective as both first and second lines of treatment in CRC. However, evidence suggests that anti-EGFR treatment is effective only in tumors lacking codon 12 and 13 mutations in KRAS. The treatment may even be detrimental when used with oxaliplatin in patients with KRAS-mutated tumors as shown in clinical trials130-134. Accordingly, anti-EGFR inhibitors are contraindicated in patients with mutant KRAS metastatic disease as per the guidelines of the American Society for Clinical Oncology135. The BRAF V600E mutation correlated with poor disease specific survival also confers resistance to anti-EGFR therapies even in the presence of wild-type KRAS127,136,137. Similarly, the presence of microsatellite instability has been shown to be predictive of failure of the standard, first-line 5-fluorouracil treatment138. Other genetic and epigenetic biomarkers also have significant implications for CRC diagnosis and treatment. These biomarkers have been previously described in detail in the literature139.
Non-coding RNAs in colorectal cancer
Only 2% of the human genome comprises protein coding genes. The remaining 98% region is transcribed into non-protein-coding RNAs. A variety of RNAs such as long non-coding RNA (lncRNA), microRNA (miRNA), small interfering RNA (siRNA), and piwi-interacting RNA (piRNA) are present in eukaryotes, which are mainly classified depending on their sizes and functions.
MicroRNAs
MicroRNAs are small (20-25 nucleotides), single-stranded, non-coding RNAs that negatively regulate gene expression140. They exert action by binding to the 3'UTR sequence of a target mRNA transcript. Binding is done by base-pairing to a partially complementary region, which leads to either transcript degradation or transcriptional inhibition 140,141. MicroRNAs can act either as tumor-suppressing miRNAs inactivating the oncogenes or as oncogenic miRNAs (oncomiRs) inactivating the tumor suppressor genes in cancer142,143. MicroRNA expression is frequently altered in cancers as they lie in unstable genomic regions. The miRNAs shown to be particularly important in CRCs are discussed in the sections that follow.
MiR-143 and miR-145
MiR-143 and miR-145 are concomitantly downregulated in most CRCs as both are found on 5q33. Their downregulation is an early event and occurs even before the APC gene aberrations144. The two miRNAs function as oncosuppressors in a coordinated fashion, that is, they either target the same genes or different genes regulating the same pathway145. MiR-143 expression is inversely related to KRAS expression and has inhibitory effects on KRAS in Lovo cells146,147. IGF-I receptor is another miR-143 target, and the overexpression of which inhibits proliferation, angiogenesis, and migration through the PI3K/AKT/HIF-1/VEGF pathway. MiR-143 overexpression also increases sensitivity to the chemotherapeutic drug, oxaliplatin148. MiR-143 and -145 also target cluster of differentiation 44 (CD44), Kruppel-like factor 5 (KLF5), and BRAF.
Lethal-7 (let-7) family and miR-18a*
Lethal-7 (let-7) family and miR-18a* have tumor suppressive roles influencing the RAS pathway. The let-7 expression is negatively correlated with RAS and c-myc expression149 and hypothesized to regulate the KRAS expression through TP53. However, no such association has been proven150. An increase in the let-7i expression is observed in relation to polyamine depletion, suggesting a role of polyamines in regulating the let-7 family. The high mobility group A2 (HMGA2), which is another let-7 target gene, is downregulated in a polyamine-dependent manner151. MiR-18a* also negatively regulates the KRAS gene. The KRAS-dependent increase in cell proliferation and anchorage dependence is observed in colon adenocarcinoma HT-29 cells transfected with anti-miR-18a*152.
MiR-200 family
The miR-200 family has a metastasis inhibitory role via the regulation of ZEB1 and ZEB2, which are E-cadherin repressors. MiR-200 downregulation is seen in invading cells of adenocarcinoma cells, which indicates a strategic decrease in EMT. However, its expression is restored when regaining the epithelial phenotype153. The miR-200c/141 cluster is also overexpressed in liver metastasis when regaining the epithelial phenotype154. MiR-200b indirectly upregulates the KRAS gene by repressing its inhibitor (i.e. PTPN12). Notably, the chemotherapeutic drug 5-FU upregulates miR-200b155.
MiR-34 family
The miR-34 family prevents metastasis by inhibiting the SNAIL expression, which is an EMT-inducing transcription factor156. MiR-34a repression is required for interleukin-6 (IL-6) induced metastasis. The IL-6 receptor (IL-6R) mediates the activation of STAT-3 transcription factor by IL-6. This activation represses miR-34a, thereby facilitating EMT. However, the activation of p53 results in the inhibition of IL-6R, which is a direct miR-34a target157. Controversially, mutation rs4938723 in the promoter region of primary miR-34b/c is associated with a decreased CRC risk158. The study has several limitations. Therefore, a more thorough analysis of the mutation on the transcriptional and translational levels can unravel the mechanisms involved.
MiR-17-92 cluster
The miR-17-92 cluster includes miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a, and miR-92a-1. This cluster is induced by transcription factors c-myc and E2F3159. All six miRNAs are significantly overexpressed in colorectal tumors. The higher expression is noted in adenocarcinoma, which indicates a role in adenoma to adenocarcinoma progression. A copy number gain at the locus is associated with the overexpression of these miRNAs except for miR-18a.
Impact of mutations
Particular CRC mutations may also regulate microRNA expression. For example, inactivating mutations in the APC increases miR-135b expression by stabilizing Î-catenin160. Moreover, epigenetic changes (e.g. DNA methylation) result in the downregulation of miRNAs. Three of the five downregulated tumor-suppressing miRNAs are restored in the CRC cell lines after treatment with DNA methyltransferase (DNMT) and histone deacetylase (HDAC) inhibitors. A comparison with the adjacent normal mucosa shows higher methylation observed in primary tumors. The miR-9-1 methylation is associated with lymph node metastases161. In another study, miR-9-1 and -34c have significantly reduced expression, while miR-34b has also been hypermethylated along with these two microRNAs162. Several SNPs in the genes coding for miR-146a, miR-196a2, miR-149, and miR-499 are tested. A significantly reduced expression of miR-499 is observed with the GG genotype for rs3746444G>A.
Therapeutic biomarkers
As previously reviewed, microRNAs serve as potential biomarkers for therapeutic outcomes163. These factors are important in predicting the response to particular therapies. MicroRNAs act as positive biomarkers indicating sensitivity to therapy or as negative biomarkers signifying resistance to treatment and assisting in choosing the appropriate treatment regimen (Table 1)164-179.
Table1.
Effect of microRNA expression on colorectal cancer treatment
| MicroRNA | Expression | Effect on treatment | Reference |
| * EGFR: Epidermal growth factor receptor. Arrows indicate upregulated or downregulated expression | |||
| Let-7 | Increased sensitivity to EGFR*-targeted treatment | 164-167 | |
| MiR-126 | Decreased sensitivity to capecitabine and oxaliplatin (XELOX) | 168-169 | |
| MiR-31 | Decreased sensitivity to 5-fluorouracil | 170 | |
| MiR-192/ miR-215 | Increased resistance to 5-fluorouracil | 171 | |
| MiR-148a | Poor sensitivity to oxaliplatin and oxaliplatin plus 5-fluorouracil | 172 | |
| MiR-21 | Poor sensitivity to 5-fluorouracil | 173,174 | |
| MiR-129 | Increased sensitivity to 5-fluorouracil | 175 | |
| MiR-19b | Increased response to 5-fluorouracil | 176 | |
| MiR-34a | Resistance to 5-fluorouracil | 177 | |
| MiR-143 | Increased response to 5-fluorouracil | 178 | |
| MiR-203 | Resistance to oxaliplatin | 179 |
Long non-coding RNAs
LncRNAs are a heterogenous group of more than 200 nucleotides. LncRNAs lack the open reading frames (ORF) larger than 100 amino acids in length180. These RNAs are categorized depending on their location and function181. They are becoming increasingly important in cancer and metastasis studies because they play important roles in chromatin remodeling and transcriptional and posttranscriptional gene expression regulation182. The role of the five most common lncRNA, namely HOTAIR, MALAT-1, CCAT-1 and -2, and H-19, in colorectal carcinogenesis is reviewed in this paper.
Hox transcript antisense intergenic RNA (HOTAIR)
The 2.2 kb long lncRNA is present on the HOXC locus. This lncRNA binds to the polycomb repressive complex 2 (PRC2) at 5' end resulting in the H3 lysine-27 (H3K27) methylation that leads to the repression of tumor suppressing gene HOXD and lysine-specific demethylase1 (LSD1) at 3' end causing H3K4 demethylation180,182. The HOTAIR expression in CRCs is lower in cancerous tissues than in their normal counterparts. The CRC cells overexpressing HOTAIR are more invasive. Moreover, its suppression decreases the invasion. A higher HOTAIR expression increases the potential of liver metastasis because it cooperates with PRC2 in maintaining the mesenchymal phenotype183.
Prostate cancer-associated ncRNA transcripts-1 (PCAT-1)
PCAT-1 is located on position 8q24 and is overexpressed up to almost 1.5 fold in cancerous versus normal tissues. This overexpression is associated with distant metastasis and influences the overall survival rate184. However, the mechanism of action of PCAT-1 in CRCs is still not understood.
Metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1)
The 6918bp to 8441bp region is located on chromosome 11q13 and is upregulated in both SW480 CRC cells and primary cancer tissues. Mutations have also been identified in this fragment185. The higher expression of MALAT-1 is correlated with cell proliferation, migration, and invasion and observed in metastatic tumors186. MALAT-1 promotes tumor metastasis in nude mice by inhibiting the polypyrimidine tract binding protein 2 (PTBP2)/splicing factor proline/glutamine-rich (SFPQ) complex and releasing the oncogene PTBP2. Furthermore, both MALAT1 and PTBP2 are overexpressed. The higher expression is associated with higher invasion and metastatic potential187. In vivo studies indicate PRKA kinase anchor protein 9 (AKAP-9) as a target gene of MALAT-1. Its upregulation is correlated with MALAT-1 tumor promoting activity188.
Colon cancer associated transcript (CCAT1)
CCAT1 is upregulated in CRCs with a 100-fold higher expression in the HCT116 cell line and more than 30-fold higher expression in tumor versus normal colon cells. This higher expression is also associated with regional and distant liver metastasis189. CCAT1-L is found on 8q24.21, upstream of MYC, and acts as an enhancer in CRC cell lines. The knockdown of CCAT1-L can reduce MYC levels. CCAT1-L assists in maintaining chromatin looping by modulating the binding of CTCF to the MYC locus190.
Colon cancer-associated transcript (CCAT2)
CCAT-2 is another lncRNA located on 8q24, which encompasses rs6983267 SNP and is involved in chromosomal instability that results in increased proliferation and metastasis in MSS tumors191. The G allele has been associated with higher susceptibility to CRC compared to the T allele with ratios of 1.4 and 1.27 for homozygotes and heterozygotes, respectively192. This difference in alleles affects its binding efficiency to the transcription factor 7-like 2 (TCF7L2), through which it upregulates MYC, miR-17-5p, and miR-20a. This difference also further enhances Wnt activity and may form a feedback loop as a downstream target of Wnt191.
H-19
The lncRNA H19 is hypomethylated at the sixth CTCF-binding site of the differentially methylated region (DMR), which results in loss of imprinting and gene expression193. H19 expression increases its precursor miR-675, which promotes colorectal malignancy by downregulating the tumor suppressor retinoblastoma protein (RB)194.
Conclusions
Colorectal cancers are one of the most prevalent and widely studied cancers in the world. The various molecular subtypes and the specific genetic and epigenetic events associated with these subtypes have been extensively investigated. This body of research has led to detailed classifications that can help identify the specific set of mutations present in a particular patient and the biomarkers that can predict treatment outcomes.
This review attempts to summarize the vast literature available on colorectal carcinogenesis. However, gaps in our knowledge of the disease process are still present. The molecular mechanisms involved in early onset sporadic cancers are yet to be identified. As the era of personalized medicine approaches, further work is still needed on the preventive, diagnostic, prognostic, and treatment-predictive signatures of diseases.
Conflict of interest statement
No potential conflicts of interest are disclosed.
References
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136: E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990; 61 : 759-67.
- Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, et al. Mutations in APC, kirsten-ras, and p53--alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci U S A. 2002;99: 9433–8. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38: 787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- East JE, Saunders BP, Jass JR. Sporadic and syndromic hyperplastic polyps and serrated adenomas of the colon: classification, molecular genetics, natural history, and clinical management. Gastroenterol Clin North Am. 2008;37:25�6. doi: 10.1016/j.gtc.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Sameer AS, Nissar S, Fatima K. Mismatch repair pathway: molecules, functions, and role in colorectal carcinogenesis. Eur J Cancer Prev. 2014;23: 246–57. doi: 10.1097/CEJ.0000000000000019. [DOI] [PubMed] [Google Scholar]
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348: 919–32. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7: 153–62. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8: 686�00. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138: 2059–72. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cummins JM, Shen D, Cahill DP, Jallepalli PV, Wang TL, et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 2004;64: 2998–3001. doi: 10.1158/0008-5472.CAN-04-0587. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ando K, Oki E, Ikawa-Yoshida A, Ida S, Kimura Y, et al. Aberrations of BUBR1 and TP53 gene mutually associated with chromosomal instability in human colorectal cancer. Anticancer Res. 2014;34: 5421–7. [PubMed] [Google Scholar]
- Shin HJ, Baek KH, Jeon AH, Park MT, Lee SJ, Kang CM, et al. Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell. 2003;4: 483–97. doi: 10.1016/S1535-6108(03)00302-7. [DOI] [PubMed] [Google Scholar]
- Derycke MS, Gunawardena SR, Middha S, Asmann YW, Schaid DJ, Mcdonnell SK, et al. Identification of novel variants in colorectal cancer families by high-throughput exome sequencing. Cancer Epidemiol Biomarkers Prev. 2013;22: 1239–51. doi: 10.1158/1055-9965.EPI-12-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SD, Britigan EM, Zasadil LM, Witte K, Audhya A, Roopra A, et al. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc Natl Acad Sci U S A. 2012;109: E2205–14. doi: 10.1073/pnas.1201911109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, et al. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63: 3511–6. [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Ishibashi M, Nezu M, Shimada H, Ochiai T, et al. Centromere protein H is up-regulated in primary human colorectal cancer and its overexpression induces aneuploidy. Cancer Res. 2005;65: 4683–9. doi: 10.1158/0008-5472.CAN-04-3613. [DOI] [PubMed] [Google Scholar]
- Ewart-Toland A, Briassouli P, De Koning JP, Mao JH, Yuan J, Chan F, et al. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat Genet. 2003;34: 403–12. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- Han D, Zhu Q, Cui J, Wang P, Qu S, Cao Q, et al. Polo-like kinase 1 is overexpressed in colorectal cancer and participates in the migration and invasion of colorectal cancer cells. Med Sci Monit. 2012;18: BR237–46. doi: 10.12659/MSM.882900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian K, Brashavitskaya O, Zih FW, Berger-Richardson D, Xu RZ, Pacholczyk K, et al. Polo-Like kinases in colorectal cancer: potential for targeted therapy. Curr Colorectal Cancer Rep. 2015;11: 187–99. doi: 10.1007/s11888-015-0275-4. [DOI] [Google Scholar]
- Katayama H, Ota T, Jisaki F, Ueda Y, Tanaka T, Odashima S, et al. Mitotic kinase expression and colorectal cancer progression. J Natl Cancer Inst 1999; 91:1160�162.
- Lin BW, Wang YC, Chang LY, Lin YJ, Yang ST, Tsou JH, et al. Overexpression of Aurora-C interferes with the spindle checkpoint by promoting the degradation of Aurora-B. Cell Death Dis 2014; 5:e1106.
- Przybylowska K, Kabzinski J, Sygut A, Dziki L, Dziki A, Majsterek I. An association selected polymorphisms of XRCC1, OGG1 and MUTYH gene and the level of efficiency oxidative DNA damage repair with a risk of colorectal cancer. Mutat Res. 2013; 745-746:6-15.
- Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108: 73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- Ertych N, Stolz A, Stenzinger A, Weichert W, Kaulfuß S, Burfeind P, et al. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat Cell Biol. 2014;16: 779–91. doi: 10.1038/ncb2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plentz RR, Wiemann SU, Flemming P, Meier PN, Kubicka S, Kreipe H, et al. Telomere shortening of epithelial cells characterises the adenoma-carcinoma transition of human colorectal cancer. Gut. 2003;52: 1304–7. doi: 10.1136/gut.52.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger L, Jones RE, Heppel NH, Williams GT, Sampson JR, Baird DM. Extensive telomere erosion in the initiation of colorectal adenomas and its association with chromosomal instability. J Natl Cancer Inst. 2013;105: 1202–11. doi: 10.1093/jnci/djt191. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Laken S, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B, et al. Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc Natl Acad Sci U S A. 2001;98: 2698–702. doi: 10.1073/pnas.051625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaslan M, Aytekin T. Loss of heterozygosity in colorectal cancer. Afr J Biotechnol. 2010;825: 7308–12. [Google Scholar]
- Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40: 600–8. doi: 10.1038/ng.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244: 217–21. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268: 1336–8. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, et al. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13: 343–6. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3: 1221–4. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- Mann B, Gelos M, Siedow A, Hanski ML, Gratchev A, Ilyas M, et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci U S A. 1999;96: 1603–8. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ER, Hoekstra E, Franken PF, Helvensteijn W, Van Deurzen CH, Van Veelen W, et al. Î-catenin signaling dosage dictates tissue-specific tumor predisposition in apc-driven cancer. Oncogene. 2013;32: 4579–85. doi: 10.1038/onc.2012.449. [DOI] [PubMed] [Google Scholar]
- Cottrell S, Bicknell D, Kaklamanis L, Bodmer WF. Molecular analysis of APC mutations in familial adenomatous polyposis and sporadic colon carcinomas. Lancet. 1992;340: 626–30. doi: 10.1016/0140-6736(92)92169-G. [DOI] [PubMed] [Google Scholar]
- Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, et al. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet. 1992;1: 229–33. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Konishi M, Kikuchi-Yanoshita R, Enomoto M, Igari T, Tanaka K, et al. Characteristics of somatic mutation of the adenomatous polyposis coli gene in colorectal tumors. Cancer Res. 1994;54: 3011–20. [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87: 159–70. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Otori K, Konishi M, Sugiyama K, Hasebe T, Shimoda T, Kikuchi-Yanoshita R, et al. Infrequent somatic mutation of the adenomatous polyposis coli gene in aberrant crypt foci of human colon tissue. Cancer. 1998;83: 896–900. doi: 10.1002/(SICI)1097-0142(19980901)83:5. [DOI] [PubMed] [Google Scholar]
- Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60: 4366–71. [PubMed] [Google Scholar]
- Christie M, Jorissen RN, Mouradov D, Sakthianandeswaren A, Li S, Day F, et al. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/Î-catenin signalling thresholds for tumourigenesis. Oncogene. 2013;32: 4675–82. doi: 10.1038/onc.2012.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, O'rourke KP, Simon J, Tschaharganeh DF, Van Es JH, Clevers H, et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell. 2015;161: 1539–52. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58: 1130–4. [PubMed] [Google Scholar]
- Watanabe K, Biesinger J, Salmans ML, Roberts BS, Arthur WT, Cleary M, et al. Integrative ChIP-seq/microarray analysis identifies a CTNNB1 target signature enriched in intestinal stem cells and colon cancer. PloS One 2014; 9:e92317.
- Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol. 2013;26: 825–34. doi: 10.1038/modpathol.2012.240. [DOI] [PubMed] [Google Scholar]
- Pretlow TP, Pretlow TG. Mutant KRAS in aberrant crypt foci (ACF): initiation of colorectal cancer. Biochim Biophys Acta. 2005;1756: 83�6. doi: 10.1016/j.bbcan.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Li W, Qiu T, Zhi W, Shi S, Zou S, Ling Y, et al. Colorectal carcinomas with KRAS codon 12 mutation are associated with more advanced tumor stages. BMC Cancer. 2015;15: 340. doi: 10.1186/s12885-015-1345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest. 2011;121: 4311–21. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidou V, Oikonomou E, Vlassi M, Avlonitis S, Katseli A, Tsipras I, et al. Tumor heterogeneity revealed by KRAS, BRAF, and PIK3CA pyrosequencing: KRAS and PIK3CA intratumor mutation profile differences and their therapeutic implications. Hum Mutat. 2014;35: 329–40. doi: 10.1002/humu.22496. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Gulhati P, Rampy BA, Han Y, Rychahou PG, Doan HQ, et al. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg. 2010;210: 767–76, 776-8. doi: 10.1016/j.jamcollsurg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suman S, Kurisetty V, Das TP, Vadodkar A, Ramos G, Lakshmanaswamy R, et al. Activation of AKT signaling promotes epithelial-mesenchymal transition and tumor growth in colorectal cancer cells. Mol Carcinog. 2014; 53 Suppl 1: E151�60.
- Chin YR, Yuan X, Balk SP, Toker A. PTEN-deficient tumors depend on AKT2 for maintenance and survival. Cancer Discov. 2014;4: 942–55. doi: 10.1158/2159-8290.CD-13-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Cai W, Zheng Y, Evers BM, She QB. ERK and AKT signaling cooperate to translationally regulate survivin expression for metastatic progression of colorectal cancer. Oncogene. 2014;33: 1828–39. doi: 10.1038/onc.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten CD. PI3K and MEK inhibitor combinations: examining the evidence in selected tumor types. Cancer Chemother Pharmacol. 2013;71: 1395–409. doi: 10.1007/s00280-013-2121-1. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10: 789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9: 724–37. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- Cooks T, Pateras IS, Tarcic O, Solomon H, Schetter AJ, Wilder S, et al. Mutant p53 prolongs NF-ÎB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell. 2013;23: 634–46. doi: 10.1016/j.ccr.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Minin G, Bellazzo A, Dal Ferro M, Chiaruttini G, Nuzzo S, Bicciato S, et al. Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol Cell. 2014;56: 617–29. doi: 10.1016/j.molcel.2014.10.013. [DOI] [PubMed] [Google Scholar]
- Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X, et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun. 2013;4: 2996. doi: 10.1038/ncomms3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WF, Wu G, Sun PC, Qiu D. P53 mutations occur more commonly than KRAS mutations in colorectal adenoma. Int J Clin Exp Med. 2015;8: 1370–5. [PMC free article] [PubMed] [Google Scholar]
- Brighenti E, Calabrese C, Liguori G, Giannone FA, Trerè D, Montanaro L, et al. Interleukin 6 downregulates p53 expression and activity by stimulating ribosome biogenesis: a new pathway connecting inflammation to cancer. Oncogene. 2014;33: 4396–406. doi: 10.1038/onc.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, Dubois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107: 1183–8. doi: 10.1016/0016-5085(94)90246-1. [DOI] [PubMed] [Google Scholar]
- Oshima M, Murai N, Kargman S, Arguello M, Luk P, Kwong E, et al. Chemoprevention of intestinal polyposis in the Apcdelta716 mouse by rofecoxib, a specific cyclooxygenase-2 inhibitor. Cancer Res. 2001;61: 1733–40. [PubMed] [Google Scholar]
- Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, et al. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30: 377–86. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- Roelofs HM, te Morsche RH, van Heumen BW, Nagengast FM, Peters WH. Over-expression of COX-2 mRNA in colorectal cancer. BMC Gastroenterol. 2014;14: 1. doi: 10.1186/1471-230X-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C, Queirós S, Galaghar A, Sousa H, Pimentel-Nunes P, Brandão C, et al. Genetic variability in key genes in prostaglandin E2 pathway (COX-2, HPGD, ABCC4 and SLCO2A1) and their involvement in colorectal cancer development. PloS One. 2014;9: e92000. doi: 10.1371/journal.pone.0092000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen J, Kim H, Piantadosi S, Liu ZF, Levitt RC, Sistonen P, et al. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med. 1994;331: 213–21. doi: 10.1056/NEJM199407283310401. [DOI] [PubMed] [Google Scholar]
- Shin SK, Nagasaka T, Jung BH, Matsubara N, Kim WH, Carethers JM, et al. Epigenetic and genetic alterations in Netrin-1 receptors UNC5C and DCC in human colon cancer. Gastroenterology. 2007;133: 1849–57. doi: 10.1053/j.gastro.2007.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford-Richens KL, Rowan AJ, Gorman P, Halford S, Bicknell DC, Wasan HS, et al. SMAD4 mutations in colorectal cancer probably occur before chromosomal instability, but after divergence of the microsatellite instability pathway. Proc Natl Acad Sci U S A. 2001;98: 9719–23. doi: 10.1073/pnas.171321498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman AM, Naranjo S, Webb E, Broderick P, Lips EH, Van Wezel T, et al. The colorectal cancer risk at 18q21 is caused by a novel variant altering SMAD7 expression. Genome Res. 2009;19: 987–93. doi: 10.1101/gr.092668.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73: 725–35. doi: 10.1158/0008-5472.CAN-12-2706. [DOI] [PubMed] [Google Scholar]
- Kozak MM, Von Eyben R, Pai J, Vossler SR, Limaye M, Jayachandran P, et al. Smad4 inactivation predicts for worse prognosis and response to fluorouracil-based treatment in colorectal cancer. J Clin Pathol. 2015;68: 341–5. doi: 10.1136/jclinpath-2014-202660. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74: 681�10. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Bak ST, Sakellariou D, Pena-Diaz J. The dual nature of mismatch repair as antimutator and mutator: for better or for worse. Front Genet. 2014;5: 287. doi: 10.3389/fgene.2014.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368: 258–61. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Papadopoulos N, Liu B, Weit YF, Carter KC, Ruben SM, et al. Mutations of two P/WS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371: 75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT, et al. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet. 2000;24: 27–35. doi: 10.1038/71643. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, et al. Reconstitution of 5'-directed human mismatch repair in a purified system. Cell. 2005;122: 693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Peltomäki P, Lothe RA, Aaltonen LA, Pylkkänen L, Nyström-Lahti M, Seruca R, et al. Microsatellite instability is associated with tumors that characterize the hereditary non-polyposis colorectal carcinoma syndrome. Cancer Res. 1993;53: 5853–5. [PubMed] [Google Scholar]
- de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010;28: 3380–7. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58: 5248–57. [PubMed] [Google Scholar]
- Bonadona V, Bonaïti B, Olschwang S, Grandjouan S, Huiart L, Longy M, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 2011;305: 2304–10. doi: 10.1001/jama.2011.743. [DOI] [PubMed] [Google Scholar]
- Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition-Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20: 269–76. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollikainen M, Hannelius U, Lindgren CM, Abdel-Rahman WM, Kere J, Peltomäki P. Mechanisms of inactivation of MLH1 in hereditary nonpolyposis colorectal carcinoma: a novel approach. Oncogene. 2007;26: 4541–9. doi: 10.1038/sj.onc.1210236. [DOI] [PubMed] [Google Scholar]
- Woods MO, Williams P, Careen A, Edwards L, Bartlett S, Mclaughlin JR, et al. A new variant database for mismatch repair genes associated with Lynch syndrome. Hum Mutat. 2007;28: 669–73. doi: 10.1002/humu.20502. [DOI] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36: 497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38: 1178–83. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- Hitchins MP. The Role of Epimutations of the Mismatch Repair Genes in the Development of Lynch Syndrome Related Cancers. In: Vogelsang M (ed.), DNA Alterations in Lynch Syndrome. Springer Netherlands. 2013:101�3.
- Kuiper RP, Vissers LE, Venkatachalam R, Bodmer D, Hoenselaar E, Goossens M, et al. Recurrence and variability of germline EPCAM deletions in Lynch syndrome. Hum Mutat. 2011;32: 407–14. doi: 10.1002/humu.21446. [DOI] [PubMed] [Google Scholar]
- Ligtenberg MJL, Kuiper RP, Geurts van Kessel A, Hoogerbrugge N. EPCAM deletion carriers constitute a unique subgroup of Lynch syndrome patients. Fam Cancer. 2013;12: 169–74. doi: 10.1007/s10689-012-9591-x. [DOI] [PubMed] [Google Scholar]
- Gylling A, Ridanpää M, Vierimaa O, Aittomäki K, Avela K, Kääriäinen H, et al. Large genomic rearrangements and germline epimutations in Lynch syndrome. Int J Cancer. 2009;124: 2333–40. doi: 10.1002/ijc.24230. [DOI] [PubMed] [Google Scholar]
- Rhees J, Arnold M, Boland CR. Inversion of exons 1-7 of the MSH2 gene is a frequent cause of unexplained Lynch syndrome in one local population. Fam Cancer. 2014;13: 219–25. doi: 10.1007/s10689-013-9688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraldsdottir S, Hampel H, Tomsic J, Frankel WL, Pearlman R, De La Chapelle A, et al. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology. 2014;147: 1308–1316. doi: 10.1053/j.gastro.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensenkamp AR, Vogelaar IP, Van Zelst-Stams WA, Goossens M, Ouchene H, Hendriks-Cornelissen SJ, et al. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. 2014;146: 643–646. doi: 10.1053/j.gastro.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, et al. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58: 3455–60. [PubMed] [Google Scholar]
- Cunningham JM, Kim CY, Christensen ER, Tester DJ, Parc Y, Burgart LJ, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69: 780–90. doi: 10.1086/323658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional Consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95: 6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuismanen SA, Holmberg MT, Salovaara R, de la Chapelle A, Peltomäki P. Genetic and epigenetic modification of MLH1 accounts for a major share of microsatellite-unstable colorectal cancers. Am J Pathol. 2000;156: 1773–9. doi: 10.1016/S0002-9440(10)65048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira L, Muñoz J, Cuatrecasas M, Quintanilla I, Leoz ML, Carballal S, et al. Prevalence of somatic mutl homolog 1 promoter hypermethylation in Lynch syndrome colorectal cancer. Cancer. 2015;121: 1395–404. doi: 10.1002/cncr.29190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä TT, Böhm JP, Friman M, Lahtinen L, Väyrynen VM, Liipo TK, et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer. 2015;112: 1966–75. doi: 10.1038/bjc.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre JN, Becker FF. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun. 1979;87: 698–705. doi: 10.1016/0006-291X(79)92015-1. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301: 89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Flatau E, Bogenmann E, Jones PA. Variable 5-methylcytosine levels in human tumor cell lines and fresh pediatric tumor explants. Cancer Res. 1983;43: 4901–5. [PubMed] [Google Scholar]
- Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11: 6883–94. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228: 187–90. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- Bariol C, Suter C, Cheong K, Ku SL, Meagher A, Hawkins N, et al. The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol. 2003;162: 1361–71. doi: 10.1016/S0002-9440(10)63932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL. Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis. 2004;19: 95–101. doi: 10.1007/s00384-003-0539-3. [DOI] [PubMed] [Google Scholar]
- Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer. 2010;9:125. doi: 10.1186/1476-4598-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchitta M, Quattrocchi A, Maugeri A, Vinciguerra M, Agodi A. LINE-1 Hypomethylation in Blood and Tissue Samples as an Epigenetic Marker for Cancer Risk: A Systematic Review and Meta-Analysis. PLoS ONE 2014; 9: e109478.
- Cui H, Horon IL, Ohlsson R, Hamilton SR, Feinberg AP. Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat Med. 1998;4: 1276–80. doi: 10.1038/3260. [DOI] [PubMed] [Google Scholar]
- Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62: 6442–6. [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96: 8681�6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yao X, Wang Y, Hu F, Wang F, Jiang L, et al. MLH1 promoter methylation frequency in colorectal cancer patients and related clinicopathological and molecular features. PloS One 2013; 8:e59064.
- Poynter JN, Siegmund KD, Weisenberger DJ, Long TI, Thibodeau SN, Lindor N, et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev. 2008;17: 3208–15. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129: 837–45. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104: 18654–9. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cho NY, Yoo EJ, Kim JH, Kang GH. CpG island methylator phenotype in colorectal cancers: comparison of the new and classic CpG island methylator phenotype marker panels. Arch Pathol Lab Med. 2008;132: 1657–65. doi: 10.5858/2008-132-1657-CIMPIC. [DOI] [PubMed] [Google Scholar]
- Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9: 305–14. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokazono K, Ueki T, Nagayoshi K, Nishioka Y, Hatae T, Koga Y, et al. A CpG island methylator phenotype of colorectal cancer that is contiguous with conventional adenomas, but not serrated polyps. Oncol Lett. 2014;8: 1937–44. doi: 10.3892/ol.2014.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roon EH van, Boot A, Dihal AA, Ernst RF, Wezel T van, Morreau H, et al. BRAF mutation-specific promoter methylation of FOX genes in colorectal cancer. Clin Epigenetics. 2013;5: 2. doi: 10.1186/1868-7083-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A-L, Dawson SN, Arends MJ, Guttula K, Hall N, Cameron EA, et al. Boosting Wnt activity during colorectal cancer progression through selective hypermethylation of Wnt signaling antagonists. BMC Cancer. 2014;14: 891. doi: 10.1186/1471-2407-14-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura N, Shinjo K, An B, Shimizu Y, Yamao K, Ohka F, et al. Aberrant TET1 methylation closely associated with CpG island methylator phenotype in colorectal cancer. Cancer Prev Res (Phila) 2015;8: 702–11. doi: 10.1158/1940-6207.CAPR-14-0306. [DOI] [PubMed] [Google Scholar]
- Tahara T, Yamamoto E, Madireddi P, Suzuki H, Maruyama R, Chung W, et al. Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology. 2014;146: 530–38. doi: 10.1053/j.gastro.2013.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons CC, Hughes LA, Smits KM, Khalid-De Bakker CA, De Bruïne AP, Carvalho B, et al. A novel classification of colorectal tumors based on microsatellite instability, the CpG island methylator phenotype and chromosomal instability: implications for prognosis. Ann Oncol. 2013;24: 2048–56. doi: 10.1093/annonc/mdt076. [DOI] [PubMed] [Google Scholar]
- Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148: 77–87. doi: 10.1053/j.gastro.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo YY, Johnston FM, Zhang DY, Juo HH, Wang H, Pappou EP, et al. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Ann Oncol. 2014;25: 2314–27. doi: 10.1093/annonc/mdu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013; 10: e1001453.
- Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World J Gastroenterol. 2012;18: 5171–80. doi: 10.3748/wjg.v18.i37.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96: 1166–9. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lièvre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, et al. KRAS mutations as an Independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26: 374–9. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- Bokemeyer C, Bondarenko I, Makhson AA, Aparicio J, De Braud F, Donea S, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the First-Line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27: 663–71. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- Peeters M, Price T, Hotko Y, Cervantes A, Ducreux M, André T, et al. 14LBA Randomized phase 3 study of panitumumab with FOLFIRI vs FOLFIRI alone as second-line treatment (tx) in patients (pts) with metastatic colorectal cancer (mCRC) Eur J Cancer Suppl. 2009;7: 10. [Google Scholar]
- Allegra CJ, Jessup JM, Somerfield MR, Hammond EH, Hayes DF, Morton RF, et al. American society of clinical oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to Anti-Epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27: 2091–6. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26: 5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer. 2015;51: 587–94. doi: 10.1016/j.ejca.2015.01.054. [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28: 3219–26. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppedè F, Lopomo A, Spisni R, Migliore L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J Gastroenterol. 2014;20: 943â€?6. doi: 10.3748/wjg.v20.i4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136: 215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisch J, Darfeuille-Michaud A, Nguyen HT. Role of microRNAs in the immune system, inflammation and cancer. World J Gastroenterol. 2013;19: 2985–96. doi: 10.3748/wjg.v19.i20.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leva GD, Garofalo M, Croce CM. MicroRNAs in Cancer. Annu Rev Pathol Mech Dis. 2014;9: 287�14. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Li X, Ji W, Sun B, Xu C, Li Z, et al. Small molecule with big role: MicroRNAs in cancer metastatic microenvironments. Cancer Lett. 2014;344: 147–56. doi: 10.1016/j.canlet.2013.10.024. [DOI] [PubMed] [Google Scholar]
- Kamatani A, Nakagawa Y, Akao Y, Maruyama N, Nagasaka M, Shibata T, et al. Downregulation of anti-oncomirs miR-143/145 cluster occurs before APC gene aberration in the development of colorectal tumors. Med Mol Morphol. 2013;46: 166–71. doi: 10.1007/s00795-013-0020-5. [DOI] [PubMed] [Google Scholar]
- Pagliuca A, Valvo C, Fabrizi E, Di Martino S, Biffoni M, Runci D, et al. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene. 2013;32: 4806–13. doi: 10.1038/onc.2012.495. [DOI] [PubMed] [Google Scholar]
- Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28: 1385–92. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li JJ, et al. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer. 2009;101: 699–706. doi: 10.1038/sj.bjc.6605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Yu J, Yin Y, He J, Wang L, Li Q, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12: 1385–94. doi: 10.4161/cc.24477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29: 903–6. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- Luu C, Heinrich EL, Duldulao M, Arrington AK, Fakih M, Garcia-Aguilar J, et al. TP53 and let-7a micro-RNA regulate K-Ras activity in HCT116 colorectal cancer cells. PloS One. 2013;8: e70604. doi: 10.1371/journal.pone.0070604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz EA, LaFleur B, Gerner EW. Polyamines are oncometabolites that regulate the LIN28/let-7 pathway in colorectal cancer cells. Mol Carcinog. 2014; 53 Suppl 1: E96-106.
- Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30: 953–9. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- Paterson EL, Kazenwadel J, Bert AG, Khew-Goodall Y, Ruszkiewicz A, Goodall GJ. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia. 2013;15: 180–91. doi: 10.1593/neo.121828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62: 1315–26. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res. 2007;56: 248–53. doi: 10.1016/j.phrs.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Siemens H, Jackstadt R, HÃnten S, Kaller M, Menssen A, Götz U, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10: 4256–71. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124: 1853–67. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Kim JW, Lee BE, Jang MJ, Chong SY, Park PW, et al. Polymorphisms of the pri-miR-34b/c promoter and TP53 codon 72 are associated with risk of colorectal cancer. Oncol Rep. 2014;31: 995–1002. doi: 10.3892/or.2013.2926. [DOI] [PubMed] [Google Scholar]
- Diosdado B, Van De Wiel MA, Terhaar Sive Droste JS, Mongera S, Postma C, Meijerink WJ, et al. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101: 707–14. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25: 469–83. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125: 2737–43. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- Vinci S, Gelmini S, Mancini I, Malentacchi F, Pazzagli M, Beltrami C, et al. Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods. 2013;59: 138–46. doi: 10.1016/j.ymeth.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Stiegelbauer V, Perakis S, Deutsch A, Ling H, Gerger A, Pichler M. MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World J Gastroenterol. 2014;20: 11727–35. doi: 10.3748/wjg.v20.i33.11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa M, Majorana A, Statello L, Maugeri M, Salito L, Barbagallo D, et al. Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol Cancer Ther. 2010;9: 3396–409. doi: 10.1158/1535-7163.MCT-10-0137. [DOI] [PubMed] [Google Scholar]
- Ruzzo A, Graziano F, Vincenzi B, Canestrari E, Perrone G, Galluccio N, et al. High let-7a microRNA levels in KRAS-mutated colorectal carcinomas May rescue anti-EGFR therapy effects in patients with chemotherapy-refractory metastatic disease. Oncologist. 2012;17: 823–9. doi: 10.1634/theoncologist.2012-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuzzo F, Sacconi A, Landi L, Ludovini V, Biagioni F, D'incecco A, et al. MicroRNA signature in metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodies. Clin Colorectal Cancer. 2014;13: 37–45. doi: 10.1016/j.clcc.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Salendo J, Spitzner M, Kramer F, Zhang X, Jo P, Wolff HA, et al. Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g. Radiother Oncol. 2013;108: 451–7. doi: 10.1016/j.radonc.2013.06.032. [DOI] [PubMed] [Google Scholar]
- Hansen TF, Christensen R dP, Andersen RF, Sørensen FB, Johnsson A, Jakobsen A. MicroRNA-126 and epidermal growth factor-like domain 7âan angiogenic couple of importance in metastatic colorectal cancer. MicroRNA-126 and epidermal growth factor-like domain 7âan angiogenic couple of importance in metastatic colorectal cancer. Results from the Nordic ACT trial. Br J Cancer. 2013;109: 1243â€?1. doi: 10.1038/bjc.2013.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TF, Sørensen FB, Lindebjerg J, Jakobsen A. The predictive value of microRNA-126 in relation to first line treatment with capecitabine and oxaliplatin in patients with metastatic colorectal cancer. BMC Cancer. 2012;12: 83. doi: 10.1186/1471-2407-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CJ, Stratmann J, Zhou ZG, Sun XF. Suppression of microRNA-31 increases sensitivity to 5-FU at an early stage, and affects cell migration and invasion in HCT-116 colon cancer cells. BMC Cancer. 2010;10: 616. doi: 10.1186/1471-2407-10-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, et al. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. 2010;9: 2265–75. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Cuatrecasas M, Balaguer F, Hur K, Toiyama Y, Castells A, et al. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PloS One. 2012;7: e46684. doi: 10.1371/journal.pone.0046684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Lei W, Fu JC, Zhang L, Li JH, Xiong JP. Targeting miR-21 enhances the sensitivity of human colon cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys Res Commun. 2014;443: 789–95. doi: 10.1016/j.bbrc.2013.11.064. [DOI] [PubMed] [Google Scholar]
- Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci U S A. 2010;107: 21098–1103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013;4: e659. doi: 10.1038/cddis.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Tanahashi T, Iima T, Yamamoto Y, Akaike Y, Nishida K, et al. Role of miR-19b and its target mRNAs in 5-fluorouracil resistance in colon cancer cells. J Gastroenterol. 2012;47: 883–95. doi: 10.1007/s00535-012-0547-6. [DOI] [PubMed] [Google Scholar]
- Akao Y, Noguchi S, Iio A, Kojima K, Takagi T, Naoe T. Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 2011;300: 197–204. doi: 10.1016/j.canlet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Borralho PM, Kren BT, Castro RE, Da Silva IB, Steer CJ, Rodrigues CM. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276: 6689–700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wan G, Spizzo R, Ivan C, Mathur R, Hu X, et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol. 2014;8: 83–92. doi: 10.1016/j.molonc.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9: 703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136: 629�1. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Hauptman N, Glavač D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14: 4655–69. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71: 6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol Northwood Lond Engl. 2013;30: 588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39: 169–75. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L, et al. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/Î-catenin signal pathway. PLoS One. 2013;8: e78700. doi: 10.1371/journal.pone.0078700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111: 736–48. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, et al. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852: 166–74. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130: 1598–606. doi: 10.1002/ijc.26170. [DOI] [PubMed] [Google Scholar]
- Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24: 513–31. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23: 1446–61. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39: 954–6. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Tang Z, Song G, Pan Y, He B, Bao Q, et al. Loss of imprinting of IGF2 correlates with hypomethylation of the H19 differentially methylated region in the tumor tissue of colorectal cancer patients. Mol Med Rep. 2012;5: 1536–40. doi: 10.3892/mmr.2012.833. [DOI] [PubMed] [Google Scholar]
- Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31: 350–8. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]


