Abstract
Nascent polypeptides are degraded by the proteasome concurrently with their synthesis on the ribosome. This process, called cotranslational protein degradation (CTPD), has been observed for years, but the underlying mechanisms remain poorly understood. Equally unclear are the identities of cellular proteins genuinely subjected to CTPD. Here we report the identification of CTPD substrates in the yeast Saccharomyces cerevisiae via a quantitative proteomic analysis. We compared the abundance of individual ribosome-bound nascent chains between a wild type strain and a mutant defective in CTPD. Of 1,422 proteins acquired from the proteomic analysis, 289 species are efficient CTPD substrates, with >30% of their nascent chains degraded cotranslationally. We found that proteins involved in translation, ribosome biogenesis, nuclear transport, and amino acid metabolism are more likely to be targeted for CTPD. There is a strong correlation between CTPD and the translation efficiency. CTPD occurs preferentially to rapidly translated polypeptides. CTPD is also influenced by the protein sequence length; longer polypeptides are more susceptible to CTPD. In addition, proteins with N-terminal disorder have a higher probability of being degraded cotranslationally. Interestingly, the CTPD efficiency is not related to the half-lives of mature proteins. These results for the first time indicate an inverse correlation between CTPD and cotranslational folding on a proteome scale. The implications of this study with respect to the physiological significance of CTPD are discussed.
Keywords: proteasome, protein degradation, protein folding, proteomics, proteostasis, ribosome, cotranslational protein degradation
Introduction
Protein homeostasis is maintained by an elaborate quality control system that regulates a delicate balance between protein synthesis, folding, and degradation. Remarkably, nascent proteins are monitored by the quality control system concurrently with their synthesis by the ribosome (1–5). The N-terminal end of a nascent polypeptide is available for folding before the other end has been synthesized. Cotranslational folding helps reduce aggregation of translation intermediates and promotes accurate folding of newly synthesized proteins. On the other hand, nascent polypeptides can be degraded by the proteasome during translation, a process called cotranslational protein degradation (CTPD).2 The term CTPD has also been used to describe the degradation of newly synthesized proteins already released from the ribosome but not yet folded. In this report, CTPD is used exclusively for the degradation of ribosome-bound nascent polypeptides, in line with the definition of bona fide cotranslational degradation.
CTPD was first inferred from early studies that applied pulse-chase experiments to measure the kinetics of degradation of radioactive isotope-labeled proteins in living cells (6–11). These studies revealed two phases of kinetics of protein degradation, with a fast rate of turnover within the first hour of chase, followed by a second kinetic of slower degradation. It was also found that the rate of protein turnover within the first hour of chase was markedly accentuated by shortening the pulse length. These observations indicate that nascent proteins are more susceptible to degradation than mature proteins and that a fraction of newly synthesized proteins has already been degraded before the chase begins. Intriguingly, the scale of degradation of newly synthesized proteins is rather substantial. Wheatley et al. (11) showed that ∼35% of labeled proteins from a 5-min pulse were degraded during a 2-h chase in mammalian cells. Extending this work, Schubert et al. (12) provided evidence that >30% of newly synthesized proteins are defective ribosomal products (DRiPs) that never achieve a native state and are rapidly degraded by the proteasome. This work led to an elaboration of the defective ribosomal product hypothesis, claiming that MHC class I-presented peptides are primarily generated from cotranslational degradation of faulty proteins (12–14). Substantial prechase degradation, accounting for 30–80% of total labeled nascent proteins, was observed in the pulse-chase experiments that analyzed the degradation of N-end rule substrates and ubiquitin-fusion degradation substrates (15, 16). These studies strongly suggest that a substantial fraction of nascent polypeptides is subjected to CTPD. Of note, the pulse-chase experiments in these studies could not distinguish CTPD from the degradation of nascent proteins already released from the ribosome. Turner and Varshavsky (17) invented the so-called ubiquitin sandwich technique and successfully demonstrated that >50% of nascent polypeptides bearing an N-terminal degron were degraded cotranslationally. Our recent study showed that ∼33% of total ribosome-bound nascent polypeptides were degraded cotranslationally in yeast cells (18).
Although it is clear that a large amount of nascent proteins is degraded cotranslationally, the protein species subjected to CTPD remain unidentified. Lack of this information hinders our understanding of the mechanisms and physiological significance of CTPD. We recently discovered that CTPD is severely impaired in the yeast mutant srp1-49 expressing a mutated version of the nuclear import factor Srp1 (also known as importin α, karyopherin α, or Kap60) (18–21). Only 4% of ribosome-bound nascent polypeptides were degraded cotranslationally in srp1-49, sharply lower than that in the WT counterpart. Srp1 directly binds nascent polypeptides emerging from the ribosome and, by collaborating with its associated protein Sts1, couples proteasomes to ribosome-nascent chain complexes (RNCs). Thus, Srp1 plays a general and critical role in CTPD, and srp1-49 can be used for identifying CTPD substrates. The difference in abundance of a ribosome-bound nascent chain between WT and srp1-49 reflects whether and how efficiently this nascent polypeptide is degraded cotranslationally. Here we report the proteome scale identification of CTPD substrates and the analysis of principles governing CTPD. The implications of the results with respect to the potential physiological functions of CTPD are discussed.
Experimental Procedures
Yeast Strains and Plasmids
The yeast strains used in this study included W303-1A (MATa ura3-1 trp1-1 leu2-3,112 his3-11 ade2-1 can1-100), JLY555 (MATα ura3-1 trp1-1 leu2-3,112 his3-11 ade2-1 can1-100 srp1-49, kindly provided by G. Fink and M. Nomura), YXY728 (a lys2Δ derivative of W303-1A), and YXY730 (a lys2Δ derivative of JLY555). The lys2Δ strains were constructed following a procedure as previously described (22). Briefly, the LYS2 deletion vector pAD2 (purchased from American Type Culture Collection) was linearized by the restriction enzyme ClaI and transformed into W303-1A and JLY555. The resulting transformants were selected on synthetic complete-ura plates to isolate Ura+ integrants, which were subsequently subjected to counterselection on 5-fluoroorotic acid plates to remove the URA3 marker and LYS2 via an intra-allele homologous recombination. The Lys− phenotype of resulting lys2Δ strains was confirmed by plating assay using synthetic complete-lys plates.
SILAC-PUNCH-P Analysis
The abundance difference of individual ribosome-bound nascent chains between WT and srp1-49 cells was quantified by a combination of SILAC (stable isotope labeling by amino acids in cell culture) and PUNCH-P (puromycin-associated nascent chain proteomics) approaches (23, 24). Yeast strains YXY728 and YXY730 were grown in 10 ml of synthetic complete medium containing either 30 μg/ml l-lysine (light) or 31.28 μg/ml l-lysine [13C615N2] (heavy) for at least 10 generations until the late log phase. (The molar concentrations of l-lysine and l-lysine [13C615N2] were identical in the medium). In a label swap experiment, YXY728 and YXY730 cells were cultured in heavy and light media, respectively. The cultures were then diluted to A600 ∼0.1 in 300 ml of fresh medium of the same composition and kept cultured to A600 ∼1.2. The cells were harvested by centrifugation and immediately frozen on dry ice. Cell pellets could be stored at −80 °C at this point. Cell pellets with equal numbers of YXY728 and YXY730 cells were mixed and grounded to fine powders with liquid nitrogen. The powders were further disrupted by glass beads in lysis buffer (20 mm HEPES, pH 7.5, 50 mm KCl, 10 mm MgCl2, and 0.1% Triton X-100) supplemented with 1 mm PMSF, protease inhibitor mixture, 50 μm MG-132, and 90 μm emetine. Emetine was used to arrest elongating ribosomes to prevent ribosomal run-off. It has been shown that emetine does not interfere with the incorporation of puromycin into nascent chains (24). Total cell extracts were clarified by centrifugation at 4 °C. Polysomes or RNCs were isolated by ultracentrifugation, following our previous procedure (18). RNC pellets were resuspended in polysome buffer (10 mm HEPES, pH 7.5, 400 mm KCl, 3 mm MgCl2) after gentle washing with 1 ml of ice-cold RNase-free water. The concentrations of RNC samples were determined by spectrophotometric measurement at A260. Labeling of ribosome-bound nascent chains with biotin-puromycin was performed as previously described (18). The RNC sample (400 μg) was incubated with 3 μm biotin-puromycin at 30 °C for 90 min. The reaction was terminated, and RNCs were disassembled by addition of 1 ml of urea/SDS denaturing buffer (50 mm Tris-HCl, pH 7.5, 8 m urea, 2% SDS, and 200 mm NaCl). Biotin-puromycin-labeled nascent chains were recovered by streptavidin-agarose beads, following the PUNCH approach (24). The recovery efficiency was examined by immunoblotting analysis. The nascent chains were subjected to on-bead digestion with protease Lys-C and separated by reverse phase chromatography, followed by tandem mass spectrometry. Spectra were searched against the Saccharomyces cerevisiae database downloaded from Uniprot using MaxQuant (version 1.5.2.8). SILAC ratios were converted to WT/mutant (WT/Mut) orientation and to log2 scale for analysis.
Calculation of CTPD Efficiency
The WT/Mut SILAC ratio (R) represents the difference in abundance of a ribosome-bound nascent chain between WT and srp1-49. In addition to cotranslational degradation, R may be affected by the difference in protein production between WT and srp1-49. Thus, R is expressed by the following equation,
 |
where Ws and Wd stand for the synthesized and degraded amounts of a nascent chain in WT cells. Likewise, Ms and Md represent the synthesis and degradation of the same nascent chain in srp1-49. The percentage of a nascent chain cotranslationally degraded in WT cells, referred to as cotranslational degradation efficiency (E), can be calculated by the following formula.
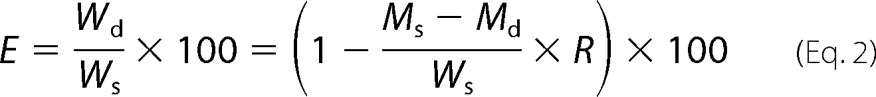 |
Because CTPD is severely inhibited in srp1-49, Md was treatedas 0 to simplify the calculation. This may yield a slightly undercalculated E value. Protein production is ∼15% lower in srp1-49 than in WT (see “Results”), i.e. Ms/Ws = 0.85. The above equation was therefore converted to: E = (1 − 0.85R) × 100.
Data Analysis
The R package was used for box plotting analysis. The p values were calculated with the Wilcoxon nonparametric statistical hypothesis test (25).
Results
Proteomic Analysis for Ribosome-bound Nascent Polypeptides Subjected to CTPD
To compare the abundance of individual ribosome-bound nascent chains between WT and srp1-49 cells, we combined the SILAC and PUNCH-P approaches (Fig. 1A). SILAC is a metabolic labeling method that depends on cellular protein synthesis to incorporate stable isotope-labeled amino acids into the entire proteome for quantitative analysis (23). PUNCH-P is a strategy that incorporates biotin-labeled puromycin into nascent chains being translated from isolated RNCs, allowing for efficient affinity purification of nascent polypeptides by streptavidin beads (24). The LYS2 gene was deleted from srp1-49 and WT strains, such that the cells depended on lysine provided in the medium for protein synthesis. WT and srp1-49 cells were grown in light and heavy media, respectively. The light medium contained l-lysine, whereas the heavy medium contained l-lysine-13C6,15N2. Each l-lysine-13C6,15N2 introduced an 8-Da mass difference between the light and heavy peptides. The relative intensity of light and heavy peptides (so-called SILAC ratio) reflects the difference in abundance of individual ribosome-bound nascent chains between WT and srp1-49. The percentage of a nascent polypeptide degraded cotranslationally in WT cells can be deduced from the SILAC ratio. To effectively eliminate experimental errors, we included a label swap experiment in which WT cells were cultured in heavy medium, whereas srp1-49 in light medium. WT and srp1-49 cells were mixed at a 1:1 ratio before lysis. RNCs were isolated by ultracentrifugation through a sucrose cushion and incubated with biotin-puromycin to label the nascent chains. No loss of nascent chains from the RNCs was detected during incubation with biotin-puromycin (data not shown). After labeling, RNCs were denatured under stringent conditions to completely break up the ribosome and release all nascent chains, which were recovered by streptavidin beads. Immunoblotting analysis indicated that virtually all biotin-puromycin-labeled nascent chains were captured by the streptavidin beads (Fig. 1B). The nascent chains were then subjected to on-bead digestion with Lys-C, a protease that cleaves proteins on the C-terminal side of lysine residues. The eluted peptides were separated by reverse phase chromatography, followed by tandem mass spectrometry. Spectra were searched against the S. cerevisiae database from Uniprot.
FIGURE 1.
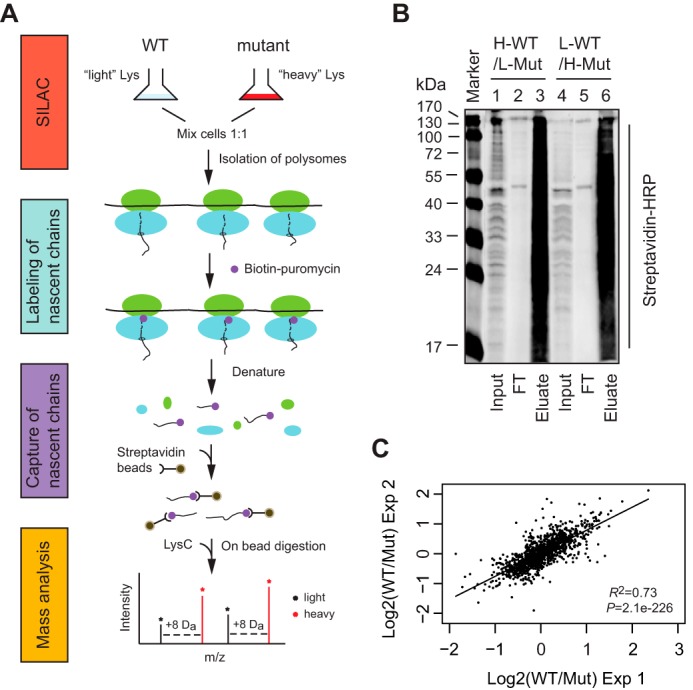
Quantitative proteomic analysis for nascent proteins subjected to cotranslational degradation. A, workflow of SILAC-PUNCH-P. B, immunoblotting analysis of biotin-puromycin-labeled nascent chains captured by streptavidin beads. Biotin-puromycin-labeled nascent chains from a pair of label-swapped samples were incubated with streptavidin beads under stringent conditions. Captured puromycylated nascent chains were eluted after extensive washing. Input (2%), flow-through (FT) (2%), and eluate (20%) were analyzed by immunoblotting with HRP-conjugated streptavidin. Lanes 1–3, WT cells labeled with heavy Lys; lanes 4–6, srp1-49 cells labeled with heavy Lys. C, scatter plotting analysis of SILAC data obtained from two independent SILAC-PUNCH-P experiments. The WT/Mut SILAC ratios of two label-swapped samples were averaged and converted to log2 scale for analysis. The proteins identified by SILAC-PUNCH-P and their SILAC ratios are listed in supplemental Table S1.
The SILAC-PUNCH-P experiments were independently performed twice. We analyzed only those proteins that were identified by both SILAC-PUNCH-P experiments. Each SILAC-PUNCH-P experiment contained two label-swapped samples: heavy WT versus light srp1-49 and light WT versus heavy srp1-49. SILAC data were analyzed with MaxQuant software (version 1.5.2.8) and expressed as WT/Mut SILAC ratios. Only those proteins that were found in both label-swapped samples and had two WT/Mut SILAC ratios in the same orientation were counted. The first SILAC-PUNCH-P experiment identified 1,543 proteins, whereas the second one collected 1,587 proteins, of which 1,422 proteins were in common. This high percentage of overlap indicates good technical reproducibility of the SILAC-PUNCH-P experiments. Moreover, scatter plotting analysis of the data obtained from two independent SILAC-PUNCH-P experiments displayed a high degree of confidence in the SILAC ratios of the 1,422 proteins (Fig. 1C). Given that not all of the 6,338 yeast genes are expressed in exponentially growing cells (26), the 1,422 proteins identified by our proteomic analysis cover a significant portion of the yeast proteome.
The WT/Mut SILAC ratio, referred to as R, presents the difference in abundance of a ribosome-bound nascent chain between WT and srp1-49. Of note, in addition to cotranslational degradation, R may be affected by the difference in protein production between WT and srp1-49. In fact, we found that the protein production in srp1-49 was ∼15%, less than that in WT (Fig. 2). Taking this into account, we formulated the following equation to calculate the cotranslational degradation efficiency (E) for each of the 1,422 proteins: E = (1 − 0.85R) × 100, where 0.85 is the relative protein production of srp1-49 versus WT. An E value means the percentage of a nascent protein degraded cotranslationally in WT cells. (The formulation of this equation was detailed under “Experimental Procedures.”) Based on their E values (supplemental Table S1), the 1,422 proteins were categorized into three groups: group 1 (E < 10%), group 2 (E = 10–30%), and group 3 (E >30%). Group 1 proteins (n = 584) are subjected to no or low level cotranslational degradation. Group 2 proteins (n = 540) are cotranslationally degraded at a medium level. Group 3 proteins (n = 298) undergo high level cotranslational degradation, with > 30% of their nascent chains degraded. When extrapolated to the entire proteome, our data suggest that ∼60% of the yeast proteome may be subjected to CTPD and ∼21% of the protein species may undergo high level CTPD. Thus, CTPD is a robust cellular process involving not only the amount of nascent proteins but also the number of protein species.
FIGURE 2.
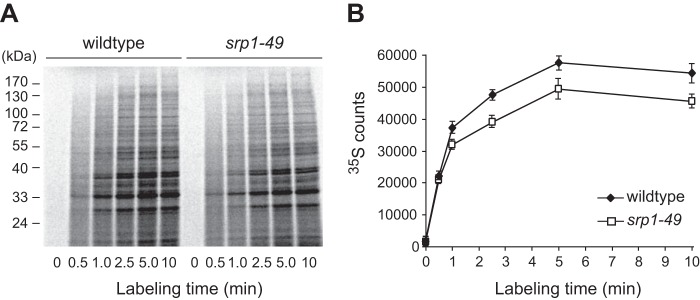
Protein production is lower in srp1-49 than in WT cells. Equal numbers of WT and srp1-49 cells were withdrawn at different time points after the addition of [35S]Met/Cys to the cell cultures. Cell extracts were resolved by SDS-PAGE (A), and the 35S intensity of each lane was quantified by a PhosphorImager and plotted against the labeling time to show the curves of protein synthesis (B). The data shown are the means of three independent experiments.
Rapidly Translated Polypeptides Are Preferred Substrates of CTPD
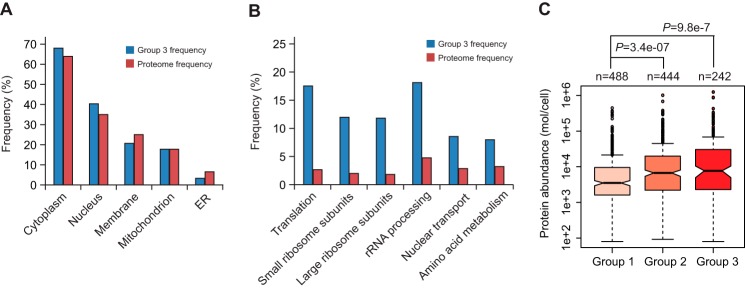
With the compilation of cotranslational degradation efficiencies of 1,422 proteins, we wanted to determine what types of proteins are prone to CTPD. We used Gene Ontology Slim Mapper to compare the distribution of group 3 proteins and the entire yeast S. cerevisiae proteome (6,338 proteins) with respect to subcellular localization and cellular processes. We found that group 3 proteins are present in all cellular compartments. The frequencies of cytoplasmic and nuclear proteins were modestly higher in group 3 than the proteome (68.6% versus 63.6%, and 40.1% versus 34.6%) (Fig. 3A). In contrast, the cluster frequencies of membrane and endoplasmic reticulum (ER) proteins were lower compared with the proteome frequencies (20.4% versus 25.4% and 3.3% versus 6.5%). These data indicate that cytoplasmic and nuclear proteins are more accessible to cotranslational degradation than membrane and ER proteins. This is not unexpected because the nascent chains of secretory and ER proteins are guided to and are protected by the ER translocation channel immediately after emerging from the ribosome. There was no difference in the frequency of mitochondrial proteins between group 3 and the proteome.
FIGURE 3.
Characterization of proteins undergoing cotranslational degradation. A, subcellular distribution of proteins subjected to high level cotranslational degradation. Gene Ontology Slim Mapper was used to compare the subcellular distribution of group 3 proteins (cluster frequency) and the entire yeast proteome (proteome frequency). B, frequency comparison between group 3 proteins and the yeast proteome in various cellular processes. C, correlation between CTPD and protein abundance. Box plotting analysis was conducted to compare the distribution of protein abundance across group 1, 2, and 3 proteins. The protein abundance data were obtained from a previous data set (42) and included in supplemental Table S2. The R package was used for this analysis. Box plots indicated the data distribution through median, 25 and 75% quartiles (filled box), and 1.5× the interquartile range (dashed line). The p values were calculated using the Wilcoxon nonparametric statistical hypothesis test.
Group 3 proteins and the proteome displayed comparable frequencies for most cellular processes classified by Gene Ontology Slim Mapper. However, we noticed that proteins involved in several cellular processes are enriched in group 3 (Fig. 3B). The most striking differences between group 3 and the proteome were the frequencies of proteins participating in translation and ribosome biogenesis, two closely related and overlapping processes. The frequency of translation proteins in group 3 was 17.4%, whereas it was only 2.7% in the proteome. The cluster frequencies for ribosomal small and big subunits and rRNA processing proteins were 11.7, 11.4, and 18.1%, but only 2.1, 1.5, and 4.8% for the proteome. Note that many of these proteins are RNA-binding proteins and participate in more than one process related to protein translation and ribosome biogenesis. In addition, the cluster frequencies were substantially higher than the proteome frequencies for proteins in nuclear transport (8.7% versus 2.8%) and amino acid metabolism (8.1% versus 3.3%). These results indicate that proteins involved in translation, ribosome biogenesis, nuclear transport, and amino acid metabolism are more likely to be targeted for cotranslational degradation. Interestingly, many of these proteins are abundant in the cell. This is consistent with the analysis for distribution of protein abundance across group 1, 2, and 3 proteins, which revealed a strong correlation between CTPD and protein abundance (Fig. 3C and supplemental Table S2). The abundance medians of group 1, 2, and 3 proteins were 3,508, 6,725, and 7,721 molecules per cell, and the mean numbers were 14,338, 26,622 and 41,570, respectively. This analysis raises the interesting possibility that CTPD may prevent aggregation of abundant proteins such as RNA-binding proteins in the cell by keeping their concentration below the threshold (more discussion below).
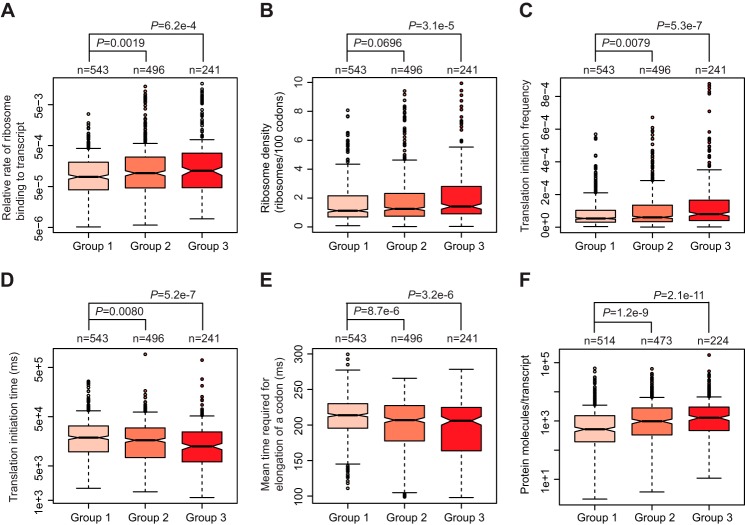
It is of interest to note that many ribosome biogenesis proteins such as ribosome proteins are rapidly translated. We decided to examine whether there is a correlation between CTPD and the translation efficiency. Taking advantage of a recent genome-wide analysis, which documented the parameters that could affect translation in S. cerevisiae (27), we acquired several key translation parameters for the proteins identified by our proteomic analysis. These parameters included relative rate of ribosome binding to transcripts,ribosome density, translation initiation frequency, and time required for translation initiation and elongation (supplemental Table S3). The relative rate of ribosome binding to transcripts, ribosome density, and translation initiation frequency were significantly higher in group 3 proteins than in group 1 proteins (Fig. 4, A–C). On the other hand, the time required for translation initiation and the mean time for elongation of one codon were significantly shorter for proteins in group 3 than in group 1 (Fig. 4, D and E). Consistently, the number of protein molecules produced per transcript was significantly larger for group 3 proteins than group 1 proteins (Fig. 4F and supplemental Table S3). The distribution of these translation parameters for group 2 proteins was between group 1 and 3 proteins. These results indicate that CTPD occurs preferentially to rapidly translated polypeptides.
FIGURE 4.
CTPD occurs preferentially to rapidly translated polypeptides. Box plotting analysis was performed to compare the distribution of relative rate of ribosome binding to transcripts (A), ribosome density (B), translation initiation frequency (C), time required for translation initiation (D), mean time for elongation of a codon (E), and number of protein molecules generated per transcript (F) across group 1, 2, and 3 proteins. The data used for plotting are shown in supplemental Table S3. The unit of time was millisecond (ms).
CTPD Is Inversely Correlated to Cotranslational Protein Folding
It has been suggested that the speed of translation elongation influences cotranslational folding of nascent polypeptides (28). Our observation that rapidly translated polypeptides are preferred substrates for CTPD strongly suggests an inverse correlation between CTPD and cotranslational protein folding. The compilation of the intrinsic sequence features of more than 50% of the yeast proteome in present databases and the CTPD efficiencies of 1,422 proteins from our proteomic analysis enabled us to examine the relationship between CTPD and cotranslational protein folding on a proteome scale. We first examined the correlation between CTPD and the protein sequence length, an important determinant of protein folding kinetics. It has been shown that long polypeptides are particularly challenged to fold cotranslationally (29). The lengths of the 1,422 proteins identified by SILAC-PUNCH-P were obtained from the S. cerevisiae genome database (supplemental Table S4). As shown in Fig. 5A, there was a strong correlation between CTPD and the protein sequence length. The length medians of group 1, 2, and 3 proteins were 487, 571, and 726 amino acids, and the mean lengths were 616, 721, and 850 amino acids, respectively. Apparently, proteins with longer sequence length are more susceptible to cotranslational degradation. We went on to investigate the effect of intrinsically disordered sequences on CTPD. To this end, we integrated our SILAC data with the data sets that compiled the positions and lengths of disordered regions of 3,273 yeast proteins (30). We were able to extract information for 958 of the 1,422 proteins (supplemental Table S5). We focused on the difference in distribution of N-terminal and internal disordered regions between group 1 and 3 proteins. As defined previously (30), the length of an N-terminal disordered region was counted as the number of residues predicted to be disordered at the N terminus of a protein, allowing for minor stretches of up to three structured residues in between the disordered residues. An internal disordered region was defined as a continuous stretch of at least 40 disordered residues in the middle of a protein. We found a significant correlation between CTPD and the N-terminal disorder length (Fig. 5B). Group 3 proteins tend to have longer N-terminal disordered regions than group 1 proteins. The average lengths were 16 and 28 amino acids for group 1 and 3 proteins. By contrast, the distribution of internal disordered regions, including the number and the length, were similar between group 1 and 3 proteins (data not shown). These results indicate that CTPD is counteracted by cotranslational protein folding and that the N-terminal folding kinetics but not the final folded structure of the nascent polypeptide affects the CTPD efficiency. Consistently, we found no significant difference in the disorder fraction and the average disorder score between group 1 and 3 proteins (Fig. 5, C and D). These two parameters are commonly used to predict overall protein structure (29, 30).
FIGURE 5.
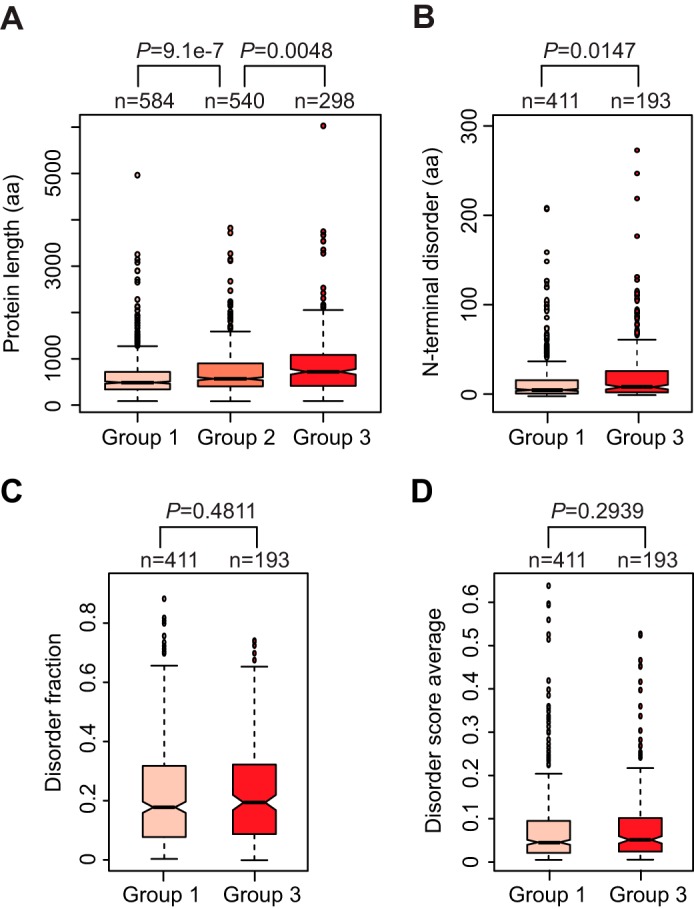
Analysis of the relationship between CTPD and intrinsic protein sequence features. A, long proteins are more susceptible to cotranslational degradation than short proteins. The lengths of 1,422 proteins were obtained from the S. cerevisiae genome database (supplemental Table S4). Distribution of protein length across groups 1, 2, and 3 was assessed by box plotting analysis. B, N-terminal disorder affects CTPD. Distribution of N-terminal disorder lengths of group 1 and 3 proteins was examined by box plotting analysis. The length of N-terminal disorder was counted as the number of disordered residues at the N terminus of a protein, allowing for minor (up to three consecutive residues) stretches of structured residues. C and D, CTPD is not related to the overall fraction of disorder of a protein. Box plotting analysis was conducted to compare the distribution of disorder fraction (C) and disorder score average (D) between group 1 and 3 proteins. Disorder fraction means the fraction of a protein predicted to be disordered, which was calculated by dividing disorder content by protein length (30). The average disorder score of a protein is the sum of the per residue scores divided by protein length. The primary data used for this analysis are shown in supplemental Table S5.
CTPD Is Not Related to the Half-lives of Mature Proteins
We next investigated the relationship between CTPD and the protein half-life. By integrating our SILAC data with a previously published data set containing the half-lives of 3,751 yeast proteins (31), we acquired the half-lives for 1,079 of the 1,422 proteins (supplemental Table S6). Scatter plotting showed no significant correlation between CTPD efficiencies and half-lives of the 1,079 proteins (Fig. 6A). Consistently, there was no statistically significant difference in half-life distribution between group 1 and 3 proteins (Fig. 6B). We also compared the distribution of CTPD efficiencies between short- and long-lived proteins. The median half-life of yeast proteins is ∼43 min, which was used to categorize short- and long-lived proteins (31). The distribution of CTPD efficiencies was similar between short- and long-lived proteins (Fig. 6C). Thus, CTPD is not related to the half-lives of mature proteins. This conclusion is in line with the lack of correlation between CTPD and internal disordered regions, which often serve as an important determinant of the half-life of a protein (32). Consistent with our observation, a recent study showed that there is no correlation between cotranslational protein ubiquitylation and the protein half-life (33).
FIGURE 6.
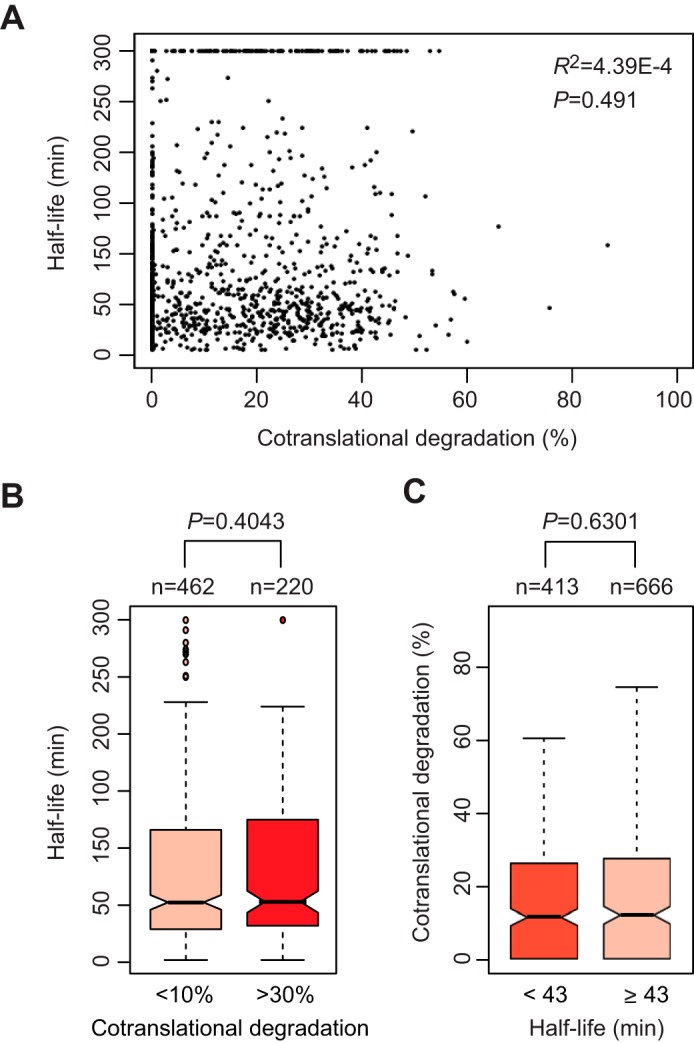
CTPD is unrelated to the protein half-life. A, scatter plotting analysis showing no significant correlation between CTPD and the protein half-life. The half-lives of 1,081 proteins were acquired from a previously published data set and integrated with our SILAC data (supplemental Table S6). A half-life of >300 min was counted as 300 min in accordance with the previous report (31). B, box plotting analysis to compare the distribution of protein half-lives across group 1 and 3 proteins. C, similar distribution of CTPD efficiencies between short- and long-lived proteins. The median half-life of yeast proteins is ∼43 min, which was used to classify short- and long-lived proteins.
Discussion
The extent, mechanism, and biological significance of CTPD are long standing and important open questions in the field of proteostasis. Although it has been shown that a substantial amount of nascent proteins are degraded cotranslationally in eukaryotic cells, the number and types of proteins subjected to CTPD remain unknown. The current study demonstrates that a large number of proteins are CTPD substrates. We estimate that ∼60% of the yeast proteome are accessible to CTPD and ∼21% protein species undergo high level CTPD. Thus, CTPD is a robust process, involving not only the amount of nascent proteins but also the number of protein species. We found that proteins involved in translation, ribosome biogenesis, nuclear transport, and amino acid metabolism are more likely to be targeted for cotranslational degradation. What are the principles that determine the susceptibility of nascent proteins to cotranslational degradation? Our bioinformatic analysis revealed a strong correlation between CTPD and the translation efficiency. CTPD occurs preferentially to rapidly translated polypeptides. It was suggested that the translation efficiency could affect cotranslational folding of nascent chains (28). Slow translation presumably provides more time for the N terminus of a nascent chain to fold before the appearance of potentially interfering C-terminal domain. This may be especially important for appropriate folding of multidomain proteins. Recent studies using engineered proteins showed that increasing the rate of translation elongation by optimizing codon usage reduced the efficiency of cotranslational protein folding (34–36). It is reasonable to speculate that rapidly translated nascent chains are prone to CTPD because of their poor cotranslational folding efficiency. This speculation is supported by the observation that proteins with long sequence length are more susceptible to CTPD than proteins with short sequence length. It has been documented that long polypeptides are particularly challenged to fold cotranslationally (29). Our study for the first time reveals a strong inverse correlation between CTPD and cotranslational protein folding on a proteome scale. It is worthy to note that CTPD is influenced by N-terminal disordered sequences but not internal disordered regions. This suggests that CTPD occurs before the nascent polypeptide has a chance to fold globally. Consistently, the CTPD efficiency is not related to the protein half-life, which is determined by the global protein structure.
This study provides insight into the biological significance of CTPD. It has been repeatedly assumed that CTPD plays a role in protein homeostasis by degrading misfolded nascent polypeptides resulting from translational errors before they fall off the ribosome, but evidence has been missing. Our finding that CTPD preferentially occurs to rapidly translated polypeptides and is inversely correlated to cotranslational folding supports the assumed function of CTPD. Accelerating translation likely increases the chance of incorporation of incorrect amino acids into nascent polypeptides because of the tradeoff between translation speed and accuracy (37). CTPD can efficiently remove the erroneous nascent chains accompanying fast translation, thereby preventing the accumulation of misfolded, defective, toxic proteins in the cell. Another potential function of CTPD is to gauge the protein concentration in the cell to prevent protein aggregation. This hypothesis is supported by the observation that many CTPD substrates including RNA-binding proteins are abundant proteins in the cell. One can image that, without CTPD, these proteins would be accumulated to an extent to which the cell can no longer tolerate. Recent studies showed that RNA-binding proteins with prion-like domains tend to form droplets and can further develop into fibrous structures (aggregation) given enough time and under the right conditions (38–41). Interestingly, aggregation of RNA-binding proteins has been observed only in solution, not in living cells. It is possible that CTPD serves to keep the concentration of these RNA-binding proteins below the threshold inside the cell. It will be of great interest and importance to further elucidate the mechanisms and biological ramifications of CTPD, an understudied and yet important area of proteostasis.
Author Contributions
Y. X. coordinated the project, designed and performed experiments, analyzed data, and wrote the manuscript. S.-W. H. designed and conducted experiments, analyzed data, and wrote the manuscript. D. J. performed experiments and analyzed data. W. H. analyzed data and reviewed and edited the manuscript. All authors analyzed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank C. Enenkel, G. Fink, N. Davis, S. Jinks-Robertson, and M. Nomura for yeast strains and plasmids. We thank P. Stemmer and N. Carruthers for mass spectrometry. The Proteomics Core of Wayne State University/Karmanos Cancer Institute is supported by National Institutes of Health Grant P30 CA022453-28).
This work was supported in part by National Science Foundation Grant MCB-0816974 and a fund from the Office of the Vice President for Research at Wayne State University (to Y. X.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S6.
- CTPD
- cotranslational protein degradation
- RNC
- ribosome-nascent chain complex
- Mut
- mutant
- ER
- endoplasmic reticulum.
References
- 1. Kramer G., Boehringer D., Ban N., and Bukau B. (2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 [DOI] [PubMed] [Google Scholar]
- 2. Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C. C., Li G.-W., Zhou S., King D., Shen P. S., Weibezahn J., Dunn J. G., Rouskin S., Inada T., Frost A., and Weissman J. S. (2012) A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hartl F. U., Bracher A., and Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 4. Pechmann S., Willmund F., and Frydman J. (2013) The ribosome as a hub for protein quality control. Mol. Cell 49, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Preissler S., and Deuerling E. (2012) Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37, 274–283 [DOI] [PubMed] [Google Scholar]
- 6. Goldberg A. L., and Dice J. F. (1974) Intracellular protein degradation in mammalian and bacterial cells. Annu. Rev. Biochem. 43, 835–869 [DOI] [PubMed] [Google Scholar]
- 7. Knowles S. E., Gunn J. M., Hanson R. W., and Ballard F. J. (1975) Increased degradation rates of protein synthesized in hepatoma cells in the presence of amino acid analogues. Biochem. J. 146, 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bradley M. O., Hayflick L., and Schimke R. T. (1976) Protein degradation in human fibroblasts (WI-38): effect of ageing, viral transformation and amino acid analogues. J. Biol. Chem. 251, 3521–3529 [PubMed] [Google Scholar]
- 9. Shakespeare V., and Buchanan J. H. (1976) Increased degradation rates of proteins in ageing human fibroblasts and in cells treated with an amino acid analog. Exp. Cell Res. 100, 1–8 [DOI] [PubMed] [Google Scholar]
- 10. Poole B., and Wibo M. (1973) Protein degradation in cultured cells. The effect of fresh medium, fluoride, and iodoacetate on the digestion of cellular protein of rat fibroblasts. J. Biol. Chem. 248, 6221–6226 [PubMed] [Google Scholar]
- 11. Wheatley D. N., Giddings M. R., and Inglis M. S. (1980) Kinetics of degradation of “short-” and “long-lived” proteins in cultured mammalian cells. Cell Biol. Int. Rep. 4, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 12. Schubert U., Antón L. C., Gibbs J., Norbury C. C., Yewdell J. W., and Bennink J. R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 [DOI] [PubMed] [Google Scholar]
- 13. Yewdell J. W., Antón L. C., and Bennink J. R. (1996) Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J. Immunol. 157, 1823–1826 [PubMed] [Google Scholar]
- 14. Yewdell J. W., and Nicchitta C. V. (2006) The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol 27, 368–373 [DOI] [PubMed] [Google Scholar]
- 15. Suzuki T., and Varshavsky A. (1999) Degradation siganls in the lysine-asparagine sequence space. EMBO J. 18, 6017–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xie Y., and Varshavsky A. (2002) UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat. Cell Biol. 4, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 17. Turner G. C., and Varshavsky A. (2000) Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117–2120 [DOI] [PubMed] [Google Scholar]
- 18. Ha S.-W., Ju D., and Xie Y. (2014) Nuclear import factor Srp1 and its associated protein Sts1 couple ribosome-bound nascent polypeptides to proteasomes for cotranslational degradation. J. Biol. Chem. 289, 2701–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yano R., Oakes M. L., Tabb M. M., and Nomura M. (1994) Yeast srp1 has homology to armadillo/plakoglobin/β-catenin and participates in apparently multiple nuclear functions including the maintenance of the nucleolar structure. Proc. Natl. Acad. Sci. U.S.A. 91, 6880–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loeb J. D., Schlenstedt G., Pellman D., Kornitzer D., Silver P. A., and Fink G. R. (1995) The yeast nuclear import receptor is required for mitosis. Proc. Natl. Acad. Sci. U.S.A. 92, 7647–7651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Enenkel C., Blobel G., and Rexach M. (1995) Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 270, 16499–16502 [DOI] [PubMed] [Google Scholar]
- 22. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., and Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 23. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 24. Aviner R., Geiger T., and Elroy-Stein O. (2014) Genome-wide identification and quantification of protein synthesis in cultured cells and whole tissues by puromycin-associated nascent chain proteomics (PUNCH-P). Nat. Protoc. 9, 751–760 [DOI] [PubMed] [Google Scholar]
- 25. Wilcoxon F. (1945) Individual comparisons by ranking methods. Biometrics Bulletin 1, 80–83 [Google Scholar]
- 26. Gandhi S. J., Zenklusen D., Lionnet T., and Singer R. H. (2011) Transcription of functionally related constitutive genes is not coordinated. Nat. Struct. Mol. Biol. 18, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siwiak M., and Zielenkiewicz P. (2010) A comprehensive, quantitative, and genome-wide model of translation. PLoS Comput. Biol. 6, e1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chaney J. L., and Clark P. L. (2015) Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys. 44, 143–166 [DOI] [PubMed] [Google Scholar]
- 29. Ouyang Z., and Liang J. (2008) Predicting protein folding rates from geometric contact and amino acid sequence. Protein Sci. 17, 1256–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van der Lee R., Lang B., Kruse K., Gsponer J., Sánchez de Groot N., Huynen M. A., Matouschek A., Fuxreiter M., and Babu M. M. (2014) Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 8, 1832–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Belle A., Tanay A., Bitincka L., Shamir R., and O'Shea E. K. (2006) Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fishbain S., Inobe T., Israeli E., Chavali S., Yu H., Kago G., Babu M. M., and Matouschek A. (2015) Sequence composition of disordered regions fine-tunes protein half-life. Nat. Struct. Mol. Biol. 22, 214–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duttler S., Pechmann S., and Frydman J. (2013) Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Brien E. P., Vendruscolo M., and Dobson C. M. (2012) Prediction of variable translation rate effects on cotranslational protein folding. Nat. Commun 3, 868. [DOI] [PubMed] [Google Scholar]
- 35. Spencer P. S., Siller E., Anderson J. F., and Barral J. M. (2012) Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 422, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu C.-H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M. S., and Liu Y. (2015) Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 59, 744–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J.-R., Chen X., and Zhang J. (2014) Codon-by-codon modulation of translational speed and accuracy via mRNA folding. PLoS Biol. 12, e1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Patel A., Lee H. O., Jawerth L., Maharana S., Jahnel M., Hein M. Y., Stoynov S., Mahamid J., Saha S., Franzmann T. M., Pozniakovski A., Poser I., Maghelli N., Royer L. A., Weigert M., et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 [DOI] [PubMed] [Google Scholar]
- 39. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A. P., Kim H. J., Mittag T., and Taylor J. P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin Y., Protter D. S., Rosen M. K., and Parker R. (2015) Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang H., Elbaum-Garfinkle S., Langdon E. M., Taylor N., Occhipinti P., Bridges A. A., Brangwynne C. P., and Gladfelter A. S. (2015) RNA controls polyQ protein phase transitions. Mol. Cell 60, 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., and Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.