Abstract
Mammalian sirtuins (SIRT1-7) are members of a highly conserved family of nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylases that regulate many biological processes including metabolism, genome stability, and transcription. Among the seven human sirtuins, SIRT7 is the least understood, to a large extent due to the lack of enzymatic activity in vitro. Here we reported that SIRT7 can be activated by DNA to hydrolyze acetyl group from lysine residues in vitro on histone peptides and histones in the chromatin context. Both N- and C- termini of SIRT7 are important for the DNA-activated deacetylase activity. The regulatory mechanism of SIRT7 is different from that of SIRT6, which also showed increased activity on chromatin substrates, but the deacetylase activity of SIRT6 on peptide substrate cannot be activated by DNA. This finding provides an improved enzymatic activity assay of SIRT7 that will promote the development of SIRT7 modulators. Further investigation into the activation mechanism of SIRT7 by DNA could provide new insights into its biological function and help the development of sirtuin activators.
The silent information regulator-2 (Sir2) family of proteins, or sirtuins, are evolutionarily conserved NAD+-dependent protein lysine deacetylases1–3. Mammals encode seven sirtuins (SIRT1-SIRT7)4, which govern important biological processes including transcriptional regulation, genome stability, stress response, metabolism, and aging5, 6. The seven mammalian sirtuins have different subcellular localizations, with SIRT1, SIRT6, and SIRT7 being mainly nuclear, SIRT2 being predominantly cytosolic, and SIRT3-5 being mainly mitochondrial7. Among the seven human sirtuins, the molecular function of SIRT7 is the least understood. SIRT7 is enriched in nucleoli where it activates rRNA gene transcription by interacting with Pol I and upstream binding transcription factor (UBF)8, 9. In response to metabolic stress, SIRT7 is released from nucleoli leading to hyperacetylation of PAF53, a subunit of Pol I complex, and decreased Pol I transcription10. SIRT7 also regulates Pol II-mediated mRNA synthesis. It can be recruited by specific transcription factors (ELK411 and Myc12) to deacetylate H3K18 acetyl modification specifically and thus suppress downstream gene transcription. SIRT7 has also been reported to regulate transcription of mitochondrial biogenesis genes by directly deacetylating and activating a transcription factor GABPβ113. SIRT7 is also involved in attenuating stress-activated protein kinase (SAPK) and p53-mediated stress responses14, 15 and regulating Pol III-mediated tRNA transcription and ribosomal biogenesis16, but the exact molecular mechanism remains unknown.
Despite all the reports that connect SIRT7’s deacetylase activity to its biological functions, the in vitro deacetylase activity of SIRT7 in many cases cannot be detected. For example, no appreciable amount of deacetylation product was observed by high pressure liquid chromatography (HPLC)-based assays using peptide substrates17. This was also observed in our laboratory (Figure 1A–B). Inspired by the fact that SIRT7 is localized mainly in the nucleus and involved in regulating transcription and protein synthesis11, 16, we postulated that DNA might affect its enzymatic activity, similar to SIRT6 that displays a nucleosome-dependent deacetylase activity18.
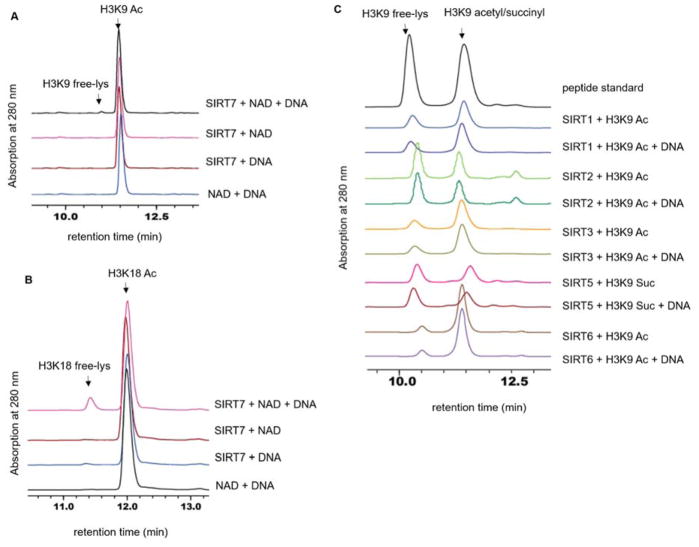
Figure 1. SIRT7 deacetylase activity is significantly enhanced by DNA.
(A) Overlaid HPLC traces showing SIRT7-catalyzed hydrolysis of H3K9 acetyl peptide with DNA. (B) HPLC traces showing SIRT7-catalyzed hydrolysis of H3K18 acetyl peptide with DNA. (C) Overlaid HPLC traces showing that other sirtuin proteins cannot be activated by DNA.
SIRT7 was expressed in E. coli and affinity purified to near homogeneity. An HPLC assay was used to monitor the activity of SIRT7 on H3K9 and H3K18 acetyl (H3K9 Ac and H3K18 Ac) peptides in the absence or presence of double-stranded DNA (dsDNA, salmon sperm DNA sheared to an average size less than 2000bp). In the absence of dsDNA, no deacetylation was observed on either peptide substrate. Strikingly, in the presence of dsDNA, SIRT7 displayed deacetylase activity on both H3K9 Ac and H3K18 Ac peptides (Figure 1A–B). SIRT7 activity increased as the mass ration of DNA to SIRT7 was increased and the activity almost reached a plateau when the ratio reaches 1:1 (Figure S1). In later studies, we generally used 3:1 DNA to SIRT7 mass ratio to make sure SIRT7 was saturated by DNA. Interestingly, all the other sirtuin proteins (except for SIRT4, which we could not acquire the recombinant protein from E. coli) could not be activated by dsDNA (Figure 1C). Even SIRT6, which is closest to SIRT7 and was shown to have a nucleosome-dependent deacetylation activity, could not be activated by dsDNA on peptide substrates. Thus, among all the human sirtuins, the activation of the deacetylase activity on peptide substrates by DNA is a unique property of SIRT7.
We further determined the turnover number (kcat) and Michaelis constant (Km) of SIRT7 in the presence of dsDNA (Table 1). Again, without dsDNA we could not detect the activity and thus could not determine the kinetic constants. Consistent with the reported activity screen11, H3K18 Ac peptide turned out to be a significantly better (16-fold) substrate of SIRT7 compared to H3K9 Ac peptide. For H3K18 Ac, SIRT7 displayed a kcat value of 0.16 min−1 and a Km value of 167 μM (kcat/Km 16 M−1s−1). In contrast, for H3K9 Ac, we could not saturate SIRT7 and thus could only obtain the kcat/Km value, which was 1.0 M−1s−1. This is the first time that we could reliably detect the enzymatic activity of SIRT7 in vitro.
Table 1.
Catalytic efficiencies of SIRT7 full-length (FL) and truncations on acetyl peptides
| kcat (min−1) | Km (μM) | kcat/Km (M−1s−1) | |
|---|---|---|---|
| H3K9 Ac, FL | NA*** | NA | NA |
| H3K9 Ac, FL + ds DNA* | ND** | ND | 1.0 |
| H3K18 Ac, FL | NA | NA | NA |
| H3K18 Ac, FL + ds DNA | 0.16 ± 0.02 | 167 ± 43 | 16 |
| H3K18 Ac, ΔN + ds DNA | ND** | ND | ~2.0 |
| H3K18 Ac, ΔC + ds DNA | NA*** | NA | NA |
| H3K18 Ac, core + ds DNA | NA | NA | NA |
dsDNA: SIRT7=3:1 (mass ratio).
ND: kcat and Km cannot be determined because V~[S] is linear, thus only kcat/Km value can be determined.
NA: product formation cannot be detected.
FL: 1-400; ΔN: 56-400; ΔC: 1-364; core: 56-364
We next investigated whether SIRT7 could be activated by DNA on histone protein substrates. We incubated calf thymus histones with recombinant SIRT7 in the presence/absence of dsDNA. The H3K18 Ac level was then blotted using an H3K18Ac-specific antibody. SIRT7 exhibited no deacetylase activity on purified histone proteins. Unexpectedly, addition of exogenous DNA did not significantly decrease the H3K18 Ac level (Figure 2A).
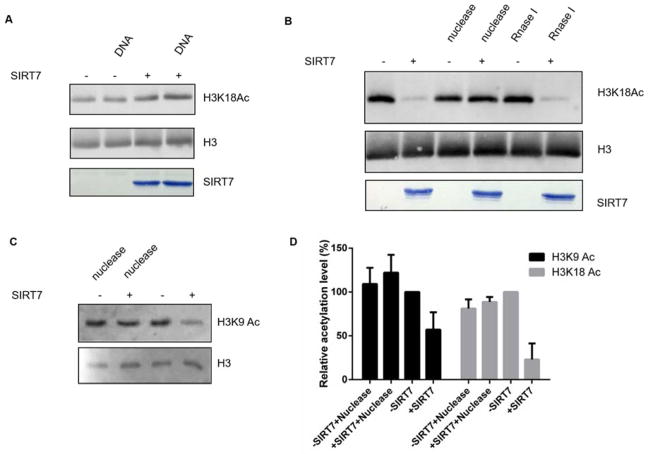
Figure 2. SIRT7 is activated by genomic DNA on chromatin substrates.
(A) SIRT7 cannot be activated by exogenous DNA on histone proteins. (B) SIRT7 can deacetylate H3K18 in the chromatin context. (C) Genomic DNA is required for SIRT7 to remove H3K9 Ac from chromatin substrate. (D) Quantification of western blot results shown in (B & C). The levels of H3K9 Ac and H3K18 Ac were normalized by the amount of histone H3 proteins. The relative acetylation level was calculated by setting the negative control (SIRT7 minus reaction) level to 100%.
A nucleosomes consists of a segment of DNA wrapped around a histone octamer. Therefore, we speculated that chromatin compositional DNA may activate SIRT7 to remove histone acetylation. We first extracted chromatin fraction from HEK 293T cells as reported19. Using the chromatin substrates, SIRT7 displayed robust NAD+-dependent H3K18 Ac deacetylase activity. Furthermore, nuclease cocktail treatment completely abolished the deacetylase activity on H3K18Ac while RNase treatment did not, suggesting that chromatin DNA is required for SIRT7’s activity on H3K18 (Figure 2B). We also tested SIRT7’s activity on H3K9 Ac with and without nuclease treatment (Figure 2C) and quantified the acetylation level (Figure 2D). Interestingly, in the context of chromatin, SIRT7 was able to remove acetyl group from H3K9. We further performed a time-course analysis of SIRT7 on H3K18 Ac in the context of chromatin (Figure S2). SIRT7 was able to hydrolyze ~50% of H3K18 Ac within 90 minutes.
We next investigated which domain(s) of SIRT7 is responsible for its activation by DNA. Sirtuin proteins contain a conserved central catalytic core flanked by highly variable N- and C-terminal extensions20. Seven human sirtuins are categorized into four classes, where SIRT6 and SIRT7 belong to class IV 4. Bioinformatics prediction of DNA-binding sites of SIRT7 suggested that the N- and C-termini that are rich in Arg and Lys residues could potentially play an important role in binding to negatively charged DNA (Figure S3 and S4)21. Thus, we hypothesized that the N- and C- termini of SIRT7 could be essential for its activation by DNA.
We obtained full length (FL, residue 1-400) and several truncated SIRT7 lacking either the N- or C-terminal extension: the N-terminal deletion (ΔN, residue 56-400), the C-terminal deletion (ΔC, residue 1-364), and the core domain (56-364) (Figure 3A–B). Using the H3K18 Ac peptide, we conducted activity assays for these four proteins. With dsDNA as the in vitro activator, we found SIRT7 ΔC and the core domain did not exhibit detectable enzymatic activity within the detection limit of the HPLC-based assay (Figure 3C). SIRT7 ΔN was only slightly activated by DNA. Kinetics studies indicated that the catalytic efficiency of SIRT7 ΔN (kcat/Km ~2.0 M−1s−1) was only 1/8 of FL SIRT7 (kcat/Km ~16.0 M−1s−1) in the presence of dsDNA (Table 1).
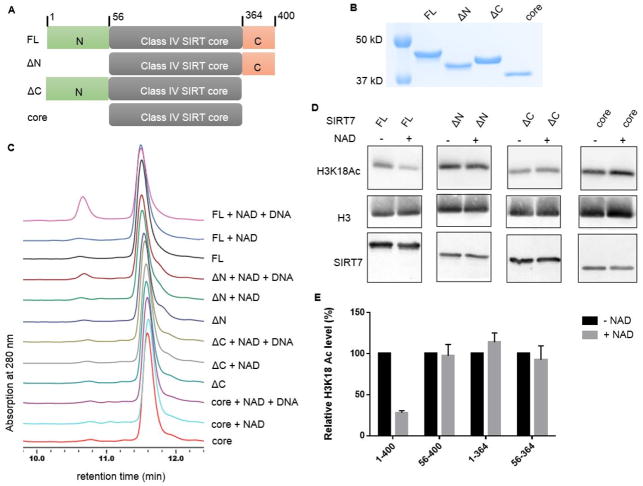
Figure 3. Both the N- and C- termini of SIRT7 are important for its activation by DNA.
(A) Scheme showing the full length (FL, residue 1-400), N-terminal deletion (ΔN, residue 56-400), C-terminal deletion (ΔC, residue 1-364), and core domain (56-364) of SIRT7. (B) Coomassie blue-stained gel showing the purity of SIRT7 deletion mutants. (C) HPLC traces showing the activity of different SIRT7 deletion mutants with and without dsDNA on H3K18 Ac peptide. (D) The deacetylase activities of SIRT7 FL and deletion mutants on H3K18 Ac on chromatin substrates. Reactions without NAD were used as negative controls. (E) Quantification of the Western blot results in (D). The H3K18 Ac level in each reaction was normalized by the amount of histone H3 protein. The relative H3K18 Ac level was calculated by setting the level of negative control to 100%.
We further examined whether SIRT7 truncations could deacetylate H3K18 Ac on chromatin substrates. As shown in Figure 3D and quantified in Figure 3E, only full length SIRT7 was able to efficiently deacetylate H3K18 Ac group while ΔN, ΔC and core domain exhibited no activity, consistent with the results obtained using the peptide substrate.
In summary, our enzymology data demonstrated that SIRT7 has extremely low basal deacetylase activity on histone peptides, but the activity can be significantly increased by dsDNA. When working on physiologically relevant histone protein substrates, exogenous DNA did not activate SIRT7’s deacetylase activity, but the chromatin compositional DNA activated SIRT7 to deacetylate H3K18 Ac. Moreover, studies with SIRT7 truncations implicated that both N- and C- termini are important for its dsDNA-activated deacetylase activity. Interestingly, the N- and C-terminal domains of SIRT7 have been previously implicated in mediating nuclear and nucleolar localization22 and interaction with Myb-binding protein 1a23, a SIRT7 inhibitor. Thus, it appears that the N- and C-terminal domains of SIRT7 play multiple roles in regulating SIRT7 physiological function. Our work demonstrates for the first time that SIRT7 enzymatic activity could be regulated by DNA molecules and identified regions of SIRT7 that are important for the activation. SIRT7 has been widely known as a transcription regulator11–13. The results presented here suggest that DNA binding and activation could serve as a molecular switch that turns on SIRT7 when it approaches histone substrates on chromatin.
SIRT7 is known to be important for regulating fatty liver formation12, 13, 24 and tumorigenesis11, 25–29. Thus, small molecule inhibitors of SIRT7 will be useful probes to further explore the biological function and therapeutic potential of SIRT7. However, so far no potent SIRT7 inhibitors have been reported. This is mainly due to lack of a reliable in vitro assay to detect SIRT7 activity. Our finding that DNA can significantly activate SIRT7 will fill in this gap and promote the development of SIRT7 inhibitors. Although controversial, sirtuin activators have been reported to provide health benefits30–34. Interestingly, the activation of SIRT1 by small molecules also involves its N-terminal domain35, similar to the activation of SIRT7 by DNA. Further investigation into the mechanism by which SIRT7 is activated by DNA may provide insights that may help the development of novel sirtuin activators.
Methods
SIRT7 purification
The open reading frames of full length human SIRT7 (1-400), ΔN (56-400), ΔC (1-364), and the core domain (56-364) were inserted into the pET28a vector. The plasmid was transformed into E. coli ArcticExpress (DE3) cells. Cells were cultured at 37°C in LB broth medium (BD biosciences catalog# 240220). Isopropyl-β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.2 mM when OD600 was between 1–1.5, and the culture was grown for 48 hours at 10 °C. Cells were harvested and then re-suspended in lysis buffer (50 mM K2HPO4, pH 7.20, 500 mM NaCl, 5% glycerol). The cells were lysed using a cell disrupter. The lysate was centrifuged at 20,000 rpm for 30 min at 4 °C. The supernatant was loaded onto a nickel column (HisTrap HP, GE healthcare life sciences) pre-equilibrated with a buffer containing 50 mM K2HPO4 (pH 7.20), 500 mM NaCl and 5% glycerol. SIRT7 was eluted with a linear gradient of imidazole (0 to 500 mM) in the buffer. The desired fractions were pooled, concentrated and buffer exchanged against the cation exchange buffer (80 mM NaCl, 50 mM K2HPO4, pH 7.20, 5% glycerol). The protein was further loaded onto a cation exchange column (HiTrap SP HP, GE healthcare life sciences) and eluted with a linear gradient of NaCl (80 mM to 1 M) in the buffer containing 50 mM K2HPO4 (pH 7.20) and 5% glycerol. Fractions were assayed for purity on SDS-PAGE and concentrated, snap frozen and stored at −80 °C.
Synthesis of acetyl peptides
Acetyl peptides corresponding to residues 4–13 and 12–24 of Histone H3 (NH2-KQTARKacSTGGWW-COOH and NH2-GGKAPRKacQLATKAWW-COOH) were synthesized by standard solid phase peptide synthesis as described below. Two Trp residues were added to the C-terminus of the peptides to facilitate the detection of peptides by monitoring the ultra-violet (UV) light absorption at 280 nm.
Peptides were synthesized by Fmoc SPPS on an automated peptide synthesizer (Focus XC from aapptec) using standard protocols. 500 mg of Wang resin (100–200 mesh, 1% DVB, 1.2 meq/g) was swollen in 10 mL dichloromethane (DCM) for 6 h. The first amino acid was coupled onto the resin for 3 h in N, N-dimethylformamide (DMF), with a 4-fold excess of amino acid, 4-fold excess of HBTU, and 10-fold excess of diisopropylethylamine (DIEA) followed by addition of 0.1 equivalent of 4-dimethylaminopyridine (DMAP) in 0.2 ml of DCM. The resin was then treated with Ac2O/pyridine (1:9 v/v) for 10 min to cap any remaining reactive functionalities on the resin. The coupling of the rest amino acids was carried out following standard protocols using 4-fold excess of amino acid, 4-fold excess of HBTU, and 10-fold excess of DIEA. Acetyl modification on lysine residues was introduced through the use of Fmoc-Lys(acetyl)-OH. The peptides were cleaved from the resin using a cocktail of trifluoroacetic acid, triisopropylsilane, water, ethanedithiol, thioanisole, and phenol (81.5:1:5:2.5:5:5 by vollume) for 2 h. The crude peptides were purified by preparative HPLC. The identities of the peptides were confirmed using LC-MS (LCQ Fleet, Thermo Scientific).
The purified free-lysine and acetyl H3K9 and H3K18 peptides were dissolved in water. The concentrations of peptides were determined at 280 nm using extinctions coefficient of the two Trp residues attached at the C-termini of the peptides.
HPLC Assay and kinetics
Activity of SIRT7 was detected using HPLC. The reactions contained 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM DTT, 2 mM NAD+, 50 μM H3K9 Ac or H3K18 Ac peptides, and 2 μM SIRT7. Reactions were incubated at 37 °C for 1 h. Salmon sperm DNA mixture (10 mg/ml) was used as the in vitro activator. Mass ratio of DNA to SIRT7 was maintained at 3:1 in the assays unless specified otherwise. The reactions were quenched with 1 volume of 10% (v/v) TFA and spun down for 10 min at 18,000 g to remove proteins. The supernatants were then analyzed by HPLC using a reverse phase analytical column (Kinetex XB-C18 100A, 75 mm × 4.60 mm, 2.6 μm, Phenomenex). The product and substrate peaks were quantified using the absorption peak areas at 280 nm from the two Trp residues added at C-terminus of the peptide substrates.
For kinetics, 2 mM NAD, 50 mM Tris (pH 8.0), 1 mM DTT, 2 μM SIRT7 (full-length) or 8 μM SIRT7 (ΔN, 56-400) was used. Peptide concentration used for H3K18 Ac and H3K9 Ac were 10, 30, 50, 100, 150, 250 and 500 μM with an incubation time of 45 minutes. The quenched reactions were then analyzed by HPLC. The product and substrate peaks were quantified as described above and converted to initial rates, which were then plotted against the peptide concentrations and fitted to the Michaelis-Menten equation using the Graphpad Prism 6 program.
Histone deacetylation assay
Calf thymus histones were obtained from Roche. In vitro deacetylation reactions were performed with 10 μg of SIRT7 and 1 μg of calf thymus histone in deacetylation buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM NAD+, 1 mM DTT), with or without 30 μg of salmon sperm DNA. The reactions were incubated at 37°C for 2h. The reactions were quenched by adding 3X Laemmli buffers and boiling for 15 minutes at 95°C. The acetylation level of histones was assessed by Western blot using H3K9 Ac (Abcam cat# ab4441)- and H3K18 Ac (Abcam cat # ab1191) -specific antibodies. To more quantitatively determine H3K18 Ac level using Western blot, we used a fluorophore-conjugated secondary antibody (Goat anti-rabbit Dylight 488, thermofisher cat # 35552). Signal intensity of all Western blot bands was quantified using ImageJ.
For the chromatin deacetylation assay, we extracted chromatin fraction from 293T cells as previously described12. 293T cells were treated with 2 μM histone deacetylases inhibitor, trichostatin A (TSA) for 2h before harvest. 10 μM SIRT7 protein was incubated with ~1 μM chromatin substrate in deacetylation buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM NAD+, 1 mM DTT). To remove DNA or RNA, 2 μL nuclease cocktail (Thermo Fisher cat. # 88702) or 4 μL Rnase I (Ambion cat. # AM 2294) was added to 30 μL of reaction mixture and incubated at 37°C for 2h. The acetylation levels of H3K18 Ac and H3K9 Ac were determined as described above.
Cell culture
Human 293T cell lines were acquired from the American Type Culture Collection (ATCC). Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) complete medium containing glucose and L-glutamine (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen).
Supplementary Material
Acknowledgments
This work is supported in part by grants from NIH GM086703 (H.L.), GM105933, CA152870 (Q.H.), and HK-RGC C7037-14G (Q.H.).
Footnotes
Supporting information. This material is available free of charge via the Internet.
References
- 1.Imai S-i, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai S-i, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 3.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 5.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haigis MC, Sinclair DA. Mammalian Sirtuins: Biological Insights and Disease Relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics. 2012;11:60–76. doi: 10.1074/mcp.A111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, Seiler J, Santiago-Reichelt M, Felbel K, Grummt I, Voit R. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol Cell. 2013;52:303–313. doi: 10.1016/j.molcel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, Struhl K, Garcia BA, Gozani O, Li W, Chua KF. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, Li P, Cheng HL, Murphy AJ, Valenzuela DM, Luo H, Kapahi P, Krauss R, Mostoslavsky R, Yancopoulos GD, Alt FW, Chua KF, Chen D. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 2013;5:654–665. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, Lee JU, Zeviani M, Romand R, Hottiger MO, Schoonjans K, Auwerx J. A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab. 2014;20:856–869. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 15.Kiran S, Oddi V, Ramakrishna G. Sirtuin 7 promotes cellular survival following genomic stress by attenuation of DNA damage, SAPK activation and p53 response. Exp Cell Res. 2015;331:123–141. doi: 10.1016/j.yexcr.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Tsai YC, Greco TM, Cristea IM. Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol Cell Proteomics. 2014;13:73–83. doi: 10.1074/mcp.M113.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil R, Barth S, Kanfi Y, Cohen HY. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res. 2013;41:8537–8545. doi: 10.1093/nar/gkt642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:W243–248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiran S, Chatterjee N, Singh S, Kaul SC, Wadhwa R, Ramakrishna G. Intracellular distribution of human SIRT7 and mapping of the nuclear/nucleolar localization signal. FEBS J. 2013;280:3451–3466. doi: 10.1111/febs.12346. [DOI] [PubMed] [Google Scholar]
- 23.Karim MF, Yoshizawa T, Sato Y, Sawa T, Tomizawa K, Akaike T, Yamagata K. Inhibition of H3K18 deacetylation of Sirt7 by Myb-binding protein 1a (Mybbp1a) Biochem Biophys Res Commun. 2013;441:157–163. doi: 10.1016/j.bbrc.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Yoshizawa T, Karim MF, Sato Y, Senokuchi T, Miyata K, Fukuda T, Go C, Tasaki M, Uchimura K, Kadomatsu T, Tian Z, Smolka C, Sawa T, Takeya M, Tomizawa K, Ando Y, Araki E, Akaike T, Braun T, Oike Y, Bober E, Yamagata K. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014;19:712–721. doi: 10.1016/j.cmet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Malik S, Villanova L, Tanaka S, Aonuma M, Roy N, Berber E, Pollack JR, Michishita-Kioi E, Chua KF. SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci Rep. 2015;5:9841. doi: 10.1038/srep09841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, Hu Y, Cai T. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep. 2015;5:9787. doi: 10.1038/srep09787. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Shen Q, Park WS, Lee JY, Borlak J, Nam SW. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 28.Wang HL, Lu RQ, Xie SH, Zheng H, Wen XM, Gao X, Guo L. SIRT7 Exhibits Oncogenic Potential in Human Ovarian Cancer Cells. Asian Pac J Cancer Prev. 2015;16:3573–3577. doi: 10.7314/apjcp.2015.16.8.3573. [DOI] [PubMed] [Google Scholar]
- 29.Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X, Zhou Y, Wang H, Pan C, Huang W. Overexpression of sirt7 exhibits oncogenic property and serves as a prognostic factor in colorectal cancer. Clin Cancer Res. 2014;20:3434–3445. doi: 10.1158/1078-0432.CCR-13-2952. [DOI] [PubMed] [Google Scholar]
- 30.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borra MT, Smith BC, Denu JM. Mechanism of Human SIRT1 Activation by Resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 33.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinclair DA, Guarente L. Small-molecule allosteric activators of sirtuins. Annu Rev Pharmacol Toxicol. 2014;54:363–380. doi: 10.1146/annurev-pharmtox-010611-134657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai H, Case AW, Riera TV, Considine T, Lee JE, Hamuro Y, Zhao H, Jiang Y, Sweitzer SM, Pietrak B, Schwartz B, Blum CA, Disch JS, Caldwell R, Szczepankiewicz B, Oalmann C, Yee Ng P, White BH, Casaubon R, Narayan R, Koppetsch K, Bourbonais F, Wu B, Wang J, Qian D, Jiang F, Mao C, Wang M, Hu E, Wu JC, Perni RB, Vlasuk GP, Ellis JL. Crystallographic structure of a small molecule SIRT1 activator-enzyme complex. Nat Commun. 2015;6:7645. doi: 10.1038/ncomms8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.