Abstract
The abundance of ribonucleotides in DNA remained undetected until recently because they are efficiently removed by the Ribonucleotides Excision Repair pathway, a process similar to Okazaki fragment processing after incision by RNase H2. All DNA polymerases incorporate ribonucleotides during DNA synthesis. How many, when and why they are incorporated has been the focus of intense work during recent years by many labs. In this review, we discuss recent advances in ribonucleotide incorporation by eukaryotic DNA polymerases that suggest an evolutionarily conserved role for ribonucleotides in DNA and review the data that indicate that removal of ribonucleotides plays an important role in maintaining genome stability.
Keywords: DNA polymerase, RNase H2, Steric gate, RER, Aicardi-Goutières syndrome, Systemic Lupus Erythematosus
Inclusion of Ribonucleotides During DNA Replication
Eukaryotic DNA replication proceeds at an enormous rate with remarkable accuracy, employing three major DNA polymerases. During embryonic development Drosophila melanogaster duplicates its genome in 8 minutes at 25 °C due in large part to the use of multiple origins of replication [1] Given the size of its genome, the rate of incorporation is about 0.5 mbp/second. As this rapid replication proceeds there are many opportunities for various types of errors. To maintain genome integrity, replicative polymerases are highly accurate in copying DNA. For DNA Pol α, the rate of incorporation of the incorrect base is about 10−4. DNA Pol δ and ε have the ability to proofread the last incorporated deoxynucleoside monophosphate (dNMP) and, if incorrect, remove it using the polymerases’ 3′-exonuclease activity. Proofreading increases their fidelity to a frequency of incorrect insertion of 10−6. Once the DNA exits the polymerase, the mismatch repair (MMR) system scans the newly synthesized strand for proper base pairing with the template, and repairs any mismatch, lowering the error rate to between 10−7 and 10−10[2].
Replication of damaged DNA requires specialized repair polymerases[3] . Replicative polymerases have a very compact binding site that can only accommodate incoming bases that form proper hydrogen bonding with the template. This tight binding pocket cannot fit template DNA containing damaged or modified bases[4, 5]. To prevent DNA synthesis from stalling when encountering damage, replicative polymerases are replaced by translesion polymerases, which have a larger active site and therefore have lower base selectivity[3]. Translesion polymerases vastly outnumber replicative polymerases, as each is specialized for polymerizing through different types of DNA damage. Together they allow completion of DNA replication and repair to maintain genome integrity but at the cost of generating mutations.
Until recently, examination of fidelity of DNA replication has mostly concentrated on how the DNA polymerases can insert the correct base, with limited concern for sugar discrimination. The abundant presence of ribonucleotides in chromosomal DNA was not detected until Kunkel’s lab, in an attempt to make mutants of the replicative polymerases to define their roles in replication, found a DNA polymerase ε (Pol ε) mutant that incorporates large numbers of ribonucleotides, and discovered that even the wild-type polymerases incorporate many ribonucleoside monophosphates (rNMPs) into DNA in vivo[6]. These mutants led to the currently accepted division of labor model of chromosomal DNA replication, which places the DNA Pol ε as the major leading strand replicase and Pol α and Pol δ as the enzymes responsible for lagging strand Okazaki Fragment (OF) production[7].
The unexpected finding that in Saccharomyces cerevisiae replicative DNA polymerases incorporate large numbers of ribonucleotides during DNA synthesis[8, 9] has opened a whole new field of research that is improving our understanding of multiple processes and will likely yield even more surprises. We describe here recent studies on incorporation of rNMPs by the major DNA replicases which provide details on how the polymerases select against rNTPs, the frequencies with which each replicative polymerase includes rNMPs, how rNMPs are removed by ribonuclease H2 (RNase H2-see Glossary) and some of the benefits of having rNMPs transiently in DNA as well as the problems when rNMPs remain in double-stranded DNA when RNase H2 is defective.
Steric Gate Limits Ribonucleotide Incorporation
DNA polymerases have a narrow active site suitable for dNTPs that would not easily accommodate the larger substituent at the 2′ position present in rNTPs (OH vs. H)[4]. Most DNA polymerases use a bulky amino acid residue called the “steric gate” located at the entrance of the active site to prevent rNMP incorporation[5, 10]. The steric gate amino acid residues are Glu for A family polymerases (see Glossary) and Tyr or Phe for members of the B, X, Y and RT families[11]. Reducing the size of the side chain of the steric gate residue increases the tolerance for ribonucleotides in the active site, in some cases to the extent of being incorporated as well as deoxyribonucleotides[11]. However, not all DNA polymerases use a bulky side chain to prevent rNMP incorporation, and crystallographic studies showed that X family members, pol β and pol λ, use the peptide backbone segment to exclude rNTPs[12-14].
The steric gate amino acid also plays a role in base discrimination and fidelity in copying the template[15]. A smaller side chain not only improves incorporation of rNMPs but decreases affinity for the correct dNTP as well. Because of its crucial role in base and sugar selectivity, replacing the steric gate tyrosine (Y645) with alanine, in yeast Pol ε results in lethality[16]. However, mutating the adjacent hydrophobic amino acid M644 has a strong effect on fidelity without impairing growth. Sugar selectivity can vary over a thirty fold range for Pol ε when M644 is replaced by Leu or Gly[8]. The former increases selectivity while the Gly substitution decreases sugar discrimination. Therefore, Pol ε is one amino acid away from either being better or worse at ribonucleotide exclusion. Although Leu is instead present at the corresponding site on both Pol α and Pol δ, Met644 is highly conserved in Pol ε from different organisms suggesting a functional value of rNMP incorporation in the leading strand[6, 8]. Polε-M644G possesses a high mutation rate and this observation was exploited in the elegant experiments that led to the conclusion that Pol ε is responsible for leading strand replication while Pol δ synthesizes the lagging strand[7, 17].
Steric gate amino acids in the Y family of translesion DNA polymerases also control sugar selectivity and occasionally have unexpected effects on base fidelity. Mutating the steric gate F35 residue of S. cerevisiae Pol η increases base fidelity at the cost of decreasing sugar selectivity[18]. The same is true for Y39 of Pol ι[19]. For other translesion polymerases the steric gate residue is important for lesion bypass [11]. Some DNA polymerases such as Terminal Deoxynucleotidyl Transferase (TdT)[20, 21] and DNA polymerase μ[20]are extraordinarily able to use rNTPs with little preference for dNTPs. When cellular concentrations of rNTPs/dNTPs (see following section) are used in vitro, as much as 80-90% of the product is composed of rNMPs[20, 21]. This may be a mechanism to accomplish DNA repair in phases of the cell cycle when the concentration of dNTPs is very low, a way to mark DNA for further repair, or a mechanism of programed mutagenesis in antibody diversification[20, 21].
The 3′-exonuclease proofreading domain of replicative DNA polymerases effectively eliminates mispaired bases [2]but it is inefficient at removing ribonucleotides[22, 23] Once a ribonucleotide has been added to the growing chain, incorporation of the next nucleotide proceeds more slowly than extension from a dNMP, possibly facilitating removal by the 3′- exonuclease activity rather than elongation[24, 25]. The abilities of several eukaryotic polymerases to incorporate, extend and bypass ribonucleotides in DNA are summarized in Table 1[2, 6, 8, 12, 14, 19, 20, 22, 26-33].
Table 1.
Base and sugar fidelity of several DNA Polymerases from the four eukaryotic families
| Polymerase | Family [2] |
Functions [2] |
Steric gate |
Base [2] Selectivity |
Sugar Discrimination |
rNMP Extensionb |
rNMP Bypass c |
|---|---|---|---|---|---|---|---|
|
Human
Pol γ |
A | mtDNA replication, BERg |
E895 | 10−5 | 1,100-77,000a [26] |
7-33% [26] |
51% [26] |
|
S. cerevisiae
Pol α |
B | Genome replication |
Y869 | 10−4 | 3,000-10,000d [6] |
100% | 50-75% [27] |
|
S. cerevisiae
Pol δ |
B | Genome replication, NERg, BER, MMRg, DSBRg |
Y613 | 10−6 | 10,000- 1,700,000d [6] |
ND | 7.4-69% [27, 28] |
|
S. cerevisiae
Pol ε |
B | Genome replication, NER, BER, MMR, DSBR |
Y645 | 10−6 | 500-6,700d [6] |
90% [8, 23] |
32-85% [27, 28] |
|
Human
Pol δ |
B | Genome replication, NER, BER, MMR, DSBR |
Y607 | 10−6 | 2000e [22] |
ND | 78-85% [22] |
|
Human
Pol ε |
B | Genome replication, NER, BER, MMR, DSBR |
Y602 | 10−6 | 210-5,300 d [29] |
37-52% [29] |
66% [29] |
|
Human
Pol β |
X | BER, MMR | Y271 | 10−3-10−4 | 2,00-8,200a [14, 20, 30] |
100% [30] |
12 [30] |
|
Human
Pol λ |
X | BER, NHEJg, V(D)J recombination |
Y505 | 10−3-10−4 | 3,000-50,000a [12, 32] |
70% [32] |
ND |
|
Human
Pol μ |
X | NHEJ, V(D)J recombination |
G433 | ND | 1.4-11a [20] |
25% [20] |
1%f [20] |
|
Human
Pol ι |
Y | TLSg, BER, SHMg |
Y39 | 10-10−4 | 1,100a [19] |
100% [19] |
ND |
|
S. cerevisiae
Pol ζ |
B | TLS, DSBR, ICLRg, SHM |
Y980 | 10−3 | 470-4,700d [33] |
ND | 80-95% [34] |
For ribonucleotide incorporation, the discrimination factor = (kcat/Km) of dNTP incorporation opposite to its cognate dNMP in template/ (kcat/Km) of rNTP incorporation opposite to its cognate dNMP in template.
Catalytic efficiency for dNTP incorporation from 3′-rNMP respect to the catalytic efficiency for dNTP incorporation from 3′-dNMP.
Catalytic efficiency of primer extension opposite a DNA template with a rNMP respect to the template with dNMP.
For ribonucleotide incorporation, the discrimination factor was obtained by dividing the percentage of dNMP product by the percentage of rNMP product, and then multiplying by the ratios of nucleotide concentrations and by the differences in enzyme concentrations and reaction times used to extend no more than 20% of the starting substrate.
Incorporation of one rNTP for approximately 2000 dNTPs.
Full RNA template.
(BER) base excision repair, (NER) nucleotide excision repair, (MMR) mismatch repair, (DSBR) double-strand break repair, (NHEJ) nonhomologous end-joining of double-strand breaks, (TLS) translesion synthesis, (SHM) somatic hypermutation, (ICLR) interstrand cross-link repair, (ND) not determined.
Abundance of rNTPs relative to dNTPs affects the frequency of incorporation of rNMPs in DNA
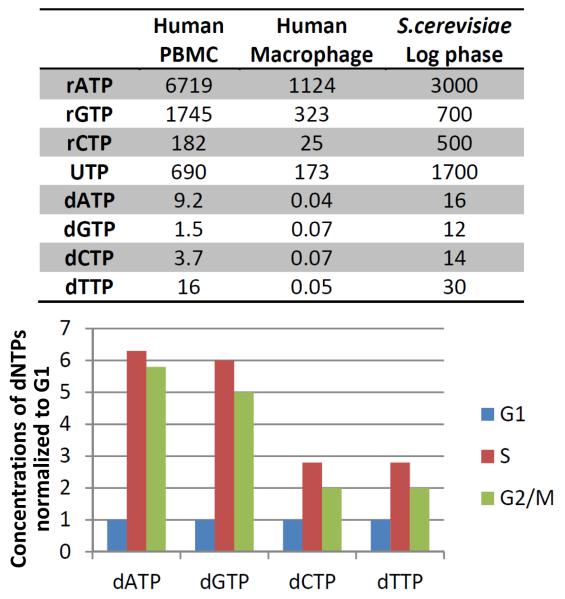
The steric gate is severely challenged in vivo due to the high concentrations of rNTPs[6, 20]. The ratio of cellular dNTPs/rNTPs varies from one organism to another, from tissue to tissue, and with stage of the cell cycle, but always in all cases rNTPs are far more abundant than dNTPs (Figure 1) . Throughout the cell cycle the concentration of rNTPs is relatively constant, however the production of dNTPs increases during the S-phase[34], suggesting fewer ribonucleotides are incorporated during DNA replication and more during repair synthesis outside the S-phase. In terminally differentiated cells, such as human macrophages[35, 36], the dNTP concentrations are very low (Figure 1). To adjust to the large pool of rNTPs, specialized DNA polymerases repairing DNA in these cells are particularly adept at incorporating ribonucleotides. Low levels of dNTPs are maintained in macrophages by SAMHD1, a dNTP triphosphohydrolase, which is degraded upon infection with human immunodeficiency virus type2 (HIV-2) resulting in increased dNTP concentrations to favor viral replication[37].
Figure 1. Concentrations of rNTPs and dNTPs in different cells.
Table (top) shows μM concentration of nucleotides in human peripheral blood mononucleocytes (PBMC)[36], human macrophages[36] and S. cerevisiae log phase cells[6]. Graph (bottom) represents concentrations of dNTPs in S. cerevisiae during the different phases of the cell cycle, normalized to G1 phase levels[34].
Ribonucleotide Excision Repair and Mismatch repair to remove rNMPs
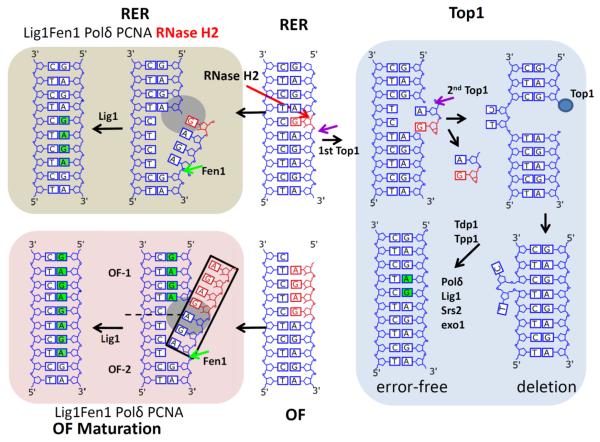
In 1993 Eder and Walder proposed involvement of RNase H2 in removal of ribonucleotides in DNA when they reported RNase H2 incision at a single rNMP in an in vitro synthetic substrate[38], however the significance of this finding was not appreciated until the discovery of the abundance of rNMPs in DNA. A role for RNase H2 in removing rNMPs in DNA in S. cerevisiae was indicated subsequently by Rydberg and Game [39]. Ribonucleotides are efficiently removed by the “Ribonucleotide Excision Repair” (RER) mechanism[40],which is initiated by RNase H2 through cleavage at the 5′-end of the ribonucleotide followed by strand displacement synthesis by Pol δ and flap removal by Fen1 and/or Dna2 and ligation by DNA ligase1 (Figure 2). Except for the first critical step carried out by RNase H2, the subsequent process is identical to OF maturation (Figure 2). RNases H were long thought to be responsible for removing OF primers. While this may occur in bacterial[41] and mitochondrial DNA replication[42], in eukaryotic genome replication Fen1 and Dna2 are the major activities removing the flap structure containing the primer[41] (Figure 2).
Figure 2. Solutions to the presence of ribonucleotides in DNA.
The vertical center depicts a single rNMP in DNA in red (top) and an Okazaki fragment (OF) that has just be synthesized with an RNA primer in red (bottom). RNase H2 cleavage at the top (red arrow) designates the site for ribonucleotide excision repair (RER), the mechanisms of which are shown in the upper left portion, including Pol δ (grey circle), PCNA (not shown), Fen1 (pale green arrow) and DNA ligase 1. In the lower left two OFs are shown. As OF-1 is extended by Pol δ, it displaces from OF-2 the “PriαR-D” (black rectangle; see text). Subsequent steps are the same as seen for RER. The right side describes error-free and error prone (deletions) repair following Top1 cleavage at an rNMP in DNA. Cleavage sites of Top1 are indicated by purple arrows. The first action of Top1 (purple arrow) results in formation of a 2′-3′ cyclic phosphate on the ribose. A second attack of Top1 (purple arrow), in this case 2 nucleotides to the 5′-side of the original event, leads to loss of a dinucleotide containing a dNMP and the rNMP with Top1 remaining attached to the duplex DNA (blue circle). Error-free repair can occur via removal of Top1 by Tdp1 and Tpp1 or Exo 1 Srs2 with Pol δ synthesizing the DNA. The DNAs shown have repetitive sequences, which allows formation of a DNA with two nucleotides of the template DNA flipped out and the gapped strand re-aligned to bring the Top1 in contact with the next nucleotide to facilitate ligation resulting in a two-nucleotide deletion. Bases with green backgrounds indicate newly synthesized DNAs.
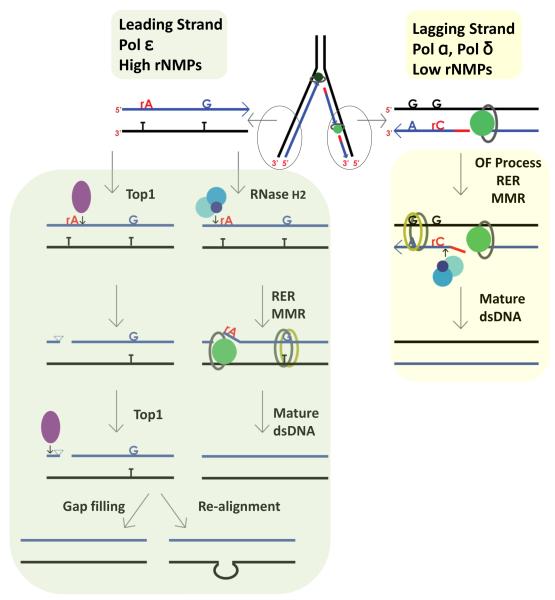
RER could occur concomitantly with or following DNA replication allowing rNMPs to serve as marks of newly synthesized DNA strands. rNMPs are more abundant in the leading strand where topological issues arise. RNase H2 incision at an rNMP could aid in releasing torsional constraints in DNA as well as creating sites to correct errors in replication. MMR provides additional means of ensuring insertion of the correct base but it requires a nick to initiate repair, a function that could be supplied by RNase H2 acting on rNMPs in leading strand DNA[43, 44] (Figure 3, Key Figure). Short lagging strand OFs contain many entry points for MMR (Figure 3, Key Figure). Replication and repair DNA synthesis requires PCNA to bring together the different processing factors and facilitate the reaction. RNase H2, like other components of the RER process, contains a PCNA-interacting peptide (PIP). Eukaryotic[45] and a few archaeal[46] RNases H2 have a PIP box suggesting they are recruited to replication and repair DNA complexes. In vitro RER requires PCNA but not the PIP box on RNase H2[40]. S. cerevisiae and human RNase H2[45, 46] activity is not stimulated by PCNA while that of Archaeoglobus fulgidus RNase HII, a single chain enzyme, is increased in vitro by PCNA[46]. This difference may reflect that the PIP box is present on the accessory RNase H2B subunit in the trimeric eukaryotic subunit which in the structure is quite remote from the catalytic center[47-49], but on the single chain A. fulgidus enzyme is much closer to the active center. It seems reasonable that RNase H2 initiates the RER process and its PIP interacts with PCNA to attract other RER components for subsequent steps of removal of rNMPs. Similarly, cleavage of the ribonucleotide by RNase H2 could attract PCNA and initiate the loading of the MMR system to scan for mismatches in the newly synthesized leading strand (Figure 3, Key Figure).
Figure 3. Key Figure. Processing of rNMPs in leading and lagging strand.
Leading strand replicase Pol ε (dark green circle associated with PCNA represented as grey oval) incorporates high number of rNMPs that are used as strand discrimination for newly synthesized DNA. In yeast Top1 can cleave rNMPs, although in a minor pathway that is only evident in the absence of RNase H2. Top1 (represented as purple filled oval) cleaves at the 3′-end of the rNMP forming a 2′-3′ cyclic phosphate that creates unligatable ends. A second Top1 cleaving a few nucleotides upstream would generate a gap that could be filled by the replicative polymerase in an error-free manner. In a sequence with a tandem repeat Top1 could induce realignment in a mutagenic way. RNase H2 (represented as three blue circles) cleaves at the 5′-end of the rNMP (red rA) initiating the RER process that includes strand displacement synthesis (represented as green circle associated with PCNA represented as grey oval), flap removal and ligation identical to OF maturation. The nick created in the DNA by RNase H2 at the rNMP can be used by the MMR system (represented as green and yellow ovals) to access mispaired nucleotides in a strand-specific manner. Lagging strand replicase Pol δ (represented as green circle associated with PCNA represented as grey oval) incorporates low number of rNMPs. The discontinuous nature of lagging strand DNA synthesis permits the loading of the MMR system before OF maturation. RER initiated by RNase H2 can occur concomitant with OF processing or independently. Black represents the template strand, blue the newly synthesized DNA and red the RNA primer of OF.
Although RER functions in association with DNA replication, in S. cerevisiae RNase H2 expression fluctuates with the cell cycle[50], peaking during the S and G2/M phase. In HeLa cells, western blot analysis indicates all three RNase H2 subunits are present throughout the cell cycle[51]. In addition to initiating RER in S-phase, ribonucleotide processing is likely also to occur during DNA repair when the cells are in G2/M, perhaps to eliminate rNMPs that escaped cleavage after replication, or to remove ribonucleotides incorporated by repair DNA polymerases.
Top1 processing of ribonucleotides in DNA
Yeasts deleted for RNase H2 are viable containing numerous rNMPs in genomic DNA[52-55], some of which are removed by alternative pathways that may induce genome instability, including mild replicative stress and check point activation, and are associated with higher mutation rates due to 2-5 base-pair deletions in short repetitive sequences[56]. These phenotypes are linked to topoisomerase 1 (Top1) acting on rNMPs[56]. Top1 resolves DNA supercoiling created during transcription or replication by making a nick in one DNA strand, which is religated after relieving the stress[56-59]. RER could also prevent supercoiling by incising at the rNMP, and perhaps that is one of the major roles of RNase H2. Absence of RNase H2 would create a need for Top1 to eliminate supercoiling incurred on leading strand DNA[60, 61]. Since lagging strand OFs are short, replication-induced supercoiling would not be an issue and may not attract Top1 (Figure 3, Key Figure). rNMP removal from lagging strand DNA could be performed by other enzymes leading to different patterns of mutations such as the single base pair deletion seen in the steric gate Pol η mutation[18], although single base pair deletions have also been observed in the leading strand in rnh201Δ-pol2G644M strains[62].
When Top1 cleaves at a ribonucleotide position, the 2′-OH group and 3′phosphate can form a 2′-3′ cyclic phosphate that is unable to be religated to the 5′-OH, creating a nick in the DNA (Figure 2 and Figure 3, Key Figure). The repair of the break can occur in either an error-free or mutagenic way. A second Top1 cleavage a few nucleotides upstream of the first releases a short oligonucleotide containing the 2′-3′ cyclic phosphate resulting in a short gap[57, 63]. The gap can be filled in employing Srs2 helicase, Exo1 nuclease and Pol δ[40, 57, 64] to correctly restore the DNA. Gap filling by a translesion polymerase would result in error-prone DNA synthesis. Moreover, if the gap is within a tandem repeat, the sequence can re-align bringing together the two ends allowing Top1-mediated ligation, creating the 2-5 bp deletions observed in association with Top1 ribonucleotide-processing (Figure 2 and Figure 3, Key Figure). The nicks introduced in DNA by Top1 and its faulty processing induces genomic instability, not the presence of ribonucleotides per se, because deleting TOP1 in the rnh201Δ strain eliminates these phenotypes while maintaining unprocessed ribonucleotides in DNA[62].
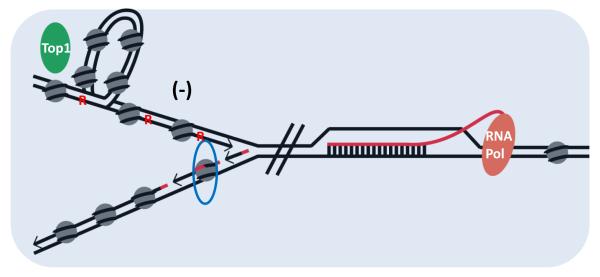
Yeast rnh201Δ pol2M644G mutant strains accumulate large numbers of rNMPs and are lethal when combined with a deletion of RNH1[33, 61]. However, because RNase H1 (see Glossary) cannot process single ribonucleotides in DNA[65], it cannot substitute for RNase H2 in the RER pathway. Consequently, the detrimental effect of combining the pol2M644G rnh201Δ and rnh1Δ mutations in the same strain may be due to abundant rNMPs in DNA together with accumulation of R-loops (see Glossary). R-loops can stall replication fork progression generating DNA damage (Figure 4). Transcription induces supercoiling in front of and behind RNA polymerases[66] providing substrates for Top1 and promoting R-loops. S. cerevisiae rnh1Δrnh201Δ strains are viable but require Top1 activity to prevent R-loop formation during transcription, most notably in rRNA genes[67, 68]. Highly transcribed genes in a wildtype RNH1 RER-defective strain also exhibit 2-5 bp deletions on the non-transcribed strand(NTS), regardless of direction of replication[69]. In this instance, even overexpression of RNase H1 fails to eliminate the deletions. High frequency of transcription-induced supercoiling may provide Top1 access to attack rNMPs in the NTS. These findings all point to the close associations between RNases H and topology.
Figure 4. Ribonucleotides both in DNA and R-loops contribute to genome instability.
In pol2M644G rnh1Δ rnh201Δ strains replication is impaired by the burden of ribonucleotides in leading strand DNA (represented as red R), and R-loops in the path of the fork which could have lethal consequences (RNA in red extruding from the RNA pol and base-paired to template DNA). Also, the block created by the R-loop in front of a replication fork will increase the supercoiling of the leading strand (−) behind the fork inducing a need for Top1 (green oval) to release the torsion. Top1 will produce multiple nicks at the ribonucleotides leading to genome instability. Nucleosome (represented by grey filled circles) assembly in the lagging strand could occur before OF maturation, leading to the incorporation of the RNA primer (represented in red) in nucleosomal DNA. The blue oval surrounding a nucleosome highlights this possibility.
Mapping ribonucleotide sites and abundance in DNA
Recently, the ability to map ribonucleotides in DNA has yielded valuable information about the complexities of the eukaryotic DNA replication process. Four different groups using distinct, but related techniques[70] have located the position of rNMPs in budding[52-54] and fission yeast[55]. All used deep sequencing and various procedures for marking the position of the rNMPs, which have been incorporated by the same DNA Pol mutants used to determine the division of labor at the replication fork. These studies mapped replication origins (some known and some new) where initiations occur in a very narrow region while converging replication forks result in broad termination zones. They found non-random distribution of rNMPs with many base pairs not having any rNMPs (no reads) while on the same scale some sites had more than 1000 reads, suggesting sequence context effects. More rNMPs were present in the leading strand confirming that wild-type Pol ε and its steric gate M644G mutant incorporate rNMPs at higher rates that their Pol δ counterparts. More surprisingly, they established that Pol α has substantial contribution to genomic DNA after OF maturation. It was thought that a majority of DNA synthesized by Pol α is removed by displacement synthesis by Pol δ and Fen1 cleavage of the flap[41]. Persistent residual rNMPs could reflect the failure to remove Pol α synthesized DNA in some of the large numbers of OFs (~105 OFs formed during replication of haploid S. cerevisiae and ~10 million OFs synthesized in diploid human DNA replication).
Nucleosomes and rNMPs in DNA replication
The replication complex plows through nucleosomes and proteins bound to DNA, such as transcription factors, ahead of the replication fork to open the DNA strands and allow synthesis. Nucleosomes rapidly form after leading strand synthesis, but the discontinuous nature of the lagging strand presents obstacles for nucleosome assembly. It is unknown if nucleosome assembly and ligation of the newly synthesized lagging strand DNA are coupled. OFs and DNA wrapped around a nucleosome plus the internucleosome spacer are similar in length. It has previously been reported that in the absence of DNA Lig1 the ends of two adjacent OFs are present at the dyad of the nucleosome[71]. Removal of the Primase-Pol α RNA-DNA (PriαR-D) product depends on Fen1 cleavage following displacement by Pol δ (Figure 2 black rectangle). Were there a delay in Pol δ synthesis or Fen1 removal of the PriαR-D region, nucleosome assembly could occur on DNA containing the PriαR-D sequence (Figure 4- blue oval), affecting complete removal of the RNA primer and leaving multiple rNMPs, which would have scored as an rNMP when mapping ribonucleotides in DNA. Interestingly, two papers described slight increases of rNMPs at the nucleosome dyad[52, 53].
Mating type switching in two distantly related Schizosaccahromyces (pombe[72]andjaponicus[73]) is characterized by the presence of a di-ribonucleotide in DNA at specific sites of the mating type locus. Di-ribonucleotide imprinting requires Pol α, suggesting the ribonucleotides are derived from the PriαR-D of OFs. Imprinting of the mating-type switching is eliminated when a region just prior to (or after) the site of imprinting is deleted. The length of the region is important, not the sequence, indicating the DNA may function as a spacer to position the site to be imprinted. Replication pausing is another important factor for imprinting. As suggested above, pausing could prevent the displacement and maturation of the OF with rNMPs at its 5′-end, and may allow nucleosome formation on a region of DNA that contains a portion of the RNA primer. It is unclear how the imprint arises and how it is unrecognized by RNase H2.
Ribonucleotides in mitochondrial DNA
Since the seventies, it has been known that mouse and human mitochondrial DNA (mtDNA) contain ribonucleotides[74] that are sensitive to alkali treatment and RNase H2[75]. These rNMPs are incorporated by mitochondrial DNA Pol γ during DNA replication, although like other polymerases, DNA Pol γ has a functional steric gate (Table1)[26]. The ribonucleotides that survive in the mtDNA are resistant to the RNase H1 present in mitochondria, indicating they are most likely present as mono- di- or tri-ribonucleotides[51, 76]. RER is not active in mitochondria due to the absence of RNase H2 in this organelle. Mitochondrial functions are not affected by the presence of these dispersed rNMPs. However, in the absence of RNase H1 failure to remove the RNA primer used to initiate mtDNA replication has catastrophic effects[42]. It will be of great interest to determine the frequency and location of rNMPs in mtDNA and assess any role they might have in mitochondrial related pathologies. S. cerevisiae mtDNA is present in strains deleted for RNH1 [50, 77] and RNH201[51, 52, 54, 77]. Yeast mtDNA is larger and more complex than metazoan mtDNA. Its replication mechanism is likely quite distinct as indicated by maintenance of mtDNA in the absence of RNases H1 and H2. Clausen et al [52] interpreted their sequences of mtDNA to be compatible with a circular mtDNA genome but is also consistent with a recombination-driven replication[78].
Ribonucleotides in DNA of Aicardi Goutières syndrome (AGS) and Systemic Lupus Erythematosus (SLE) patients with mutations in RNase H2
Unlike in yeasts where the presence of ribonucleotides in DNA is not detrimental per se, it has been suggested that the embryonic lethality at E9.5 of mice lacking RNase H2 is due to abundant rNMPs in DNA[51, 79] (estimated to be 1. 3 million or approximately one rNMP per 7,500 nucleotides[51]) This conclusion is important for understanding patients with AGS[80] and SLE[81] who have mutations in RNase H2. AGS is a neuroinflammatory autoimmune disease resulting from mutations in seven genes encoding nucleotide-processing enzymes, with more than 50% of the patients having biallelic mutations in one of the three subunits of RNase H2[82]. A recent study reporting a higher frequency of rare variance in RNase H2 genes in a cohort of SLE patients compared with normal controls established a genetic association between RNase H2 and SLE[81]. The hallmark of defects in RNase H2, rNMPs in DNA, was suggested to be responsible for the observed activation of autoimmunity. While rNMPs in DNA may be the culprit, RNA/DNA hybrid accumulation might contribute to the observed phenotypes since RNase H2 represents greater than 90% of the hybrid degradative activity in eukaryotic cells. Somewhat surprisingly, RNA/DNA hybrids were found at higher than normal levels in cell lines from AGS patients with mutations in RNASEH2A, RNASEH2B, SAMHD1 and TREX1, suggesting hybrids as possible causes of AGS[83]. The debate between consequences of rNMPs in DNA and failure to resolve RNA/DNA hybrids continues.. Two recent studies in S. cerevisiae examining recombination in diploid strains reported genome instability in the absence of RNase H2 but reached opposite conclusions as to the contributions of the two RNase H2 activities to the observed instability [59, 84]. Clearly more work lies ahead that will be aided by a RNase H2-RED (Ribonucleotide Excision Defective) mutant that lacks RER but can hydrolyze RNA/DNA hybrids[85].
Concluding remarks
All DNA polymerases incorporate ribonucleotides due to imperfect exclusion of rNTPs at the catalytic center and high ratio rNTPs/dNTPs in all cells. DNA Pol ε incorporates more rNMPs as it synthesizes the leading strand than do Pol δ and Pol α during lagging strand formation. Evidence suggests that ribonucleotides in leading strand are important for subsequent steps in DNA metabolism. Their processing, initiated by RNase H2 nicking at the rNMPs, may release torsional stress and provide an entry point to the MMR system, contributing to DNA integrity. Over one million rNMPs are present in DNA of mouse embryonic cells with no RNase H2 resulting in death at E9.5. Patients with AGS and SLE autoimmune syndromes have mutations that decrease RNase H2 activity and accumulate rNMPs. RNase H2 provides the initial step in removal of rNMPs, but also processes RNA/DNA hybrids, products of RNA transcription and reverse transcription. Still remains to be determined the contribution of each activity to the phenotypes observed in mouse and human. The RNase H2 RED mutant would be valuable to ascribe the role of rNMP processing in eukaryotes.
TRENDS BOX.
Ribonucleotides are incorporated in DNA during DNA synthesis and are removed by the Ribonucleotide Excision Repair pathway, which is initiated by RNase H2.
Ribonucleotides are markers for the newly synthesized DNA strand and strand-discrimination signals for the mismatch repair system.
Recently, ribonucleotide mapping in DNA allowed mapping of replication origins and showed that Pol substantially contributes to genomic DNA after Okazaki fragment maturation.
In the absence of RNase H2, Top1 can initiate the removal of ribonucleotides in DNA in error-free or error-prone processes.
Deleting RNase H2 causes high levels of rNMPs in DNA, which is tolerated in yeast but causes embryonic death in mice. Biallelic mutations in RNase H2 in humans cause a neuroinflammatory autoimmune disease, Aicardi-Goutières Syndrome, possibly due to accumulation of cytosolic nucleic acids.
OUTSTANDING QUESTIONS BOX.
Is there a role for ribonucleoside monophosphate (rNMP) incorporation during DNA replication and repair synthesis other than to serve as strand-discrimination signal for mismatch repair (MMR) In addition, is nicking by RNase H2 at rNMPs involved in resolving supercoiling?
What is the timing of the RER process? Is it associated with the replication machinery during S-phase or can it work in trans at other times of the cell cycle and what might be the substrates?
Is the role for the PCNA-Interacting-Peptide of RNase H2 in bringing the RER and MMR systems to the rNMP after incision by RNase H2?
Is there an alternative pathway to remove rNMPs in the lagging strand in the absence of RNase H2? If there is one, is it mutagenic?
What cytosolic nucleic acids might accumulate when RNase H2 is defective in AGS and SLE patients and do these nucleic acids result from rNMPs in DNA and/or RNA/DNA hybrids?
Acknowledgements
We thank Andrei Chabes (Umeå University Medical Biochemistry and Biophysics SE-901 87 Umeå, SWEDEN) for providing comments on concentrations of dNTPs and rNTPS in S. cerevisiae, Baek Kim (Department of Pediatrics Emory School of Medicine Atlanta, GA) for comments on NTP concentrations in mammalian cells, David Clark (NICHD NIH) for discussion of nucleosomes and Ariadne Cerritelli for help with figures. This work was supported by the Intramural Program of NIH.
Glossary
- Eukaryotic DNA polymerase families
Based on primary amino acid sequence homology, DNA polymerases can be grouped into several families. There are four families of DNA polymerases in eukaryotes. Polymerases, α, δ and ε for chromosomal DNA replication and γ for mitochondrial DNA belong to the A and B families. They are highly selective and make few mistakes during DNA synthesis because of their narrow active site and proofreading activity. Translesion and specialized polymerases belong mostly to the X and Y families and are error-prone because they have a wider active site to accommodate bulky adducts on damaged DNA and lack proofreading activity.
- RNases H
Ribonucleases H (RNases H) are enzymes that hydrolyze RNA of RNA/DNA duplexes, sometimes described as hybrids and other times as R-loops. Embedded ribonucleotides in duplex DNA can be single or several consecutive rNMPs that are incorporated during DNA synthesis. RNA/DNA hybrids can form during transcription and reverse transcription. There are two types of RNases H in eukaryotic cells. RNase H1 is a monomeric enzyme that cleaves RNA/DNA hybrids and rNMPs in DNA that are at least 4 nucleotides long. RNase H2 is a heterotrimeric complex that process hybrids and also single ribonucleotides in DNA and participates in the Ribonucleotide Excision Repair process.
- R-loops
R-loops are triple stranded complexes in which RNA base pairs with one DNA strand and the non-base paired single-stranded DNA forms the ‘loop’.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Spradling A, Orr-Weaver T. Regulation of DNA Replication During Drosophila Development. Ann Rev Genet. 1987;21:373–403. doi: 10.1146/annurev.ge.21.120187.002105. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA. Evolving Views of DNA Replication (In)Fidelity. Cold Spring Harbor Sym. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodman MF, Woodgate R. Translesion DNA Polymerases. Cold Spring Harbor Perspect Biol. 2013 doi: 10.1101/cshperspect.a010363. cshperspect.a010363v1 5/10/a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joyce CM. Choosing the right sugar: How polymerases select a nucleotide substrate. Proc Natl Acad Sci. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astatke M, et al. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, et al. Division of Labor at the Eukaryotic Replication Fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams JS, Kunkel TA. Ribonucleotides in DNA: Origins, repair and consequences. DNA Rep. 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel PH, Loeb LA. Multiple Amino Acid Substitutions Allow DNA Polymerases to Synthesize RNA. J Biol Chem. 2000;275:40266–40272. doi: 10.1074/jbc.M005757200. [DOI] [PubMed] [Google Scholar]
- 11.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry-US. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JA, et al. A Novel Mechanism of Sugar Selection Utilized by a Human X-Family DNA Polymerase. J Mol Biol. 2010;395:282–290. doi: 10.1016/j.jmb.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bebenek K, Pedersen LC, Kunkel TA. Structure–Function Studies of DNA Polymerase λ. Biochemistry-US. 2014;53:2781–2792. doi: 10.1021/bi4017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavanaugh NA, et al. Molecular insights into DNA polymerase deterrents for ribonucleotide insertion. J Biol Chem. 2011;286:31650–31660. doi: 10.1074/jbc.M111.253401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, et al. Structural Factors That Determine Selectivity of a High Fidelity DNA Polymerase for Deoxy-, Dideoxy-, and Ribonucleotides. J Biol Chem. 2012;287:28215–28226. doi: 10.1074/jbc.M112.366609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavlov YI, Shcherbakova PV, Kunkel TA. In Vivo Consequences of Putative Active Site Mutations in Yeast DNA Polymerases α, ε, δ, and ζ. Genetics. 2001;159:47–64. doi: 10.1093/genetics/159.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pursell ZF, et al. Yeast DNA Polymerase ε Participates in Leading-Strand DNA Replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donigan KA, et al. Unlocking the steric gate of DNA polymerase η leads to increased genomic instability in Saccharomyces cerevisiae. DNA Rep. 2015;35:1–12. doi: 10.1016/j.dnarep.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donigan KA, et al. The Steric Gate of DNA Polymerase ι Regulates Ribonucleotide Incorporation and Deoxyribonucleotide Fidelity. J Biol Chem. 2014;289:9136–9145. doi: 10.1074/jbc.M113.545442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nick McElhinny SA, Ramsden DA. Polymerase Mu Is a DNA-Directed DNA/RNA Polymerase. Mol Cell Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulé J-B, Rougeon F, Papanicolaou C. Terminal Deoxynucleotidyl Transferase Indiscriminately Incorporates Ribonucleotides and Deoxyribonucleotides. J Biol Chem. 2001;276:31388–31393. doi: 10.1074/jbc.M105272200. [DOI] [PubMed] [Google Scholar]
- 22.Clausen AR, et al. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ. DNA Rep. 2013;12:121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams JS, et al. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase ε. DNA Rep. 2012;11(8):649–656. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnin A, et al. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type φ29 DNA polymerase1. J Mol Biol. 1999;290:241–251. doi: 10.1006/jmbi.1999.2900. [DOI] [PubMed] [Google Scholar]
- 25.Lin TC, et al. 3′–5′ Exonucleolytic Activity of DNA Polymerases: Structural Features That Allow Kinetic Discrimination between Ribo- and Deoxyribonucleotide Residues. Biochemistry-US. 2001;40:8749–8755. doi: 10.1021/bi0105936. [DOI] [PubMed] [Google Scholar]
- 26.Kasiviswanathan R, Copeland WC. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J Biol Chem. 2011;286:31490–31500. doi: 10.1074/jbc.M111.252460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watt DL, et al. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Rep. 2011;10:897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clausen AR, et al. Structure-function analysis of ribonucleotide bypass by B family DNA replicases. Proc Natl Acad Sci. 2013;110:16802–16807. doi: 10.1073/pnas.1309119110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Göksenin AY, et al. Human DNA polymerase ε is able to efficiently extend from multiple consecutive ribonucleotides. J Biol Chem. 2012;287:42675–42684. doi: 10.1074/jbc.M112.422733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavanaugh NA, Beard WA, Wilson SH. DNA polymerase β ribonucleotide discrimination insertion, misinsertion, extension, and coding. J Biol Chem. 2010;285:24457–24465. doi: 10.1074/jbc.M110.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gosavi RA, et al. The catalytic cycle for ribonucleotide incorporation by human DNA Pol λ. Nucleic Acids Res. 2012;40:7518–7527. doi: 10.1093/nar/gks413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makarova AV, et al. Ribonucleotide incorporation by yeast DNA polymerase ζ. DNA Rep. 2014;18:63–67. doi: 10.1016/j.dnarep.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazzaro F, et al. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chabes A, et al. Survival of DNA Damage in Yeast Directly Depends on Increased dNTP Levels Allowed by Relaxed Feedback Inhibition of Ribonucleotide Reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen LA, et al. Pre-steady state kinetic analysis of HIV-1 reverse transcriptase for non-canonical ribonucleoside triphosphate incorporation and DNA synthesis from ribonucleoside-containing DNA template. Antivir Res. 2015;115:75–82. doi: 10.1016/j.antiviral.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy EM, et al. Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J Biol Chem. 2010;285:39380–39391. doi: 10.1074/jbc.M110.178582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahouassa H, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 39.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparks JL, et al. RNase H2-Initiated Ribonucleotide Excision Repair. Mol Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balakrishnan L, Bambara RA. Okazaki Fragment Metabolism. Cold Spring Harb Perspect Biol. 2013;5:pii: a010173. doi: 10.1101/cshperspect.a010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmes JB, et al. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc Natl Acad Sci. 2015;112:9334–9339. doi: 10.1073/pnas.1503653112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghodgaonkar MM, et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lujan SA, et al. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chon H, et al. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 2009;37:96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bubeck D, et al. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011;39:3652–3666. doi: 10.1093/nar/gkq980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaban NM, et al. The Structure of the Mammalian RNase H2 Complex Provides Insight into RNA·DNA Hybrid Processing to Prevent Immune Dysfunction. J Biol Chem. 2010;285:3617–3624. doi: 10.1074/jbc.M109.059048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Figiel M, et al. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi- Goutières syndrome defects. J Biol Chem. 2011;286(12):10540–10550. doi: 10.1074/jbc.M110.181974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reijns MAM, et al. The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J Biol Chem. 2011;286:10530–10539. doi: 10.1074/jbc.M110.177394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arudchandran A, et al. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells. 2000;5:789–802. doi: 10.1046/j.1365-2443.2000.00373.x. [DOI] [PubMed] [Google Scholar]
- 51.Reijns MAM, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clausen AR, et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struc Mol Biol. 2015;22:185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reijns MAM, et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 2015;518:502–506. doi: 10.1038/nature14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koh KD, et al. Ribose-seq: Global mapping of ribonucleotides embedded in genomic DNA. Nat Meth. 2015;12:251–257. doi: 10.1038/nmeth.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daigaku Y, et al. A global profile of replicative polymerase usage. Nat Struc Mol Biol. 2015;22(3):192–198. doi: 10.1038/nsmb.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim N, et al. Mutagenic Processing of Ribonucleotides in DNA by Yeast Topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sparks JL, Burgers pp.M. Error-free and mutagenic processing of topoisomerase 1-provoked damage at genomic ribonucleotides. EMBO J. 2015;34:1259–1269. doi: 10.15252/embj.201490868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho JE, et al. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA Rep. 2013;12:205–211. doi: 10.1016/j.dnarep.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Connell K, Jinks-Robertson S, Petes TD. Elevated Genome-Wide Instability in Yeast Mutants Lacking RNase H Activity. Genetics. 2015;201:963–975. doi: 10.1534/genetics.115.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu H, Dröge pp. Replication-induced supercoiling: a neglected DNA transaction regulator? Trends Biochem Sci. 2014;39:219–220. doi: 10.1016/j.tibs.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 61.Williams JS, et al. Evidence that processing of ribonucleotides in DNA by topoisomerase 1 is leading-strand specific. Nat Struc Mol Biol. 2015;22:291–297. doi: 10.1038/nsmb.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams JS, et al. Topoisomerase 1-Mediated Removal of Ribonucleotides from Nascent Leading-Strand DNA. Molecular Cell. 2013;49(5):1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang SYN, Ghosh S, Pommier Y. Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites. J Biol Chem. 2015;290:14068–14076. doi: 10.1074/jbc.M115.653345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Potenski CJ, et al. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El Hage A, et al. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Devel. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.French SL, et al. Distinguishing the Roles of Topoisomerases I and II in Relief of Transcription-Induced Torsional Stress in Yeast rRNA Genes. Mol Cell Biol. 2011;31:482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cho J-E, Kim N, Jinks-Robertson S. Topoisomerase 1-dependent deletions initiated by incision at ribonucleotides are biased to the non-transcribed strand of a highly activated reporter. Nucleic Acids Res. 2015;43:9306–9313. doi: 10.1093/nar/gkv824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jinks-Robertson S, Klein HL. Ribonucleotides in DNA: Hidden in plain sight. Nat Struc Mol Biol. 2015;22(3):176–178. doi: 10.1038/nsmb.2981. [DOI] [PubMed] [Google Scholar]
- 71.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zech J, et al. The DNA-Binding Domain of Mrc1 (Claspin) Acts to Enhance Stalling at Replication Barriers. PLoS ONE. 2015;10:e0132595. doi: 10.1371/journal.pone.0132595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu C, Bonaduce MJ, Klar AJS. Defining the Epigenetic Mechanism of Asymmetric Cell Division of Schizosaccharomyces japonicus Yeast. Genetics. 2013;193:85–94. doi: 10.1534/genetics.112.146233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grossman LI, Watson R, Vinograd J. The Presence of Ribonucleotides in Mature Closed-Circular Mitochondrial DNA. Proc Natl Acad Sci. 1973;70:3339–3343. doi: 10.1073/pnas.70.12.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keller W, Crouch R. Degradation of DNA RNA Hybrids by Ribonuclease H and DNA Polymerases of Cellular and Viral Origin. Proc Natl Acad Sci. 1972;69:3360–3364. 65. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cerritelli SM, et al. Failure to Produce Mitochondrial DNA Results in Embryonic Lethality in Rnaseh1 Null Mice. Mol Cell. 2003;11(3):807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 77.El Hage A, et al. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10:e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bendich AJ. The End of the Circle for Yeast Mitochondrial DNA. Molecular Cell. 39:831–832. doi: 10.1016/j.molcel.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 79.Hiller B, et al. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Crow YJ, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 81.Günther C, et al. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J Clinl Invest. 2015;125:413–424. doi: 10.1172/JCI78001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crow YJ, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 83.Lim YW, et al. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi–Goutières syndrome. 2015;4 doi: 10.7554/eLife.08007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conover HN, et al. Stimulation of Chromosomal Rearrangements by Ribonucleotides. Genetics. 2015;201:951–961. doi: 10.1534/genetics.115.181149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chon H, et al. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41:3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]