Abstract
The saliva of ixodid ticks contains a mixture of bioactive molecules that target a wide spectrum of host defense mechanisms to allow ticks to feed on the vertebrate host for several days. Tick salivary proteins cluster in multigenic protein families, and individual family members display redundancy and pluripotency in their action to ameliorate or evade host immune responses. It is now clear that members of different protein families can target the same cellular or molecular pathway of the host physiological response to tick feeding. Here we present and discuss our hypothesis that the redundancy and pluripotency has evolved in tick salivary immunomodulators in order to evade immune recognition by the host, while retaining the immunomodulatory potential of their saliva.
Keywords: Multigenic protein families, pluripotency, redundancy, tick salivary proteins, immunomodulation, silent antigens
Multigenic protein families in ticks – many questions, few answers
Ticks are obligatory blood-feeding ecto-parasitic arthropods. There are two major tick families: Argasidae (soft ticks) and Ixodidae (hard ticks). Hard ticks remain attached to their hosts for up to two weeks, during which time they are exposed to host homeostatic and defense mechanisms [1]. To complete their blood meal, ticks need to evade host immune surveillance and/or suppress the host immune response. The evasion of host immune system is mediated mainly via tick salivary secretion that is injected into the tick-feeding cavity [2]: ixodid tick saliva is a highly complex natural mixture of low and high molecular weight bioactive molecules. The pharmacological effects of tick saliva and its individual components have been studied for almost half century and research in this field could be divided into the times before and after transcriptomes, but this journey was outlined elsewhere [3] and its description outreaches the scope of this article. Transcriptomic studies have shown that secreted salivary proteins cluster in multigenic protein families (see Glossary) containing as many as 200 individual members differing by only a few amino acids [4, 5]. Only a small number of family members has been functionally characterized at the protein level, recombinant or native. Therefore, the role of individual family members remains to be elucidated. Moreover, a significant portion of the identified transcripts fall into the category of hypothetical proteins, without any relationship or similarity to known proteins from any living organism. Therefore, their (potentially important) role in the tick lifecycle is currently unknown.
The existence of multigenic salivary protein families raises many questions. Which host’s homeostatic functions do individual tick proteins affect? Do members of a given tick protein family target different, similar or identical host physiological processes? Why is such a large number of functionally and structurally similar proteins produced, and what is the mechanism of their gene expression regulation? Although we can hypothesize that multigenic families exist to meet the need for large numbers of immunomodulatory proteins to block specific host defense mechanisms, is their effect additive and what are the immunological consequences? Indeed, expressing small quantities of many different antigens (at once or sequentially throughout tick feeding) can diminish immune recognition due to low-dose tolerance and/or antigenic competition effects and impair immune cell activation [6]. As research has progressed in this field, it is now clear that tick salivary proteins display both pluripotency and redundancy in their actions. Recent publications show that the high level of redundancy, identified at mRNA level, translates into the proteomic profile of the saliva [7, 8]. In this short opinion article we suggest that multigenic families of tick salivary secreted proteins evolved to reduce the immunogenicity of immunomodulatory tick effectors by lowering the amount of each individual antigen in tick salivary secretion.
Redundancy and pluripotency in the action of tick salivary immunomodulators
In order to remain attached to their host and successfully complete their long (up to two weeks) feeding course, ticks block the actions of many host immune system components. From an evolutionary point of view, an efficient mechanism must exist to evolve high numbers of proteins with novel functions in the 'arms race' between ticks and their multiple and diverse hosts. There has been extensive research on how proteins with novel functions evolve after gene duplication events [9, 10]. According to phylogenetic analyses, gene duplication is usually followed by rapid evolution driven by positive selection rather than by the accumulation of neutral mutations [10, 11]. The concepts of redundancy and pluripotency in the action of tick salivary immunomodulators represent strategies that fulfill the need for many different low-immunogenicity effectors, while simultaneously reflect evolutionary mechanisms and constraints such as the requirement for providing an evolutionary advantage [12]. Moreover, a single pluripotent tick salivary protein may exert multiple effects on the host either by targeting more than one mechanism or pathway in the same immune cell type or by targeting different functions in different cell types. For instance, certain tick inhibitors of vertebrate proteases usually target more than one protease involved in host immunity (Figure 1A). The evolutionary mechanisms by which protease inhibitors gain new specificities are well described in general [13], as well as for tick salivary protease inhibitors of the Kunitz family [14, 15], where a positive evolutionary pressure on inhibitor functionality is known to exist. An additional element in host immunomodulation by tick saliva is redundancy (or overlap) in actions, i.e., when a certain component of host immunity is targeted by more than one salivary protein (Figure 1A, B). Characterization of secreted tick salivary proteins has revealed at least two types of functional redundancy: (i) different tick proteins (from the same or different multigenic protein families) target an identical host cellular mechanism (Figure 1B), or (ii) different tick proteins affect different parts of the same cellular pathway to produce an identical final outcome and phenotype (Figure 1C). A combination of redundancy and pluripotency of secreted tick salivary proteins enables tick saliva to interfere with a wide range of host defense mechanisms (such as hemostasis, inflammation, or antigen-dependent acquired immunity) while evading the host’s response to compromised homeostasis due to the intrusion of tick mouthparts into the host skin. In the following sections, we illustrate the aforementioned concepts with specific examples. Redundancy in targeting hemostasis, such as coagulation and platelet aggregation, has been already well described [16, 17], so here we focus only on proteins with immunomodulatory features.
Figure 1. Redundancy and pluripotency in the action of tick salivary proteins on host cellular processes.
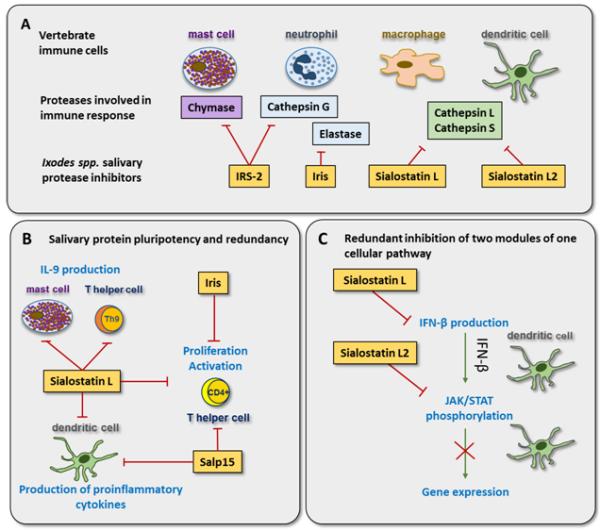
Different members of different tick protein families target the same host immune function, or the same protein targets different modules of the host immune response. (A) By inhibiting different immune cell-derived proteases with specific inhibitors, Ixodes spp. ticks can block multiple defense mechanisms. The anti-inflammatory serpin IRS-2 (Ixodes ricinus serpin-2) inhibits mast cell chymase and neutrophil cathepsin G. Both proteases are known to be involved in promoting inflammation. The second of the three major neutrophil serine proteases involved in inflammatory responses, elastase, is inhibited by another serpin, Iris (Ixodes ricinus immunosuppressor). Sialostatin L and L2 are inhibitors of the intracellular cysteine proteases cathepsin L and cathepsin S that contribute to NLRP3 inflammasome activation and the production of the pro-inflammatory cytokines interleukin-1β (IL-1β) and IL-18 in macrophages and dendritic cells (DCs). (B) The cystatin from Ixodes scapularis, sialostatin L, was shown to inhibit production of proinflammatory cytokines from DCs, the proliferation of T cells, and the production of IL-9 from Th9 cells and mast cells. Another member of an I. scapularis protein family, Salp (Salivary protein) 15, inhibited proliferation of T cells and the production of pro-inflammatory cytokines from DCs. Finally, the tick salivary serpin Iris also inhibited T cell proliferation. (C) Redundant inhibition of different modules of the interferon-β (IFN-β) signaling pathway by two members of the same multigenic family. Sialostatin L, unlike sialostatin L2, inhibits IFN-β production by DCs. Sialostatin L2, however, inhibits the phosphorylation of STAT-3 in the JAK/STAT signaling pathway and subsequent expression of several genes downstream from IFN-β-dependent DC activation.
Pluripotency and redundancy - some practical examples
Thorough functional characterization of several individual recombinant tick salivary-secreted proteins has revealed their pluripotent immunomodulatory activities (Figure 1B). One example is the Ixodes scapularis salivary cystatin sialostatin L (SialoL), which is an inhibitor of human cathepsins with highest affinity for cathepsin L. Early SialoL studies in a mouse model of acute inflammation revealed its anti-inflammatory potential and inhibitory effect on cytotoxic T cells [18, 19]. A follow-up study showed that SialoL also inhibited proliferation of CD4+ T helper cells, dendritic cell (DC) maturation, and the production of proinflammatory cytokines [19]. It was later shown that SialoL diminished interleukin-9 (IL-9) production in Th9 lymphocytes and mast cells [20, 21]. Finally, SialoL impaired production of interferon-β (IFN-β) in DCs activated by Borrelia burgdorferii spirochetes [22]. When this latter study is evaluated in light of the data from another study [23], it appears that two members of the same multigenic family of I. scapularis cystatins (namely sialostatin L and sialostatin L2) target different modules of the same pathway. Specifically, the activity of sialostatin L2 (SialoL2) differed from that of SialoL: unlike SialoL, SialoL2 did not affect IFN-β production, but instead it attenuated STAT phosphorylation in spleen DCs, thus inhibiting JAK/STAT signaling downstream of IFN-β-dependent signaling. As a result, SialoL2 also inhibited IFN-β-dependent expression of the neutrophil-attracting chemokines MIP-1α and IP-10 in DCs [23] (Figure 1C). In summary, these two sialostatins demonstrate that: (i) a single protein can exert several functions, and (ii) members of the same multigenic family, despite sequence similarity, can differ in their activities, but have complementary effects (Figure 1B, C).
A second example of a pluripotent tick immunomodulator is the salivary protein (Salp) 15 (Figure 1B), a glycoprotein that belongs to another large protein family present in several Ixodes species [24]. Similar to SialoL, Salp15 displays an effect on both CD4+ T cells and DCs [25] and it carries out several activities; the most prominent is the direct binding of Salp15 to the CD4 receptor and subsequent inhibition of T cell activation, proliferation, and IL-2 production [26, 27]. In DCs, Salp15 interacts with adhesion and receptor molecule DC-SIGN, leading to the inhibition of proinflammatory cytokine production (IL-6 and tumor necrosis factor (TNF)) [28]. Inhibition of CD4+ T cell proliferation has also been observed with another Ixodes species-specific protein, the serpin Iris (Ixodes ricinus immunosuppressor) [29]. Therefore, SialoL, Salp15 and Iris represent another type of redundancy, in which specific host functions are targeted by phylogenetically distinct proteins (Figure 1B). The concept of pluripotency and redundancy of tick salivary proteins is supported by many other examples: two related lipocalins from Ornithodoros savignyi, tick salivary gland protein (TSGP) 2 and TSGP3, bind and inhibit complement by inhibiting C5 convertase and also bind the neutrophil chemoattractant leukotriene B4 [30]. Three lipocalins from Rhipicephalus appendiculatus, Ra-HBP-1, 2 and 3, display histamine binding features [31], while two members of the Salp16-like family from Ixodes persulcatus, Salp16-Iper1 and 2, inhibit neutrophil migration and reactive oxygen species production [32], similar to two disintegrin members from the ADAMTS (a disintegrin and metalloprotease with thrombospondin motifs) family (ISL 929 and ISL 1373) isolated from I. scapularis [33]. Since they are present in many ixodid species, Salp16-like and ADAMTS protein families represent another example of functional redundancy among different protein family members and members of a single protein family. It is important to note that not every observed tick salivary protein effect on host immunity has a known mechanistic explanation. Therefore, some effects of a given recombinant tick protein may be indirect rather than direct, i.e., they may be caused by modulating upstream components of the same molecular pathway or via pathway crosstalk. For instance, the I. ricinus salivary serpin IRS-2 (Ixodes ricinus serpin-2) was shown to inhibit T cell differentiation into Th17 cells [34]. This effect, however, resulted from an upstream inhibition of DC-derived production of the pro-inflammatory cytokine IL-6 [34]. More detailed mechanistic studies are therefore necessary to determine the primary in vivo targets for each tick salivary protein in question.
When testing tick salivary recombinant proteins in-vitro or ex-vivo, especially candidate immunodulators, high concentrations of pure recombinant proteins are often needed to achieve a detectable effect. Such high protein concentrations are not usually observed in pure tick saliva, raising the question of whether these proteins really are responsible for the physiological immunomodulatory effects of tick saliva observed both in vivo and in vitro. The observed redundancy in tick salivary immunodulators could explain this need for high protein concentrations in vitro. Since each protein family contains many members with high sequence similarity, it is likely that many of these members share the same function. Therefore, although each protein is present only at low concentrations in the tick saliva, they may act in concert and display an additive (or even synergistic) functional effect that is quantitatively equivalent to a higher concentration of a single protein. Salivary proteins in recombinant form are usually biologically active at concentrations as high as 1-6 μM [22, 28, 34]. In the complex salivary mixture, this effective concentration could be achieved by combining numerous redundant proteins (Figure 2A). The functional characterization of five anti-complement multigenic family members from I. ricinus, IxAC-B1 – IxAC B5, supports this hypothesis [35]: all five proteins inhibit the alternatively activated complement pathway by preventing C3 convertase complex formation in spite of their differing primary structure [35] and, also their epitope structures (Figure 2A). In contrast, some other tick salivary activities are detected at much lower recombinant protein concentrations. This might be explained by the hypothesis that a certain host function, while targeted by these proteins, is not blocked by numerous other tick salivary effectors (and thus assistance in their action is not provided by other salivary effectors) or that the mechanism in question is the primary target of a given salivary effector (the protein is a key modulator of the specific host homeostatic mechanism or pathway). For example, the I. ricinus serpin Iris significantly inhibited TNF production in human peripheral blood mononuclear cells at concentrations as low as 25nM [36], but the contact phase of coagulation and fibrinolysis was affected at much higher concentrations (3-6 uM) [37]. The serpin superfamily, expressed in Ixodes spp. salivary glands, contains over 40 members [38]; the Iris reactive center loop with methionine at its main active site, the P1 site, however, is rather unique among them. We can, therefore, speculate that the inhibition of TNF production represents a major Iris function while its anti-hemostatic activity plays only a minor role in concert with other tick inhibitors more specialized for hemostasis inhibition.
Figure 2. Redundancy in a single multigenic family and antigenic variability.
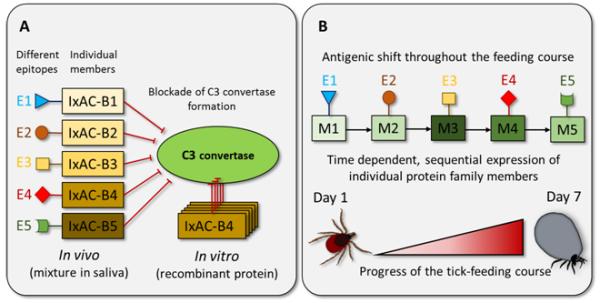
(A) Five members of the anti-complement protein multigenic family IxAC-B1-5 affect the same process: they bind to properdin and thus inhibit the stabilization of the C3 convertase complex which, in consequence, inhibits the activation of the alternative pathway of complement. Since there is a divergence in primary structure of individual members of IxAC-B family, it is likely that there is epitope variability and, therefore, different antigenicity across family members, while all members exert the same function on the host. Indeed, the study of Couvreur and colleagues supports such a suggestion, as the sera, raised against IxAC-B1 did not recognize other members of the family [35]. The observed functional (but not antigenical) identity results in a lower amount of protein required for each family member to achieve the inhibition of host complement, which in consequence leads to lower immunogenicity and helps to evade effective recognition by the host immune system (amount effect). On the other hand, when a single recombinant family member (e.g. IxAC-B4, Figure 2A) is tested in a functional bioassay, its concentration would have to be higher to achieve an observable effect, according to our hypothesis. E1-E5 – Epitope 1-5. (B) The sequential expression of members from one multigenic family throughout feeding results in continuous antigenic shift, while the targeted host process remains blocked. Due to this change in epitope exposure, an effective and timely antigen-specific response cannot be mounted against the tick salivary immunomodulators. Similar to other parasites such as Plasmodium spp., Trypanosoma spp., or Schistosoma spp., the antigen switch takes place at the parasite-host interface, which - in the case of ticks - is between the blood and tick saliva. Both simultaneous and sequential expression of several antigenically different family members can result in a silent antigen phenomenon. E1-E5 – Epitope 1-5, M1-M5 – member of one hypothetical multigenic family 1-5.
Antigenic variation
The genetic mechanism that drives multigenic family evolution in ticks is a combination of multiple gene duplications and subsequent mutations [15, 39, 40]. This mechanism, ubiquitous in eukaryotes [41], was also proposed as a hypothesis for multigenic evolution in ticks [42]. This was subsequently supported by a phylogenetic analysis [30] and confirmed experimentally by the analysis of synonymous vs. non-synonymous mutation rates in salivary proteins [43]. The latter analysis revealed significantly higher non-synonymous mutation rates in putative secreted proteins, when compared to other protein groups. During the evolution, newly emerging functional proteins/mutants either retain their original function or gain novel features/activities via a process of positive selection rather than by the accumulation of neutral mutations [10]. Although retaining their original function, divergent (in primary structure) proteins can change their epitope structure and immunogenicity by altering a few amino acids. A similar model was introduced in an attempt to experimentally decrease epitope immunogenicity of hirudin, a medically interesting anticoagulant of parasitic origin [44, 45]. Another participant in the race to evade and/or exhaust the host immune system is antigenic variability, the simultaneous or sequential change in multiple antigens upon host immune system exposure. The mechanism of immune system exhaustion by continuous antigen switching is known to occur in Trypanosoma brucei, the causative agent of sleeping sickness, which can sequentially express around 2000 different surface antigens [46]. Every newly expressed antigen stimulates a massive immune reaction that eliminates the majority of parasites, but some of the surviving individuals with altered surface antigen expression escape from the host’s antigen-specific reaction and start a new round of multiplication. This results in the typical wave-like course of the disease [47]. Similarly, the malarial protozoa Plasmodium spp. also use the antigenic diversity to evade immune recognition and elimination by the host immune system [48]. Among non-protozoan pathogens, antigenic variability is exploited by Schistosoma spp. [49] and it also plays an important role in pathogenesis of certain bacteria [50]. Interestingly, antigenic variability mechanisms have evolved several times independently, i.e., the molecular and genetic mechanisms underpinning antigenic variability in parasites and pathogens are different. However, all these parasites have in common their contact with blood and the host immune system. All the aforementioned examples relate to endoparasites, and thus the variable antigens are expressed on their surface (which is in contact with host immunity). The equivalent in blood-feeding ectoparasites, such as ixodid ticks, should be the antigenic variability of the salivary proteins that are found at the actual interface between the host immune system and the tick. Indeed, as described in previous sections, similar mechanism of antigenic variation do seem to exist in ectoparasites, as suggested by several transcriptomic analyses of tick salivary glands throughout feeding. Several multigenic protein family members are expressed sequentially during the progression of tick feeding; at each time point during feeding, a different multigenic family member is expressed in tick salivary glands and secreted into the host [43, 51, 52] (Figure 2B). The concept of antigenic variability, as a conserved mechanism to evade host immune recognition and responses, may therefore be applied to blood-feeding ectoparasites as well and may represent a ubiquitous mechanism exploited by parasites from very distant phylogenetic groups.
Recombinant protein vs. saliva – the difference in immunogenicity
In our study from 2008, we describe a discrepancy in the immunogenicity of a recombinant tick salivary protein that was artificially injected into the host at high concentrations vs. when it was delivered naturally by multiple exposures of the host to tick saliva due to natural tick feeding [53]. Specifically, the tick salivary protein SialoL2 was highly immunogenic in recombinant form, since guinea pig vaccination with high amounts of the protein and subsequent exposure of the guinea-pigs to ticks resulted in a strong anti-tick response and significantly higher tick rejection. However, natural exposure of guinea pigs to tick saliva by repeated tick feeding did not lead to SialoL2 recognition by their sera unless the animals were previously artificially vaccinated/sensitized with recombinant protein [53]. The observed discrepancy can have several causes. First, immunogenicity is correlated with antigen concentration, therefore an exposure to low levels of an antigen can lead to immunotolerance. Similarly, time-dependent expression of different multigenic protein family members in tick salivary glands limits the time and intensity of the exposure of a given tick antigen to the host immune system and thus reduces the likelihood of a host developing immunity to a specific protein (Figure 2A, B). Salivary proteins are often of relatively low molecular weight, which is also an important factor that, in general, negatively correlates with antigen immunogenicity. Furthermore, the salivary proteins in native state could undergo posttranslational modifications that diminish their immunogenicity. To summarize, the low molecular weight, post-translational modifications, time-limited exposure of a single antigen, and low antigen concentration due to the presence of several members of the same protein family in tick saliva (displaying the same function, but not antigenicity), impairs antibody production and T-cell activation. This could explain why immunomodulatory proteins in tick saliva do not elicit humoral responses despite their immunogenic potential in the recombinant form.
Concluding remarks
Several important questions persist in tick-host interaction research and the functional characterization of tick secreted salivary proteins, especially in testing them as candidate immunomodulators. Why do these proteins form large multigenic families? What is the reason for sequential expression of individual family members throughout tick feeding? What is the purpose of redundancy and pluripotency in salivary protein function and what impact does it have on host humoral recognition and immune response? What are the implications of performing functional characterization of tick salivary proteins in recombinant form? In this article we have attempted to answer these questions by presenting the evidence base for the herein proposed hypothesis. We hypothesize that the “raison d’être” of multigenic families among tick salivary secreted proteins lies in their ability to provide sufficient amount of members with the same function, but different antigenicity. Combined with the pluripotency of some key salivary effectors, we can identify a very powerful network of salivary proteins that are functionally interconnected by the pluripotency and redundancy in their actions (Figure 3). This, in turn, leads to an efficient suppression and modulation of important anti-tick defenses of the host, while the antigen-specific immune responses are diminished due to the presence of silent antigens; these silent antigens do not elicit strong immune response, because of their low amount in the tick saliva and low molecular weight. Our hypothesis brings possible explanation to (and is supported by) the fact that researchers need to use relatively high concentrations of pure recombinant tick candidate immunomodulators in bioassays (3-6 μM) in order to see an observable effect [34, 37]. Moreover, it brings out the suggestion that some pluripotent proteins (such as Iris, SialoL or Salp15 and other not yet characterized proteins) could be considered as key factors, because they can target more than one branch of immune reaction (Figure 3). We hypothesize that the functional network of tick salivary immunomodulators could be disrupted by targeting these proteins with vaccines, leading to stronger host resistance to ticks and the pathogens that ticks transmit. To prove or disprove this hypothesis is one of the many possible future directions in anti-tick vaccine development (see Outstanding Questions Box).
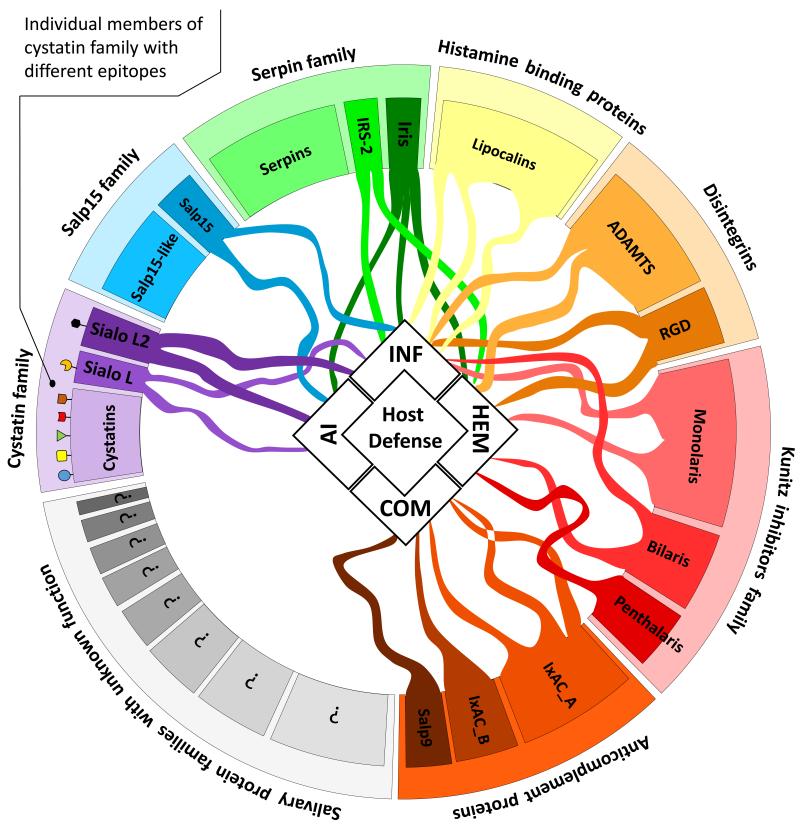
Figure 3. A perplexed network of actions in the vertebrate host is mediated by tick salivary proteins.
Pluripotency and redundancy in the action of tick salivary effectors drive the formation of a complex network of molecular interactions between ticks and the hosts they parasitize. Host anti-tick defenses can be schematically divided into four branches – hemostasis (HEM), inflammation (INF), complement (COM) and antigen specific acquired immune response (AI). Tick saliva contains multigenic protein families with known or unknown function that can together target all host’s defense mechanisms (the effect on host defense mechanisms is shown as a 'tentacle' in the figure). There is redundancy in their actions both in the frame of a single family or among the members of different families. The individual members of single family can exert the same effect on the host, while expressing different antigenic epitopes (as shown on the example of cystatin family), which leads to immune system evasion. Some pluripotent proteins (e.g. Iris, Sialo L or Salp15 in the figure and other -not yet characterized- proteins) could be considered as key factors, because they can target more than one branch of immune reaction. We hypothesize that by targeting these proteins with vaccines, the functional network of tick salivary immunomodulators could be disrupted, leading to stronger host resistance to ticks and the pathogens that ticks transmit. RGD – proteins containing integrin binding motif (Arg-Gly-Asp); ADAMTS – a disintegrin and metalloprotease with thrombospondin motifs; IRS – Ixodes ricinus serpin; Iris – Ixodes ricinus immunosuppressor; Salp – Salivary protein; Sialo – Sialostatin; IxAC – Ixodes anti-complement.
Outstanding questions box.
Are there some crucial tick salivary components that serve as cornerstones, without which the tick feeding would be significantly disrupted?
What is the mechanism behind the regulation of sequential expression of individual protein family members? Is it dependent on tick feeding status?
How can we incorporate the antigenic shuffle throughout the tick feeding progress into anti-tick vaccine development?
Are tick-borne pathogens adapted to the change in salivary proteins expression and how?
Despite the fact that a larger body of experimental evidence from bioassays and/or in silico work on transcriptomic and proteomic datasets is still required to validate these hypotheses, there is already sufficient accumulated knowledge and data to at least direct the way of designing and developing anti-tick vaccines and to better understand the interface, at which pathogen acquisition and/or transmission occur upon tick feeding.
Trends box.
A major aim for tick research is the development of anti-tick or transmission-blocking vaccines.
Given the redundancy and pluripotency of tick salivary proteins, the biggest challenge in this area of research is unraveling the key immunomodulatory molecules to be targeted for anti-tick vaccine development.
Antigenic variation throughout the tick feeding should be also considered as an important factor in the development of both anti-tick and transmission blocking vaccines.
In addition to vaccine development, the discovery of powerful immunomodulatory proteins in tick salivary glands presents another important direction in tick research that might enrich the current repertoire of biotherapeutic molecules.
Acknowledgments
We thank Nextgenediting (www.nextgenediting.com) for providing editorial assistance. This work was supported by the Grant Agency of the Czech Republic (GACR grant P502/12/2409 to MK), the Czech Academy of Sciences (grant no. RVO60077344 to the Biology Center—Institute of Parasitology), the 7th Framework Program of the European Union (EU FP7; Marie Curie Reintegration grant PIRG07-GA-2010-268177 to MK) and internal grant of the Faculty of Science at the University of South Bohemia in České Budějovice. This publication reflects only the authors' views and the European Union is not liable for any use that may be made of the information contained herein. MK and JHFP were supported by the National Institutes of Health (R01 AI093653), and JHFP was supported by start-up funds provided by the University of Maryland-Baltimore, School of Medicine. MK received support from the Czech Academy of Sciences (Jan Evangelista Purkyně Fellowship).
Because this is a governmental work, the work is in the public domain in the United States.
Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and be use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. Rights can be established outside of the United States subject to a government use license.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Glossary
- Antigenic variation
A mechanism developed independently by different parasitic species to evade or exhaust the host immune system. A parasite systematically switches the expression of antigens present at the host-parasite interface. The genetic mechanism underlying this phenomenon varies among different parasites. In ticks, the antigens present at the host-parasite interface are on the tick salivary proteins. Accordingly, different antigens are expressed either once or sequentially throughout tick feeding. The phenomena of silent antigens and antigenic variation in tick saliva are most likely interconnected.
- Gene duplication
One of the dominant mechanisms in gene evolution. Gene duplication occurs due to recombination, and is followed either by positive or negative (purifying) selection. Positive selection results in a protein with a novel function, while negative selection eliminates the duplicated gene.
- Immunogenicity
The ability of an antigen to induce humoral or cellular antigen-specific immune responses. Several factors influence the immunogenicity: the type of molecule (proteins are usually the best antigens), molecular weight (larger proteins display higher immunogenicity), and concentration (low and high concentrations of antigens can induce immunotolerance or anergy, respectively).
- Multigenic family
A group of phylogenetically closely-related proteins that evolved from a single gene ancestor by the process of multiple gene duplications and subsequent evolution due to the pressure of positive selection. Individual members of one multigenic family can exert identical, similar, or completely different functions.
- Pluripotency
In the context of this review, pluripotency means that a single salivary protein displays more than one function. For instance, a protease inhibitor can target more proteases with variable specificity and thus inhibit more physiological processes. Pluripotency can be misidentified, if two different observed effects of a given tick protein actually result from the inhibition of the same upstream process.
- Redundancy
Several redundancy scenarios in tick salivary protein function can be anticipated and observed: two or more proteins from one or several different multigenic families (i) target an identical cellular mechanism, or (ii) target different modules of the same cellular or physiological pathway to exert the same final effect on host physiology.
- Silent antigen
An antigen, present on a tick salivary protein that can efficiently raise an antibody response when injected at high concentrations in recombinant form, but is unable to elicit such a response as a part of the physiological salivary mixture. This might be due to the lower concentration of the antigen in tick saliva. The concentration of a given effector in tick saliva is low due to the combination of functionally identical but antigenically different salivary proteins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sonenshine DE, Roe RM. Biology of ticks. Oxford University Press; 2014. [Google Scholar]
- 2.Kotal J, et al. Modulation of host immunity by tick saliva. Journal of proteomics. 2015 doi: 10.1016/j.jprot.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chmelar J, et al. Sialomes and Mialomes: A Systems-Biology View of Tick Tissues and Tick-Host Interactions. Trends in parasitology. 2015 doi: 10.1016/j.pt.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karim S, et al. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PloS one. 2011;6:e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarz A, et al. De novo Ixodes ricinus salivary gland transcriptome analysis using two next-generation sequencing methodologies. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:4745–4756. doi: 10.1096/fj.13-232140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy KP, et al. Janeway's immunobiology. Garland Science; 2012. [Google Scholar]
- 7.Schwarz A, et al. A systems level analysis reveals transcriptomic and proteomic complexity in Ixodes ricinus midgut and salivary glands during early attachment and feeding. Molecular & cellular proteomics : MCP. 2014;13:2725–2735. doi: 10.1074/mcp.M114.039289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francischetti IM, et al. An insight into the salivary transcriptome and proteome of the soft tick and vector of epizootic bovine abortion, Ornithodoros coriaceus. Journal of proteomics. 2008;71:493–512. doi: 10.1016/j.jprot.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes AL. The evolution of functionally novel proteins after gene duplication. Proceedings. Biological sciences / The Royal Society. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 10.Kondrashov FA, et al. Selection in the evolution of gene duplications. Genome biology. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proceedings. Biological sciences / The Royal Society. 2012;279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto SP, Yong P. The evolution of gene duplicates. Advances in genetics. 2002;46:451–483. doi: 10.1016/s0065-2660(02)46017-8. [DOI] [PubMed] [Google Scholar]
- 13.Christeller JT. Evolutionary mechanisms acting on proteinase inhibitor variability. The FEBS journal. 2005;272:5710–5722. doi: 10.1111/j.1742-4658.2005.04975.x. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz A, et al. Understanding the evolutionary structural variability and target specificity of tick salivary Kunitz peptides using next generation transcriptome data. BMC evolutionary biology. 2014;14:4. doi: 10.1186/1471-2148-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai SX, et al. Evolution, expansion and expression of the Kunitz/BPTI gene family associated with long-term blood feeding in Ixodes scapularis. BMC evolutionary biology. 2012;12:4. doi: 10.1186/1471-2148-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chmelar J, et al. Tick salivary secretion as a source of antihemostatics. Journal of proteomics. 2012;75:3842–3854. doi: 10.1016/j.jprot.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazimírová M. Bioactive compounds in ticks acting on host thrombohemostasis. In: Viroj W, editor. Thrombohemostatic disease research. Nova Biomedical Books; 2007. p. vi.p. 202. [Google Scholar]
- 18.Kotsyfakis M, et al. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick Ixodes scapularis. The Journal of biological chemistry. 2006;281:26298–26307. doi: 10.1074/jbc.M513010200. [DOI] [PubMed] [Google Scholar]
- 19.Sa-Nunes A, et al. The immunomodulatory action of sialostatin L on dendritic cells reveals its potential to interfere with autoimmunity. Journal of immunology. 2009;182:7422–7429. doi: 10.4049/jimmunol.0900075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horka H, et al. The tick salivary protein sialostatin L inhibits the Th9-derived production of the asthma-promoting cytokine IL-9 and is effective in the prevention of experimental asthma. Journal of immunology. 2012;188:2669–2676. doi: 10.4049/jimmunol.1100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein M, et al. Tick Salivary Sialostatin L Represses the Initiation of Immune Responses by Targeting IRF4-Dependent Transcription in Murine Mast Cells. Journal of immunology. 2015;195:621–631. doi: 10.4049/jimmunol.1401823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieskovska J, et al. Tick sialostatins L and L2 differentially influence dendritic cell responses to Borrelia spirochetes. Parasites & vectors. 2015;8:275. doi: 10.1186/s13071-015-0887-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieskovska J, et al. Tick salivary cystatin sialostatin L2 suppresses IFN responses in mouse dendritic cells. Parasite immunology. 2015;37:70–78. doi: 10.1111/pim.12162. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, et al. Genetic diversity of Salp15 in the Ixodes ricinus complex (Acari: Ixodidae) PloS one. 2014;9:e94131. doi: 10.1371/journal.pone.0094131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramamoorthi N, et al. The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature. 2005;436:573–577. doi: 10.1038/nature03812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg R, et al. Cutting edge: CD4 is the receptor for the tick saliva immunosuppressor, Salp15. Journal of immunology. 2006;177:6579–6583. doi: 10.4049/jimmunol.177.10.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anguita J, et al. Salp15, an Ixodes scapularis salivary protein, inhibits CD4(+) T cell activation. Immunity. 2002;16:849–859. doi: 10.1016/s1074-7613(02)00325-4. [DOI] [PubMed] [Google Scholar]
- 28.Hovius JW, et al. Salp15 binding to DC-SIGN inhibits cytokine expression by impairing both nucleosome remodeling and mRNA stabilization. PLoS pathogens. 2008;4:e31. doi: 10.1371/journal.ppat.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leboulle G, et al. Characterization of a novel salivary immunosuppressive protein from Ixodes ricinus ticks. The Journal of biological chemistry. 2002;277:10083–10089. doi: 10.1074/jbc.M111391200. [DOI] [PubMed] [Google Scholar]
- 30.Mans BJ, Ribeiro JM. Function, mechanism and evolution of the moubatin-clade of soft tick lipocalins. Insect biochemistry and molecular biology. 2008;38:841–852. doi: 10.1016/j.ibmb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paesen GC, et al. Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure. Molecular cell. 1999;3:661–671. doi: 10.1016/s1097-2765(00)80359-7. [DOI] [PubMed] [Google Scholar]
- 32.Hidano A, et al. Suppressive effects of neutrophil by Salp16-like salivary gland proteins from Ixodes persulcatus Schulze tick. Insect molecular biology. 2014;23:466–474. doi: 10.1111/imb.12101. [DOI] [PubMed] [Google Scholar]
- 33.Guo X, et al. Inhibition of neutrophil function by two tick salivary proteins. Infection and immunity. 2009;77:2320–2329. doi: 10.1128/IAI.01507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palenikova J, et al. Ixodes ricinus salivary serpin IRS-2 affects Th17 differentiation via inhibition of the interleukin-6/STAT-3 signaling pathway. Infection and immunity. 2015;83:1949–1956. doi: 10.1128/IAI.03065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Couvreur B, et al. Variability and action mechanism of a family of anticomplement proteins in Ixodes ricinus. PloS one. 2008;3:e1400. doi: 10.1371/journal.pone.0001400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prevot PP, et al. Exosites mediate the anti-inflammatory effects of a multifunctional serpin from the saliva of the tick Ixodes ricinus. The FEBS journal. 2009;276:3235–3246. doi: 10.1111/j.1742-4658.2009.07038.x. [DOI] [PubMed] [Google Scholar]
- 37.Prevot PP, et al. Anti-hemostatic effects of a serpin from the saliva of the tick Ixodes ricinus. The Journal of biological chemistry. 2006;281:26361–26369. doi: 10.1074/jbc.M604197200. [DOI] [PubMed] [Google Scholar]
- 38.Mulenga A, et al. Ixodes scapularis tick serine proteinase inhibitor (serpin) gene family; annotation and transcriptional analysis. BMC genomics. 2009;10:217. doi: 10.1186/1471-2164-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mans BJ, et al. Evolution of hematophagy in ticks: common origins for blood coagulation and platelet aggregation inhibitors from soft ticks of the genus Ornithodoros. Molecular biology and evolution. 2002;19:1695–1705. doi: 10.1093/oxfordjournals.molbev.a003992. [DOI] [PubMed] [Google Scholar]
- 40.Mans BJ, et al. Comparative sialomics between hard and soft ticks: implications for the evolution of blood-feeding behavior. Insect biochemistry and molecular biology. 2008;38:42–58. doi: 10.1016/j.ibmb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersson DI, et al. Evolution of new functions de novo and from preexisting genes. Cold Spring Harbor perspectives in biology. 2015;7 doi: 10.1101/cshperspect.a017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mans BJ, Neitz AW. Adaptation of ticks to a blood-feeding environment: evolution from a functional perspective. Insect biochemistry and molecular biology. 2004;34:1–17. doi: 10.1016/j.ibmb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Kotsyfakis M, et al. Tissue- and time-dependent transcription in Ixodes ricinus salivary glands and midguts when blood feeding on the vertebrate host. Scientific reports. 2015;5:9103. doi: 10.1038/srep09103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asgari S, et al. Rational design of stable and functional hirudin III mutants with lower antigenicity. Biologicals : journal of the International Association of Biological Standardization. 2015 doi: 10.1016/j.biologicals.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 45.Cai Y, et al. Computational design, functional analysis and antigenic epitope estimation of a novel hybrid of 12 peptides of hirudin and reteplase. Journal of molecular modeling. 2015;21:2774. doi: 10.1007/s00894-015-2774-2. [DOI] [PubMed] [Google Scholar]
- 46.McCulloch R, Field MC. Quantitative sequencing confirms VSG diversity as central to immune evasion by Trypanosoma brucei. Trends in parasitology. 2015;31:346–349. doi: 10.1016/j.pt.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tabel H, et al. T cells and immunopathogenesis of experimental African trypanosomiasis. Immunological reviews. 2008;225:128–139. doi: 10.1111/j.1600-065X.2008.00675.x. [DOI] [PubMed] [Google Scholar]
- 48.Guizetti J, Scherf A. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cellular microbiology. 2013;15:718–726. doi: 10.1111/cmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeMarco R, et al. Protein variation in blood-dwelling schistosome worms generated by differential splicing of micro-exon gene transcripts. Genome research. 2010;20:1112–1121. doi: 10.1101/gr.100099.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer GH, et al. 'Nothing is permanent but change'- antigenic variation in persistent bacterial pathogens. Cellular microbiology. 2009;11:1697–1705. doi: 10.1111/j.1462-5822.2009.01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chmelar J, et al. Insight into the sialome of the castor bean tick, Ixodes ricinus. BMC genomics. 2008;9:233. doi: 10.1186/1471-2164-9-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ribeiro JM, et al. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect biochemistry and molecular biology. 2006;36:111–129. doi: 10.1016/j.ibmb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 53.Kotsyfakis M, et al. Cutting edge: Immunity against a “silent” salivary antigen of the Lyme vector Ixodes scapularis impairs its ability to feed. Journal of immunology. 2008;181:5209–5212. doi: 10.4049/jimmunol.181.8.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]