Abstract
Non-viral gene delivery methods represent a potential safe and effective approach for treating myriad diseases. For many gene therapy applications, delivering multiple exogenous genes and controlling the time profile that these genes are expressed would be advantageous. Polymeric non-viral gene carriers are versatile and can be readily tailored for particular therapeutic applications, have the ability to carry multiple large genes within each particle, and can be more easily manufactured than viruses used for gene delivery. A layer-by-layer (LbL) theranostic-enabling nanoparticle was developed to incorporate two plasmid types which have differing expression time-profiles. Temporally controlling the expression of exogenous DNA enables superior control over the microenvironment and could lead to better control over differentiation pathways and cell fate.
Keywords: gold nanoparticle, layer-by-layer, gene delivery, poly(beta-amino ester), cell engineering
Introduction
Gene therapy is capable of curing or treating many genetic disorders and cancer.1 Polymeric gene delivery techniques have advantages over viral methods in that they are chemically customizable, are able to be manufactured on a large scale, have low potential for insertional mutagenesis, and are less immunogenic. However, the balance between safety and efficacy remains elusive, as there are not any U.S. FDA approved viral or non-viral gene therapies available.
Delivering DNA in conjunction with biocompatible gold nanoparticles (AuNP)s enables a therapeutic function (gene/thermotherapy) as well as a diagnostic function (imaging). AuNPs are known to be effective clinical X-ray contrast agents with extended imaging times and sharper contrast than traditional iodine-based agents.2 AuNPs are also able to be imaged via photoacoustics3 and can be viewed by transmission electron microscopy (TEM)4. While the imaging properties of AuNPs are well known, they are not intrinsically able to deliver DNA. Our current research focuses on polymer coatings of AuNPs to promote gene delivery, making the polymer-coated AuNPs a theranostic-enabling platform. There are additional advantages of AuNPs as well. Photothermal therapy, in which AuNPs are excited by incident electromagnetic radiation at their surface plasmon resonance wavelength, has been shown to effectively thermally ablate tumors in mammalian in vivo models.5 AuNPs are also attractive due to their monodispersity,6 their ability to be highly charged and suitable for layer-by-layer (LbL) polyelectrolyte coating processes, their ability to be easily chemically modified via thiol groups,7 and their optical properties.
LbL assembly involves the sequential addition of oppositely charged polyelectrolytes onto a given substrate.8–11 DNA loading can be modified by the number and the order of the DNA layers.12 If DNA layers were able to be transcribed and translated at different time points, it would improve temporal control over the cellular microenvironment. Stem cell differentiation is one type of potential application that could benefit from such control over expression time profiles, as exposure to growth factors at different time points can heavily influence differentiation pathways.13,14 As a proof of principle, this work demonstrates the ability to control the expression time profiles of two proteins using a single theranostic-enabling AuNP LbL platform.
Materials and methods
Gold nanoparticle synthesis and characterization
The AuNPs used in this study were synthesized via a modified Frens method6 and previously reported.4 Briefly, a 0.01% tetrachloroauric acid trihydrate, HAuCl4 (Fisher Scientific), in ultra pure distilled water was vigorously boiling when a 1% sodium citrate tribasic dihydrate (Sigma Aldrich) solution (in water) was injected to reduce the gold ions.
11-MUA Stabilization
To maintain monodispersity and create a stably charged anionic coating on the AuNPs, 11-mercaptoundecanoic acid (MUA) was conjugated to the AuNPs via a thiol bond as previously reported and was previously shown to be superior for ionic complexation LbL processes compared to citrate-stabilized AuNPs.4 The 11-MUA at 1 mM in ethanol was added to the citrate stabilized AuNPs and allowed to thiolate for 48 hours. The solution was then washed twice in ethanol and re-solvated in a 0.45% solution of sodium citrate. The resulting conjugated AuNPs were then diluted to 0.31 nM in preparation for the layering process.
Polymer synthesis and characterization
To synthesize the hydrolytically degradable cationic poly(β-amino ester) (PBAE) polyelectrolyte layer, diacrylate (1,4-butanediol diacrylate) and amine small molecules (4-amino-1-butanol) were reacted neat in a 1.2:1 molar ratio15 and were allowed to react for 24 hours (>100 RPMs). Subsequently, the linear polymers were end-capped for 1 hour at 1000 RPM via an excess of 1-(3-aminopropyl)-4-methylpiperazine in anhydrous tetrahydrofuran (THF), resulting in 1-(3-aminopropyl)-4-methylpiperazine end-modified poly(1,4-butanediol diacrylate-co-4-amino-1-butanol). This cationic PBAE polymer is referred to as B4-S4-E7 or 447 (Figure S1). Polymer 447 was then ether-purified to remove any unreacted monomers and then solvated to 100 mg/mL in anhydrous dimethyl sulfoxide (DMSO) and was stored at −20°C until use. We have previously reported NMR spectra on 447 and verified its chemical structure [1].
The polymer molecular weight was quantified using gel permeation chromatography (Waters®, Breeze 2 software). The solvent constituted 94% THF, 5% DMSO, 1% piperidine and a trace amount of butylated hydroxytoluene at a flow rate of 1 mL min−1.
Multilayer particle formation
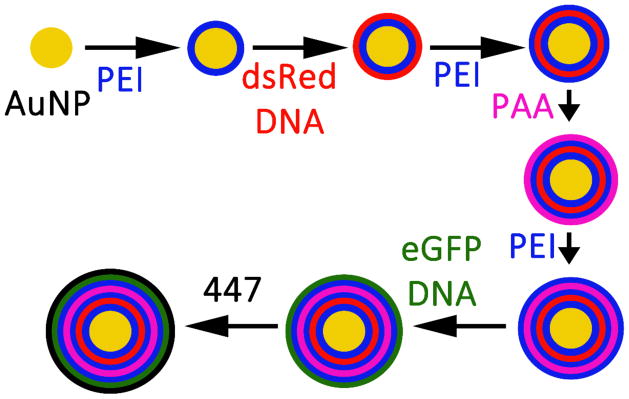
The multilayer nanoparticles were designed to have 3 polymer layers (polyethylenimine (PEI), poly(acrylic acid) (PAA), and PBAE) between the two functional gene layers (the dsRed and eGFP pDNA layers) so that the outer eGFP layer would be transcribed and translated first, followed by the expression of the more inner dsRed layer at a later point in time. PEI was chosen because it is not-degradable in water, which is important for the inner layers, is highly cationic, and has been demonstrated to be effective for gene delivery. Poly(acrylic acid) was chosen because it is highly anionic and has been used for LbL processes in the past.16 PBAE was chosen as a last layer due to its high efficacy as a cationic gene delivery polymer and based on our earlier research on PBAEs as coating materials to improve intracellular delivery.4,17 The LbL process is shown in Scheme 1. 80 μL of 25 kDa branched PEI was added to 112 μL of 0.31 nM (1.9×1011 particles mL−1) aqueous AuNPs which had been MUA-conjugated. The AuNP-PEI solution was shaken at room temperature for 30 minutes at 500 RPM, and then washed twice at 4°C via centrifugation for 10 minutes at 10 krcf. The supernatant was replaced with 150 mM sodium acetate (NaAc, pH=5) up to 192 μL for the first wash, and then up to 112 μL for the second wash. Subsequent layers were deposited by adding 80 μL of polyelectrolyte suspended in 150 mM NaAc, followed by the same washing procedure. PEI was added at 10 mg mL−1 (Layers 1, 3, 5), dsRed-Max-N1 (dsRed; Addgene) plasmid DNA (pDNA) at 0.5 mg mL−1 (Layer 2), PAA (Mw ~1800 Da; Sigma Aldrich) at 0.5 mg mL−1 (Layer 4), enhanced Green Fluorescent Protein (eGFP-N1, referred to as eGFP; Clontech) pDNA at 0.5 mg mL−1 (Layer 6), and PBAE 447 at 2.5 mg mL−1 (Layer 7). To reduce acidity 25 mM NaAc was used instead of 150 mM NaAc for the final wash of the eGFP pDNA layer. In addition, PBAE 447 was solvated in 25 mM NaAc and the final layer was not washed as we have previously observed excess 447 is necessary for transfection.4
Scheme 1.
LbL process of 7 layering steps of the LbL inorganic/polymeric nanoparticles beginning with the MUA-conjugated AuNPs. While a single gold core is shown for clarity, typically 3–4 inorganic gold nanoparticles are co-encapsulated per larger LbL inorganic/polymeric nanoparticle.
Particle diameter
Nanoparticle tracking analysis (NTA) was performed with a NanoSight NS500 at each of the 7 layering stages (n=2). Particles were diluted up to a total of 500 μL in 150 mM NaAc and mixed thoroughly immediately prior to loading into the NanoSight. Transmission electron microscopy (TEM) was accomplished on the complete LbL formulation to confirm the diameter ascertained via NTA (n=3). ImageJ (NIH) was also used to quantify diameters of the TEM images based on the area of the particles.
Particle zeta potential
Dynamic light scattering (DLS) analysis was performed with a Zetasizer (Malvern) following the completion of each of the 7 layers to measure the zeta potential (n=2 per layer). Particles were diluted up to a total of 700 μL in UltraPure™ Distilled Water (Invitrogen).
Nucleic acid loading
Based on our previously reported decomplexation process,4 the nucleic acid content was quantified via a calibration curve using a fluorescence plate reader (Synergy2; 485/528 nm excitation/emission); the stock nucleic acid decomplexation solution (fluorescence dye (YO-PRO®-1: 4.37 μL at 10 μM; phosphate buffered saline (PBS): 395.89 μL at 10x (1500 mM); heparin: 36.95 μL at 3.55 mg/mL) was added to 112 μL of the LbL solution (150 mM NaAc) to a final volume of 192 μL (final concentrations: salt (PBS and NaAc): 653 mM; heparin: 300 μg/mL). Previously reported data shows that the salt and heparin concentration used was sufficient to measure the nucleic acid content present in the layered formulation.4 The nucleic acid content was assessed after the dsRed layer and after both the dsRed and eGFP layers were complexed. The eGFP content was quantified via mass balance by subtracting the total nucleic acid content from the dsRed content. The uncomplexed nucleic acid content was also assessed in the supernatants of the washing steps following the addition of the dsRed layer and following the addition of the eGFP layer as well (Figure S2). The efficiency of layering was calculated by multiplying 100 by the fraction of nucleic acid retained in the LbL formulation to the total nucleic acid content used for layering.
Cell culture and transfection
Human glioblastoma cells18 (GB319) cultured in T-75 flasks were cultured at 37°C and 5% CO2 atmosphere using DMEM:F12 (1:1), 1x antibiotic-antimycotic and 10% fetal bovine serum (FBS). The cells were seeded at 10,000 cells/well in 96-well tissue culture treated polystyrene plates the day before transfection and were confluent at the time of transfection. Transfection was performed by diluting 192 μL of the final LbL formulation up to 256 μL using 25 mM NaAc. From the diluted 256 μL, 20 μL was delivered per well and allowed to incubate with the seeded cells in the 96-well plates for 2 hours at 37°C before the media was replaced with complete media. The full layered nanoparticles were incubated using 10% and 50% serum by volume for the 2 hour time period to assess any differences, as the 50% serum concentration more closely resembles in vivo conditions. The glioblastoma cells in the plates were cultured in the same conditions as in the flasks (37°C and 5% CO2 atmosphere using DMEM:F12 (1:1), 1x antibiotic-antimycotic and 10% FBS) throughout the 21 day period.
As a positive control, Lipofectamine® 2000 (Lipo2k) was also incubated with the cells in like manner. Lipo2k was mixed with dsRed and eGFP pDNA using ratios of 0.5:1, 1.5:1, 2.5:1, and 3.5:1 (Lipo2k μL: pDNA μg) at the same dsRed:eGFP pDNA ratio and dosages as the LbL formulation. Total dosages of dsRed:eGFP pDNA of 100 and 200 ng were also assessed at the same Lipo2k:pDNA (μL:μg; n=4) ratios.
Protein expression and relative metabolic activity
Fluorescence microscopy images were obtained with a Carl Zeiss fluorescence microscope on days 2, 5, 9, 14, and 21 post-transfection (5x magnification). The eGFP and dsRed channel exposure times were 300 and 600 ms, respectively.
Flow cytometry was performed with a BD Accuri™ C6 flow cytometer on days 2, 5, 9, 14, and 21 post-transfection as well; a HyperCyt sampler was used to automate data collection. Cells were detached via incubation at 37°C with trypsin-ethylenediaminetetraacetic acid (0.05%) for five minutes, followed by immediate quenching with 2% FBS in PBS. FSC-H vs. SSC-H was used to isolate the singlet population; expression of eGFP and dsRed were analyzed using FL1-H vs. FL3-H (FloJo). Because cellular uptake is required for successful transcription and translation of both the eGFP DNA and the dsRed DNA, the presence of the expressed fluorescent proteins by fluorescence microscopy and flow cytometry confirms cellular uptake.
An MTS-based Promega assay CellTiter 96® was used to determine the relative metabolic activity of transfected cells as compared to untreated controls at 24 hours for 10% and 50% serum conditions which is an indicator of viability.
Statistics
Figure 3: 10% serum: n=12; 50% serum: n=4. Figure S2, S4, and S5 were n≥4, ≥4, =4, respectively. When comparing the positive control (Figure 3 vs. Figure S5) to the LbL formulation at 2 days post-transfection, a two-tailed Student’s t-test was used. All error bars in figures and the text are reported as standard errors of the mean (SEM) unless otherwise indicated.
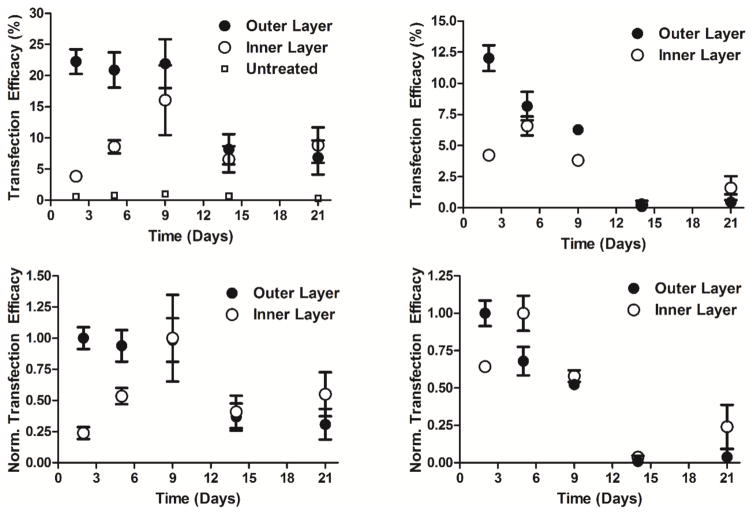
Figure 3.
Transfection efficacies (top) and normalized transfection efficacies (bottom) in time for the outer (eGFP) and inner (dsRed) pDNA layers using 10% (left) and 50% (right) serum containing media. Transfection is of human glioblastoma GB319 cells. (10% serum: n=12; 50% serum: n=4).
Results
Gold nanoparticle characterization and 11-MUA stabilization
TEM analysis confirmed that the stabilized AuNPs were monodisperse as well as uniform in size at 17 ± 2 nm (Mean ± StDev) as we have previously found.4 SPR results showed a red-shift of the wavelength from 520 nm for solely citrate stabilized AuNPs to 526 nm for 11-MUA conjugated nanoparticles.4
Polymer characterization
The number-average (Mn) and weight-average (Mw) molecular weights of PBAE 447 were 11.9 kDa and 35.5 kDa, respectively, with a polydispersity index of 2.98.
Particle diameter and zeta potential
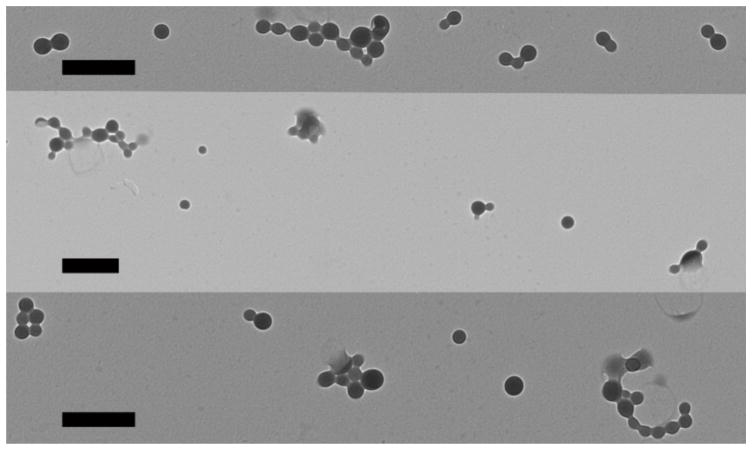
The TEM of the complete LbL inorganic/polymeric nanoparticle formulations with all 7 layers is shown in Figure 1. The arithmetic and geometric means of cores per LbL inorganic/polymeric nanoparticle were 4.3 and 2.7, respectively, demonstrating 3–4 inorganic gold nanoparticles per larger LbL inorganic/polymeric nanoparticle. According to the area of the particles assessed by ImageJ, the dry inorganic nanoparticle core diameters were 104 ± 0.8 nm (StDev) in diameter and the larger dry LbL inorganic/polymeric nanoparticle diameters were 210 ± 10 nm (StDev) between the 3 images. The dry nanoparticle diameters according to TEM were similar in size but slightly smaller than the hydrodynamic nanoparticle diameters as measured in aqueous buffer by NTA. The hydrodynamic diameters of the LbL inorganic/polymeric nanoparticle were approximately 300 nm for all 7 layers (Figure 2; left).
Figure 1.
TEM of LbL inorganic/polymeric nanoparticles. Scale bars are 500 nm.
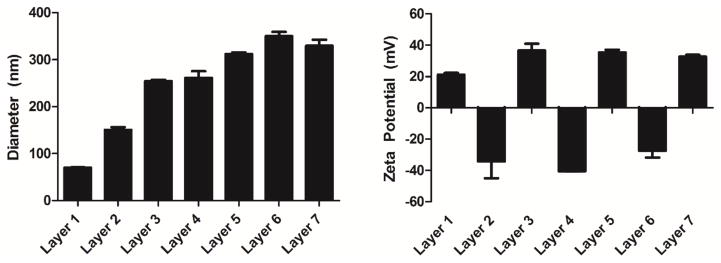
Figure 2.
Hydrodynamic diameter (left) and zeta potential (right) of LbL inorganic/polymeric nanoparticles following the addition of each of the 7 sequential layers. (n=2)
According to NTA, the hydrodynamic radius increased with each subsequent layer for the first 5 layers. There was no statistical difference in diameter between layers 5–7. The largest increase in diameter occurred at the first DNA layer similarly to what was observed previously and is believed to be due to the plasmid DNA molecules being relatively large (~3 MDa) and multivalent with negative charge.4 The zeta potential of the LbL inorganic/polymeric nanoparticles reversed at each of the 7 stages of layering, validating that coatings of each polyelectrolyte layer were achieved as designed (Figure 2; right).
Nucleic acid loading
The inner (dsRed) and outer (eGFP) pDNA layers contained DNA in an approximate 2:1 mass ratio. When the LbL inorganic/polymeric nanoparticles were used for cellular experiments, 208 ± 9 ng/well dsRed pDNA and 120 ± 10 ng/well eGFP pDNA, respectively, were contained in the LbL inorganic/polymeric nanoparticle formulations, for a total of 328 ± 9 ng of pDNA used per cellular well in a 96-well plate. The layering efficiency of the inner and outer layers were 6.6 ± 0.6% and 4 ± 1% (Figure S2). The dsRed pDNA nucleic acid content in the supernatant of the first and second washings following dsRed pDNA layering accounted for 79 ± 2% and 4 ± 1% respectively (Figure S2). The eGFP pDNA nucleic acid content in the supernatant of the first and second washings following eGFP pDNA layering accounted for 77 ± 3% and 4 ± 2% respectively (Figure S2). The mass ratio of polymer 447 to total pDNA was 48:1.
Expression and relative metabolic activity
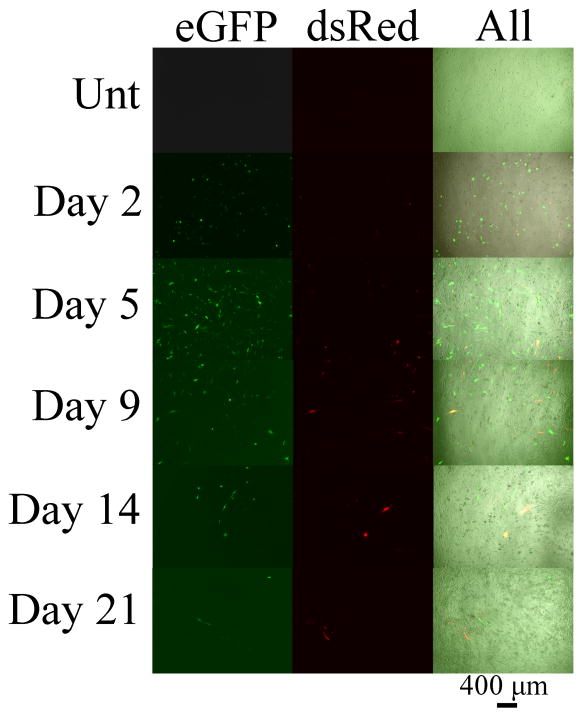
In the 10% serum condition (Figure 3; left), the transfection levels of the outer (eGFP) pDNA layer of the LbL inorganic/polymeric nanoparticles was 22 ± 2% positively transfected human cells (Figure 3; top) and remained similar out to day 9 and then decreased to approximately 7% thereafter. In contrast, the inner (dsRed) pDNA layer (Figure 3; top) had low expression initially and steadily increased to a maximum at 16 ± 6% positively transfected human cells on day 9. The expression thereafter was similar to the outer eGFP levels near 7%. Figure S3 shows the dot plots of the dsRed and eGFP fluorescence channels (FloJo) for the 10% serum condition on days 2, 5, 9, 14, and 21. Qualitatively, it can be seen that the eGFP and dsRed expression maxima are on days 2 and 9, respectively. The fluorescence microscope images (shown in Figure 4) also qualitatively match these findings.
Figure 4.
Fluorescence microscopy images of the GB319 cells in 10% serum transfected with the LbL inorganic/polymeric nanoparticles on days 2, 5, 9, 14, and 21 post-transfection. The fluorescence channels on the left, middle and right correspond to eGFP, dsRed, and all channels (brightfield, eGFP, and dsRed), respectively. The scale bar is 400 μm.
In the 50% high serum condition during transfection (Figure 3; right), the outer (eGFP) layer was higher at 12 ± 1% initially and steadily decreased to near 0% on day 14 (Figure 3; top). The inner (dsRed) layer had similar kinetics to the 10% serum condition, except the peak expression happened sooner, on day 5, and the transfection efficacy was lower, with a maximum at 6.5 ± 0.8% (Figure 3; top).
Figure 3 (bottom) shows the normalized transfection levels for the LbL inorganic/polymeric nanoparticles in the 10% serum condition (bottom left) and the 50% serum condition (bottom right), which more clearly shows the kinetics of exogenous protein expression over time as well as the maximum expression levels. The outer layers for the 10 and 50% serum conditions were highest on day 2 whereas the inner layers for the 10% and 50% serum conditions peaked on days 9 and 5, respectively. The relative metabolic activities (untreated = 100%) of the LbL inorganic/polymeric nanoparticles incubated in 10% and 50% serum at 24 hours post-transfection were 93 ± 4% and 88 ± 2%, respectively, as shown in Figure S4. This demonstrates good cellular viability and minimal cytotoxicity of the LbL inorganic/polymeric nanoparticles.
Generally speaking, as the dosage increased for the Lipo2k formulations both the transfection efficacies of eGFP and dsRed increased and the relative metabolic activities decreased (Figure S5). At comparable dosages to the LbL inorganic/polymeric nanoparticles (eGFP: 120 ± 10 ng; dsRed: 208 ± 9 ng; see Nucleic acid loading section), the 3.5:1, 2.5:1, 1.5:1, and 0.5:1 (Lipo2k μL: μg pDNA) had relative metabolic activities of 30 ± 2%, 53 ± 1%, 79 ± 1%, and 96 ± 1%, respectively. The transfection levels for the same Lipo2k:pDNA ratios for eGFP and dsRed were 21 ± 5%, 22 ± 4%, 27 ± 4%, 22 ± 1% and 28 ± 2%, 25 ± 2%, 32 ± 3%, 13 ± 1%, respectively. The highest transfection level achieved with the Lipo2k positive control at the same dosage as the LbL inorganic/polymeric nanoparticles was the 1.5:1 formulation which was similar statistically to the LbL inorganic/polymeric nanoparticles in 10% serum for both eGFP (p-value = 0.53) and dsRed (p-value = 0.14) transfection (Figure S5), but had lower cellular metabolic activity, indicating cytotoxicity (79 ± 1%).
While holding the eGFP and dsRed mass ratio for the Lipo2k similar to the LbL inorganic/polymeric nanoparticles (0.34 ng : 0.66 ng), the highest transfection level was found to be using a pDNA dosage of 200 ng and a Lipo2k μL: μg pDNA ratio of 2.5:1. The eGFP and dsRed transfection levels were 28 ± 1% and 32 ± 3% which were statistically similar to the LbL inorganic/polymeric nanoparticles in 10% serum as well for both eGFP (p-value = 0.38) and dsRed (p-value = 0.13). However, the optimized Lipo2k formulation (200 ng; 2.5:1) also had a much lower relative metabolic activity of 65 ± 1% (Figure S5).
Discussion
In these experiments we have determined that we can fabricate LbL inorganic/polymeric nanoparticles composed of both theranostic inorganic nanoparticle cores and polyelectroyte coatings that are useful for intracellular gene delivery. The layer of plasmid DNA in the multilayered nanoparticle is able to tune the kinetics of expression with the outer layer being expressed quickly and inner DNA layer experiencing delayed expression over a time span of several days. As expected, subsequent layering of polyelectrolytes increased the diameter of the nanoparticles compared to uncoated gold nanoparticles and there was reversal of the zeta potential following each successive layering and washing step. This biophysical characterization demonstrates that successful ionic complexation of each layer was achieved for these hybrid nanoparticles. In this method, 3–4 AuNP cores were encapsulated within each LbL inorganic/polymeric nanoparticle. While this may be able provide stronger X-ray/CT contrast or higher per particle drug loading, it may also reduce cellular uptake compared to coating of a single AuNP core. Alternative fabrication methods, such as those that utilize microfluidics and/or a reduced concentration of AuNPs cores, may enable LbL inorganic/polymeric nanoparticle construction with only single AuNP cores. Nonetheless, the LbL inorganic/polymeric nanoparticles described here were able to be successfully internalized by human cells leading to successful gene delivery of multiple genes.
Because the backbones of the two plasmids used in this study are very similar, including the same plasmid size and promoter, differences observed in the exogenous gene expression profiles over time are attributable to the physical placement of these different plasmids within the LbL inorganic/polymeric nanoparticles. The inner plasmid is likely secluded from transcription and translation for a longer time period within the non-degradable inner polymer layers of PEI and PAA, whereas the eGFP plasmid is closer to the outside of the particle and most strongly associated with the poly(beta-amino ester), the polymer that coats it and that is degradable in water. Based on the particle sizing data (Figure 2), the dsRed pDNA layer is thicker than the eGFP pDNA layer, leading to there being approximately twice as much dsRed pDNA as eGFP pDNA. The dsRed pDNA was secluded sufficiently that the kinetics of expression of these two plasmids were independent and separate in time.
When investigating both types of serum media conditions, the outer layer transfected the cells quickly, reaching its maximum expression at the earliest time point measured (day 2) and then decreasing over time. The inner layer had a delay in peak gene expression depending on the local environment, with lower serum leading to a longer delay (peak expression at day 9) and increased serum leading to a shorter delay (peak expression at day 5). In all cases, little toxicity was observed with the LbL inorganic/polymeric nanoparticles. In the high serum condition, it is possible that the nanoparticle decomplexation process may be enhanced by the presence of additional serum proteins, resulting in quick nanoparticle disassembly and inner plasmid release to the cytoplasm. We have also recently found that for poly(beta-amino ester)-based nanoparticles, not all uptake mechanisms (i.e., clathrin, caveolae, and macropinocytosis) are equally effective for the eventual transcription and translation of exogenous plasmids.20 It is possible that the LbL nanoparticles in the presence of a higher concentration of serum proteins may experience more serum adsorption to the outer nanoparticle surface in a manner that may change the cellular uptake properties of the nanoparticle.21 Future mechanistic studies may be able to test these hypotheses and elucidate design criteria to make related LbL inorganic/polymeric nanoparticles even more effective at non-viral gene delivery. Because the design of these hybrid nanoparticles are flexible, additional nucleic acids may be included and by varying the number of layers, temporal release may be able to be further tuned.
There are several potential advantages of delivering, with temporal control, two plasmids types within the same nanoparticle as compared to multiple doses of single nanoparticles. Within a tissue engineered construct or in vivo setting, multiple doses at a desired spatial and temporal location may not be possible. Moreover, it has been demonstrated that when polymeric gene delivery nanoparticles, each encoding different plasmids, are added in combination to cells, mosaic expression is observed.25 Yet, when the different plasmids are co-encapsulated in the same nanoparticles, co-expression of these plasmids in the same cells is observed instead.25 Thus, co-encapsulation helps to ensure co-expression, and in the case of LbL inorganic/polymeric nanoparticles, kinetically controlled co-expression. An alternative approach for kinetic control of gene expression that has been recently demonstrated is the utilization of engineered genetic circuits and synthetic biology for control.22–24 In contrast, the method that we present here is orthogonal as the kinetic control is mediated by biomaterial properties and physical sequestration. These two approaches could potentially be used in combination to enable further enhanced control of the kinetics of exogenous gene expression, including temporal release following an environmental trigger as well as control independent of the state of a cell.
Using an LbL approach, we were able to successfully develop a theranostic-enabling platform technology which is capable of loading two different types of plasmids within the same nanoparticle, safely and effectively non-virally delivering them to human cells, and then temporally controlling the exogenous gene expression between the two plasmids. The outer layer of the nanoparticle is composed of a biodegradable polymer that mediates quick and efficient transfection of the outermost plasmid later. Over time as the LbL inorganic/polymeric nanoparticle disassembles, the inner most plasmid layer becomes active to be transcribed and translated with its own kinetics. This coating approach could also be extended to other types of charged nanoparticles, including those with additional imaging modalities. Such control over the timing of non-viral exogenous gene expression, mediated by theranostic nanoparticles, may be of significant interest as a research tool for controlling the cellular microenvironment and as a potential therapeutic.
Supplementary Material
Acknowledgments
Dr. Alfredo Quiñones-Hinojosa is thanked for the use of the GB319 cells. This work was supported by the National Institutes of Health (R01EB016721). We would like to acknowledge the Microscope Facility at JHU for allowing usage of their facility, and the National Science Foundation Graduate Research Fellowship given to CJB (DGE-0707427).
Abbreviations
- AuNP
gold nanoparticle
- DMSO
dimethyl sulfoxide
- DLS
dynamic light scattering
- dsRed
dsRed-Max-N1
- eGFP
eGFP-N1
- FBS
fetal bovine serum
- GB319
primary human glioblastoma
- LbL
layer-by-layer
- Lipo2k
Lipofectamine™ 2000
- NaAc
sodium acetate
- Mn
number-average molecular weight
- Mw
weight-average molecular weight
- MUA
11-mercaptoundecanoic acid
- NTA
nanoparticle tracking analysis
- PAA
polyacrylic acid
- PBAE
poly(β-amino ester)
- PBS
phosphate buffered saline
- pDNA
plasmid DNA
- PEI
polyethylenimine
- SEM
standard error of the mean
- StDev
standard deviation
- SN1
the 1st supernatant washing after a nucleic acid layer
- SN2
the 2nd supernatant washing after a nucleic acid layer
- TEM
transmission electron microscopy
- THF
tetrahydrofuran
Footnotes
Supplementary material can be found online.
References
- 1.Bishop CJ, Kim J, Green JJ. Biomolecule Delivery to Engineer the Cellular Microenvironment for Regenerative Medicine. Annals of Biomedical Engineering. 2014;42(7):1557–1572. doi: 10.1007/s10439-013-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hainfeld JF, Slatkin DN, Focella TM, Smilowitz HM. Gold nanoparticles: a new X-ray contrast agent. British Journal of Radiology. 2006;79(939):248–253. doi: 10.1259/bjr/13169882. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Q, Iwakuma N, Sharma P, Moudgil BM, Wu C, McNeill J, Jiang H, Grobmyer SR. Gold nanoparticles as a contrast agent for in vivo tumor imaging with photoacoustic tomography. Nanotechnology. 2009;20(39) doi: 10.1088/0957-4484/20/39/395102. [DOI] [PubMed] [Google Scholar]
- 4.Bishop CJ, Tzeng SY, Green JJ. Degradable polymer-coated gold nanoparticles for co-delivery of DNA and siRNA. Acta Biomaterialia. 2014;11:393–403. doi: 10.1016/j.actbio.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Sayed IH, Huang XH, El-Sayed MA. Selective laser photo-thermal therapy of epithelial carcinoma using anti-EGFR antibody conjugated gold nanoparticles. Cancer Letters. 2006;239(1):129–135. doi: 10.1016/j.canlet.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 6.Frens G. Controlled Nucleation for Regulation of Particle-Size in Monodisperse Gold Suspensions. Nature-Physical Science. 1973;241(105):20–22. [Google Scholar]
- 7.Zhang ZW, Jia J, Lai YQ, Ma YY, Weng J, Sun LP. Conjugating folic acid to gold nanoparticles through glutathione for targeting and detecting cancer cells. Bioorganic and Medicinal Chemistry. 2010;18(15):5528–5534. doi: 10.1016/j.bmc.2010.06.045. [DOI] [PubMed] [Google Scholar]
- 8.Elbakry A, Zaky A, Liebkl R, Rachel R, Goepferich A, Breunig M. Layer-by-Layer Assembled Gold Nanoparticles for siRNA Delivery. Nano Letters. 2009;9(5):2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 9.Flessner RM, Yu Y, Lynn DM. Rapid release of plasmid DNA from polyelectrolyte multilayers: A weak poly(acid) approach. Chemical Communications. 2011;47(1):550–552. doi: 10.1039/c0cc02926b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poon Z, Lee JB, Morton SW, Hammond PT. Controlling in Vivo Stability and Biodistribution in Electrostatically Assembled Nanoparticles for Systemic Delivery. Nano Letters. 2011;11(5):2096–2103. doi: 10.1021/nl200636r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saurer EM, Flessner RM, Sullivan SP, Prausnitz MR, Lynn DM. Layer-by-Layer Assembly of DNA- and Protein-Containing Films on Microneedles for Drug Delivery to the Skin. Biomacromolecules. 2010;11(11):3136–3143. doi: 10.1021/bm1009443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blacklock J, Mao GZ, Oupicky D, Mohwald H. DNA Release Dynamics from Bioreducible Layer-by-Layer Films. Langmuir. 2010;26(11):8597–8605. doi: 10.1021/la904673r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Indrawattana N, Chen GP, Tadokoro M, Shann LH, Ohgushi H, Tateishi T, Tanaka J, Bunyaratvej A. Growth factor combination for chondrogenic induction from human mesenchymal stem cell. Biochemical and Biophysical Research Communications. 2004;320(3):914–919. doi: 10.1016/j.bbrc.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 14.Worster AA, Brower-Toland BD, Fortier LA, Bent SJ, Williams J, Nixon AJ. Chondrocytic differentiation of mesenchymal stem cells sequentially exposed to transforming growth factor-beta 1 in monolayer and insulin-like growth factor-I in a three-dimensional matrix. Journal of Orthopaedic Research. 2001;19(4):738–749. doi: 10.1016/S0736-0266(00)00054-1. [DOI] [PubMed] [Google Scholar]
- 15.Bishop CJ, Ketola TM, Tzeng SY, Sunshine JC, Urtti A, Lemmetyinen H, Vuorimaa-Laukkanen E, Yliperttula M, Green JJ. The Effect and Role of Carbon Atoms in Poly(beta-amino ester)s for DNA Binding and Gene Delivery. Journal of the American Chemical Society. 2013;135(18):6951–6957. doi: 10.1021/ja4002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinnane CR, Wark K, Such GK, Johnston AP, Caruso F. Peptide-functionalized, low-biofouling click multilayers for promoting cell adhesion and growth. Small. 2009;5(4):444–8. doi: 10.1002/smll.200801012. [DOI] [PubMed] [Google Scholar]
- 17.Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Letters. 2009;9(6):2402–6. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-viral gene delivery nanoparticles based on poly(beta-amino esters) for treatment of glioblastoma. Biomaterials. 32(23):5402–10. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ketola TM, Hanzlikova M, Leppanen L, Ravina M, Bishop CJ, Green JJ, Urtti A, Lemmetyinen H, Yliperttula M, Vuorimaa-Laukkanen E. Independent versus Cooperative Binding in Polyethylenimine-DNA and Poly(L-lysine)-DNA Polyplexes. Journal of Physical Chemistry B. 2013;117(36):10405–10413. doi: 10.1021/jp404812a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Sunshine JC, Green JJ. Differential Polymer Structure Tunes Mechanism of Cellular Uptake and Transfection Routes of Poly(beta-amino ester) Polyplexes in Human Breast Cancer Cells. Bioconjugate Chemistry. 2014;25(1):43–51. doi: 10.1021/bc4002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albanese A, Walkey CD, Olsen JB, Guo H, Emili A, Chan WC. Secreted biomolecules alter the biological identity and cellular interactions of nanoparticles. ACS Nano. 2014;8(6):5515–26. doi: 10.1021/nn4061012. [DOI] [PubMed] [Google Scholar]
- 22.Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. Proc Natl Acad Sci U S A. 2012;109(15):5850–5. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130(2):363–72. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 24.Brophy JA, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014;11(5):508–20. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhise NS, Shmueli RB, Gonzalez J, Green JJ. A novel assay for quantifying the number of plasmids encapsulated by polymer nanoparticles. Small. 2012;8(3):367–73. doi: 10.1002/smll.201101718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.