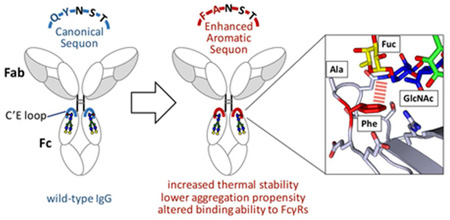
Abstract
Monoclonal antibodies (mAbs) exhibiting highly selective binding to a protein target constitute a large and growing proportion of the therapeutics market. Aggregation of mAbs results in the loss of their therapeutic efficacy and can result in deleterious immune responses. The CH2 domain comprising part of the Fc portion of Immunoglobulin G (IgG) is typically the least stable domain in IgG-type antibodies and therefore influences their aggregation propensity. We stabilized the CH2 domain by engineering an enhanced aromatic sequon (EAS) into the N-glycosylated C’E loop and observed a 4.8 °C increase in the melting temperature of the purified IgG1 Fc fragment. This EAS-stabilized CH2 domain also conferred enhanced stability against thermal and low pH induced aggregation in the context of a full-length monoclonal IgG1 antibody. The crystal structure of the EAS-stabilized (Q295F/Y296A) IgG1 Fc fragment confirms the design principle, i.e., the importance of the GlcNAc1•F295 interaction and surprisingly reveals that the core fucose attached to GlcNAc1 also engages in an interaction with F295. Inhibition of core fucosylation confirms the contribution of the fucose-Phe interaction to the stabilization. The Q295F/Y296A mutations also modulate the binding affinity of the full-length antibody to Fc receptors by decreasing the binding to low affinity Fc gamma receptors (FcγRIIa, FcγRIIIa and FcγRIIIb), while maintaining wild-type binding affinity to FcRn and FcγRI. Our results demonstrate that engineering an EAS into the N-glycosylated reverse turn on the C’E loop leads to stabilizing N-glycan–protein interactions in antibodies and that this modification modulates antibody-Fc receptor binding.
Graphical Abstract
Monoclonal antibodies (mAbs) are used extensively in scientific research, and are becoming increasingly important for diagnosing and treating diseases.(1) Despite their widespread applications, antibodies, like all proteins, remain susceptible to chemical and biological degradation processes. For example, mAbs undergoing significant structural fluctuations during purification, processing and storage can aggregate if at sufficient concentration.(2) Local mAb unfolding can expose peptide substructures that have a high propensity for intermolecular β-sheet formation, enabling mAb misassembly.(2) Aggregates of mAbs generally no longer recognize their target, and even a minor population of aggregates can sometimes lead to an unwanted immune response that can neutralize a therapeutic antibody.(2, 3) Therapeutic antibodies that sample alternative conformations can also be degraded by proteases in the body. Therefore, it is common practice in the pharmaceutical industry to select lead antibodies both based on their biological activity and their stability to minimize endoproteolysis and aggregation.(4) Engineering strategies that can reliably stabilize mAb native states both during storage and in vivo are valuable.
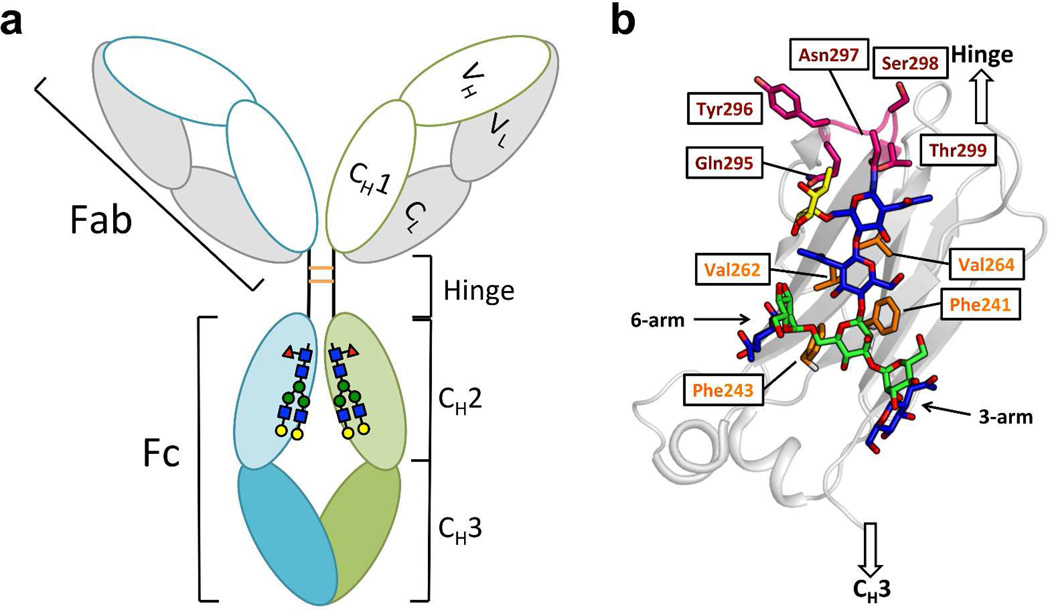
The immunoglobulin G (IgG) glycoproteins account for about 75% of the total immunoglobulins in human plasma. IgGs are Y-shaped macromolecules comprising two Fab (“Fragment antigen-binding”) and one Fc (“Fragment crystallizable”) substructures (Figure 1a). Two identical light chains and two identical heavy chains linked by disulfide bonds make up each IgG (Figure 1a). Each IgG heavy chain contains a variable domain (VH) and three constant domains (CH1, CH2 and CH3). The horseshoe-shaped IgG Fc is a homodimer composed of the CH2 and CH3 domains of the two heavy chains, which are covalently linked via disulfide bonds between the hinge regions that bridge the Fab and Fc fragments (Figure 1a). The interaction of the Fc substructure with the neonatal Fc receptor (FcRn) is responsible for the long serum half-lives of antibodies. Furthermore, the interaction of the Fc substructure with Fc gamma receptors (FcγRs) is necessary for obtaining the desired Fc-mediated effector functions, while interaction of the Fc with the C1q component of complement is necessary for clearing pathogens or aberrant cells.(1) The Fc region of IgG is also valuable for extending the serum half-lives of proteins-of-interest by generating therapeutic protein–Fc fusions (e.g., abatacept, alefacept, and etanercept).(1, 5, 6)
Figure 1. Domain organization and glycosylation site of IgG antibodies.
(a) IgG is composed of the Fab region, the hinge region and the Fc region. The Fab comprises the light chain (grey) and VH and CH1 domains (white) of the heavy chain. The Fc region comprises the CH2 (light green and light cyan) and CH3 (green and cyan) domains of the heavy chain. Each CH2 domain is N-glycosylated at Asn297. The heavy chains are disulfide linked (orange) in the hinge region. (b) Details of the glycan-protein interactions in the CH2 domain (grey ribbon). The N-glycan is presented with GlcNAc colored in blue, mannose colored in green and the core fucose colored in yellow. The N-glycan interacts with the hydrophobic residues (side chains highlighted in orange) on the inner surface of each CH2 domain. The C’E loop (backbone and side chains) is colored in magenta.
In most human IgGs, CH2 is the least stable domain, as reflected by its low melting temperature (Tm).(7) Given this circumstance, it is not surprising that, in many cases, IgG antibody aggregation is mediated by the CH2 domain.(2, 8–10) The conformational instability of the CH2 domain is also established to contribute to the aggregation of therapeutic protein-of-interest–Fc fusions.(6) Thus, stabilizing the native state of the CH2 domain should lower the rate of conformational excursions or unfolding of the Fc region and in turn, reduce the aggregation propensity of antibodies and therapeutic protein–Fc fusions.
A key feature of the Fc region of all IgG subclasses is that each CH2 domain contains a single Asn297 side chain amide N-linked glycan (or “N-glycan”) in the loop between strands C‘ and E (hereafter called the “C’E loop”).(11) These N-glycans are sequestered within the space between the two CH2 domains, bridging them while shielding the hydrophobic inner surface of the two CH2 domains from solvent (Figures 1a and 1b). The glycan-glycan and glycan-protein interactions involving the CH2 domains maintain the relative glycan orientation and are thought to influence Fc-FcγR interactions.(7, 12) The glycan-protein interactions are established to be vital for the stability of the CH2 domains, as deglycosylation greatly destabilizes the CH2 domain and abolishes binding to low-affinity FcγRs.(1, 13, 14) It has also been shown by NMR analysis that the termini of both the α1–3 and α1–6 arms of the N-glycan are highly dynamic, rendering the termini accessible to glycan-modifying enzymes.(15) Terminal glycan remodeling is known to influence the receptor binding abilities of the Fc.(15–17)
A common strategy to minimize the aggregation propensity of a protein-of-interest is to identify and remove the aggregation “hot spots” by altering the protein’s primary sequence. Such a strategy has been used previously on the Fc fragment to reduce antibody aggregation under thermal stress.(8, 9) CH2 domain sequences facilitating N-glycan conformational flexibility can lead to exposure of the hydrophobic inner surface of the CH2 domain, contributing to the aggregation of IgG antibodies and therapeutic protein–Fc fusions.(7) We hypothesize that strengthening the interactions between the glycan and the CH2 domain should stabilize the CH2 domain, lowering the concentration of alternatively folded CH2 domains, and thereby reducing CH2 domain aggregation propensity. We posit that the interactions between the glycan and CH2 domain can be engineered to be more stabilizing by replacing the N-glycosylated C’E loop with an enhanced aromatic sequon (EAS)—a portable structural module that features stabilizing aromatic side chain-glycan interactions while at the same time enhancing N-glycosylation efficiency by oligosaccharyltransferase.(18–23) A 4.8 °C increase in the melting temperature of a purified IgG1 Fc fragment was realized using this strategy. Moreover, this EAS-stabilized CH2 domain also conferred enhanced stability against thermal and low pH stress-mediated aggregation in the context of a full-length monoclonal IgG1 antibody. The crystal structure of the EAS-stabilized (Q295F/Y296A) IgG1 Fc fragment supports the engineering strategy, i.e., the importance of the GlcNAc1•F295 interaction and surprisingly, along with fucosylation inhibition data, reveals that the core fucose attached to GlcNAc1 also engages in a stabilizing interaction with F295. The Q295F/Y296A mutations also modulate the binding affinity of the full-length antibody to Fc receptors by decreasing the binding to low affinity Fc gamma receptors (FcγRIIa, FcγRIIIa and FcγRIIIb), while maintaining wild-type binding affinity to FcRn and FcγRI.
RESULTS AND DISCUSSION
Design
Enhanced aromatic sequons are sequences that include an aromatic residue prior to an Asn-Xxx-Ser/Thr sequence, or “sequon”, harbored in a reverse turn conformation that allows the glycan and the aromatic side chain to interact upon enzymatic N-glycosylation. The best studied EAS features an aromatic residue at the n-2 position relative to the N-glycosylated Asn that is integrated into a type I β-turn with a G1 β-bulge (a five-residue EAS; e.g., Phen-2-Alan-1-Asnn-Glyn+1-Thrn+2). Another EAS that is well studied is one where the aromatic residue is 3 residues N-terminal to a sequon in a type II β-turn within a larger six-residue loop (a six-residue EAS, e.g., Phen-3-Argn-2-Sern-1-Asnn-Glyn+1-Thrn+2). The EAS glycan-aromatic side chain interaction generally stabilizes proteins by ≈ 1–2 kcal/mol.(18–20, 22, 24) An extensive study of five- and six-residue EASs in the Pin WW domain, whose folding thermodynamics and kinetics have been extensively studied,(25–28) showed that the stabilizing carbohydrate-aromatic interactions are driven by the hydrophobic effect and CH-π interactions, powered largely by dispersion forces in this context.(20)
In the IgG1 Fc structures available in the Protein Data Bank (PDB), the C’E loop of the CH2 domains, which contains the N-glycosylated Asn297 residue, comprises one of two sequences, resulting in two different turn structures. Almost all the IgG1 Fc fragments have type I β-turns with a G1 β-bulge in the C’E loop(29–35); the exception being those IgG1 variants isolated from the serum of a patient with multiple myeloma (Cri, allotype G1m3)(36–39), which have a type IV β-turn (Table S1 and Figures S1 and S2). A type I β-turn with a G1 β-bulge, as noted above, is known to be amenable to conversion to an EAS. Thus, simply replacing Gln295 with a Phe residue should enable the aromatic-glycan interactions in the EAS. Although the Tyr296 side chain is usually in a conformation that projects away from the rest of the protein, and is therefore generally solvent exposed, Tyr296, together with the C’E loop, are flexible.(31) Moreover, this residue has been predicted to increase the aggregation propensity of antibodies.(8) Thus, to prevent Tyr296 from interacting with the Phe residue of the EAS or enhancing aggregation, Tyr296 was mutated to an Ala residue. This EAS sequence, with Phe at the n-2 position and Ala at the n-1 position relative to the N-glycosylated Asn residue in the context of a type I β-turn with a G1 β-bulge, has been shown to be effective in stabilizing the N-glycosylated rat ortholog of the immune protein CD2, muscle acylphosphatase, and the Pin WW domain. (18, 19, 24)
Introduction of the Enhanced Aromatic Sequon Stabilizes the CH2 domain
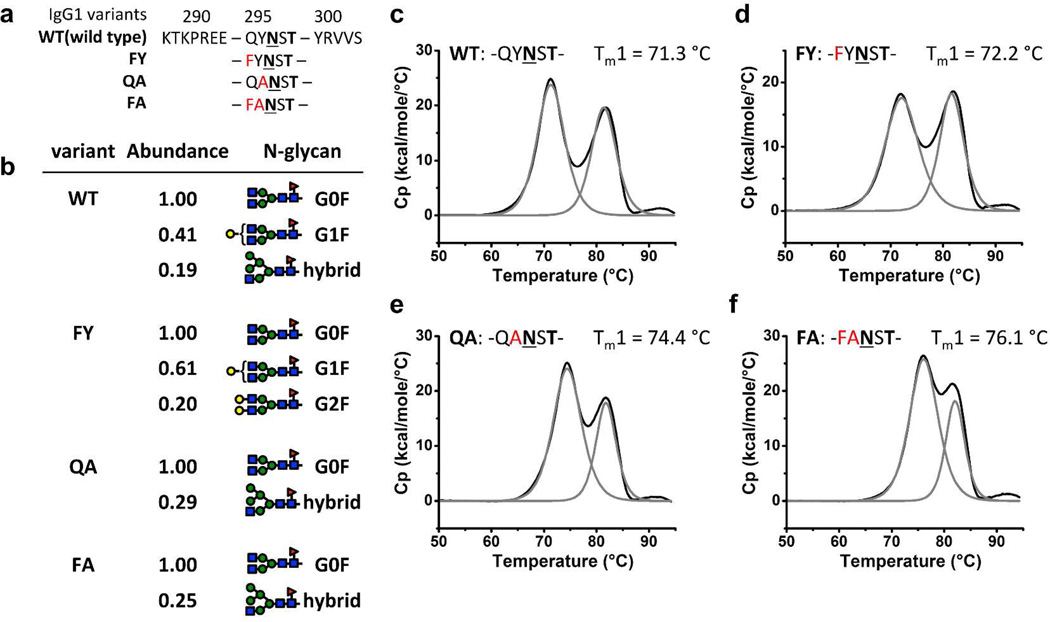
We overexpressed the above-described double mutant with Phe and Ala in the n-2 and n-1 positions, respectively, relative to the Asn that is N-glycosylated (hereafter called the “FA variant”), as well as the wild-type IgG1 Fc fragment in HEK293F cells. Single mutant variants, including variant FY (with Phe and Tyr in the n-2 and n-1 positions, respectively) and variant QA (with Gln and Ala in the n-2 and n-1 positions, respectively), were also biosynthesized so that we could parse the individual contributions of each mutation (Figure 2a). The wild-type (WT) and mutant IgG1 Fc fragments were purified from the harvested conditioned media by Protein A affinity chromatography followed by size-exclusion chromatography. The purity of the Fc fragment variants was very high, as discerned by SDS-PAGE (Figure S3).
Figure 2. Sequences, isoforms and thermal stabilities of Fc variants.
(a) Protein primary sequences of the IgG1 Fc variants around the C’E loop (Gln295-Thr299). Asn297 is shown in bold and underlined and Thr299 is shown in bold. Mutations are shown in red. (b) ESI-MS analysis of the purified Fc variant glycoforms. Relative abundance of glycoforms for each variant is normalized to the most abundant glycoform observed, and only those glycoforms that have relative abundance above 0.15 are shown. G# refers to the number of Gal on the two arms and F to fucosylation of the initial GlcNAc residue. (c–f) DSC analysis of the stability of Fc variants WT, FY, QA and FA. The baseline-adjusted DSC trace (black line) showed a two-peak feature for each variant which could be deconvoluted (grey lines) to the melting of CH2 (lower temperature, Tm1) and CH3 (higher temperature, Tm2) domains. The protein primary sequence of the C’E loop and Tm1 for each variant are shown above the DSC curves.
We confirmed that the mutant sequences of the purified Fc fragments were those expected and that they were N-glycosylated, using dithiothreitol treatment followed by an electrospray ionization (ESI) mass spectrometry (MS) analysis (Figures 2b, S4, and S5). The most abundant glycoform for all Fc variants was G0F (see Figures 2b and S5 for glycoform nomenclature and abundances), consistent with the glycan analysis for the recombinant WT IgG Fc by positive ion matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) MS of the enzymatically released glycans.(1, 40, 41) The second most abundant glycoform for the WT and FY variant is G1F. However, the second most abundant glycoform for variants QA and FA is a hybrid glycoform, suggesting less processing of the N-glycan of these two variants in the Golgi compartment (Figures 2b, S5). This observation of reduced processing of the N-glycans in the most stable CH2 domains (see below) could be explained by the lower accessibility of the N-glycan to glycan processing enzymes and has been observed before.(21, 42)
Differential scanning calorimetry (DSC) was used to examine the thermal stabilities of the purified Fc fragments. The thermal unfolding of the WT Fc fragment showed two endothermic peaks at 71.3 °C (Tm1) and 82.5 °C (Tm2), which correspond to the unfolding of the CH2 and CH3 domains, respectively (Figure 2c).(7–9) The thermal unfolding of the FA variant also showed two endothermic peaks, but Tm1 of the FA variant (76.1 °C) was substantially elevated compared to the WT, whereas Tm2 was unchanged (82.5 °C; cf. Figure 2f to Figure 2d–e). The Tm1 values of the FY and QA variants were both higher than that of WT Fc and lower than the FA variant, while the Tm2 of the FY and QA variants were unchanged compared to the WT Fc (Figures 2d–e). Comparing the Tm1 of the WT Fc and the QA variant shows that removing the aromatic side chain of Tyr296 by mutating it to an Ala residue increased Tm1 by ∼3 °C, possibly because the aromatic side chain of Tyr296 being solvent exposed is destabilizing. The next step of installing the EAS by introducing an aromatic Phe residue to replace Gln295 (comparing variants QA and FA) further increases the Tm1 by 1.7 °C. This increase likely originates from the Phe-glycan interactions in the EAS. Replacing Gln295 with Phe in the wild-type Fc (variant FY) slightly increases the Tm1 by 0.9 °C, 0.8 °C less than the effect observed for the same mutation on the QA variant (QA to FA), showing that there is some cooperativity between the stabilizing effects of the Q295F and Y296A mutations.
The WT and FA variants each display a slightly different distribution of glycoforms, with FA having less-processed N-glycans. However, it is unlikely that these differences in glycoforms result in the observed stabilization of the CH2 domain in variant FA (ΔTm1 = 4.8 °C) because prior studies have shown that an Fc with less-processed hybrid N-glycans was less stable than an Fc with complex N-glycans.(40) The stabilization of the C’E loop by EAS incorporation that we observed by DSC compares favorably with that achieved by past efforts at stabilizing CH2.(8, 9) Moreover, it is reasonable to assume that the Q295F/Y296A mutations in the variant FA could work cooperatively with other stabilizing mutations in IgG antibodies.(8, 9) It is also important to note that the C’E loop sequences and structures of the Fc region in IgG1, IgG2, IgG3 and IgG4 are very similar, suggesting that comparable stabilization could be achieved by engineering the FA variant of an EAS into other IgG subclasses (Figure S6).(43, 44)
Structure of the Enhanced Aromatic Sequon in the Fc Fragment CH2 Domain
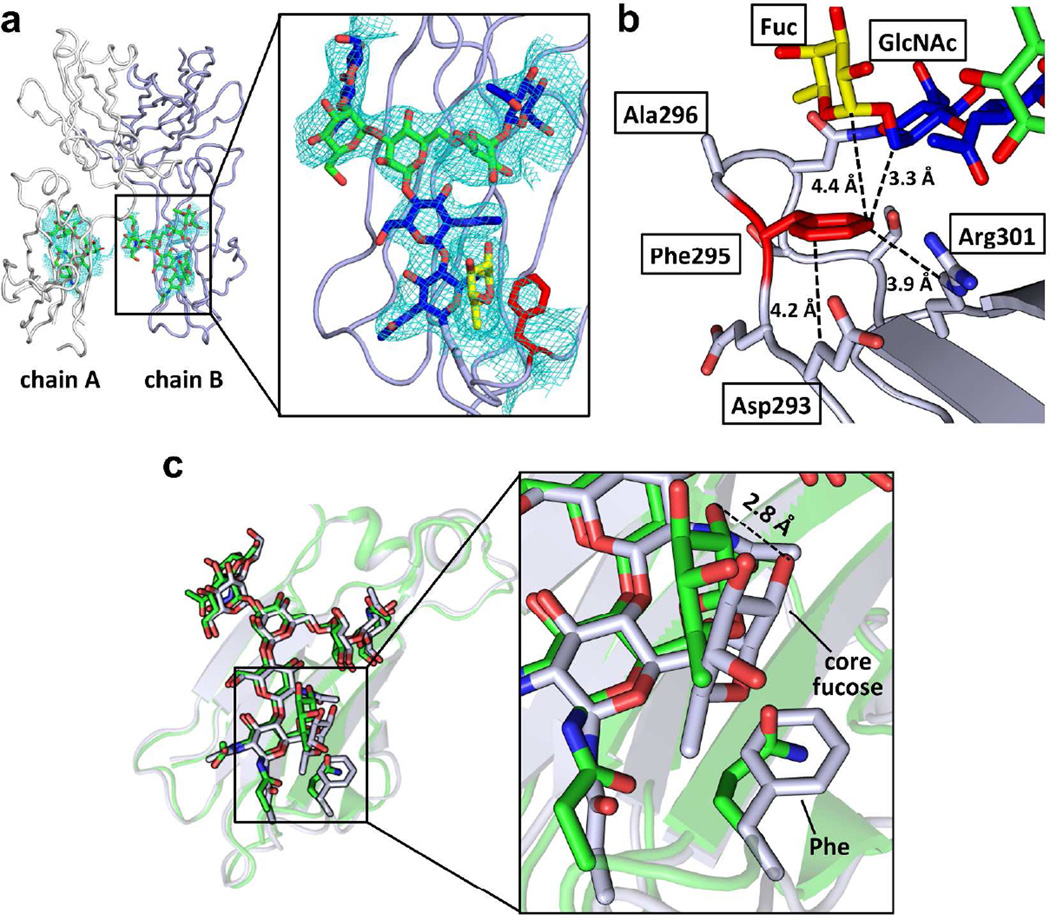
To further understand how the EAS stabilizes the CH2 domain, we solved the crystal structure of the FA variant of the Fc fragment (Figure 3, Table S2, PDBID:4QGT). The two identical polypeptide chains that form the Fc homodimer (chains A and B) are generally well-defined as judged from the electron density (Figures 3a,b). Chain B engages in more crystal contacts, and thereby has a slightly lower average B-value (71 Å2) than Chain A (77 Å2). Tighter crystal packing also explains why the Asn297 glycan on chain B (average B-value = 101 Å2) is substantially more ordered than the same glycan on chain A (average B-value = 238 Å2). Therefore, our analysis of the EAS focused on chain B.
Figure 3. Crystal structure of the Fc variant FA.
(a) Overview (left) of the crystal structure of Fc variant FA (PDBID:4QGT) and a close-up view of the N-glycan and the engineered region of chain B of the CH2 domain (right). The protein part of Fc is shown in ribbon format (chain A colored in white and chain B colored in light purple) and N-glycans are depicted in stick format. In the close-up view, the GlcNAc of the N-glycan is colored in blue, the mannose in green and the core fucose in yellow. The GlcNAc1 and the core fucose are interacting with the engineered Phe (red) in the Fc. The 2Fo-Fc map contoured at 1 sigma for the N-glycan and Phe is shown. (b) The engineered Phe residue packs with the aliphatic portion of the side chains of Asp293 and Arg301 and with GlcNAc1 and the core fucose of the N-glycan. Distances are shown in Å. (c) Alignment of the CH2 domains of the wild type (3AVE, green) and EAS-stabilized (grey) FA variant Fc chain B structures. The N-glycan and the side chain of Gln295 in the wild-type and the Phe295 in the EAS-stabilized Fc structures are shown in stick format. The N-glycans of the wild type Fc and the EAS-stabilized Fc align very well, with the largest difference being the orientation of the core fucose. In the EAS-stabilized Fc, this fucose is shifted closer to the engineered Phe295 residue by about 2 Å (see inset).
As expected from the relatively minor engineered modification made, the overall structure of the FA variant chain B (protein backbone) aligns very well with the WT Fc chain B structures (PDBID 3AVE; RMS=0.53 Å) (Figures 3c and S7, grey and green structures, respectively). The backbone conformations of the C’E loops with and without the EAS have very similar structures and backbone dihedral angles (Table S3). A slight shift in the position of Ser298 and the side chain of Asn297 is observed (Figures 3c and S7). The N-linked glycans also align well, but with a substantial difference in the core fucose moiety, which is shifted 2 Å towards the engineered Phe295 relative to the corresponding fucose on the wild-type Fc (Figure 3c). The engineered Phe side chain in the FA variant packs well with the protein backbone and the aliphatic portions of the Asp293 and Arg301 side chain (Figure 3b). Unlike the aromatic-glycan interactions observed previously in type I β-bulge turn EASs,(20) which consist mainly of face-to-face packing between the Phe side chain and the α-face of GlcNAc1 of the N-glycan, Phe295 in the FA variant of the Fc fragment interacts primarily with the C6-H of GlcNAc1 and C1-H of the invariant core fucose, probably engaging in hydrophobic packing and CH-π interactions (Figure 3b). These new aromatic-glycan interactions have not been observed before because the previously studied EASs in type I β-turns with a G1 β-bulge did not have fucosylated N-glycans.
To further probe the role of Phe-fucose interaction in the stabilization of the FA variant, we expressed and purified the WT and FA Fc fragments with core fucosylation pharmacologically inhibited (2-deoxy-2-fluoro-L-fucose at 200 µM) during expression. Using dithiothreitol treatment followed by an ESI-MS analysis, we found that the most abundant glycoform for both WT and FA variants was G0, showing that the core fucosylation was mostly inhibited (≈ 30% residual core fucosylation; Figure S8a). The stabilization in the FA variant compared to WT Fc is significantly less with core fucosylation inhibition (ΔTm1 = 3.2 °C; Figure S8b–c) than was observed without core fucosylation inhibition (ΔTm1 = 4.8 °C), suggesting that the Phe-fucose interactions contribute to the stabilization of the CH2 domain in the FA variant, as probed by DSC (also consistent with the reduction in glycoform elaboration in the EAS CH2 domain noted above).
A CH2 EAS Stabilizes a Full-Length Antibody against Thermal Aggregation
To be useful in the context of antibody engineering, the mutations that we introduced into the Fc fragment must be able to stabilize full-length antibodies against thermal denaturation and aggregation. Therefore, to examine whether the stabilization of the CH2 domain in the Fc fragment by the FA variant translates to full-length antibody stabilization, we expressed and purified the wild type, QA, and FA variants of the antibody 5J8, a human IgG1 monoclonal antibody that recognizes a conserved epitope on the hemagglutinin receptor binding site of the H1N1 influenza virus (Figure S9).(45, 46)
DSC was used to evaluate whether the CH2 domain is stabilized in the QA and FA variants of antibody 5J8. The DSC trace of wild-type 5J8 antibody could be deconvoluted into the melting of CH2, Fab, and CH3 domains, with CH2 being the first to unfold (Figures 4a and S10). The unfolding of the CH2 domain results in the apparent shoulder visible at about 71 °C in the DSC trace of 5J8. This shoulder is missing in the DSC trace of both the QA and FA variants, consistent with the stabilization of the CH2 domain against thermal denaturation in these two variants (Figure 4a).
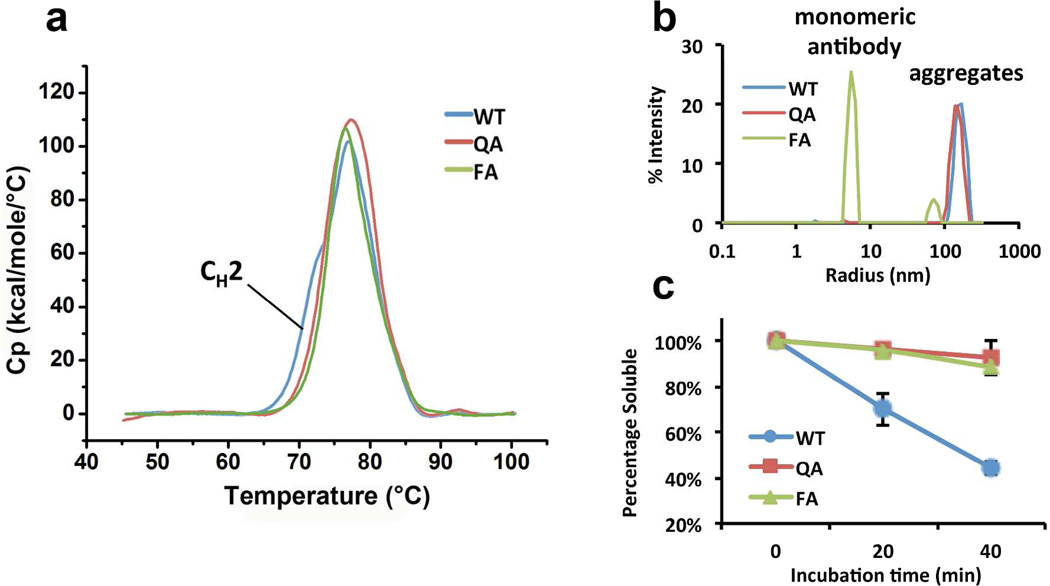
Figure 4. Stabilities of 5J8 variants.
(a) Baseline-adjusted DSC curves of 5J8 antibody variants WT (blue), QA (red) and FA (green). The apparent shoulder on the WT curve, reflecting the melting of the CH2 domain (Figure S9), is not present in the curves of QA and FA variants, indicating the stabilization of the CH2 domain in these two variants. (b) DLS signal of 5J8 antibody variants WT (blue), QA (red) and FA (green) upon dilution in pH 3.5 buffer. WT and QA exhibit high signal at the region of Rh = 100–300 nm for the aggregates while the signal for the monomeric FA antibody (Rh = 7 nm) is almost completed suppressed. The FA variant exhibits much less signal for the aggregates while maintaining a strong signal for the monomeric antibody. (c) Soluble protein percentage of 5J8 antibody variants WT (blue), QA (red) and FA (green) after 0, 20 and 40, min of thermal stress at 70 °C.
Next, we analyzed the global stability of these 5J8 antibody variants by measuring the extent to which they aggregate at low pH or at high temperature. It has been shown that antibodies aggregate with different mechanisms under different kinds of stress, with low pH stress producing relatively small aggregates, whereas heat stress produces large insoluble aggregates.(10, 47, 48) To stress 5J8 using low pH buffer, we diluted 5J8 antibodies from a concentration of 10 mg/mL in PBS buffer into 10 mM sodium acetate buffer (pH 3.5) at a final concentration of 0.5 mg/mL and then monitored their aggregation using dynamic light scattering (DLS), a highly sensitive method frequently used to study antibody aggregation.(49, 50) DLS analysis reveals that the WT and QA variants of 5J8 formed aggregates with hydrodynamic radii (Rh) of 0.1–0.3 µm immediately upon dilution into pH 3.5 buffer (Figure 4b). In contrast, the EAS-stabilized FA variant of 5J8 was resistant to aggregation (Figure 4b), with the predominant peak corresponding to monomeric antibody. Thus, the combined Gln295Phe and Tyr296Ala mutations protected 5J8 from acid-induced aggregation almost completely. The intensity-weighted particle size distribution calculated from the DLS signal shown in Figure 4b tends to over-represent the population of large aggregates, because large aggregates scatter light to a much greater extent than smaller particles. Thus, to quantify the extent of acid-mediated aggregation of the 5J8 variants, we measured the protein concentration in solution before and after centrifugation at 20,000 × g for 30 min. Centrifugation was sufficient to completely sediment the aggregates, as demonstrated by examining the supernatant by DLS (Figure S11). This approach revealed that 6% ± 3% of the total antibody aggregated at low pH for WT 5J8 vs. 2% ± 3% for the QA variant of 5J8 (Figure S12). Prolonged incubation of the aggregate-free supernatant of 5J8 antibodies in pH 3.5 buffer at 37 °C (up to 3 days) did not cause further aggregation. Also, we note that the mechanical process of diluting these samples without changing the pH, by using PBS buffer, does not result in the generation of new aggregates for any of the three variants (cf. Figures S13 and 4b).
To determine whether the FA variant could protect 5J8 against aggregation due to heat stress, we incubated the WT, QA and FA variants of 5J8 at 70 °C in PBS buffer for 20 and 40 min. Since the aggregates formed were too large to characterize by DLS, we pelleted the insoluble aggregates and measured the concentration of soluble antibody in the supernatant. After 20 min, only 70% of the wild-type 5J8 antibody was in the soluble form. In contrast, 96% of the mutant QA and FA 5J8 antibody variants remained soluble (Figure 4c). After 40 min of heat stress at 70 °C, only 44% of the wild-type 5J8 antibody remained in the supernatant, whereas 92% and 88% of the QA and FA variants, respectively, remained soluble (Figure 4c). Taken together, these data show that introducing the Gln295Phe and Tyr296Ala mutations into a full-length IgG1 antibody globally stabilizes the antibody against aggregation, in part by EAS-associated stabilization of the Fc.
The EAS Modulates Antibody Interactions with Fc Receptors
The Fc region (Figure 1a) is responsible for the long half-lives of antibodies and therapeutic protein–Fc fusions, via its interaction with FcRn.(51–53) The Fc region also mediates effector functions, such as antibody-dependent cell-mediated cytotoxicity (ADCC) and antibody-dependent cell-mediated phagocytosis (ADCP) via its interactions with FcγRs.(54) Mutations in the C’E loop (Q295A and Y296F) have previously been shown to mildly decrease the binding to FcγRs.(55) To determine how the Q295F/Y296A mutations affect Fc receptor binding by the 5J8 antibody, we measured the energetics of the binding between the WT, QA and FA variants of 5J8 and several important Fc receptors using bio-layer interferometry.
The Y296A mutation in the QA variant of 5J8 moderately perturbs binding to the Fc receptors that were examined: it decreases the affinities of 5J8 for FcγRI, FcγRIIIa and FcγRIIIb by 5- to 6-fold each, and for FcRn and FcγRIIa by less than 2-fold (Table 1). Similarly, the Q295F/Y296A double mutation in the FA variant of 5J8 has little effect on binding of 5J8 to FcRn or FcγRI, but decreases the affinity for FcγRIIIa by 9-fold. However, binding of the FA variant of 5J8 to FcγRIIa and FcγRIIIb is substantially impaired; in fact, the on- and off-rates could not be accurately measured for these variants (Table 1).
Table 1.
Summary of binding parameters kon (1/M s), koff (1/s) and Kd (M). The Kd reported is the ratio koff / kon.
| Fc receptor | variant | kon | koff | Kd |
|---|---|---|---|---|
| FcRn | WT | 4.4 × 104 | 2.2 × 10−3 | 5.1 × 10−8 |
| QA | 3.8 × 104 | 3.5 × 10−3 | 9.3 × 10−8 | |
| FA | 4.6 × 104 | 2.2 × 10−3 | 4.7 × 10−8 | |
| FcγRI | WT | 5.0 × 105 | 2.2 × 10−4 | 4.4 × 10−10 |
| QA | 3.1 × 105 | 7.3 × 10−4 | 2.0 × 10−9 | |
| FA | 4.9 × 105 | 3.3 × 10−4 | 6.9 × 10−10 | |
| FcγRIIa | WT | 1.1 × 106 | 2.9 × 10−1 | 2.7 × 10−7 |
| QA | 7.7 × 105 | 3.0 × 10−1 | 3.9 × 10−7 | |
| FA | - | - | - | |
| FcγRIIIa | WT | 4.6 × 105 | 5.0 × 10−2 | 1.1 × 10−7 |
| QA | 2.8 × 105 | 2.0 × 10−1 | 6.1 × 10−7 | |
| FA | 4.1 × 105 | 4.3 × 10−1 | 1.0 × 10−6 | |
| FcγRIIIb | WT | 4.3 × 105 | 7.1 × 10−1 | 1.6 × 10−6 |
| QA | 6.3 × 104 | 5.2 × 10−1 | 8.3 × 10−6 | |
| FA | - | - | - | |
The most likely reason why the FA variant antibody maintains its ability to bind to FcRn and FcγRI is simply because the binding sites used for interacting with these two Fc receptors should be minimally perturbed by the two mutations in the C’E loop.(34) FcRn binds Fc at the CH2/CH3 interface and the high affinity receptor FcγRI binds the Fc region at the FG loop.
In contrast, Y296 interacts extensively with residues in FcγRIIIa, as found in the crystal structure of the Fc fragment in complex with FcγRIIIa,(32) thus accounting for the somewhat diminished binding of the FA variant antibody to FcγRIIIa. The origin of the diminished binding of the Q295F/Y296A mutations of 5J8 to FcγRIIa and FcγRIIIb is less clear. While the mutated residues are not directly involved in the binding of the Fc fragment to these Fc receptors, it is possible that the C’E loop needs to maintain a certain level of flexibility to adopt the optimal conformations for the polypeptide chain and the N-glycan upon binding to some low-affinity FcγRs. It is thus possible that the C’E loop prefers a conformation that is not optimal for FcγR binding in the FA variant antibody. For example, upon the binding of Fc to FcγRIIIb, the C’E loop adopts a conformation in which Y296 interacts with the core fucose;(38) such an interaction would be in competition with the EAS aromatic-glycan interactions in the FA variant. Further investigation is needed to better understand the mechanism of tuning low-affinity FcγR binding by manipulating the protein primary sequence in or close to the C’E loop.
Although the FA variant of 5J8 loses its ability to bind to some FcγRs, we note that the ability to modulate the Fc receptor-mediated effector functions of antibodies is often desirable in circumstances in which neither FcγR binding nor immune activation is desired; examples include immunomodulatory antibodies(11, 56) and conjugate-based therapy.(57) Two common ways to abolish the binding to FcγRs are to use aglycosylated Fc (for example, MPDL3280A, Genentech/Roche, phase III) or IgG subclasses other than IgG1, such as IgG2 (e.g., tremelimumab, Pfizer, phase III) and IgG4 (e.g., Nivolumab, Bristol-Myers Squibb; and Pembrolizumab, Merck; both approved).(56–58) Unfortunately, the CH2 domain of non-glycosylated Fcs is greatly destabilized compared to glycosylated Fcs, and engineering antibodies from IgG1 to other subclasses may encounter problems such as increased aggregation (IgG2 and IgG4) and light chain exchange when using IgG4.(59, 60) Engineered antibodies containing the FA variant Fc fragment could be a viable alternative, given the stabilizing influence imparted by integrating an EAS, its low aggregation propensity, and its altered Fc receptor affinity. Interestingly the FcγR-binding profile of the FA variant antibody resembles that of IgG4, which binds strongly with FcRn and FcγRI, but poorly with other FcγRs.(61) Thus the FA variant antibody might diminish or abolish some Fc-mediated effector functions while enhancing the stability of the CH2 domain and protecting the antibody (or Fc-fusion protein) from aggregation.
CONCLUSIONS
In summary, we utilized an EAS to engineer the N-glycosylated C’E loop in the Fc fragment of human IgG1 and thereby successfully stabilized the CH2 domain. The stabilization effect has two sources: removing the side chain of the solvent exposed Tyr296 and introducing new aromatic-glycan interactions via the EAS. The engineered C’E loop adopts a type I β-turn with a G1 β-bulge conformation, which enables protein-glycan interactions between the Phe side chain on the protein and the N-glycan. In this protein, GlcNAc1 and the core fucose on the N-glycan interact with the aromatic side chain of Phe, which is different from the face-to-face packing observed in previous EASs, showing the portability and adaptability of the EAS to different glycoforms. These interactions not only stabilized the CH2 domain within the Fc fragment, but also improved the global stability of an intact antibody against thermal and acid-mediated aggregation. Finally, the stabilized FA variant antibody was able to bind FcRn and FcγRI with wild-type affinity, but exhibited reduced affinity to other FcγRs. Our results show that simply manipulating the sequence space close to the N-glycosylated Asn in the Fc is useful both for creating stabilized antibodies and modulating the binding of these antibodies to the receptors that mediate their effector functions.
METHODS
Protein Expression and Purification
The Fc region of human IgG1 (residues 98–330) was cloned into the phCMV3 vector and the human CD5 antigen leader sequence(62, 63) was added to the N-terminus of the fusion protein. All mutations were introduced by QuikChange site-directed mutagenesis per the manufacturer’s instructions (Agilent Technologies). The plasmids for IgG1 Fc or for full-length IgG 5J8(45, 46) variants were transiently transfected into HEK293F cells (Invitrogen) using 293-fectin (Invitrogen) according to the manufacturer’s protocol. In the experiments in which core fucosylation was inhibited, 2-deoxy-2-fluoro-L-fucose (CARBOsynth, cat. no. MD06089) was added to the medium at a final concentration of 200 µM. Six days post-transfection, the conditioned media containing the recombinant IgG1 Fc and 5J8 antibodies were harvested and centrifuged at 6000 × g for 30 min to pellet the cells, before filtering with a 0.22 µm filter to remove residual cells. The supernatants were run over a 5 mL HiTrap Protein A column (GE Healthcare), and proteins were eluted with 0.1 M Glycine pH 2.7, and equilibrated to pH 6.6–7.0 with 1 M Tris-HCl pH 9.0, and further purified by size-exclusion chromatography (Superdex 200, GE Healthcare) in phosphate buffered saline (PBS; pH 7.4). Purified proteins were analyzed for purity by SDS-PAGE employing Coomassie staining for visualization.
Differential Scanning Calorimetry
The thermal stabilities of the wild-type (WT) and variant Fc fragments as well as the 5J8 antibody variants were assessed by differential scanning calorimetry (DSC) using a Microcal VP-DSC or a VP-capillary DSC system (Malvern Instruments). Purified Fc fragment and 5J8 antibody variants were analyzed at a concentration of 1 mg mL−1 in PBS at scan rates of 1.0 °C (Fc fragments) or 1.5 °C (5J8 antibodies) per min in triplicate. The data were analyzed using DSC Data Analysis in Origin®, by subtraction of the reference data, normalization to the protein concentration and DSC cell volume, and interpolation of a progression baseline. The peaks were deconvoluted using a non-2-state model to fit the data.(64, 65)
ESI-MS characterization
Electrospray ionization mass spectrometry (ESI-MS) analysis was performed using an Agilent 1100 LC coupled to an Agilent 1100 single quad ESI mass spectrometer. Liquid chromatography was performed with a Zorbax 300SB-C8 column (Agilent).
Crystallization and Data Collection
Purified human Fc fragment containing the EAS (FA variant) was purified by size-exclusion chromatography in 50 mM NaCl, 20 mM Tris-HCl pH 7.2 and concentrated to 8–12 mg mL−1 before being screened manually for crystallization. The Fc fragment protein formed crystals at 20 °C in a crystallization buffer consisting of 30% (w/v) PEG 1000, and 100 mM HEPES, pH 7.5. The crystals were cryoprotected by brief immersion in mother liquor supplemented with 10% ethylene glycol prior to flash-cooling in liquid nitrogen. Data were collected at beamline 11-1 at the Stanford Synchrotron Radiation Lightsource (SSRL), and processed with HKL-2000.(66) The crystal diffracted to 3.0 Å, and the diffraction data were indexed in space group C2. Data processing statistics are summarized in Table S2.
Structure Determination and Refinement
The structure of the human Fc fragment containing the EAS was determined by molecular replacement using Phaser(67) using a previously published structure of WT human Fc (PDBID: 3AVE) as the search model. Model building was carried out using Coot-0.7(68) and refinement was implemented with the Phenix program.(69) The final Rcryst and Rfree values are 29.8% and 32.6%, respectively. These high R values reflect the high B values and the partial disorder in some of the Fc structure. See Table S1 for final refinement statistics. Glycan structure and geometry were monitored throughout the refinement with the PDB CArbohydrate REsidue check (PDBCARE) online tool (www.glycosciences.de) as described previously.(70) All structures in the figures were rendered by using Pymol. The Protein Data Bank reference for the structure of the FA variant of the Fc fragment is 4QGT.
Thermal Stress-induced Aggregation of 5J8 Antibodies
Solutions of 5J8 antibodies were prepared at a concentration of 0.5 mg/mL in PBS in a 1.5 mL microcentrifuge tube (Axygen) and centrifuged at 20,000 × g for 30 min to remove any soluble aggregates. Monomeric 5J8 antibody solutions were incubated at 70 °C for 20 min or 40 min. The heated samples were briefly cooled on ice before centrifuging at 20000 × g for 30 min to remove aggregates and collecting the supernatant. Soluble protein concentration was determined by comparing the UV absorbance at 280 nm before and after heat treatment.
Low pH Stress-induced Aggregation of 5J8 Antibodies and Dynamic Light Scattering Analysis
5J8 antibodies were concentrated to 10 mg mL−1 in PBS. Protein samples were prepared at 0.5 mg mL−1 by diluting the 10 mg mL−1 stock solution into PBS or 10 mM sodium acetate buffer, pH 3.5, for determination of effective hydrodynamic radius (Rh). Dynamic light scattering (DLS) measurements were performed using a DynaPro NanoStar instrument (Wyatt Technology Corporation) at 25 °C. Data were analyzed by using the DYNAMICS software package (Wyatt Technology Corporation) following the manufacturer’s instructions.
Kd determination with Fc receptors
Dissociation constant (Kd) values were determined by biolayer interferometry using an Octet RED instrument (ForteBio, Inc.) as previously described.(45, 71) Briefly, 5J8 antibody variants at 50 µg mL−1 in kinetics buffer (PBS or 50 mM sodium phosphate, 100 mM NaCl buffer, pH 6.0, 0.01% bovine serum albumin, and 0.002% Tween 20) were immobilized onto anti-human Fab-CH1 biosensors and incubated with three concentrations of Fc receptors. Recombinant Fc gamma receptors were from R&D Systems (Cat. No. 1257FC050, 1330CD050CF, 4325FC050, 1597FC050CF). Recombinant FcRn was from Novoprotein (Cat. No. 50828338). All binding data were collected at 25°C. The experiments comprised 5 steps: 1. Baseline acquisition (60 s); 2. Antibody loading onto biosensor (180 s); 3. Second baseline acquisition (120 s); 4. Association of Fc receptors for the measurement of kon (60–180 s); 5. Dissociation of Fc receptors for the measurement of koff (60–180s). The kon and koff values of each Fc receptor were measured in real time to determine the Kd values for each 5J8 antibody variant tested.
Supplementary Material
Acknowledgments
This work was supported by the Skaggs Institute for Chemical Biology, the Lita Annenberg Hazen Foundation, and by National Institutes of Health grants GM051105 (J.W.K.), and AI084817 (I.A.W.). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209).
Footnotes
ASSOCIATED CONTENT
Supporting information. Supplementary Figures and Tables are supplied as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
Competing financial interests: Authors E.T.P. and J.W.K. are listed as inventors on a provisional patent (US20130034547 A1) that is currently unlicensed.
REFERENCES
- 1.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8:226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 2.Wu H, Kroe-Barrett R, Singh S, Robinson AS, Roberts CJ. Competing aggregation pathways for monoclonal antibodies. FEBS Lett. 2014;588:936–941. doi: 10.1016/j.febslet.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 3.Schellekens H. Bioequivalence and the immunogenicity of biopharmaceuticals. Nat Rev Drug Discov. 2002;1:457–462. doi: 10.1038/nrd818. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan A, Clementel V, Woods R, Harn N, Bowen MA, Mo W, Popovic B, Bishop SM, Dall’Acqua W, Minter R, Jermutus L, Bedian V. Engineering a therapeutic IgG molecule to address cysteinylation, aggregation and enhance thermal stability and expression. MAbs. 2013;5:255–262. doi: 10.4161/mabs.23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kontermann RE. Strategies for extended serum half-life of protein therapeutics. Curr Opin Biotechnol. 2011;22:868–876. doi: 10.1016/j.copbio.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Fast JL, Cordes AA, Carpenter JF, Randolph TW. Physical instability of a therapeutic Fc fusion protein: domain contributions to conformational and colloidal stability. Biochemistry. 2009;48:11724–11736. doi: 10.1021/bi900853v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voynov V, Chennamsetty N, Kayser V, Helk B, Forrer K, Zhang H, Fritsch C, Heine H, Trout BL. Dynamic fluctuations of protein-carbohydrate interactions promote protein aggregation. PLoS One. 2009;4:e8425. doi: 10.1371/journal.pone.0008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chennamsetty N, Helk B, Voynov V, Kayser V, Trout BL. Aggregation-prone motifs in human immunoglobulin G. J Mol Biol. 2009;391:404–413. doi: 10.1016/j.jmb.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 9.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci U S A. 2009;106:11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brummitt RK, Nesta DP, Chang L, Kroetsch AM, Roberts CJ. Nonnative aggregation of an IgG1 antibody in acidic conditions, part 2: nucleation and growth kinetics with competing growth mechanisms. J Pharm Sci. 2011;100:2104–2119. doi: 10.1002/jps.22447. [DOI] [PubMed] [Google Scholar]
- 11.Liu T, Zhang Y, Liu Y, Wang Y, Jia H, Kang M, Luo X, Caballero D, Gonzalez J, Sherwood L, Nunez V, Wang D, Woods A, Schultz PG, Wang F. Functional human antibody CDR fusions as long-acting therapeutic endocrine agonists. Proc Natl Acad Sci U S A. 2015;112:1356–1361. doi: 10.1073/pnas.1423668112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson QM, Barb AW. A perspective on the structure and receptor binding properties of immunoglobulin G Fc. Biochemistry. 2015;54:2931–2942. doi: 10.1021/acs.biochem.5b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. MAbs. 2011;3:568–576. doi: 10.4161/mabs.3.6.17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lux A, Yu X, Scanlan CN, Nimmerjahn F. Impact of immune complex size and glycosylation on IgG binding to human FcgammaRs. J Immunol. 2013;190:4315–4323. doi: 10.4049/jimmunol.1200501. [DOI] [PubMed] [Google Scholar]
- 15.Barb AW, Prestegard JH. NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat Chem Biol. 2011;7:147–153. doi: 10.1038/nchembio.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subedi GP, Barb AW. The Structural Role of Antibody N-Glycosylation in Receptor Interactions. Structure. 2015;23:1573–1583. doi: 10.1016/j.str.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subedi GP, Hanson QM, Barb AW. Restricted motion of the conserved immunoglobulin G1 N-glycan is essential for efficient FcgammaRIIIa binding. Structure. 2014;22:1478–1488. doi: 10.1016/j.str.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Culyba EK, Price JL, Hanson SR, Dhar A, Wong CH, Gruebele M, Powers ET, Kelly JW. Protein native-state stabilization by placing aromatic side chains in N-glycosylated reverse turns. Science. 2011;331:571–575. doi: 10.1126/science.1198461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price JL, Powers DL, Powers ET, Kelly JW. Glycosylation of the enhanced aromatic sequon is similarly stabilizing in three distinct reverse turn contexts. Proc. Natl. Acad. Sci. USA. 2011;108:14127–14132. doi: 10.1073/pnas.1105880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Enck S, Price JL, Powers DL, Powers ET, Wong CH, Dyson HJ, Kelly JW. Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins. J Am Chem Soc. 2013;135:9877–9884. doi: 10.1021/ja4040472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray AN, Chen W, Antonopoulos A, Hanson SR, Wiseman RL, Dell A, Haslam SM, Powers DL, Powers ET, Kelly JW. Enhanced Aromatic Sequons Increase Oligosaccharyltransferase Glycosylation Efficiency and Glycan Homogeneity. Chem Biol. 2015;22:1052–1062. doi: 10.1016/j.chembiol.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price JL, Culyba EK, Chen W, Murray AN, Hanson SR, Wong CH, Powers ET, Kelly JW. N-glycosylation of enhanced aromatic sequons to increase glycoprotein stability. Biopolymers. 2012;98:195–211. doi: 10.1002/bip.22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price JL, Shental-Bechor D, Dhar A, Turner MJ, Powers ET, Gruebele M, Levy Y, Kelly JW. Context-dependent effects of asparagine glycosylation on Pin WW folding kinetics and thermodynamics. J Am Chem Soc. 2010;132:15359–15367. doi: 10.1021/ja106896t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson SR, Culyba EK, Hsu TL, Wong CH, Kelly JW, Powers ET. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Natl Acad Sci U S A. 2009;106:3131–3136. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jager M, Nguyen H, Crane JC, Kelly JW, Gruebele M. The folding mechanism of a beta-sheet: the WW domain. J Mol Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 26.Jager M, Zhang Y, Bieschke J, Nguyen H, Dendle M, Bowman ME, Noel JP, Gruebele M, Kelly JW. Structure-function-folding relationship in a WW domain. Proc Natl Acad Sci U S A. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koepf EK, Petrassi HM, Sudol M, Kelly JW. WW: An isolated three-stranded antiparallel beta-sheet domain that unfolds and refolds reversibly; evidence for a structured hydrophobic cluster in urea and GdnHCl and a disordered thermal unfolded state. Protein Sci. 1999;8:841–853. doi: 10.1110/ps.8.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araya CL, Fowler DM, Chen W, Muniez I, Kelly JW, Fields S. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci U S A. 2012;109:16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva-Martin N, Bartual SG, Ramirez-Aportela E, Chacon P, Park CG, Hermoso JA. Structural basis for selective recognition of endogenous and microbial polysaccharides by macrophage receptor SIGN-R1. Structure. 2014;22:1595–1606. doi: 10.1016/j.str.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 31.Matsumiya S, Yamaguchi Y, Saito J, Nagano M, Sasakawa H, Otaki S, Satoh M, Shitara K, Kato K. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J Mol Biol. 2007;368:767–779. doi: 10.1016/j.jmb.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 32.Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, Umana P, Benz J. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between Fc gamma RIII and antibodies lacking core fucose. Proc. Natl Acad Sci USA. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank M, Walker RC, Lanzilotta WN, Prestegard JH, Barb AW. Immunoglobulin G1 Fc domain motions: implications for Fc engineering. J Mol Biol. 2014;426:1799–1811. doi: 10.1016/j.jmb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu J, Chu J, Zou Z, Hamacher NB, Rixon MW, Sun PD. Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding. Proc Natl Acad Sci U S A. 2015;112:833–838. doi: 10.1073/pnas.1418812112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crispin M, Bowden TA, Coles CH, Harlos K, Aricescu AR, Harvey DJ, Stuart DI, Jones EY. Carbohydrate and domain architecture of an immature antibody glycoform exhibiting enhanced effector functions. J Mol Biol. 2009;387:1061–1066. doi: 10.1016/j.jmb.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 36.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 37.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J. Mol. Biol. 2003;325:979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 38.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 39.Ramsland PA, Farrugia W, Bradford TM, Sardjono CT, Esparon S, Trist HM, Powell MS, Tan PS, Cendron AC, Wines BD, Scott AM, Hogarth PM. Structural basis for Fc gammaRIIa recognition of human IgG and formation of inflammatory signaling complexes. J Immunol. 2011;187:3208–3217. doi: 10.4049/jimmunol.1101467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowden TA, Baruah K, Coles CH, Harvey DJ, Yu X, Song BD, Stuart DI, Aricescu AR, Scanlan CN, Jones EY, Crispin M. Chemical and structural analysis of an antibody folding intermediate trapped during glycan biosynthesis. J Am Chem Soc. 2012;134:17554–17563. doi: 10.1021/ja306068g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mimura Y, Ashton PR, Takahashi N, Harvey DJ, Jefferis R. Contrasting glycosylation profiles between Fab and Fc of a human IgG protein studied by electrospray ionization mass spectrometry. J Immunol Methods. 2007;326:116–126. doi: 10.1016/j.jim.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 42.Hang I, Lin CW, Grant OC, Fleurkens S, Villiger TK, Soos M, Morbidelli M, Woods RJ, Gauss R, Aebi M. Analysis of site-specific N-glycan remodeling in the endoplasmic reticulum and the Golgi. Glycobiology. 2015;25:1335–1349. doi: 10.1093/glycob/cwv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies AM, Rispens T, Ooijevaar-de Heer P, Gould HJ, Jefferis R, Aalberse RC, Sutton BJ. Structural determinants of unique properties of human IgG4-Fc. J Mol Biol. 2014;426:630–644. doi: 10.1016/j.jmb.2013.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teplyakov A, Zhao Y, Malia TJ, Obmolova G, Gilliland GL. IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface. Mol Immunol. 2013;56:131–139. doi: 10.1016/j.molimm.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 45.Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE, Jr, Ward AB, Wilson IA. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. 2013;87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE., Jr A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joubert MK, Luo Q, Nashed-Samuel Y, Wypych J, Narhi LO. Classification and characterization of therapeutic antibody aggregates. J Biol Chem. 2011;286:25118–25133. doi: 10.1074/jbc.M110.160457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vazquez-Rey M, Lang DA. Aggregates in monoclonal antibody manufacturing processes. Biotechnol Bioeng. 2011;108:1494–1508. doi: 10.1002/bit.23155. [DOI] [PubMed] [Google Scholar]
- 49.Ahrer K, Buchacher A, Iberer G, Jungbauer A. Thermodynamic stability and formation of aggregates of human immunoglobulin G characterised by differential scanning calorimetry and dynamic light scattering. J Biochem Biophys Methods. 2006;66:73–86. doi: 10.1016/j.jbbm.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Nobbmann U, Connah M, Fish B, Varley P, Gee C, Mulot S, Chen J, Zhou L, Lu Y, Sheng F, Yi J, Harding SE. Dynamic light scattering as a relative tool for assessing the molecular integrity and stability of monoclonal antibodies. Biotechnol. Genet. Eng. Rev. 2007;24:117–128. doi: 10.1080/02648725.2007.10648095. [DOI] [PubMed] [Google Scholar]
- 51.Ghetie V, Hubbard JG, Kim JK, Tsen MF, Lee Y, Ward ES. Abnormally short serum half-lives of IgG in beta 2-microglobulin-deficient mice. Eur J Immunol. 1996;26:690–696. doi: 10.1002/eji.1830260327. [DOI] [PubMed] [Google Scholar]
- 52.Ghetie V, Ward ES. Multiple roles for the major histocompatibility complex class I- related receptor FcRn. Annu Rev Immunol. 2000;18:739–766. doi: 10.1146/annurev.immunol.18.1.739. [DOI] [PubMed] [Google Scholar]
- 53.Roopenian DC, Christianson GJ, Sproule TJ, Brown AC, Akilesh S, Jung N, Petkova S, Avanessian L, Choi EY, Shaffer DJ, Eden PA, Anderson CL. The MHC class I-like IgG receptor controls perinatal IgG transport, IgG homeostasis, and fate of IgG-Fc-coupled drugs. J Immunol. 2003;170:3528–3533. doi: 10.4049/jimmunol.170.7.3528. [DOI] [PubMed] [Google Scholar]
- 54.Borrok MJ, Jung ST, Kang TH, Monzingo AF, Georgiou G. Revisiting the role of glycosylation in the structure of human IgG Fc. ACS Chem Biol. 2012;7:1596–1602. doi: 10.1021/cb300130k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, Fox JA, Presta LG. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem. 2001;276:6591–6604. doi: 10.1074/jbc.M009483200. [DOI] [PubMed] [Google Scholar]
- 56.Stewart R, Hammond SA, Oberst M, Wilkinson RW. The role of Fc gamma receptors in the activity of immunomodulatory antibodies for cancer. J Immunother. Cancer. 2014;2:29. [Google Scholar]
- 57.Kaneko E, Niwa R. Optimizing therapeutic antibody function: progress with Fc domain engineering. BioDrugs. 2011;25:1–11. doi: 10.2165/11537830-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 58.Jung ST, Kelton W, Kang TH, Ng DT, Andersen JT, Sandlie I, Sarkar CA, Georgiou G. Effective phagocytosis of low Her2 tumor cell lines with engineered, aglycosylated IgG displaying high FcgammaRIIa affinity and selectivity. ACS Chem Biol. 2013;8:368–375. doi: 10.1021/cb300455f. [DOI] [PubMed] [Google Scholar]
- 59.Ito T, Tsumoto K. Effects of subclass change on the structural stability of chimeric, humanized, and human antibodies under thermal stress. Protein Sci. 2013;22:1542–1551. doi: 10.1002/pro.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salfeld JG. Isotype selection in antibody engineering. Nat Biotechnol. 2007;25:1369–1372. doi: 10.1038/nbt1207-1369. [DOI] [PubMed] [Google Scholar]
- 61.Jefferis R. Isotype and glycoform selection for antibody therapeutics. Arch Biochem Biophys. 2012;526:159–166. doi: 10.1016/j.abb.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 62.Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 63.Yang Q, Li C, Wei Y, Huang W, Wang LX. Expression, glycoform characterization, and antibody-binding of HIV-1 V3 glycopeptide domain fused with human IgG1-Fc. Bioconjug Chem. 2010;21:875–883. doi: 10.1021/bc9004238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukada H, Takahashi K, Sturtevant JM. Differential scanning calorimetric study of the thermal unfolding of Taka-amylase A from Aspergillus oryzae. Biochemistry. 1987;26:4063–4068. doi: 10.1021/bi00387a048. [DOI] [PubMed] [Google Scholar]
- 65.Sturtevant JM. Biochemical Applications of Differential Scanning Calorimetry. Ann. Rev. Phys. Chem. 1987;38:463–488. [Google Scholar]
- 66.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 67.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.