Abstract
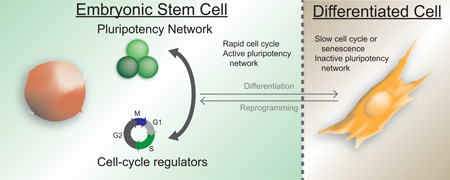
Pluripotent stem cells, defined by an unlimited self-renewal capacity and an undifferentiated state, are best typified by embryonic stem cells. These cells have a unique cell cycle compared to somatic cells as defined by a rapid progression through the cell cycle and a minimal time spent in G1. Recent reports indicate that pluripotency and cell cycle regulation are mechanistically linked. In this review, we discuss the reciprocal co-regulation of these processes, how this co-regulation may prevent differentiation, and how cellular reprogramming can re-establish the unique cell cycle regulation in induced pluripotent stem cells.
Graphical abstract
Introduction
The proper development of a metazoan organism consisting of a variety of specialized cell types requires the strict co-regulation of the differentiation and cell cycle machineries. As a cell acquires its fully differentiated state, concomitant exit from the cell cycle ensures the integrity of the genome and prevents tumorigenesis. At the opposite end of this spectrum, pluripotent stem cells persist in a state of rapid proliferation. These cells have a unique cell cycle consisting of a short G1 phase, which in part serves to impede differentiation [1–3]. Once the purview of developmental biologists, the fundamental question of how the cell cycle and differentiation are linked has become critical to a broad swath of disciplines including regenerative medicine, cancer biology, and aging. This review will examine recent findings on the dynamic regulation between the pluripotency and cell cycle networks.
Reciprocal regulation of cell cycle and pluripotency networks: Pluripotency regulation of the cell cycle
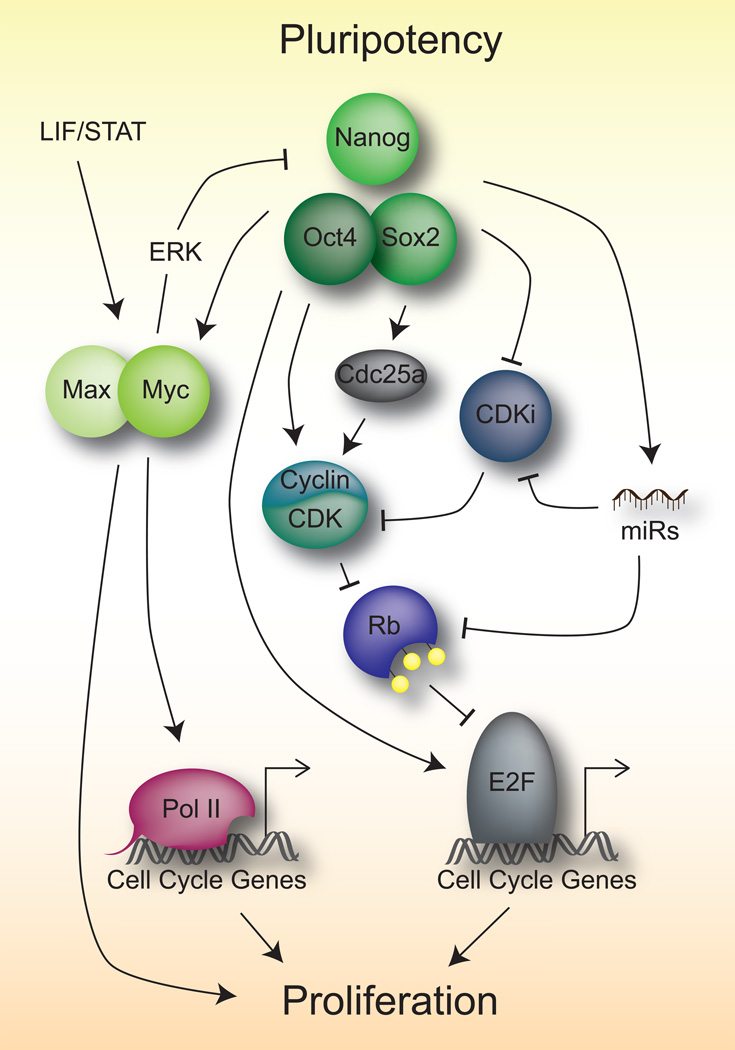
The pluripotent network consists of a core set of transcription factors, including Oct4 (Pou5f1), Sox2, and Nanog, which serve to establish the undifferentiated state and the self-renewing capacity of embryonic stem (ES) cells [reviewed in 4,5]. While it is clear that a major role of these core transcription factors is the activation of the greater pluripotency network [6], an emerging emphasis on crosstalk with the cell cycle machinery has recently been identified (Figure 1, Table 1). Early studies of the core pluripotency network identified Myc as a target of Oct4 and Nanog in ES cells that is central to the maintenance of pluripotency [7–9]. Myc then binds to and regulates many cell cycle genes in ES cells [10,11]. It does so in part by overcoming paused Pol II at target genes allowing for successful transcriptional elongation [12,13]. The dependency of Myc, and PI3K signaling, which also promotes pluripotency [14], can be relieved by growth in media containing GSK3β and MEK1/2 inhibitors (2i conditions) [15].
Figure 1.
Means of pluripotency control of the cell cycle
Table 1.
Molecular Pathways which regulate pluripotency and the cell cycle in ES cells
| Pathway | Role | Members | Ref |
|---|---|---|---|
| Core Pluripotency Network |
Maintain the undifferentiated state and self-renewal of pluripotent cells |
Oct4 (Pou5f1), Sox2, Nanog | [4,5] |
| Myc | Upregulate expression of genes to promote rapid proliferation. Often oncogenic driver of cancer |
c-Myc | [11] |
| Rb/E2F | Establish G1/S restriction point to regulate proliferation with growth signals |
Pocket Proteins (Rb, p107, p130) E2F transcription factors (E2F1- 5), DP1, CyclinD/E, Cdk2/4, Cdk- inhibitors (p16Ink4a, p15Ink4b p19Arf, p21Cip1, and p27Kip1) |
[37] |
| p53 | Genomic stability, stress response, growth arrest, apoptosis |
p53 (Trp53, TP53), Mdm2, p21Cip1, ATM, Chk2 |
[53– 55,70,71] |
| LIF/Stat3 | ES cell self-renewal, Myc activation |
LIF receptor (LIFR), gp130, Stat3, Jak |
[7] |
| PI3K/Akt | Growth factor signaling, apoptosis, and cell survival |
Receptor tyrosine kinases, PI3K, Akt |
[15,62] |
| MAPK/ERK | Extracellular signaling to nuclear transcription factors |
FGF, Ras, Raf, Mek, Erk | [46] |
| Hippo | Organ size control, cell proliferation |
Mst1/2, Lats1/2, Yap/Taz | [43] |
| ESC cell cycle regulating (ESCC) miRs |
Pluripotent gene activation, rapid cell cycle in ES cells |
miR290–295 cluster, miR302 | [25,27] |
Pluripotency and cell cycle control also converge on the Rb/E2F pathway (Table 1), one of the major regulators of the cell cycle, which is indeed critically involved in the regulation of the cell cycle in ES cells [16,17]. Rb, and its family members p107 and p130, comprise the family of “pocket proteins” which canonically repress E2F activity by an E2F-binding pocket domain. Through this pathway mitogen signaling can affect the activity of Cyclin/CDK complexes which, through phosphorylation of the pocket proteins, can relieve inhibition of the E2F family of transcription factors to initiate DNA replication [reviewed in 18,19]. ES cells are characterized by high CDK activity, subsequent phosphorylation of all three pocket proteins, and high E2F activity. Indeed Myc can directly regulate E2F activity [11]. Oct4 can also directly regulate the expression of E2F3a, which is partly responsible for the high proliferative rates in ES cells [20]. In addition, Nanog can upregulate CDKs and the CDK activator, Cdc25a [21]. To further enhance high CDK activity, several CDK inhibitors (including p16Ink4a, p15Ink4b, p19Arf, p21Cip1, and p27Kip1) are repressed in part by core pluripotency members [19,22,23]. The core pluripotency network also upregulates miRNAs, particularly of the miR290–295 cluster, miR302, and miR590 (Table 1), which in turn repress CDK inhibitors, pocket proteins, pro-differentiation miRNAs, and apoptosis [24–28]. Beyond transcriptional regulation and post-transcriptional regulation by miRNAs, post-translational modifications of key pathway members are also utilized by the cell to enforce high proliferation in ES cells. For example, the F-box protein Fbw7 (Fbxw7), a component of the SCF-type ubiquitin ligase complex, targets c-Myc for degradation and is therefore downregulated in ES cells to maintain high c-Myc protein stability [29,30]. In addition, the O-GlcNAcylation of a RINGB, a member of the polycomb repressive complex 1 (PRC1), removes PRC1 from regulatory DNA elements of cell cycle genes to promote differentiation [31].
One complication of fast cell proliferation is the potentially increased accumulation of genetic mutations due to error-prone DNA synthesis. Oct4 has been shown to directly bind to and inhibit Cdk1 resulting in a lengthening of G2 phase which allows more time for the DNA repair machinery to correct de novo mutations [32]. Similarly, a miR-590/Acvr2a/Rad51b axis also serves to balance the needs of the cell to maintain fast proliferation and resolve DNA damage. This occurs through the expression of miR-590, which becomes upregulated in ES cells during damage and mediates the upregulation of Rb and p21 to slow the cell cycle, while Activin signaling induces expression of the DNA-damage repair gene Rad51 [28].
Reciprocal regulation of cell cycle and pluripotency networks: Cell cycle regulation of pluripotency
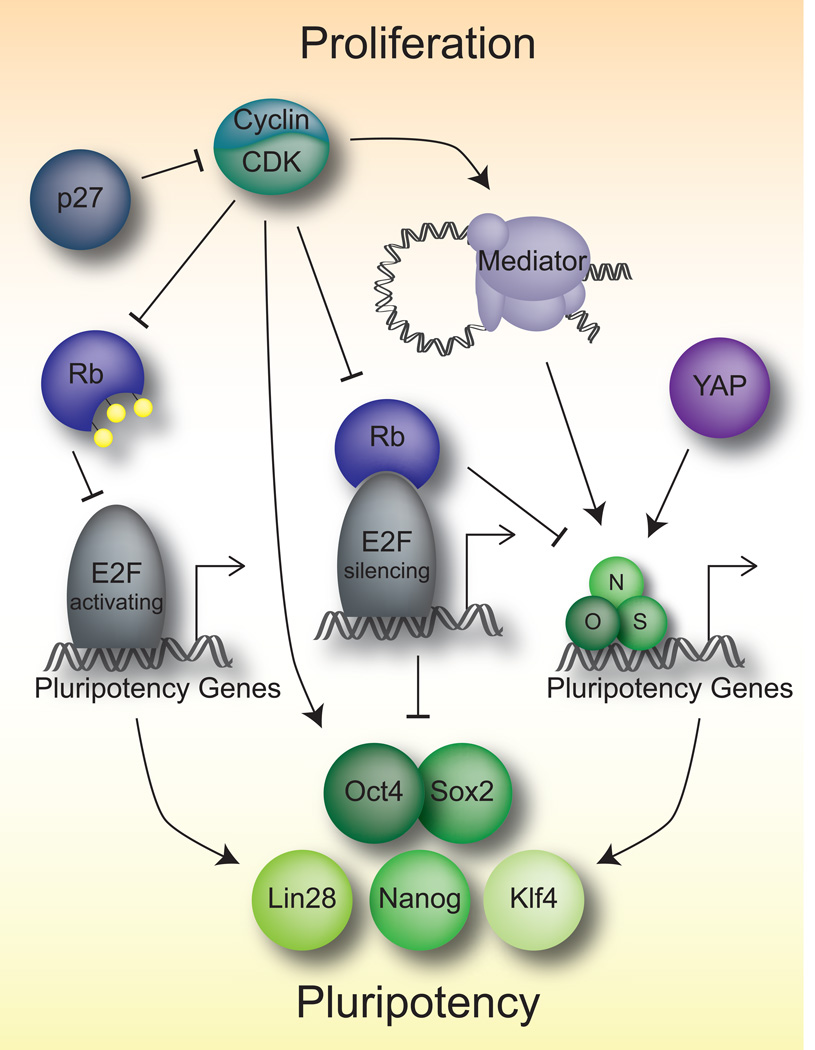
As the core pluripotency network can control the cell cycle, there are multiple means by which cell cycle regulators control pluripotency (Figure 2). Indeed there are several examples of how the high CDK activity in ES cells may influence the pluripotency network. Loss of CDK1 in human ES cells results in a reduction of pluripotency gene expression, including the core factors OCT4, KLF4, and LIN28, and subsequently increases differentiation [33]. Additionally, these cells show increased DNA damage and ensuing apoptosis [33,34]. Similar results were found performing chemical CDK2-inhibition in human ES cells [35]. Sox2 can be phosphorylated by Cdk2, although this is dispensable for the maintenance of pluripotency [36]. Mediator, which is controlled by Cdk8, plays an important role in the activation of genes containing Oct4, Sox2, and Nanog bound at their enhancers by looping them to promoter regions using cohesion [4]. Rb and associated proteins can silence members of the core pluripotency network in differentiated tissues, therefore this high Cdk activity serves to block this repression on pluripotency [37–39]. Similarly, Cdk inhibitors such as p27Kip1 also silence some pluripotency factors and are themselves repressed in ES cells [23,40]. This repression of Cdk inhibitors is E2F4-dependent, a reflection of the complex transcriptional role of the E2Fs, where E2F4–6 are transcriptional inhibitors rather than transcriptional activators and S-phase inducers [18,40]. Not surprisingly, activating E2Fs are implicated in the regulation of many pathways in ES cells, including the core pluripotency network genes Oct4, Sox2, and Nanog, as well as the Wnt and FGF pathways [39,41]. On the other hand, knockdown of E2F2 alone, reduced the proliferation of ES cells, but pluripotency was not effected [42]. This could reflect a specialization amongst the activating E2Fs to separate cell cycle and pluripotency functions. In addition, the Yes-associated protein (YAP), a member of the Hippo pathway which regulates organ size and cell cycling in somatic tissues, is indeed active in ES cells and can bind to many of the targets of the core pluripotency network [43].
Figure 2.
Means of cell cycle control of pluripotency
A rapid cell cycle to inhibit differentiation
Not only does the cell cycle play an integral role in the maintenance of pluripotency, but rapid proliferation may also serve to inhibit differentiation, thereby maintaining the undifferentiated state [1]. By the action of the core pluripotency network, multiple lineage-specific transcription factors are repressed [4,44]. Similarly, recent advances have indicated that the rapid cycling of ES cells maintains pluripotency by resisting differentiation [3,45] and that slowing of the cell cycle aids differentiation [1,45]. The high CDK activity in ES cells results in inactivation of the pocket proteins, which have been implicated in differentiation as well [reviewed in 18]. Myc, while keeping ES cells in a proliferative state, also directly represses the FGF/ERK pathway to inhibit differentiation (Figure 1) [7,46]. In silico techniques have shed light on the interactions between cell cycle networks and differentiation, which could be the source of future mechanistic studies [47].
Cell cycle control in the reprogramming of induced pluripotent stem (iPS) cells
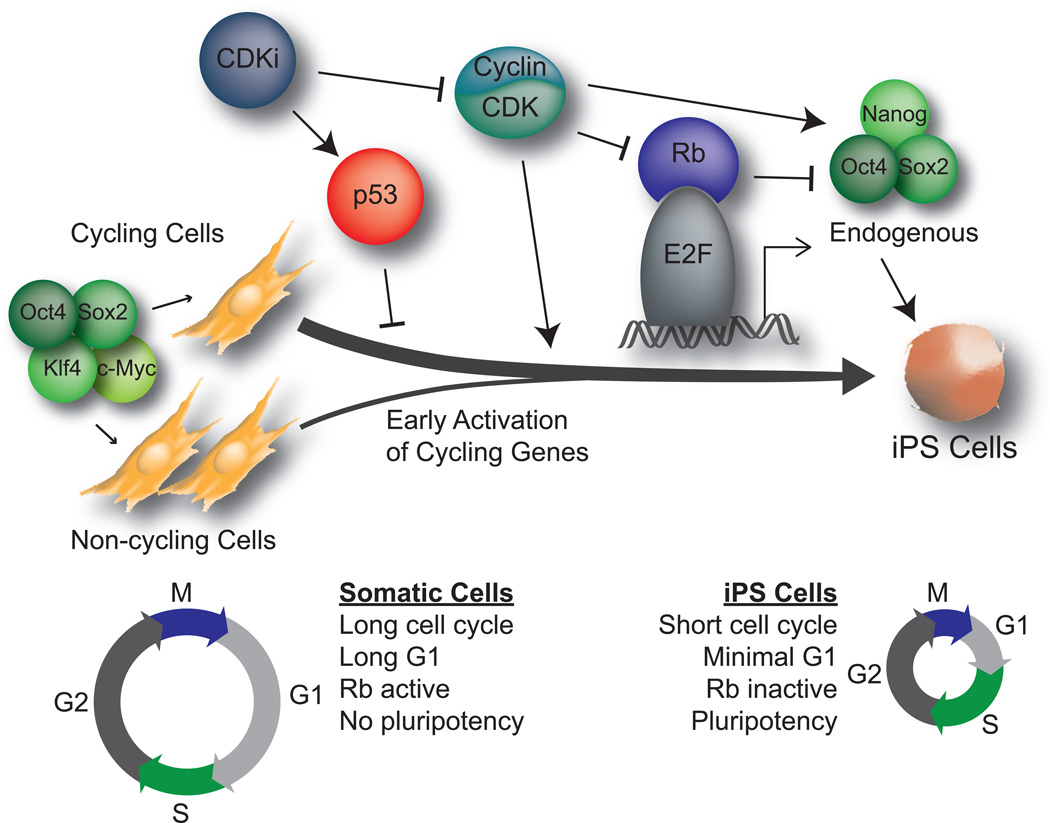
When Yamanaka and colleagues first successfully reprogrammed somatic cells to a pluripotent state, they observed that iPS cells grew at a rate similar to ES cells [48]. An integrative genomic analysis of human iPS cell reprogramming indicates that cell cycle genes are upregulated as early as day 5 of reprogramming [49]. Indeed, fully reprogrammed iPS cells acquire the minimal G1 phase typical of ES cells, and accelerated proliferation in the starting cell population aids in the efficiency of reprogramming [2,45]. Using nuclear transfer as a rapid means of reprogramming, Gurdon and colleagues observed a phenomena they termed “mitotic advantage”. They detected that mitotic nuclei are reprogrammed at rates significantly higher than interphase nuclei and that this is due in part to the de-ubiquitination of histones during mitosis resulting in gene derepression [50]. Similar studies have shown that enriching for proliferating cells enhances reprogramming [51]. This has been further supported by single cell studies that show that rapidly dividing cells are in a privileged state, whereby they account for the majority of successfully reprogrammed cells [52].
The mechanisms that underlie this increase in proliferation as cells reprogram have been the focus of multiple studies (Figure 3). Cells with Cdk inhibition are refractive to reprogramming [53–55], and the Cdk2-mediated phosphorylation of Sox2, while not required for pluripotency does indeed aid reprogramming [36]. The ability of the p53 pathway (Table 1) to restrict reprogramming is in part through increasing the proliferation of p53-null cells, although proliferation-independent roles for p53 activity cannot be strictly excluded [56,57]. The process of reprogramming stresses the cells, e. g. through DNA damage, induction of apoptotic programs, and oncogene expression [55], and reducing the checkpoint activity of p53 and Cdk inhibitors promotes successful reprogramming [58]. Loss of members of the Rb pathway also increases reprogramming [37,40]. However, the effects of Rb loss on reprogramming involves a direct repression of the core pluripotency network, rather than perturbations of the cell cycle, indicating that at least this critical tumor suppressor may possess separate functions to both regulate proliferation and pluripotency [37]. Along with CDK regulation, modulating the proper levels of CDK partner Cyclins is fundamental to proper reprogramming [59,60]. As in ES cells, Myc activity increases the rate of proliferation and reprogramming by helping to establish a pluripotent cell cycle in reprogrammed cells [10–12,61].
Figure 3.
Cell cycle control in reprogramming to pluripotency
Concluding comments
There are multiple molecular connections between the cell cycle and pluripotency. It appears plausible that these links are critical to establishing and maintaining the undifferentiated state, while setting the stage for later differentiation. The two processes of cell cycle regulation and pluripotency appear to exist in a circular relationship in ES cells where disruption of one will affect the other, resulting in generally two outcomes: cell death and/or differentiation. Indeed there are many proteins that coordinately link both processes, such as Rb and p53 [37,38,57]. Many signaling pathways can also simultaneously induce rapid proliferation and the core pluripotency gene network [15,62]. In ES cells it does appear, though, that there may be conditions in which cell cycle regulation and pluripotency may be uncoupled [37,63]. For example, Rb regulates the induction of pluripotency in a manner that is distinct from its cell cycle functions [37]. Perhaps the apparent connection of these two processes in ES cells reflects a fine balance that is achieved in the culture conditions that effectively pause these cells in their developmental state. This however, may not be true during normal development where a hierarchy between the cell cycle and pluripotency is still unclear. To address this question more studies should be performed in rodent and non-human primate models of early development, rather than ES cells that exist in a developmentally paused state, which may acquire the balanced co-regulation of proliferation and pluripotency to maintain ex vivo self-renewal. Furthermore, reprogramming techniques, such as nuclear transfer, cell fusion, or in vitro reprogramming with defined factors can further illuminate the mechanistic interrelatedness and potential hierarchy of cell cycle regulation and pluripotency.
It is perhaps not surprising that the genes and signaling pathways that simultaneously regulate proliferation and pluripotency in ES cells are often misregulated in cancer. After all, one of the main hallmarks of cancer is uncontrolled cell division. In addition, cancer has many characteristics reminiscent of ES and iPS cells including self-renewal, the ability to bypass senescence, oncogene activation, tumor suppressor repression, and tumor formation [64,65]. Of note, many cancer types are refractive to reprogramming to iPS cells, and examples of cancer cell-derived iPS cells are few [66–69]. This observation is counterintuitive because inhibition of tumor suppressors and activation of oncogenes facilitates reprogramming [66–69]. It is possible that these cancer cells are unable to acquire the unique ES cell cycle with a short G1 phase. Therefore, a greater elucidation of the crosstalk between cell cycle and pluripotency pathways can not only advance the fields of developmental biology and regenerative medicine, but may also provide novel insights into the biology of cancerous transformation.
Highlights.
The pluripotent state has a unique cell cycle relative to somatic cells
Extensive reciprocal crosstalk exists to maintain both pluripotency and a rapid cell cycle
Unique cell cycle maintains pluripotency by inhibiting differentiation
iPS cell reprogramming re-establishes theunique cell cycle of ES cells
Acknowledgments
The authors thank members of the Wernig and Sage laboratories for critical comments on the manuscript. This work was supported by the Lucile Packard Foundation for Children’s Health (M.S.K., J.S.), and the NIH (grant CA114102 to J.S.). M.W. is a New York Stem Cell Foundation-Robertson Investigator and a Tashia and John Morgridge Faculty Scholar, and J.S. is the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michael S. Kareta, Email: mkareta@stanford.edu.
Julien Sage, Email: julsage@stanford.edu.
Marius Wernig, Email: wernig@stanford.edu.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1. Li VC, Kirschner MW. Molecular ties between the cell cycle and differentiation in embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:9503–9508. doi: 10.1073/pnas.1408638111. ** The authors utilized various methods to inhibit the cell cycle and observed a more rapid establishment of terminally differnetiated gene networks.
- 2.Ghule PN, Medina R, Lengner CJ, Mandeville M, Qiao M, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Reprogramming the pluripotent cell cycle: restoration of an abbreviated G1 phase in human induced pluripotent stem (iPS) cells. J Cell Physiol. 2011;226:1149–1156. doi: 10.1002/jcp.22440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coronado D, Godet M, Bourillot PY, Tapponnier Y, Bernat A, Petit M, Afanassieff M, Markossian S, Malashicheva A, Iacone R, et al. A short G1 phase is an intrinsic determinant of naive embryonic stem cell pluripotency. Stem Cell Res. 2013;10:118–131. doi: 10.1016/j.scr.2012.10.004. * Using the Fluorescence Ubiquitination Cell Cycle Indicator (FUCCI) reporter system the authors demonstrated that naïve ES cells are more prone to differentiate in G1.
- 4.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackett JA, Surani MA. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 8.Hishida T, Nozaki Y, Nakachi Y, Mizuno Y, Okazaki Y, Ema M, Takahashi S, Nishimoto M, Okuda A. Indefinite self-renewal of ESCs through Myc/Max transcriptional complex-independent mechanisms. Cell Stem Cell. 2011;9:37–49. doi: 10.1016/j.stem.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 9.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, Adelman K. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell. 2015;58:311–322. doi: 10.1016/j.molcel.2015.02.003. * Using GRO-seq the authors demonstrated that cell cycle genes are often subject to Pol II pausing in naïve ES cells, and that perturbation of Pol II pausing decreases proliferation.
- 14.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci U S A. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hishida T, Nakachi Y, Mizuno Y, Katano M, Okazaki Y, Ema M, Takahashi S, Hirasaki M, Suzuki A, Ueda A, et al. Functional compensation between Myc and PI3K signaling supports self-renewal of embryonic stem cells. Stem Cells. 2015;33:713–725. doi: 10.1002/stem.1893. [DOI] [PubMed] [Google Scholar]
- 16.Conklin JF, Baker J, Sage J. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat Commun. 2012;3:1244. doi: 10.1038/ncomms2254. [DOI] [PubMed] [Google Scholar]
- 17.Wirt SE, Adler AS, Gebala V, Weimann JM, Schaffer BE, Saddic LA, Viatour P, Vogel H, Chang HY, Meissner A, et al. G1 arrest and differentiation can occur independently of Rb family function. J Cell Biol. 2010;191:809–825. doi: 10.1083/jcb.201003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julian LM, Blais A. Transcriptional control of stem cell fate by E2Fs and pocket proteins. Front Genet. 2015;6:161. doi: 10.3389/fgene.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh AM, Dalton S. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 2009;5:141–149. doi: 10.1016/j.stem.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanai D, Ueda A, Akagi T, Yokota T, Koide H. Oct3/4 directly regulates expression of E2F3a in mouse embryonic stem cells. Biochem Biophys Res Commun. 2015;459:374–378. doi: 10.1016/j.bbrc.2015.02.105. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Neganova I, Przyborski S, Yang C, Cooke M, Atkinson SP, Anyfantis G, Fenyk S, Keith WN, Hoare SF, et al. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J Cell Biol. 2009;184:67–82. doi: 10.1083/jcb.200801009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J, Go Y, Kang I, Han YM, Kim J. Oct-4 controls cell-cycle progression of embryonic stem cells. Biochem J. 2010;426:171–181. doi: 10.1042/BJ20091439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell. 2005;16:2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Melton C, Li YP, Shenoy A, Zhang XX, Subramanyam D, Blelloch R. miR-294/miR-302 promotes proliferation, suppresses G1-S restriction point, and inhibits ESC differentiation through separable mechanisms. Cell Rep. 2013;4:99–109. doi: 10.1016/j.celrep.2013.05.027. ** The authors demonstrate that miR-294 and miR-302 function in Rb-dependent and Rb-independent role to promote a rapid transition through the G1/S checkpoint to maintain the rapid cycling in ES cells.
- 26.Zhang Z, Hong Y, Xiang D, Zhu P, Wu E, Li W, Mosenson J, Wu WS. MicroRNA-302/367 cluster governs hESC self-renewal by dually regulating cell cycle and apoptosis pathways. Stem Cell Reports. 2015;4:645–657. doi: 10.1016/j.stemcr.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Q, Wang G, Chen Y, Li G, Yang D, Kang J. A miR-590/Acvr2a/Rad51b axis regulates DNA damage repair during mESC proliferation. Stem Cell Reports. 2014;3:1103–1117. doi: 10.1016/j.stemcr.2014.10.006. * The authors mapped out an interaction between miR-590 and DNA repair pathways as a means to ensure genomic integrity during rapid ES proliferation.
- 29.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeishi S, Nakayama KI. Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br J Cancer. 2014;111:1054–1059. doi: 10.1038/bjc.2014.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maury JJ, El Farran CA, Ng D, Loh YH, Bi X, Bardor M, Choo AB. RING1B O-GlcNAcylation regulates gene targeting of polycomb repressive complex 1 in human embryonic stem cells. Stem Cell Res. 2015;15:182–189. doi: 10.1016/j.scr.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 32. Zhao R, Deibler RW, Lerou PH, Ballabeni A, Heffner GC, Cahan P, Unternaehrer JJ, Kirschner MW, Daley GQ. A nontranscriptional role for Oct4 in the regulation of mitotic entry. Proc Natl Acad Sci U S A. 2014;111:15768–15773. doi: 10.1073/pnas.1417518111. * The authors demonstrated a role for Oct4 in regulating preventing premature mitotic entry by direct Cdk1 inhibitition.
- 33.Neganova I, Tilgner K, Buskin A, Paraskevopoulou I, Atkinson SP, Peberdy D, Passos JF, Lako M. CDK1 plays an important role in the maintenance of pluripotency and genomic stability in human pluripotent stem cells. Cell Death Dis. 2014;5:e1508. doi: 10.1038/cddis.2014.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huskey NE, Guo T, Evason KJ, Momcilovic O, Pardo D, Creasman KJ, Judson RL, Blelloch R, Oakes SA, Hebrok M, et al. CDK1 inhibition targets the p53-NOXA-MCL1 axis, selectively kills embryonic stem cells, and prevents teratoma formation. Stem Cell Reports. 2015;4:374–389. doi: 10.1016/j.stemcr.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kallas A, Pook M, Trei A, Maimets T. Assessment of the Potential of CDK2 Inhibitor NU6140 to Influence the Expression of Pluripotency Markers NANOG, OCT4, and SOX2 in 2102Ep and H9 Cells. Int J Cell Biol. 2014;2014:280638. doi: 10.1155/2014/280638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouyang J, Yu W, Liu J, Zhang N, Florens L, Chen J, Liu H, Washburn M, Pei D, Xie T. Cyclin-Dependent Kinase-Mediated Sox2 Phosphorylation Enhances the Ability of Sox2 to Establish the Pluripotent State. J Biol Chem. 2015 doi: 10.1074/jbc.M115.658195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kareta MS, Gorges LL, Hafeez S, Benayoun BA, Marro S, Zmoos AF, Cecchini MJ, Spacek D, Batista LF, O'Brien M, et al. Inhibition of pluripotency networks by the rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell. 2015;16:39–50. doi: 10.1016/j.stem.2014.10.019. * The authors revealed a role for Rb in the direct repression of pluripotency networks in a cell cycle-independent manner.
- 38.Kitajima S, Kohno S, Kondoh A, Sasaki N, Nishimoto Y, Li F, Abdallah Mohammed MS, Muranaka H, Nagatani N, Suzuki M, et al. Undifferentiated State Induced by Rb-p53 Double Inactivation in Mouse Thyroid Neuroendocrine Cells and Embryonic Fibroblasts. Stem Cells. 2015;33:1657–1669. doi: 10.1002/stem.1971. [DOI] [PubMed] [Google Scholar]
- 39.O'Connor MD, Wederell E, Robertson G, Delaney A, Morozova O, Poon SS, Yap D, Fee J, Zhao Y, McDonald H, et al. Retinoblastoma-binding proteins 4 and 9 are important for human pluripotent stem cell maintenance. Exp Hematol. 2011;39:866–879. e861. doi: 10.1016/j.exphem.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Collado M, Villasante A, Matheu A, Lynch CJ, Canamero M, Rizzoti K, Carneiro C, Martinez G, Vidal A, et al. p27(Kip1) Directly Represses Sox2 during Embryonic Stem Cell Differentiation. Cell Stem Cell. 2012;11:845–852. doi: 10.1016/j.stem.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeo HC, Beh TT, Quek JJ, Koh G, Chan KK, Lee DY. Integrated transcriptome and binding sites analysis implicates E2F in the regulation of self-renewal in human pluripotent stem cells. PLoS One. 2011;6:e27231. doi: 10.1371/journal.pone.0027231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki DE, Nakahata AM, Okamoto OK. Knockdown of E2F2 inhibits tumorigenicity, but preserves stemness of human embryonic stem cells. Stem Cells Dev. 2014;23:1266–1274. doi: 10.1089/scd.2013.0592. [DOI] [PubMed] [Google Scholar]
- 43.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz S, Panopoulos AD, Herrerias A, Bissig KD, Lutz M, Berggren WT, Verma IM, Izpisua Belmonte JC. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chappell J, Sun Y, Singh A, Dalton S. MYC/MAX control ERK signaling and pluripotency by regulation of dual-specificity phosphatases 2 and 7. Genes Dev. 2013;27:725–733. doi: 10.1101/gad.211300.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Re A, Workman CT, Waldron L, Quattrone A, Brunak S. Lineage-specific interface proteins match up the cell cycle and differentiation in embryo stem cells. Stem Cell Res. 2014;13:316–328. doi: 10.1016/j.scr.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 49.Cacchiarelli D, Trapnell C, Ziller MJ, Soumillon M, Cesana M, Karnik R, Donaghey J, Smith ZD, Ratanasirintrawoot S, Zhang X, et al. Integrative Analyses of Human Reprogramming Reveal Dynamic Nature of Induced Pluripotency. Cell. 2015;162:412–424. doi: 10.1016/j.cell.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Halley-Stott RP, Jullien J, Pasque V, Gurdon J. Mitosis gives a brief window of opportunity for a change in gene transcription. PLoS Biol. 2014;12:e1001914. doi: 10.1371/journal.pbio.1001914. ** The authors revealed that mitotic nuclei reprogram significantly faster than interphase nuclei after transfer into amphibian oocytes.
- 51.Roccio M, Schmitter D, Knobloch M, Okawa Y, Sage D, Lutolf MP. Predicting stem cell fate changes by differential cell cycle progression patterns. Development. 2013;140:459–470. doi: 10.1242/dev.086215. [DOI] [PubMed] [Google Scholar]
- 52. Guo S, Zi X, Schulz VP, Cheng J, Zhong M, Koochaki SH, Megyola CM, Pan X, Heydari K, Weissman SM, et al. Nonstochastic reprogramming from a privileged somatic cell state. Cell. 2014;156:649–662. doi: 10.1016/j.cell.2014.01.020. * Utilizing live-cell imaging the authors showed that there is a priveledged class of rare somatic cells which have a rapid proliferation rate, and that these cells account for a large majority of cells that successfully reprogram to iPS cells.
- 53.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdelalim EM, Tooyama I. The p53 inhibitor, pifithrin-alpha, suppresses self-renewal of embryonic stem cells. Biochem Biophys Res Commun. 2012;420:605–610. doi: 10.1016/j.bbrc.2012.03.041. [DOI] [PubMed] [Google Scholar]
- 58.Lin YC, Murayama Y, Hashimoto K, Nakamura Y, Lin CS, Yokoyama KK, Saito S. Role of tumor suppressor genes in the cancer-associated reprogramming of human induced pluripotent stem cells. Stem Cell Res Ther. 2014;5:58. doi: 10.1186/scrt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McLenachan S, Menchon C, Raya A, Consiglio A, Edel MJ. Cyclin A1 is essential for setting the pluripotent state and reducing tumorigenicity of induced pluripotent stem cells. Stem Cells Dev. 2012;21:2891–2899. doi: 10.1089/scd.2012.0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edel MJ, Menchon C, Menendez S, Consiglio A, Raya A, Izpisua Belmonte JC. Rem2 GTPase maintains survival of human embryonic stem cells as well as enhancing reprogramming by regulating p53 and cyclin D1. Genes Dev. 2010;24:561–573. doi: 10.1101/gad.1876710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lavagnolli T, Gupta P, Hormanseder E, Mira-Bontenbal H, Dharmalingam G, Carroll T, Gurdon JB, Fisher AG, Merkenschlager M. Initiation and maintenance of pluripotency gene expression in the absence of cohesin. Genes Dev. 2015;29:23–38. doi: 10.1101/gad.251835.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu CC, Wu HJ, Wang CH, Lin CH, Hsu SC, Chen YR, Hsiao M, Schuyler SC, Lu FL, Ma N, et al. Akt suppresses DLK for maintaining self-renewal of mouse embryonic stem cells. Cell Cycle. 2015;14:1207–1217. doi: 10.1080/15384101.2015.1014144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li VC, Ballabeni A, Kirschner MW. Gap 1 phase length and mouse embryonic stem cell self-renewal. Proc Natl Acad Sci U S A. 2012;109:12550–12555. doi: 10.1073/pnas.1206740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daley GQ. Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harb Symp Quant Biol. 2008;73:171–174. doi: 10.1101/sqb.2008.73.041. [DOI] [PubMed] [Google Scholar]
- 65. Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–677. doi: 10.1016/j.cell.2014.01.005. * By the use of an reprogrammable mouse system, the authors showed that tumors generated from premature reprogramming termination share similarities to human Wilms tumors, indicating that some tumors may arise by reprogramming-like processes.
- 66.Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, Wernig M, Jaenisch R, Brummelkamp TR. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J, Hoffman JP, Alpaugh RK, Rhim AD, Reichert M, Stanger BZ, Furth EE, Sepulveda AR, Yuan CX, Won KJ, et al. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013;3:2088–2099. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, Nagano H, Sekimoto M, Doki Y, Mori M. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci U S A. 2010;107:40–45. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stricker SH, Feber A, Engstrom PG, Caren H, Kurian KM, Takashima Y, Watts C, Way M, Dirks P, Bertone P, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654–669. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]