Abstract
The dynamic interaction of nucleosome binding proteins with their chromatin targets is an important element in regulating the structure and function of chromatin. Histone H1 variants and High Mobility Group (HMG) proteins are ubiquitously expressed in all vertebrate cells, bind dynamically to chromatin, and are known to affect chromatin condensation and the ability of regulatory factors to access their genomic binding sites. Here, we review the studies that focus on the interactions between H1 and HMGs and highlight the functional consequences of the interplay between these architectural chromatin binding proteins. H1 and HMG proteins are mobile molecules that bind to nucleosomes as members of a dynamic protein network. All HMGs compete with H1 for chromatin binding sites, in a dose dependent fashion, but each HMG family has specific effects on the interaction of H1 with chromatin. The interplay between H1 and HMGs affects chromatin organization and plays a role in epigenetic regulation.
Introduction
Chromatin dynamics play a key role in the ability of regulatory factors to access their target sites and in various processes that ultimately affect gene expression. A major group of nuclear proteins known to affect chromatin structure and function are named “chromatin architectural proteins”. Architectural proteins are defined as structural proteins, devoid of enzymatic activity, that bind to nucleosomes without apparent DNA sequence specificity and change the local and global architecture of chromatin.
Two major groups of proteins are known to function as architectural proteins in all vertebrate cells. The first group is the linker H1 protein family, the focus of this special BBA issue. Histone H1 is the most abundant family of chromatin binding proteins; most nuclei contain sufficient protein to bind to all nucleosomes. The second major group of architectural proteins is the High Mobility Group (HMG) protein superfamily [1–7]}. In general, H1 proteins promote and stabilize the formation of compact chromatin structures [8–11], while HMG proteins promote chromatin decompaction and the formation of distorted DNA structures. In the nucleus, H1 and HMG protein variants are mobile, they move rapidly throughout the entire nuclear space, interact transiently with nucleosomes and their chromatin interactions seem to be interdependent [11–14]. H1 and HMGs were the first nuclear proteins shown to affect the structure of the chromatin fiber and their properties and chromatin interactions have been the subject of several reviews [1, 4, 5, 10–12, 15–17]. In this review we focus on the interplay between H1 and HMG architectural proteins.
HMG proteins
The HMG superfamily is composed of three families: HMGN, HMGB, and HMGA proteins [2, 6]. The structural features and biological properties of these proteins have been described in several reviews and in a special issue of BBAGRM [5, 15, 18].
Each HMG family has a distinct protein structure and a distinct DNA or chromatin binding motif [1, 2, 4]. The HMG box is the functional motif of the HMGB proteins, the AT hook is the functional motif of the HMGA family, and the nucleosomal binding domain (NBD) is the functional motif of the HMGN family. Through these functional motifs HMGs bind to specific structures in DNA or chromatin, with low if any specificity for the DNA sequence. All non-chromatin-bound HMG are highly disordered proteins, a structural characteristic they share with linker H1 variants, endowing them with the ability to form multiple protein-protein interactions [19]. All HMGs promote chromatin decompaction and generate a chromatin configuration that alters various DNA-dependent activities such as transcription, replication and the repair of damaged DNA. HMGs affect genomic functions not only by directly binding to chromatin but also by interacting with regulatory factors that affect gene expression. All three families are ubiquitously present in all vertebrate cells; in addition, proteins containing the functional motifs of HMGA and HMGB were found in most eukaryotes and have also been detected in bacteria.
The human HMGN family consists of 5 members with a similar structure: a bipartite nuclear localization signal, a conserved, positively charged nucleosome binding domain, and a C-terminal regulatory domain. The nucleosome binding domain contains the invariant octapeptide RRSARLSA which serves as the signature motif of this protein family [3, 20]. As elaborated below, this octapeptide serves as the anchoring point of HMGNs on the nucleosome [20]. Genome wide, HMGN proteins preferentially bind to DNase I hypersensitive sites, the hallmark of regulatory chromatin sites [21–23]. HMGNs have been shown to modulate the global and local structure of chromatin [24], and the levels of histone modifications [25, 26], factors which may be involved in their ability to affect gene expression [27].
HMGB proteins contain two HMG boxes, and a negatively charged C-terminal domain [28]. Through the HMG box these proteins bend DNA in a sequence-independent fashion. In addition, HMGB proteins were shown to bind preferentially to pre-bent DNA or DNA with distorted geometry [4].
The hallmark of the HMGA proteins is the “AT hook”, a palindromic sequence containing the invariant tripeptide GRP, flanked by arginine residues. Most HMGA proteins contain several AT-hooks. The AT hook binds preferentially to the minor groove of short stretches of AT-rich DNA [17].
HMG-H1 interactions
The precisely organization of H1 in nucleosomes is not known; however it has been established that the globular domain of histone H1 contacts the DNA near the nucleosome dyad axis and adjacent linker DNA, and thus stabilizes DNA wrapping around the histone octamer [29–32]. While the binding of H1 to the surface of nucleosomes is directed by the globular domain, the chromatin-condensing functions of the protein are primarily provided by the highly basic C-terminal domain which is thought to interact primarily with the negatively charged linker DNA [33]. Yet, the interactions of the C-terminal tail of H1 with linker DNA are not solely determined by charge, since fluorescence recovery after photobleaching (FRAP) and NMR results suggest that specific residues in this domain, rather than the distribution of positively charged residues, are major factors in regulating its interaction with linker DNA [30, 34, 35]. The binding of histone H1 to the nucleosomes plays a pivotal role in the stabilization of the compact 30 nm structure of the chromatin fiber [31, 32]. Differences among H1 variants in the amino acid sequence of the C-terminal domain, and variations in the position and orientation of the globular domains of H1 within the nucleosome may contribute to the heterogeneity of chromatin structure and also affect gene expression.
A distinguishing feature of all HMGNs is that they recognize specifically the generic structure of the nucleosome core particle. They bind better to nucleosomes than to free histones or purified DNA. Under physiological conditions nucleosomes bind two molecules of HMGNs. Significantly, even though most cells contain several HMGN variants, both in vivo and in vitro analyses indicated that the HMGN nucleosome complexes contain two molecules of the same HMGN variant; complexes containing two different variants (i.e. one HMGN1 and one HMGN2) are not detected [36]. The position of HMGN variants in nucleosomes was mapped by DNase I, by hydroxyl radical footprinting, by site specific cross-linking, and by a combination of methyl-transverse relaxation optimized nuclear magnetic resonance spectroscopy (methyl-TROSY) and mutational analysis [37–40]. In the HMGN-nucleosome complex, the invariant octapeptide RRSARLSA located in the NBD of HMGN, binds to a negatively charged patch formed by the H2A-H2B dimer. The N terminal region of the HMGN NBD contacts histone H2B and the DNA approximately 25 base-pairs away from the end of the 147 base pair nucleosomal core DNA, while the C-terminal region of the NBD contacts the DNA near its nucleosomal exit/entry. The C-terminal domain of the HMGN protein contacts the DNA in the two major grooves flanking the nucleosome dyad axis and is in close proximity to the N-terminal tail of H3, which protrudes beyond the periphery of the nucleosomal DNA [37, 40].
The overlap between the location of the HMGN and the globular H1 near the dyad axis of the nucleosomes may play a major role in the interplay between these proteins in chromatin [15, 37]. It is possible that negatively charged residues in the C-terminal domain of certain HMGNs, which are positioned near the linker DNA, may interfere with the interactions of H1 at this site [38]. Since H1 is known to facilitate and promote chromatin compaction HMGNs may decompact chromatin by interfering with the binding of H1 to nucleosomes. The chromatin-decondensing activity of HMGN can be also attributed to its interaction with the acidic patch of the H2A-H2B dimer and with the N-terminal of H3 since internucleosomal interaction have been shown to be facilities by the tail of H3 and by interactions between the N-terminal of H4 and the H2A.H2B acidic patch. We speculate that H1 could also interact with histone tails and if such interactions do indeed occur they could be affected by the presence of HMGNs and perhaps other HMG proteins, in particular members of the HMGB family which have a highly acidic C-terminal tail.
In addition, it has been demonstrated that the HMGN5 variant, which has a particularly long negatively charged C-terminal domain, targets the positively charged C-terminal region of histone H5 in the nucleoplasm, beyond the nucleosomal context [24]. Likely, similar interaction could occur between H1 and other HMGN variants.
HMGB proteins and histone H1 interact both in the context of chromatin and in the nucleoplasm. HMGB variants bind to purified DNA, to specific DNA structures such as four-way junctions, and to chromatin. Competition between HMGB1/-B2 proteins and linker histones for four-way junctions and cisplatin-modified DNA has been clearly demonstrated [41, 42]. Although their organization in isolated nucleosomes or in chromatin is not fully understood, HMGB1/B2 seems to preferentially bind to linker DNA at the entry/exit of nucleosomes, next to the nucleosome dyad axis [43, 44]. Since these are also major sites of H1-nucleosome interaction it is likely that this positional overlap plays an important role in the HMGB-H1 interplay in chromatin.
As chemical cross-linking and gel-filtration experiments with purified proteins have shown, an equimolar H1-HMGB1 complex is formed, which persists at physiological ionic strength [45]. According to NMR spectroscopy data, H1 linker histone binds, predominantly through its basic C-terminal domain, to the acidic tail of HMGB1, thereby disrupting intramolecular interaction of the HMGB tail with the DNA-binding faces of the HMG boxes. A potential consequence of this interaction is enhanced DNA binding by HMGB1, with a concomitantly lower affinity of H1 for DNA. In the context of chromatin, this might facilitate displacement of H1 by HMGB1 [45]. Indeed, mononucleosomes devoid of H1 but containing near stoichiometric amounts of HMGB1 are preferentially released upon micrococcal nuclease digestion [46]. Oxidation of HMGB1, which leads to the formation of disulfide bridges, abolishes its ability to compete with H1 [47].
HMGA1 proteins bind preferentially to the narrow minor groove of A/T-rich regions such as the highly repetitive alpha-satellite DNA. In addition, HMGA proteins recognize and bind to DNAs with unusual structural features, such as four-way, three-way or distorted DNA [17, 48]} and have been shown to bind specifically to isolated nucleosomes [49]. As discussed above, H1 also displays affinity for distorted DNA structures [41].
HMGA proteins co-localize with histone H1 at scaffold attachment regions (SAR), which are believed to be cis-acting regulatory elements located at the stem of large loops (domains) of gene-containing DNA [50]. It has been suggested that competition between HMGA1 and histone H1 for binding to AT-rich SAR elements affects chromatin compaction thereby impacting gene transcription [51]. Likely, competition between H1 and HMGAs extends beyond just the AT-rich elements in SARs, since their distribution in various fraction of micrococcal nuclease-digested chromatin is drastically different [51].
Mobile Architectural Elements: Chromatin Network
In the nucleus, H1 variants and all HMG proteins bind dynamically to chromatin. Photobleaching analysis of live cells expressing H1-GFP and HMG-GFP revealed that these architectural proteins are highly mobile and that a specific molecule binds only temporarily to a specific nucleosome. The proteins move throughout the nucleus in a “stop-and-go” mode and reside on a specific nucleosome for only a short time [11, 52]. The mobility of architectural proteins provides an opportunity for exchange of one protein for another through competition for overlapping binding sites. HMG proteins have a higher mobility and a shorter chromatin residence time than that of histone H1 [14, 52]. Given that the binding site of each member of the HMG protein family overlaps to some extent with those of linker H1 [13], it could be expected that HMGs would affect the chromatin interactions of H1, in living cell.
Indeed, using fluorescence recovery after photobleaching (FRAP) to monitor the binding of GFP-H1 to chromatin, it was shown that an increase in the content of one H1 decreases the chromatin residence time of the remaining variant. Furthermore, upregulation of any member of the HMG protein superfamily also decreased the H1 chromatin residence time [13, 14, 53]. The effect was dose-dependent and not observed with HMG mutant proteins that do not bind to chromatin [14]. For example, neither HMGN1 with two serines mutated in their nucleosomal binding domain, nor HMGB1 lacking either the A or B box, affected the binding of H1 to chromatin as measured by FRAP [13, 53].
Members of each HMG family compete for chromatin binding sites with members of the same family, but not with members of a different family, Thus, upregulation of HMGN1 levels reduced the chromatin binding of HMGN2 but not that of HMGB1, while upregulation of HMGB1 did not affect the chromatin residence time of an HMGN or an HMGA variant [13]. Loss of one HMGN variant led to a compensatory increase in the chromatin binding of a remaining variant from the same family [21] suggesting functional compensation within a family.
The absence of competition between HMG variants from different families, suggest that each HMG family has distinct binding sites on chromatin. Yet, since all HMG chromatin binding sites overlap to some extent with those of H1, and since all HMGs affected the binding of H1 to chromatin it is possible that different HMG variants synergistically impact the binding of H1 to chromatin. Indeed, an HMG mix comprised of members from different families affected the mobility of H1 to a larger degree than an HMG mix containing equal amounts of HMG proteins from one family [13, 14]. Thus, HMG proteins might function within a synergistic network and their combined action modulates the binding of H1 to chromatin.
The dynamic competition between HMGs and H1 seems to be functionally significant. Thus, HMGA1 is enriched at origin of replications where H1 levels could be reduced [54]. Likewise, HMGN1 and HMGN2 preferentially localize to DNAse I-hypersensitive promoters sites [21–23], and HMGB proteins were predicted to be preferentially associated with cis-regulatory modules (CRMs) [55], these chromatin regulatory sites are thought to be relatively depleted of histone H1 [56].
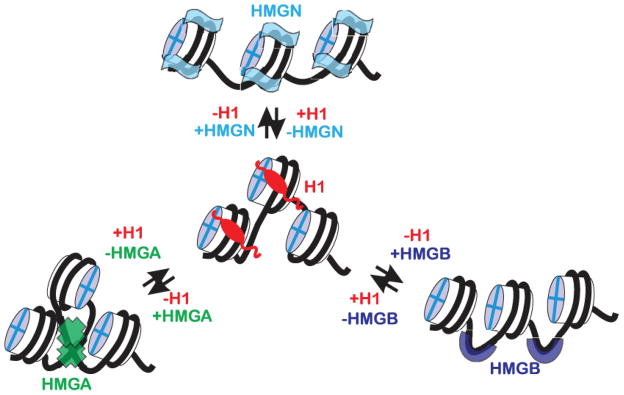
The emerging picture suggests that all HMGs and histone H1 variants form a network of dynamic interactions on the surface of the nucleosome [11](Figure 1). Histone H1 and HMGs move rapidly through the nucleoplasm and reside for a short time on the nucleosome; the residence tome of H1 on a specific site is significantly longer than that of the HMGs [9, 13, 57, 58]. As elaborated elsewhere [11], the various components within this network compete for binding sites and some of their chromatin-modifying activities could be interchangeable. The network of competitive interactions establishes the overall binding mode of each member of the network at each site of the genome. Although some of these competitive interactions can be tested individually, new integrative and system biology tools will be required to better understand how the steady state equilibrium between H1 and HMGs is established and how it changes in response to various internal or extracellular signals. Obviously, this network could include additional members since most nuclear proteins bind dynamically to chromatin [52].
Figure 1. Dynamic interplay between H1 and HMG Proteins in Chromatin.
Shown is a schematic depiction of the network of competitive interactions between all H1 variants and all the members of the HMG protein superfamily. All the members of the three families of HMG proteins synergistically affect the binding of H1 to nucleosomes. Each HMG family has a distinct set of chromatin binding sites. Members of an HMG family compete with other but do not compete with members of another HMG family for nucleosome binding sites.
Functional significance of HMG-H1 interactions
The reversible folding of the chromatin fiber into compact structures and the occupancy of nucleosomes at chromatin regulatory sites are known to play key roles in DNA related activities such as transcription, replication, and repair. At the most fundamental level, chromatin compaction and heterochromatinization inhibits DNA related activities such as transcription, replication, and repair while chromatin decompaction enhances these activities. Histone H1 and HMGN proteins have opposite effects on the structure and activity of the chromatin fiber. H1 promotes the formation and stabilization of compact heterochromatin while HMGs facilitate chromatin decompaction and enhance the ability of regulatory factors to reach their binding sites. Thus, it could be expected that the interplay between these architectural proteins would affect chromatin function.
Given that the interplay between H1 and HMGs occurs within the context of a multicomponent network of interactions, it is difficult to establish precisely the biological significance of a specific interaction between any specific HMG and H1 variant. Furthermore, since HMG variants are known to interact with specific regulatory factors, it is likely that changes in the H1-HMG interplay have specific effect in a gene or tissue specific context. Below is a summary of some of the biological effects that have been attributed to changes in the interaction between H1 and HMGs.
Several studies suggest that the H1-HMG interplay affect gene expression. Indeed HMGNs are enriched, while linker histones are depleted from actively transcribed genes [22, 59–62]. In SV40 minichromosomes, HMGN1 reduced the H1 mediated chromatin compaction and Pol II-mediated transcriptional repression [63]. The connection between chromatin compaction and transcription repression was solidified by showing that a mutant HMGN1 lacking the C-terminal acidic domain was unable to induce unfolding of the chromatin [64], and also unable to stimulate transcription. Likewise, in chromatin templates reconstituted by Drosophila extracts, H1 inhibits, while HMGN2 stimulates in vitro transcription [65, 66].
In vitro chromatin assembly studies have shown that histone H1 can inhibit chromatin remodeling by ATP-dependent SWI/SNF [67]. Phosphorylation of H1, which can alter the interaction of H1 with chromatin, rescues chromatin remodeling by SWI/SNF, ACF and other complexes [68]. Given that HMGNs increases accessibility to chromatin it is possible that HMGN might facilitate chromatin remodeling. However, HMGN1 did not affect the rate of SWI/SNF-dependent remodeling of H1-depleted mononucleosomes or nucleosomal arrays [69] and HMGN2 represses remodeling by reducing the binding of ACF to chromatin [70]. It would be interesting to study SWI/SNF or ACF remodeling activities on chromatin templates assembled in the presence of both H1 and HMGNs.
Fluorescence microscopy analyses of transfected cells revealed that upregulation of the HMGN5 variant lead to visible reduction in heterochromatin [24]. Because HMGNs binds to the acidic patch of the nucleosome core [38], some of the decondensation could be due to interference with chromatin condensation mediated by the core histone tails [71]. However, it is likely that the effects are mainly due to a reduction in the chromatin condensing activities of H1. As elaborated above, FRAP analyses of living cells showed that upregulation of HMGNs decreases the H1 chromatin residence time [53] and in vitro analyses with reconstituted chromatin and isolated proteins revealed that HMGN5 counteracts the chromatin-condensing activity of H1 [24, 72].
Loss of heterochromatin may have functional implications beyond changes in transcription. In most vertebrate nuclei, a layer of heterochromatin is positioned at the nuclear periphery, in contact with the nuclear lamina, which is known to provide mechanical support to the nuclear membrane. A recent study revealed that HMGN5 mediated chromatin decompaction reduced the mechanical sturdiness of the nucleus [73]. Analyses of various cells and transgenic mice overexpressing HMGN5 indicated that the HMGN5-mediated chromatin decompaction weakened the ability of the nuclear lamina to support nuclear sturdiness. Significantly, lamina disruption and reduced nuclear sturdiness were not due to alteration in gene expression. Although not directly proven, it is likely that HMGN5 up-regulation decompacts chromatin by interfering with the heterochromatin stabilization by H1. Thus, the H1-HMGN interplay may have direct effect on the physical properties of the nucleus.
Alterations in chromatin compaction could also facilitate cellular reorganizations necessary for efficient migration [74]. Increased chromatin condensation and H1 reorganization is observed in response to induction of directed cell migration. Conversely, chromatin decondensation inhibited the rate of cell migration, in a transcription-independent manner. Global chromatin condensation facilitates nuclear movement and reshaping, which are important for cell migration [75]. Taken together, the data suggest that chromatin compaction, which in part is modulated by the interplay between HMGs and H1, affects the mechanical integrity and the stiffness of the nucleus, a structural role for chromatin is not related to gene expression [73].
Numerous studies indicated that both HMGBs and HMGA impact chromatin structure and transcription. However, very few experiments addressed directly the possibility that these effects could in part be due to, or affected by an interplay between these HMGs and H1. Overexpression of HMGB1 or HMGB2 in cultured cells stimulates transcription from minichromosome templates. This transcriptional upregulation was accompanied by increased content of HMGB, a decreased content of H1, and by structural changes in the minichromosome templates [76]. The DDDDE sequence in acidic C-tail of HMGB1 was shown to be essential for transcriptional stimulation, most likely because it facilitates the binding of HMGB1 to linker DNA [77]. In addition, HMGB1 was shown to affect chromatin remodeling by facilitating the binding of the ISWI-containing remodeling factors ACF and CHRAC to chromatin [78], however it is not clear whether this process is associated with changes in H1.
HMGA proteins have been shown to antagonize histone H1-mediated repression of transcription at regions with unique properties. The transcription of the chromatin templates containing scaffold-associated regions (SARs) were preferentially repressed by H1 as compared to non-SAR control templates. HMGA protein relieved the transcription repression in a dose-dependent manner. In addition, upon micrococcal nuclease digestion of H1-depleted nuclei, HMGA1-associated nucleosomes which are positioned at transcription start sites are the first to be released from chromatin [51]. Furthermore, HMGA proteins assist to the assembly of enhanceosomes thereby facilitating transcriptional activation [79]. HMGAs contain multiple DNA-binding domains and multiple protein–protein interaction surfaces and potentially could interact with a wide range of nuclear factors, including ATP-dependent remodeling complexes [80]. It may be relevant that the AT-hook motif is an essential component of subunits of ATP-dependent chromatin remodeling complexes such as yeast Rsc-1 and Rsc-2 [81], and Drosophila NURF [82]. HMGA proteins have been shown to facilitate recruitment of chromatin remodeling complexes to gene regulatory regions; however, it is not clear whether this recruitment is related to changes in H1, which are usually depleted from regulatory sites.
Outlook
The relative abundance, ubiquitous distribution, and sequence conservation of H1 and HMG proteins suggests that they have an important biological function. Yet, despite numerous studies, the exact biological function of these architectural proteins remains one of the most perplexing aspects of chromatin biology. The ubiquitous presence of most of the variants of each family in most cells argues that each variant performs a specific function, yet numerous types of experiments show redundancy in chromatin binding. As elaborated above, a major obstacle in deciphering the biological function of nucleosome-binding architectural proteins is that they function within a dynamic network in which their chromatin binding and chromatin modifying activities are interdependent and a change in one component may trigger compensatory adjustments to establish a new steady state.
The increasing availability of mouse and other genetically modified animal models provide new insights into the biological function of individual architectural proteins. The animal models combined with genome wide studies and bioinformatics approaches provide novel insights into the mechanisms whereby the interplay between various members of the network of chromatin binding proteins, including the interplay between H1 and HMG proteins, modulate epigenetic regulatory process and impact the cellular phenotype. A major challenge is to determine how the interplay between specific H1 and HMG variants affects the structure and biological function of the genome.
Highlights.
HMGs and H1 proteins bind dynamically to chromatin and compete for nucleosome binding sites.
HMGs and H1 function as members of a protein network that fine tunes chromatin function
HMGs and H1 tend to have opposite effects on chromatin compaction and function.
Acknowledgments
The literature on H1 and HMGs is relatively large and we could not acknowledge and quote all the pertinent references. We do apologize to the authors which we did not quote. This research was supported by project #ZIA BC011154 and ZIA BC004496 from the Intramural Research Programs of the CCR, NCI, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Molecular and cellular biology. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustin M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends in biochemical sciences. 2001;26:152–153. doi: 10.1016/s0968-0004(00)01777-1. [DOI] [PubMed] [Google Scholar]
- 3.Bustin M. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends in biochemical sciences. 2001;26:431–437. doi: 10.1016/s0968-0004(01)01855-2. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Current opinion in genetics & development. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Reeves R. Nuclear functions of the HMG proteins. Biochimica et biophysica acta. 2010;1799:3–14. doi: 10.1016/j.bbagrm.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustin M. High mobility group proteins. Biochimica et biophysica acta. 2010;1799:1–2. doi: 10.1016/j.bbagrm.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Progress in nucleic acid research and molecular biology. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 8.Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–14787. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 9.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 10.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 11.Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Molecular cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nature structural & molecular biology. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Molecular and cellular biology. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catez F, Hock R. Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochimica et biophysica acta. 2010;1799:15–27. doi: 10.1016/j.bbagrm.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Postnikov Y, Bustin M. Regulation of chromatin structure and function by HMGN proteins. Biochimica et biophysica acta. 2010;1799:62–68. doi: 10.1016/j.bbagrm.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas JO, Stott K. H1 and HMGB1: modulators of chromatin structure. Biochemical Society transactions. 2012;40:341–346. doi: 10.1042/BST20120014. [DOI] [PubMed] [Google Scholar]
- 17.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 18.Ueda T, Yoshida M. HMGB proteins and transcriptional regulation. Biochimica et biophysica acta. 2010;1799:114–118. doi: 10.1016/j.bbagrm.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. The Journal of biological chemistry. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 20.Ueda T, Catez F, Gerlitz G, Bustin M. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Molecular and cellular biology. 2008;28:2872–2883. doi: 10.1128/MCB.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng T, Zhu ZI, Zhang S, Postnikov Y, Huang D, Horsch M, Furusawa T, Beckers J, Rozman J, Klingenspor M, Amarie O, Graw J, Rathkolb B, Wolf E, Adler T, Busch DH, Gailus-Durner V, Fuchs H, Hrabe de Angelis M, van der Velde A, Tessarollo L, Ovcherenko I, Landsman D, Bustin M. Functional compensation among HMGN variants modulates the DNase I hypersensitive sites at enhancers. Genome research. 2015 doi: 10.1101/gr.192229.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng T, Zhu ZI, Zhang S, Leng F, Cherukuri S, Hansen L, Marino-Ramirez L, Meshorer E, Landsman D, Bustin M. HMGN1 modulates nucleosome occupancy and DNase I hypersensitivity at the CpG island promoters of embryonic stem cells. Molecular and cellular biology. 2013;33:3377–3389. doi: 10.1128/MCB.00435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuddapah S, Schones DE, Cui K, Roh TY, Barski A, Wei G, Rochman M, Bustin M, Zhao K. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Molecular and cellular biology. 2011;31:700–709. doi: 10.1128/MCB.00740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, Bustin M. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Molecular cell. 2009;35:642–656. doi: 10.1016/j.molcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim JH, Catez F, Birger Y, West KL, Prymakowska-Bosak M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Molecular cell. 2004;15:573–584. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. The EMBO journal. 2005;24:3038–3048. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kugler JE, Deng T, Bustin M. The HMGN family of chromatin-binding proteins: dynamic modulators of epigenetic processes. Biochimica et biophysica acta. 2012;1819:652–656. doi: 10.1016/j.bbagrm.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochemical Society transactions. 2001;29:395–401. doi: 10.1042/bst0290395. [DOI] [PubMed] [Google Scholar]
- 29.Hayes JJ, Pruss D, Wolffe AP. Contacts of the globular domain of histone H5 and core histones with DNA in a “chromatosome”. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7817–7821. doi: 10.1073/pnas.91.16.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou BR, Feng H, Kato H, Dai L, Yang Y, Zhou Y, Bai Y. Structural insights into the histone H1-nucleosome complex. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu RM, Zhu P, Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 32.Zhou BR, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y. Structural Mechanisms of Nucleosome Recognition by Linker Histones. Molecular cell. 2015 doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. Journal of molecular biology. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 34.Th’ng JP, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. The Journal of biological chemistry. 2005;280:27809–27814. doi: 10.1074/jbc.M501627200. [DOI] [PubMed] [Google Scholar]
- 35.Raghuram N, Carrero G, Th’ng J, Hendzel MJ. Molecular dynamics of histone H1. Biochem Cell Biol. 2009;87:189–206. doi: 10.1139/O08-127. [DOI] [PubMed] [Google Scholar]
- 36.Postnikov YV, Trieschmann L, Rickers A, Bustin M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. Journal of molecular biology. 1995;252:423–432. doi: 10.1006/jmbi.1995.0508. [DOI] [PubMed] [Google Scholar]
- 37.Alfonso PJ, Crippa MP, Hayes JJ, Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. Journal of molecular biology. 1994;236:189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- 38.Kato H, van Ingen H, Zhou BR, Feng H, Bustin M, Kay LE, Bai Y. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12283–12288. doi: 10.1073/pnas.1105848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shick VV, Belyavsky AV, Mirzabekov AD. Primary organization of nucleosomes. Interaction of non-histone high mobility group proteins 14 and 17 with nucleosomes, as revealed by DNA-protein crosslinking and immunoaffinity isolation. Journal of molecular biology. 1985;185:329–339. doi: 10.1016/0022-2836(85)90407-3. [DOI] [PubMed] [Google Scholar]
- 40.Trieschmann L, Martin B, Bustin M. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5468–5473. doi: 10.1073/pnas.95.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zlatanova J, van Holde K. Binding to four-way junction DNA: a common property of architectural proteins? FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1998;12:421–431. [PubMed] [Google Scholar]
- 42.Zlatanova J, van Holde K. Linker histones versus HMG1/2: a struggle for dominance? BioEssays : news and reviews in molecular, cellular and developmental biology. 1998;20:584–588. doi: 10.1002/(SICI)1521-1878(199807)20:7<584::AID-BIES10>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 43.An W, van Holde K, Zlatanova J. The non-histone chromatin protein HMG1 protects linker DNA on the side opposite to that protected by linker histones. The Journal of biological chemistry. 1998;273:26289–26291. doi: 10.1074/jbc.273.41.26289. [DOI] [PubMed] [Google Scholar]
- 44.Nightingale K, Dimitrov S, Reeves R, Wolffe AP. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. The EMBO journal. 1996;15:548–561. [PMC free article] [PubMed] [Google Scholar]
- 45.Cato L, Stott K, Watson M, Thomas JO. The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. Journal of molecular biology. 2008;384:1262–1272. doi: 10.1016/j.jmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 47.Polanska E, Pospisilova S, Stros M. Binding of histone H1 to DNA is differentially modulated by redox state of HMGB1. PloS one. 2014;9:e89070. doi: 10.1371/journal.pone.0089070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hill DA, Pedulla ML, Reeves R. Directional binding of HMG-I(Y) on four-way junction DNA and the molecular basis for competitive binding with HMG-1 and histone H1. Nucleic acids research. 1999;27:2135–2144. doi: 10.1093/nar/27.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeves R, Nissen MS. Interaction of high mobility group-I (Y) nonhistone proteins with nucleosome core particles. The Journal of biological chemistry. 1993;268:21137–21146. [PubMed] [Google Scholar]
- 50.Saitoh Y, Laemmli UK. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 51.Zhao K, Kas E, Gonzalez E, Laemmli UK. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. The EMBO journal. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Molecular and cellular biology. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO reports. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie D, Chen CC, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome research. 2010;20:804–815. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Croston GE, Kerrigan LA, Lira LM, Marshak DR, Kadonaga JT. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 57.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 58.Harrer M, Luhrs H, Bustin M, Scheer U, Hock R. Dynamic interaction of HMGA1a proteins with chromatin. Journal of cell science. 2004;117:3459–3471. doi: 10.1242/jcs.01160. [DOI] [PubMed] [Google Scholar]
- 59.Karpov VL, Preobrazhenskaya OV, Mirzabekov AD. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5′ region. Cell. 1984;36:423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- 60.Kimura T, Mills FC, Allan J, Gould H. Selective unfolding of erythroid chromatin in the region of the active beta-globin gene. Nature. 1983;306:709–712. doi: 10.1038/306709a0. [DOI] [PubMed] [Google Scholar]
- 61.Kamakaka RT, Thomas JO. Chromatin structure of transcriptionally competent and repressed genes. The EMBO journal. 1990;9:3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Postnikov YV, Shick VV, Belyavsky AV, Khrapko KR, Brodolin KL, Nikolskaya TA, Mirzabekov AD. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic acids research. 1991;19:717–725. doi: 10.1093/nar/19.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding HF, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Molecular and cellular biology. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trieschmann L, Postnikov YV, Rickers A, Bustin M. Modular structure of chromosomal proteins HMG-14 and HMG-17: definition of a transcriptional enhancement domain distinct from the nucleosomal binding domain. Molecular and cellular biology. 1995;15:6663–6669. doi: 10.1128/mcb.15.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 66.Paranjape SM, Krumm A, Kadonaga JT. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes & development. 1995;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 67.Hill DA, Imbalzano AN. Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry. 2000;39:11649–11656. doi: 10.1021/bi001330z. [DOI] [PubMed] [Google Scholar]
- 68.Horn PJ, Carruthers LM, Logie C, Hill DA, Solomon MJ, Wade PA, Imbalzano AN, Hansen JC, Peterson CL. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nature structural biology. 2002;9:263–267. doi: 10.1038/nsb776. [DOI] [PubMed] [Google Scholar]
- 69.Hill DA, Peterson CL, Imbalzano AN. Effects of HMGN1 on chromatin structure and SWI/SNF-mediated chromatin remodeling. The Journal of biological chemistry. 2005;280:41777–41783. doi: 10.1074/jbc.M509637200. [DOI] [PubMed] [Google Scholar]
- 70.Rattner BP, Yusufzai T, Kadonaga JT. HMGN proteins act in opposition to ATP-dependent chromatin remodeling factors to restrict nucleosome mobility. Molecular cell. 2009;34:620–626. doi: 10.1016/j.molcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC. The role of the nucleosome acidic patch in modulating higher order chromatin structure. Journal of the Royal Society, Interface/the Royal Society. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malicet C, Rochman M, Postnikov Y, Bustin M. Distinct properties of human HMGN5 reveal a rapidly evolving but functionally conserved nucleosome binding protein. Molecular and cellular biology. 2011;31:2742–2755. doi: 10.1128/MCB.05216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, Bustin M. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nature communications. 2015;6:6138. doi: 10.1038/ncomms7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerlitz G, Bustin M. Efficient cell migration requires global chromatin condensation. Journal of cell science. 2010;123:2207–2217. doi: 10.1242/jcs.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerlitz G, Bustin M. The role of chromatin structure in cell migration. Trends in cell biology. 2011;21:6–11. doi: 10.1016/j.tcb.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ogawa Y, Aizawa S, Shirakawa H, Yoshida M. Stimulation of transcription accompanying relaxation of chromatin structure in cells overexpressing high mobility group 1 protein. The Journal of biological chemistry. 1995;270:9272–9280. doi: 10.1074/jbc.270.16.9272. [DOI] [PubMed] [Google Scholar]
- 77.Ueda T, Chou H, Kawase T, Shirakawa H, Yoshida M. Acidic C-tail of HMGB1 is required for its target binding to nucleosome linker DNA and transcription stimulation. Biochemistry. 2004;43:9901–9908. doi: 10.1021/bi035975l. [DOI] [PubMed] [Google Scholar]
- 78.Bonaldi T, Langst G, Strohner R, Becker PB, Bianchi ME. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. The EMBO journal. 2002;21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Merika M, Thanos D. Enhanceosomes. Current opinion in genetics & development. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 80.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 81.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Molecular cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 82.Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Molecular cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]