Abstract
Background and aims
Proton pump inhibitors (PPIs) are among the top 10 most widely used drugs in the world. PPI use has been associated with an increased risk of enteric infections, most notably Clostridium difficile. The gut microbiome plays an important role in enteric infections, by resisting or promoting colonisation by pathogens. In this study, we investigated the influence of PPI use on the gut microbiome.
Methods
The gut microbiome composition of 1815 individuals, spanning three cohorts, was assessed by tag sequencing of the 16S rRNA gene. The difference in microbiota composition in PPI users versus non-users was analysed separately in each cohort, followed by a meta-analysis.
Results
211 of the participants were using PPIs at the moment of stool sampling. PPI use is associated with a significant decrease in Shannon's diversity and with changes in 20% of the bacterial taxa (false discovery rate <0.05). Multiple oral bacteria were over-represented in the faecal microbiome of PPI-users, including the genus Rothia (p=9.8×10−38). In PPI users we observed a significant increase in bacteria: genera Enterococcus, Streptococcus, Staphylococcus and the potentially pathogenic species Escherichia coli.
Conclusions
The differences between PPI users and non-users observed in this study are consistently associated with changes towards a less healthy gut microbiome. These differences are in line with known changes that predispose to C. difficile infections and can potentially explain the increased risk of enteric infections in PPI users. On a population level, the effects of PPI are more prominent than the effects of antibiotics or other commonly used drugs.
Keywords: INTESTINAL BACTERIA, ENTERIC INFECTIONS, PROTON PUMP INHIBITION
Significance of this study.
What is already known on this subject
Proton pump inhibitors (PPI) use is associated with increased risk of enteric infections, in particular with a 65% increase in incidence of Clostridium difficile infection.
PPI is one of the most commonly used drugs.
Changes in the gut microbiome can resist or promote the colonisation of enteric infections.
What are the new findings
PPI use is associated with decreased bacterial richness and profound changes in the gut microbiome: 20% of the identified bacteria in this study showed significant deviation.
Oral bacteria and potential pathogenic bacteria are increased in the gut microbiota of PPI users.
On the population level we see more microbial alterations in the gut associated with PPI use than with antibiotics or other drug use.
How might it impact on clinical practice in the foreseeable future?
Given the widespread use of PPI, the morbidity and mortality associated with enteric infections, and the increasing number of studies investigating the microbiome, healthcare practitioners and researchers should take into consideration the influence of PPI on the gut microbiome.
Background and aims
Proton pump inhibitors (PPIs) are among the top 10 most widely used drugs in the world. In 2013, 7% of the population of the Netherlands used omeprazole. In the same year, esomeprazole was the second largest drug in terms of revenue in the USA.1 2 PPIs are used to treat GORD and to prevent gastric and duodenal ulcers.3 4 Of the general population, 25% report having heartburn at least once a month, explaining the large demand for PPIs.4 Nevertheless, PPIs are frequently prescribed or taken for long periods without evidence-based indication.5 6
PPI use has been associated with increased risk of enteric infections.5 7–9 A meta-analysis of 23 studies, comprising almost 300 000 patients, showed a 65% increase in the incidence of Clostridium difficile-associated diarrhoea among patients who used PPI.9 In healthcare-related settings, PPI use also increases the risk of recurrent C. difficile infections.5 Another meta-analysis of 11 280 patients, from six studies evaluating Salmonella, Campylobacter and other enteric infections, also found an increased risk due to acid suppression, with a greater association with PPI than with H2-receptor antagonists.8 Recently, the Dutch National Institute for Public Health and the Environment noticed a marked increase in the occurrence of campylobacteriosis associated with increased PPI use in the Netherlands.7
The gut microbiome plays an important role in these enteric infections.10–13 Gut microbiota can resist or promote the microbial colonisation of the gut by C. difficile and other enteric infections through several mechanisms that either directly inhibit bacterial growth or enhance the immune system.10 11 Moreover, substituting the gut microbiota of patients with C. difficile-associated diarrhoea with a healthy microbiome through faecal transplantation has been proven to cure C. difficile infection.14 The increased incidence of enteric infections in PPI users and the importance of the gut microbiome composition in the development of these infections led us to investigate the influence of PPI use on the gut microbiome.
Methods
Cohorts
We studied the effect of PPI use on the gut microbial composition in three independent cohorts from the Netherlands. These cohorts together comprise 1815 adult individuals, including healthy subjects and patients with GI diseases. Cohort 1 consists of 1174 individuals who participate in the general population study LifeLines-DEEP in the northern provinces of the Netherlands.15 Cohort 2 consists of 300 patients with IBD from the department of Gastroenterology and Hepatology University Medical Center Groningen (UMCG), the Netherlands. Cohort 3 consists of 189 patients with IBS and 152 matched controls from Maastricht University Medical Center (MUMC), the Netherlands.
Medication use
Current medication use at the time of stool collection of Cohort 1 participants was extracted from a standardised questionnaire.16 Two medical doctors reviewed all the medication for 1174 participants. PPI use was scored if participants used omeprazole, esomeprazole, pantoprazole, lansoprazole, dexlansoprazole or rabeprazole. To exclude other possible drug effects on the gut microbiome, medication use was scored in eight categories, allowing for later correction of parameters or exclusion of certain participants. These categories were medication that: (1) changes bowel movement or stool frequency, (2) lowers triglyceride levels, (3) lowers cholesterol levels, (4) anti-diabetic medication (oral and insulin), (5) systemic anti-inflammatory medication (excluding NSAIDs), (6) topical anti-inflammatory medication, (7) systemic antibiotics, including antifungal and antimalarial medication, and (8) antidepressants including serotonin-specific reuptake inhibitors (SSRIs), serotonin-norepinephrine reuptake inhibitors (SNRIs), mirtazapine, and tricyclic antidepressants (TCAs). The definitions of these categories are described in the online supplementary appendix. Analysis of drugs used in Cohort 2 was based on the IBD-specific electronic patient record in the UMCG. Current PPI use, as well as current IBD medication (mesalazines, thiopurines, methotrexate, steroids, tumour necrosis factor (TNF)-α inhibitors and other biologicals) were scored at the time of sampling by the gastroenterologist treating the patient with IBD. Current PPI consumption in the IBS case-control Cohort 3 was based on self-reported questionnaires. Pseudonymised data for all three cohorts was provided to the researchers.
Gut complaints and other clinical characteristics
Information on age, gender and body mass index (BMI) was available for all three cohorts. In Cohort 1, gut complaints were investigated using an extensive questionnaire that included defecation frequency and the Bristol Stool Scale. Possible IBS and functional diarrhoea or constipation were determined using self-reported ROME III criteria. The patients with IBD in Cohort 2 were diagnosed based on accepted radiological, endoscopic and histopathological evaluation. All the IBD cases included in our study fulfilled the clinical criteria for IBD. IBS in Cohort 3 was diagnosed by a gastroenterologist according to the ROME III criteria.
Stool and oral cavity mucus sample collection
A total of 1815 stool samples and 116 oral cavity mucus samples were collected. Cohorts 1 and 2 used identical protocols to collect the stool samples. Participants of cohort 1 and 2 were asked to collect one stool sample at home. Stool samples were frozen within 15 min after stool production in the participants’ home freezer and remained frozen until DNA isolation. A research nurse visited all participants to collect the stool samples shortly after production and they were transported and stored at –80°C. Participants of cohort 3 were asked to bring a stool sample to the research facility within 24 h after stool production. These samples were immediately frozen upon arrival at −80°C.
Oral cavity mucus samples were collected from 116 additional healthy volunteers using buccal swabs.
DNA isolation and analysis of microbiota composition
Microbial DNA from stool samples was isolated with the Qiagen AllPrep DNA/RNA Mini Kit (cat. # 80204). DNA isolation from oral cavity swabs was performed using the UltraClean microbial DNA isolation kit (cat.# 12224) from MoBio Laboratories (Carlsbad, California, USA). To determine the bacterial composition of the stool and oral cavity mucus samples, sequencing of the variable region V4 of the 16S rRNA gene was performed using Illumina MiSeq. DNA isolation is described in the Methods section of the online supplementary appendix.
Taxonomy determination
Bacterial taxonomy was determined by clustering the sequence reads with UCLUST (V.1.2.22q) with a distance threshold of 97%, using Greengenes (V.13.8) as the taxonomy reference database. Sequencing and the determination of taxonomy are described in the Methods section of the online supplementary appendix.
Statistical analysis
In each cohort, differentially abundant taxa in the gut microbiome between PPI users and non-PPI users were analysed using the multivariate statistical framework MaAsLin.17 MaAsLin performs boosted, additive, general linear models between metadata and microbial abundance data. After running the association studies in the individual cohorts, we performed a meta-analysis of the three cohorts, using the weighted Z-score method. The Cochran's Q test was used to check for heterogeneity. The significance cut-off for the Cochran's Q test was determined by Bonferroni correction for the 92 significant results: p<5.43×10−4. Differences in richness (the number of species within a sample), principal coordinate analyses (PCoA) and Shannon diversity analysis were determined using the QIIME microbiome analysis software.18 The Wilcoxon test and Spearman's correlations were used to identify differences in Shannon's diversity and relations between the PCoA scores of PPI users and non-PPI users, while the χ2 test, Fisher's exact test, Spearman's correlation and Wilcoxon-Mann-Whitney test (WMW test) were used to determine differences in age, gender, BMI, antibiotics use, and gut complaints between PPI users and non-users. In all the microbiome analyses, multiple test corrections were based on the false discovery rate (FDR). An FDR value of 0.05 was used as a statistically significant cut-off.
In addition to the PPI effect, we also tested the influence of other commonly used drugs in Cohort 1. Using MaAsLin with similar settings to those described above, we tested the microbial changes associated with the use of other drugs, with and without correction for PPI, and the changes when including these common drugs as a correcting factor in the PPI versus non-PPI analysis.
Significant results were graphically represented in cladograms using GraPhlAn.19 More details on the statistical analysis can be found in the Methods section (see online supplementary appendix).
Correction for factors influencing the gut microbiota
Differentially abundant taxa were corrected for several parameters, which were identified by statistical analysis of cohort phenotypes or univariate MaAsLin runs and subsequently added as cofactors to the additive linear model. Analyses in the general population Cohort 1 were corrected for age, gender, BMI, antibiotics use, sequence read depth and ROME III diagnosis (IBS-Constipation (IBS-C), IBS-Diarrhoea (IBS-D), IBS-Mixed (IBS-M), IBS-Undetermined (IBS-U), functional bloating, functional constipation, functional diarrhoea or none). The analysis of patients with IBD in Cohort 2 was corrected for age, gender, BMI, antibiotics use, sequence read depth, diagnosis (Crohn's disease or UC) combined with disease location (colon, ileum or both) and IBD medication (use of mesalazines, steroids, thiopurines, methotrexate or anti-TNF antibodies). The analysis of the IBS case-control Cohort 3 was corrected for age, gender, BMI, sequence read depth and IBS status according to the ROME III criteria. In the meta-analysis, all microbiome data were corrected for age, gender, BMI, antibiotics use and sequence read depth.
Results
PPI use is associated with older age and higher BMI
PPI were used by 211 (11.6%) of the 1815 participants: 8.4% of the general population (Cohort 1), 20.0% of the patients with IBD (Cohort 2) and 15.2% of the participants of case-control Cohort 3. Women use PPI more often than men: 9.2% versus 7.4%, albeit this was not statistically significant (p=0.61, χ2 test). PPI users were generally older: 51.6 (SD 13.4) years of age versus 44.4 (SD 14.7) years of age (p=2.50×10-11 WMW test) and have a higher BMI of 26.9 (SD 5.0) versus 24.9 (SD 4.2) for non-users (p=1.89×10−8, WMW test). Antibiotics were concomitantly used by 2% of the 99 PPI users of Cohort 1 and 33% of the 60 PPI users of Cohort 2. There was no overlap between PPI users and antibiotics users in Cohort 3. Based on our data, we included age, gender, BMI and antibiotics as cofactors in the microbiome analyses. Table 1 provides an overview of the characteristics per cohort and the use of PPI.
Table 1.
Characteristics of the three independent cohorts in this study
| Cohort 1: LifeLines-DEEP (general population) |
Cohort 2: patients with IBD UMCG |
Cohort 3: IBS case-control study MUMC |
||||
|---|---|---|---|---|---|---|
| PPI users (n=99) |
Non-PPI users (n=1075) | PPI users (n=60) |
Non-PPI users (n=240) | PPI users (n=52) |
Non-PPI users (n=289) | |
| Average (SD)* | Average (SD)* | Average (SD)* | Average (SD)* | Average (SD)* | Average (SD)* | |
| Age | 51.94 (13.59) | 44.79 (13.58) | 50.87 (14.49) | 42.45 (14.57) | 51.94 (14.27) | 44.57 (18.24) |
| BMI | 27.73 (5.10) | 25.05 (4.03) | 26.14 (5.53) | 25.58 (4.72) | 26.24 (4.10) | 24.16 (4.11) |
| Gender (% male) | 36.36 | 42.05 | 61.67 | 39.17 | 30.77 | 33.56 |
| Reads per sample | 48879 (43 001) | 55 884 (40057) | 51 081 (43 990) | 52 970 (37787) | 43 807 (28604) | 65842 (119296) |
| Antibiotics (%) | 2.02 | 1.02 | 31.67 | 16.67 | 0.00 | 1.73 |
| IBD (%) | 0.00 | 0.00 | 100.00 | 100.00 | 0.00 | 0.00 |
| IBS (%) | 34.34 | 25.77 | 0.00 | 0.00 | 90.38 | 49.48 |
| Diarrhoea (%) (IBS-D and functional diarrhoea together) | 7.07 | 4.47 | – | – | 28.4 | 17.3 |
| Average bowel movements per day | 1.36 (0.53) | 1.38 (0.61) | – | – | 1.60 (0.81) | 1.92 (1.11) |
| Anti-TNF-a (%) | – | – | 38.33 | 28.75 | – | – |
| Mesalazine (%) | –- | –- | 26.67 | 39.58 | – | – |
| Methotrexate (%) | – | – | 16.67 | 5.42 | – | – |
| Steroids (%) | – | – | 30.00 | 20.42 | – | – |
| Thiopurines (%) | –- | – | 21.67 | 37.08 | – | – |
*Unless otherwise stated
IBD, Inflammatory Bowel Disease; IBS, Irritable Bowel Syndrome; SD, standard deviation, BMI, body mass index.
MUMC, Maastricht University Medical Center; PPI, proton pump inhibitor, TNF-a, tumour necrosis factor; UMCG, University Medical Center Groningen.
Composition of the gut microbiota
The predominant phylum in each cohort was Firmicutes with abundances of 76.7%, 73.8% and 77.4% in Cohorts 1, 2 and 3, respectively. Information on the composition of the gut microbiome for all three cohorts and on all taxonomic levels is provided in online supplementary figures S1, S2 and table S1. Independent of PPI use, the overall high-level bacterial composition of the gut was homogeneous in all three cohorts (by phylum, class and order level, Spearman's correlations: r>0.94; p<1.6×10−13).
Reduced diversity of the gut microbiome associated with PPI use
In all three cohorts we identified a lower species richness and lower Shannon diversity, although not significant (Cohort 1, p=0.85 ; Cohort 2, p=0.16; Cohort 3, p=0.53), however in combined analysis of all three data sets we identified, moderate but significant decrease in gut α diversity of PPI users was observed in the meta-analysis of all 1815 gut microbiome samples: Shannon index (p=0.01) and species richness (p=0.02) (see online supplementary figures S3 and S4).
Meta-analysis: differences in gut microbiome associated with PPI use
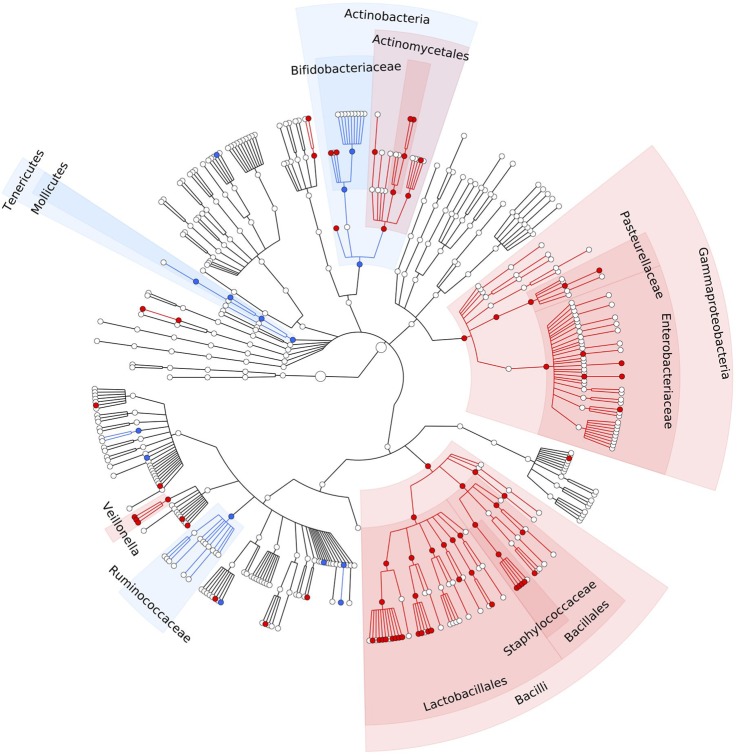
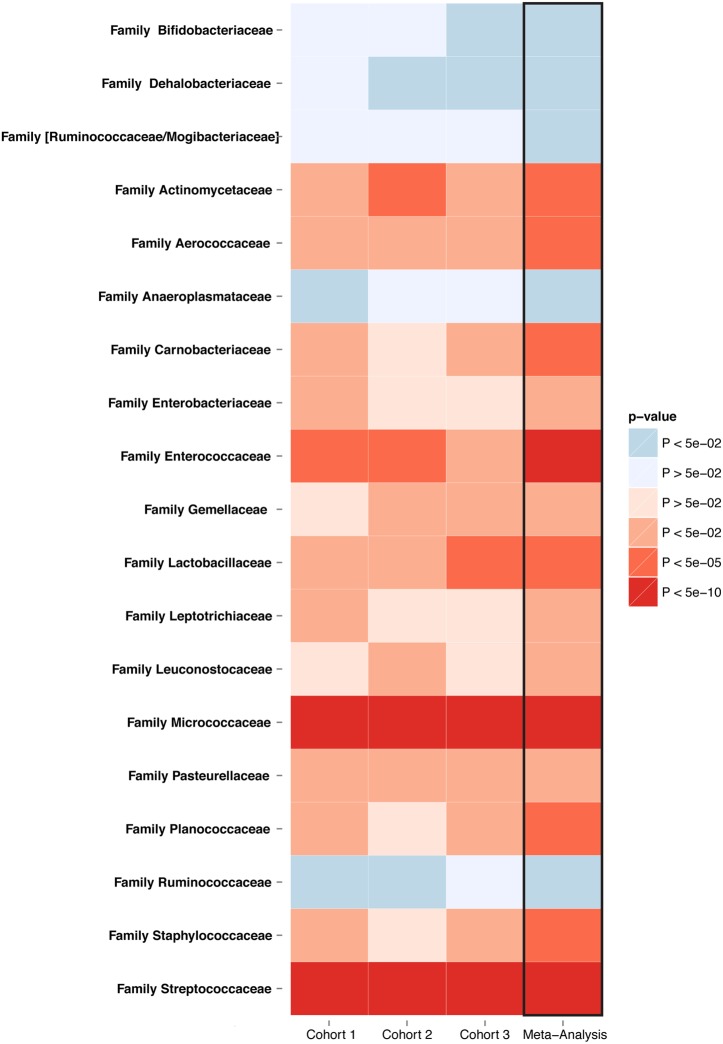
The meta-analysis across all three cohorts showed statistically significant alterations in 92 of the 460 bacterial taxa abundance (FDR<0.05). These changes are depicted in a cladogram in figure 1 and in a heatmap in figure 2, and in online supplementary figure S5. Details of each taxon, including the individual direction, coefficient, p value and FDR for each cohort, as well as the meta-analysis, are provided in online supplementary tables S2 and S3. Cochran's Q test was used to check for heterogeneity. None of the 92 reported associations were significantly heterogeneous at the Bonferroni corrected p value cut-off (p<5.43×10−4) (see online supplementary table S2).
Figure 1.
PPI-associated statistically significant differences in the gut microbiome. Meta-analysis of three independent cohorts comprising 1815 faecal samples, showing a cladogram (circular hierarchical tree) of 92 significantly increased or decreased bacterial taxa in the gut microbiome of PPI users compared with non-users (FDR<0.05). Each dot represents a bacterial taxon. The two innermost dots represent the highest level of taxonomy in our data: the kingdoms Archea and Bacteria (prokaryotes), followed outwards by the lower levels: phylum, class, order, family, genus and species. Red dots represent significantly increased taxa. Blue dots represent significantly decreased taxa. FDR, false discovery rate; PPI, proton pump inhibitor.
Figure 2.
Significantly altered families in PPI users consistent in three cohorts. Meta-analysis of three independent cohorts comprising 1815 faecal samples. The heatmap shows 19 families significantly increased or decreased associated with PPI use in the gut microbiome for each cohort and for the meta-analysis (meta-analysis FDR<0.05). FDR, false discovery rate; PPI, proton pump inhibitor.
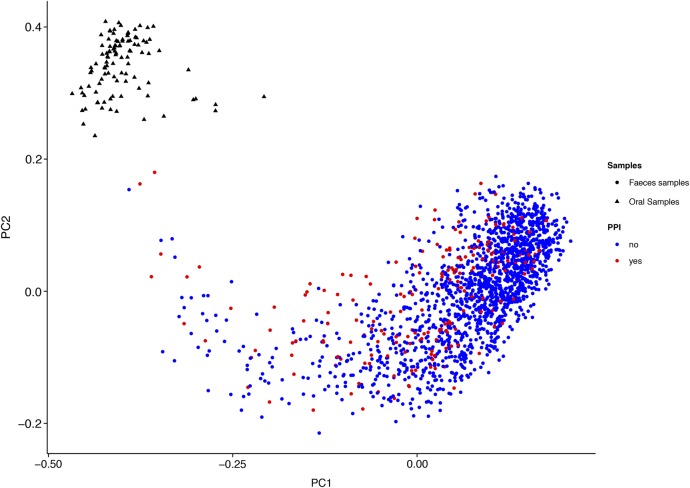
The overall difference of the gut microbiome associated with PPI use was also observed in the PCoA of all the data sets together (figure 3 and see online supplementary figure S6). The same PCoA with separate colours for each cohort has been added in online supplementary figure S7. Notably, we observed statistically significant differences between PPI users and non-users in two principal coordinates (PCoA1: p=1.39×10−20, PCoA3: p=0.0004, Wilcoxon test).
Figure 3.
Principal coordinate analysis of 1815 gut microbiome samples and 116 oral microbiome samples. The gut microbiome of PPI users is significantly different from non-PPI users in the first coordinate (PCoA1: p=1.39×10−20, Wilcoxon test). For Principal Coordinate 1 there is a significant shift of the gut microbiome of PPI users towards the oral microbiome. PCoA, principal coordinate analysis; PPI, proton pump inhibitor.
Similar changes in three independent cohorts were associated with PPI use
The order Actinomycetales, families Streptococcoceae, Micrococcoceae, genus Rothia and species Lactobacillus salivarius were increased in participants using PPI in each cohort. None of the individual cohorts contained any significantly decreased taxa (FDR<0.05). In the general population (Cohort 1), 41 of the 829 bacterial taxa were significantly increased, including the class Gammaproteobacteria, the family Enterococcoceae and the genera Streptococcus, Veillonella and Enterococcus (FDR<0.05) (see online supplementary table S4). In patients with IBD (Cohort 2), PPI use was associated with an increase of 12 of the 667 bacterial taxa, including the family Lactobacillaceae as well as the genera Streptococcus and Lactobacillus (FDR<0.05) (see online supplementary table S5). In IBS case-control Cohort 3, 18 of the 624 taxa were significantly increased, including the order Lactobacillales (FDR<0.05) (see online supplementary table S6).
Oral cavity bacteria are more abundant in the gut microbiome of PPI users
We hypothesised that the changes in the gut microbiome associated with PPI use are caused by reduced acidity of the stomach and the subsequent survival of more bacteria that are ingested with food and oral mucus. Indeed, some of the statistically significantly increased bacteria in PPI users (eg, Rothia dentocariosa, Rothia mucilaginosa, the genera Scardovia and Actinomyces and the family Micrococcaceae) are typically found in the oral microbiome.20 By analysing 116 oral microbiome samples from participants in Cohort 1, we could compare the overall composition of bacteria in the oral microbiome with the composition of the gut microbiome. We observed a statistically significant shift in Principal Coordinate 1 in the gut microbiome samples of the PPI users towards the oral samples, compared with non-PPI users (p=1.39×10−20, Wilcoxon test) (figure 3). In online supplementary figure S8, the over-representation of oral cavity bacteria in the guts of PPI users is depicted in a cladogram.
PPI use is independent of bowel movement frequency and stool consistency
Some of the significantly increased taxa were more abundant in the small intestine.11 To ensure that the changes observed in microbiota composition were not due to diarrhoea and/or more frequent bowel movements, we checked in our general population whether clinical symptoms of diarrhoea were more often present in PPI users. Neither diarrhoeal complaints (IBS-D and functional diarrhoea, p=0.22, Fisher's exact test), stool consistency as defined by the Bristol Stool Scale (r=0.027, p=0.36, Spearman's correlation) nor the defecation frequency (r=−0.001, p=0.98, Spearman's correlation) of the participants in Cohort 1 were related to PPI use.
PPI, antibiotics and other commonly used drugs
In Cohort 1, 16 taxa were associated with antibiotics and other commonly used drug categories besides PPI (see online supplementary table S7). After correction for PPI use, only six taxa remained associated with certain drugs: statins, fibrates and drugs that change bowel movements. All 92 alterations in bacterial taxa associated with PPI use remained statistically significant if we correct the microbiome analyses for antibiotics and other commonly used drugs.
Conclusions
We show that PPI use is consistently associated with profound changes in the gut microbiome. In our study, these changes were more prominent than changes associated with either antibiotics or other commonly used drugs. While PPIs have proven to be useful in the prevention and treatment of ulcers and GORD, they have also been associated with an increased risk of C. difficile, Salmonella spp, Shigella spp, Campylobacter spp, and other enteric infections.4 5 7–9 The increased risk of acquiring one of these enteric infections is likely due to changes in the PPI user's gut microbiome. Gut microbiota can resist or promote colonisation of C. difficile and other enteric infections through mechanisms that either directly inhibit bacterial growth or enhance the immune system.10–13 In the case of C. difficile, spores might be able to germinate more easily because of metabolites synthesised by certain gut bacteria.12 13
We hypothesised that PPIs change the gut microbiome through their direct effect on stomach acid. This acidity forms one of the main defenses against the bacterial influx that accompanies ingesting food and oral mucus. PPIs reduce the acidity of the stomach, allowing more bacteria to survive this barrier. We have shown here that species in the oral microbiome are more abundant in the gut microbiome of PPI users. Moreover, a study looking into the effect of PPIs on the oesophageal and gastric microbiome in oesophagitis and Barret's oesophagus showed similar bacterial taxa associated with PPI use, including increased levels of Enterobacteriaceae, Micrococcaceae, Actinomycetaceae and Erysipelotrichaceae.21 Gastric bypass surgery compromises the stomach acid barrier and leads to gut microbiome changes similar to the PPI-associated alterations in this study, thereby supporting our hypothesis.22
We looked at the role of the gut microbiome in C. difficile infections, which cause 12.1% of all nosocomial infections and were responsible for half a million infections and associated with 29 000 deaths in the USA in 2011.23 24 Virulent strains of C. difficile can only colonise a susceptible gut, after which toxins are produced and spores are shed. This leads to a wide spectrum of symptoms varying from mild diarrhoea to fulminant relapsing diarrhoea and pseudomembranous colitis.25 Recent human, animal and in vitro studies show an overlap between the specific alterations in the gut microbiota associated with PPI use found in this study and bacterial changes that lead to increased susceptibility to C. difficile. The reduced α diversity in PPI-users is associated with increased susceptibility to C. difficile infection.13 27 28 The PPI-associated decreases of the family Ruminococcoceae and the genus Bifidobacterium, as well as the PPI-associated increases of the class Gammaproteobacteria, the families Enterobacteriaceae, Enterococcoceae, Lactobacillaceae and the genera Enterococcus and Veillonella, have been consistently linked to increased susceptibility to C. difficile infection (table 2).10 13 26–32
Table 2.
Taxa and microbiome aspects associated with PPI use and increased risk of C. difficile infection
| Taxa or microbiome aspect | Direction in PPI users that increases the risk of C. difficile infection | References of role on risk of C. difficile infection |
|---|---|---|
| α diversity | Reduced | Buffie et al13
Nature. 2015 Chang et al27 Journal of Infectious Diseases. 2008 Antharam et al28 Journal of Clinical Microbiology. 2013 |
| k__Bacteria p__Firmicutes c__Clostridia o__Clostridiales f__Ruminococcaceae |
Decreased | Reeves et al26
Gut Microbes. 2011 Antharam et al28 Journal of Clinical Microbiology. 2013. Buffie et al.13 Nature 2015 (Extended Figure 3d and 3e) Schubert et al31 Mbio. 2014. Rea et al29 Journal of Clinical Microbiology. 2011. |
| k__Bacteria p__Actinobacteria c__Actinobacteria o__Bifidobacteriales f__Bifidobacteriaceae g__Bifidobacterium |
Decreased | Buffie et al10
Nature Reviews Immunology. 2013 Rea et al29 Journal of Clinical Microbiology. 2011 Baines et al30 Journal of Antimicrobial Chemotherapy. 2013 |
| k__Bacteria p__Firmicutes c__Bacilli o__Lactobacillales f__Enterococcaceae g__Enterococcus |
Increased | Antharam et al28
Journal of Clinical Microbiology. 2013 Schubert et al31 Mbio. 2014 Rea et al29 Journal of Clinical Microbiology. 2011 (Figure 4) Buffie et al13 Nature. 2015 (Extended Figure 3d and 3e) Baines et al30 Journal of Antimicrobial Chemotherapy. 2013 |
| k__Bacteria p__Firmicutes c__Bacilli o__Lactobacillales f__Lactobacillaceae, g__Lactobacillus, s__delbrueckii, s__plantarum and s__reuteri |
Increased | Reeves et al26
Gut Microbes. 2011 Antharam et al28 Journal of Clinical Microbiology. 2013 Rea et al 29 Journal of Clinical Microbiology. 2012 Buffie et al10 Nature Reviews Immunology. 2013 Buffie et al13 Nature. 2015 |
| k__Bacteria p__Firmicutes c__Clostridia o__Clostridiales f__Veillonellaceae g__Veillonella |
Increased | Antharam et al28 The Journal of Clinical Microbiology. 2013 |
| k__Bacteria p__Proteobacteria c__Gammaproteobacteria o__Enterobacteriales f__Enterobacteriaceae g__Escherichia s__coli |
Increased | Antharam et al28
Journal of Clinical Microbiology. 2013 Reeves et al26 Gut Microbes. 2011 Schubert et al31 Mbio. 2014 Peterfreund et al32 PLOS ONE. 2012 |
PPI, proton pump inhibitor.
k__, kingdom; p__, phylum; c__, class; o__, order; f__, family; g__, genus; s__, species
associations are in bold
The Ruminococcaceae family is significantly decreased in patients with C. difficile and enriched in healthy controls.28 29 31 Moreover, mice that have been treated with a mixture of antibiotics that do not become clinically ill after a challenge with C. difficile have higher levels of Ruminococcaceae.26 Within the Ruminococcaceae family, the Faecalibacterium genus was significantly increased in patients who recovered from C. difficile illness, whereas it was severely decreased in patients with C. difficile with active disease.31 Last, a decreased Ruminococcus torques operational taxonomic unit (OTU) was significantly associated with C. difficile infection in another study, although their OTU picking was done using a different reference database and associations were performed using OTU level, making direct comparisons with our study difficult.13
Species of the Bifidobacterium genus: Bifidobacterium longum, Bifidobacterium lactis, Bifidobacterium pseudocatenulatum, Bifidobacterium breve, Bifidobacterium pseudolongum, Bifidobacterium adolescentis and Bifidobacterium animalis lactis have been shown to inhibit or prevent C. difficile infection.10 The administration of antibiotics that enhance the susceptibility to C. difficile in an in vitro model of the gut also significantly reduce the genus Bifidobacterium.30 Moreover, active C. difficile diarrhoea is associated with decreased Bifidobacteria in elderly patients.29
The class Gammaproteobacteria and the family Enterobacteriaceae are significantly increased in PPI users. Gammaproteobacteria are enriched in patients with C. difficile compared with healthy controls.28 Within the class Gammaproteobacteria, the family Enterobacteriaceae dominate the murine gut microbiome after administration of clindamycin. Those mice that became clinically ill after the administration of an antibiotic cocktail containing clindamycin and a C. difficile challenge, had profoundly increased levels of Enterobacteriaceae in their gut microbiome, while mice that did not become clinically ill had a gut microbiome that predominantly consisted of Firmicutes.26 The family Enterobacteriaceae is also increased in hamsters that were treated with clindamycin and subsequently infected with C. difficile.32
The Enterococcus genus, which is also more abundant in PPI users, is significantly enriched in C. difficile-infected patients compared with healthy controls.28 31 An Enterococcus faecalis OTU and an Enterococcus avium OTU are significantly associated with increased susceptibility to C. difficile infections in mice.13 Moreover, an Enterococcus avium OTU is also significantly associated with C. difficile in humans.13 The administration of the antibiotic ceftriaxone led to an increase in the genus Enterococcus and enhanced the susceptibility to C. difficile in an in vitro model of the gut.30
The increased abundance of the family Lactobacillaceae in PPI users was associated with increased risk of C. difficile infection in several studies. Mice treated with a cocktail of antibiotics (consisting of kanamycin, gentamycin, colistin, metronidazole and vancomycin), cefoperazone or a combination of clindamycin and cefoperazone have higher levels of Lactobacillaceae in their gut.26 Mice treated with cefoperazone and clindamycin that developed C. difficile infection after being challenged with the pathogen also had a higher level of Lactobacillaceae.26 Within the Lactobacillaceae family, the Lactobacillus genus is significantly enriched in patients with C. difficile infection compared with healthy controls.28 Lactobacillus spp in the gut microbiome are also associated with active C. difficile diarrhoea in patients.29 In contrast to these studies, the Lactobacillus species Lactobacillus delbrueckii, Lactobacillus plantarum and a Lactobacillus reuteri OTU increased colonisation resistance to C. difficile.10 13 However, in concordance with increased risk, a Lactobacillus johnsonii OTU enhanced C. difficile infection.13
Last, the Veillonella genus that is increased in PPI users is significantly enriched in patients with C. difficile compared with healthy controls.28
The prevention of healthcare-associated C. difficile infections is a priority in the USA and reduction targets for 2020 have been established.5 33 A recent study looking into the effect of PPI on the risk of developing recurrent C. difficile infections found that of 191 PPI users admitted to a hospital, only 47.1% had an evidence-based indication for PPI use.5 Moreover, PPI use was discontinued in only 0.6% of the cases.5 The US Food and Drug Administration already recommends limiting PPI use to a minimum dose and duration.34 Despite these recommendations, PPIs are still often overprescribed.5 6 The risk of unnecessary antibiotics use is already addressed.35 However, limiting the unnecessary use of PPI should also be considered in preventing C. difficile and other enteric infections.
The microbiome is being intensively studied in various diseases and conditions including IBD, IBS, obesity, old age, non-alcoholic steatohepatitis and non-alcoholic fatty liver disease (NAFLD).36 PPI users are over-represented in these groups as they more likely to have GI complaints or experience GORD, either due to their health condition or their associated lifestyle. Prominent microbiome studies looking into obesity, IBD and NAFLD include results that researchers have contributed to the condition under study, but we show they are also associated with PPI use.17 37 It could well be that some of the observed effects should rather have been attributed to the use of PPI. Future microbiome studies in humans should therefore always take the effect of PPI on the gut microbiome into account.
This paper reports the largest study to date investigating the influence of PPI on the gut microbiome. The profound alterations seen in the gut microbiome could be linked to the increased risk of C. difficile and other enteric infections. Given the widespread use of PPI, the morbidity and mortality associated with enteric infections, and the increasing number of studies investigating the microbiome, healthcare practitioners and microbiome researchers should be fully aware of the influence of PPI on the gut microbiome.
Supplementary Material
Acknowledgments
The authors thank all the participants of the IBS, IBD and Lifelines-DEEP studies for contributing samples; Astrid Maatman, Tiffany Poon, Wilma Westerhuis, Daan Wiersum, Debbie van Dussen, Martine Hesselink and Jackie Dekens for logistics support; Timothy Tickle, Curtis Huttenhower, Alexandra Sirota, Chengwei Luo, Dirk Gevers and Aleksander Kostic for their help in training the first authors; Hendrik van Dullemen and Rinze ter Steege for including patients with IBD; Marten Hofker and Eelke Brandsma for contributing to the scientific discussion and Jackie Senior and Kate Mc Intyre for editing the manuscript.
Footnotes
Contributors: AZ, RKW, CW and DJ designed the study. FI, EFT, SAJ, ZM, LV, MCC and GD acquired the data; FI, MJB and AVV analysed and interpreted the data; FI drafted the manuscript; MJB and AVV performed the statistical analysis; AZ, RKW, CW, DJ, GD, LF, JF, HJMH and RJX critically revised the manuscript; AZ, RKW, CW, DJ, LF and JF obtained funding; AZ, RKW, CW and DJ supervised the study.
Funding: Sequencing of the LifeLines-DEEP and IBS MUMC cohorts was funded by the Top Institute Food and Nutrition grant GH001 to CW. CW is further supported by an ERC advanced grant ERC-671274, AZ holds a Rosalind Franklin fellowship (University of Groningen) and MCC holds a postdoctoral fellowship from the Spanish Fundación Alfonso Martín Escudero. RKW, JF and LF are supported by VIDI grants (016.136.308, 864.13.013 and 917.14.374) from the Netherlands Organisation for Scientific Research (NWO).
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: The institutional review boards of the UMCG and the MUMC (MUMC http://www.clinicaltrials.gov, NCT00775060).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.The Dutch Foundation for Pharmaceutical Statistics (SFK) Data and Facts on 2013. 2014.
- 2.Drugs.com. Top 100 sales in the United States in 2013 2013.
- 3.Olbe L, Carlsson E, Lindberg P. A proton-pump inhibitor expedition: the case histories of omeprazole and esomeprazole. Nat Rev Drug Discov 2003;2:132–9. 10.1038/nrd1010 [DOI] [PubMed] [Google Scholar]
- 4.Moayyedi P, Talley NJ. Gastro-oesophageal reflux disease. Lancet 2006;367:2086–100. 10.1016/S0140-6736(06)68932-0 [DOI] [PubMed] [Google Scholar]
- 5.McDonald EG, Milligan J, Frenette C, et al. . Continuous proton pump inhibitor therapy and the associated risk of recurrent clostridium difficile infection. JAMA Intern Med 2015;175:784–91. 10.1001/jamainternmed.2015.42 [DOI] [PubMed] [Google Scholar]
- 6.Kelly OB, Dillane C, Patchett SE, et al, The Inappropriate prescription of oral proton pump inhibitors in the hospital setting: a prospective cross-sectional study. Dig Dis Sci 2015;60:2280–6. 10.1007/s10620-015-3642-8 [DOI] [PubMed] [Google Scholar]
- 7.Bouwknegt M, Pelt W Van, Kubbinga ME, et al. . Potential association between the recent increase in campylobacteriosis incidence in the Netherlands and proton-pump inhibitor use—an ecological study. Eurosurveillance 2014;19:1–6. [DOI] [PubMed] [Google Scholar]
- 8.Leonard J, Marshall JK, Moayyedi P. Systematic review of the risk of enteric infection in patients taking acid suppression. Am J Gastroenterol 2007;102:2047–56. 10.1111/j.1572-0241.2007.01275.x [DOI] [PubMed] [Google Scholar]
- 9.Janarthanan S, Ditah I, Adler DG, et al. . Clostridium difficile-Associated Diarrhea and Proton Pump Inhibitor Therapy: a Meta-Analysis. Am J Gastroenterol 2012;107:1001–10. 10.1038/ajg.2012.179 [DOI] [PubMed] [Google Scholar]
- 10.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamada N, Chen GY, Inohara N, et al. . Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013;14:685–90. 10.1038/ni.2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britton RA., Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 2014;146:1547–53. 10.1053/j.gastro.2014.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buffie CG, Bucci V, Stein RR, et al. . Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2014;517:205–8. 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Nood E, Vrieze A, Nieuwdorp M, et al. . Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013;368:407–15. 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- 15.Tigchelaar EF, Zhernakova A, Dekens JA. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open 2015;5:e006772 10.1136/bmjopen-2014-006772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholtens S, Smidt N, Swertz M a., et al. . Cohort Profile: LifeLines, a three-generation cohort study and biobank. Int J Epidemiol 2015;44:1172–80. 10.1093/ije/dyu229 [DOI] [PubMed] [Google Scholar]
- 17.Morgan XC, Tickle TL, Sokol H, et al. . Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79 10.1186/gb-2012-13-9-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, et al. . correspondEnce QIIME allows analysis of high- throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat Publ Gr 2010;7:335–6. 10.1038/nmeth0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asnicar F, Weingart G, Tickle TL, et al. . Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 2015;3:e1029 10.7717/peerj.1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N, Haake S, Mannon P, et al. . Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 2012;13:R42 10.1186/gb-2012-13-6-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amir I, Konikoff FM, Oppenheim M, et al. . Gastric microbiota is altered in oesophagitis and Barrett's oesophagus and further modified by proton pump inhibitors. Environ Microbiol 2014;16:2905–14. 10.1111/1462-2920.12285 [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, DiBaise JK, Zuccolo a, et al. . Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA 2009;106:2365–70. 10.1073/pnas.0812600106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lessa FC, Mu Y, Bamberg WM, et al. . Burden of Clostridium difficile Infection in the United States. N Engl J Med 2015;372:825–34. 10.1056/NEJMoa1408913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magill SS, Edwards JR, Bamberg W, et al. . Multistate point-prevalence survey of health care–associated infections. N Engl J Med 2014;370:1198–208. 10.1056/NEJMoa1306801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 2009;7:526–36. 10.1038/nrmicro2164 [DOI] [PubMed] [Google Scholar]
- 26.Reeves AE, Theriot CM, Bergin IL, et al. . The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2011;2:145–58. 10.4161/gmic.2.3.16333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang JY, Antonopoulos DA, Kalra A, et al. . Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 2008;197:435–8. 10.1086/525047 [DOI] [PubMed] [Google Scholar]
- 28.Antharam VC, Li EC, Ishmael A, et al. . Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 2013;51:2884–92. 10.1128/JCM.00845-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rea MC, O'Sullivan O, Shanahan F, et al. . Clostridium difficile carriage in elderly subjects and associated changes in the intestinal microbiota. J Clin Microbiol 2012;50:867–75. 10.1128/JCM.05176-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baines SD, Crowther GS, Todhunter SL, et al. F and MHW . Mixed infection by Clostridium difficile in an in vitro model of the human gut. J Antimicrob Chemother 2013;68:1139–43. 10.1093/jac/dks529 [DOI] [PubMed] [Google Scholar]
- 31.Schubert AM, Rogers M a M, Ring C, et al. . Microbiome data distinguish patients with clostridium difficile infection and non- C. difficile -associated diarrhea from healthy. MBio 2014;5:e01021–14. 10.1128/mBio.01021-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterfreund GL, Vandivier LE, Sinha R, et al. . Succession in the Gut Microbiome following Antibiotic and Antibody Therapies for Clostridium difficile. PLoS ONE 2012;7 10.1371/journal.pone.0046966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Department H and HS. Request for Comments on the Proposed 2020 Targets for the National Action Plan To Prevent Health Care-Associated Infections: Road Map To Elimination (Phase I: Acute Care Hospital) Measures. 2014. https://www.federalregister.gov/articles/2014/02/25/2014-04069/request-for-comments-on-the-proposed-2020-targets-for-the-national-action-plan-to-prevent-health
- 34.U.S. Food and Drug Administration. FDA Drug Safety Communication: Clostridium difficile-associated diarrhea can be associated with stomach acid drugs known as proton pump inhibitors (PPIs) 2012. http://www.fda.gov/drugs/drugsafety/ucm290510.htm
- 35.Blaser M. Antibiotic overuse: Stop the killing of beneficial bacteria. Nature 2011;476:393–4. 10.1038/476393a [DOI] [PubMed] [Google Scholar]
- 36.Mehal WZ. The Gordian Knot of dysbiosis, obesity and NAFLD. Nat Rev Gastroenterol Hepatol 2013;10:637–44. 10.1038/nrgastro.2013.146 [DOI] [PubMed] [Google Scholar]
- 37.Goodrich JK, Waters JL, Poole AC, et al. . Human Genetics Shape the Gut Microbiome. Cell 2014;159:789–99. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.