Abstract
The p53 protein has been extensively studied for its role in suppressing tumorigenesis, in part through surveillance and maintenance of genomic stability. p53 has been associated with the induction of a variety of cellular outcomes including cell cycle arrest, senescence, and apoptosis. This occurs primarily, but not exclusively, through transcriptional activation of specific target genes. By contrast, the participation of p53 in normal developmental processes has been largely understudied. This review focuses on possible functions of p53 in cerebellar development. It can be argued that a better understanding of such mechanisms will provide needed insight into the genesis of certain embryonic cancers including medulloblastomas, and thus lead to more effective therapies.
Keywords: p53, Sonic Hedgehog pathway, cerebellum development, medulloblastomas
Towards Better Animal Models of Human Embryonic Tumors
p53 is a quintessential tumor suppressor. Over three decades of research have focused on this so-called “Guardian of the Genome” [1]. Indeed, the TP53 gene is found to be often mutated in a majority of human cancers [2]. This supports the notion that dysfunctional p53 contributes to tumor progression. More recently, developmental biologists have turned their attention to p53, particularly studying its possible roles in stem cell biology and induced pluripotency. These relatively new findings, in turn, are likely to impact the understanding of the genesis of embryonic cancers.
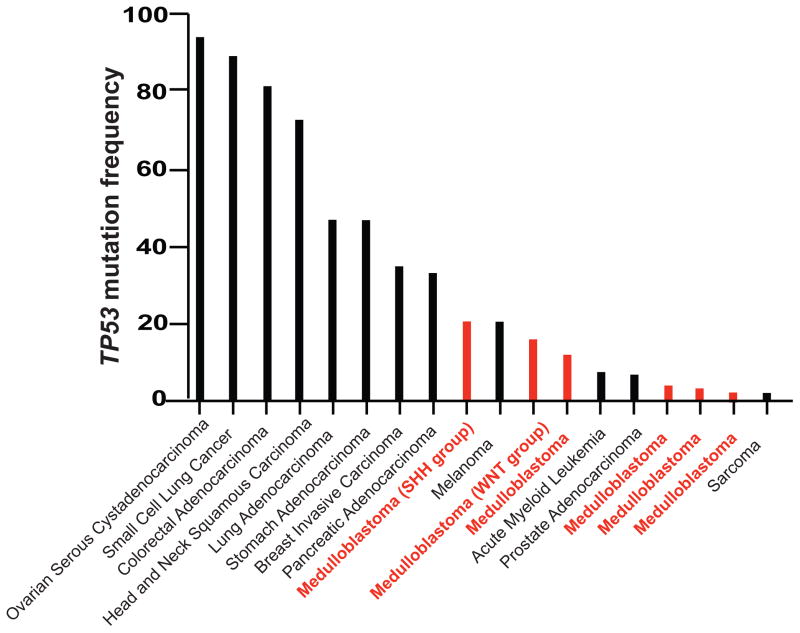
Current animal models attempt to address subtypes of one such tumor of developmental origin, namely medulloblastoma (see Glossary). Most of these models have been engineered to have deletion or loss of p53. As compared to other human cancers, multiple studies have shown that medulloblastomas are less likely to have mutated p53 (Figure 1) [3–9]. Thus, the p53 null animal models do not genetically mimic human medulloblastoma where p53 mutation is not as common as that which is seen in other types of cancers. This review addresses the importance of a more profound understanding of p53 function in development and the ways in which this can be translated into the design of more accurate animal models that better recapitulate the genetics of the human disease. With this outlook, it will may be possible to achieve more effective treatment modalities for various malignancies.
Figure 1. Unlike Other Cancer Types, Most Human Medulloblastomas Do Not Harbor Sustained TP53 Mutations.
The frequency of mutations in the gene encoding human p53 is plotted for various cancers and compared to those found in human medulloblastomas. Several published studies of human medulloblastoma are compared to data of other human cancers, derived from cBioPortal. (3,4,5,6,9, 72)
Possible Roles of the Tumor Suppressor p53 in Development
The role of p53 role as a transcription factor and the molecular determinants of its transcriptional program continue to be focus of ongoing studies. In response to a variety of cellular insults, including DNA damage and oncogenic stress, p53 triggers distinct outcomes, including cell cycle arrest, senescence, or apoptosis [10]. A key outstanding question for the field, however, is what are the possible roles of p53 in development.
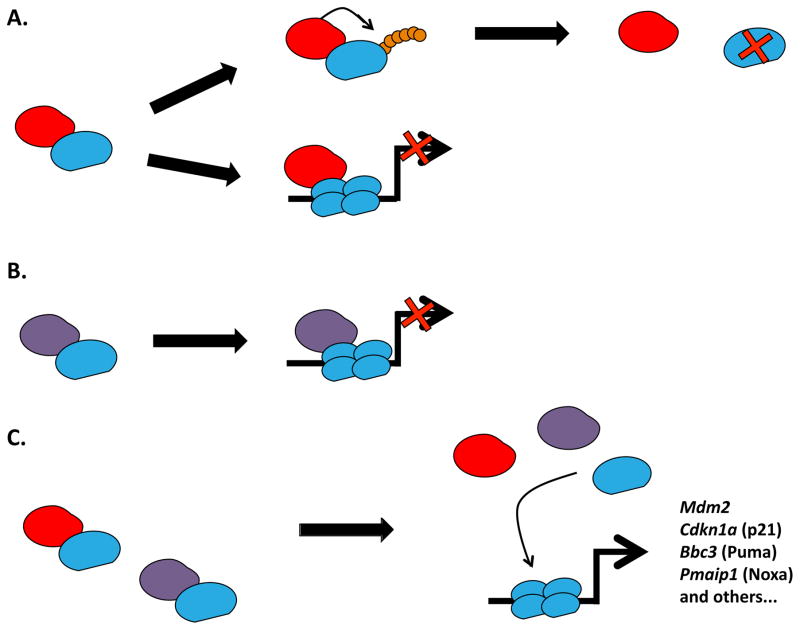
Although a suggested role for p53 in development can be gleaned from a variety of studies, definitive demonstrations of p53 contributions to vertebrate development have been elusive. For instance, Mdm2−/− mice are embryonic lethals at implantation. This phenotype can be fully rescued by p53 loss [11–13]. Indeed, Mdm2/MDM2 is a key negative regulator of p53 and thus plays critical roles in the control of p53 protein stability as well as its downstream function as a transcription factor. p53 in turn, regulates MDM2 gene expression. The repressive functions of Mdm2 disable the transcriptional activation of p53 target genes and thus, the induction of cell cycle arrest, senescence (via Cdkn1a) or apoptosis (via Bbc3 and Pmaip1). As a target gene of p53, increased Mdm2 levels either bring p53 levels back to normal, or, inhibit p53 function after cellular stress (Figure 2). This occurs despite the fact that Trp53−/− mice develop normally for the most part, although published data show a dramatic increase in spontaneous tumor formation (primarily lymphomas and sarcomas) in the mutant mice [14]. p53 mRNA has also been reported to be expressed throughout embryogenesis with a highly heterogeneous pattern [15]. This raises the question as to whether tissue-specific expression of p53 is observable even during homeostasis [16], and if there is an actual role for p53 during cellular differentiation [17–19]. There are data suggesting that p53 finely regulates stem cell differentiation during either embryogenesis or adult homeostasis. And, this fits well with its defined tumor-suppressive role when considering any potential threat represented by an uncontrolled stem-like behaving cell [20,21].
Figure 2. p53 is Negatively Regulated by Mdm2 and Mdm4.
The classical model of the p53 pathway revolves around its core elements: p53 and its main repressor Mdm2. During homeostasis, protein levels of p53 are kept low by ongoing proteosomal degradation triggered via the ubiquitin ligase activity of Mdm2 (A). Although Mdm4 does not target p53 for degradation, both Mdm4 and Mdm2 inhibit p53-dependent transcriptional activation of its target genes by directly binding to p53 (A and B). Following cellular stress such as DNA damage or oncogenic activation (C), both p53-Mdm2 and p53-Mdm4 interactions are disrupted. The resulting free p53 can fully function as a transcription factor in its homotetrameric form and activate the transcription of target genes involved in different cellular outcomes such as cell cycle arrest (Cdkn1a) or apoptosis (Bbc3 and Pmaip1).
Several studies highlight a role for p53 in stem cell maintenance, homeostasis, and differentiation. For example, p53 has been shown to block induced pluripotent stem cell reprogramming by causing cell cycle arrest or apoptosis in mouse embryonic fibroblasts [22]. p53 has also been reported to affect stem cell self-renewal and quiescence by decreasing the frequency of symmetric cell division in mouse mammary stem cells [23] and through transcriptional activation of certain target genes such as Gfi-1 [24]. Furthermore, there is evidence to suggest that cell “stemness” might be affected by indirect down-regulation of key factors such as c-Kit via p53, through a mechanism of upregulation of micro-RNA miR-34 in human colorectal cancer cell lines [25]. Thus, these studies have revealed a function for p53 beyond its well-characterized tumor suppressive role [26][15]. We summarize below the current data describing the potential role of p53 in development, with a specific emphasis on the cerebellum.
Development of the Mouse Cerebellum: A Primer
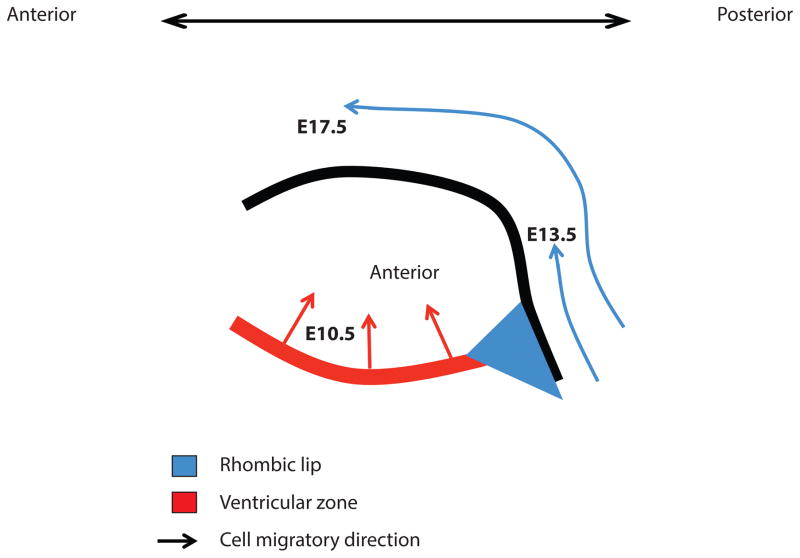
The cerebellum has a simple laminar structure that results from fine developmental processes starting around embryonic day (E) 10.5 and ending around postnatal day (P) 21. The cerebellar primordium arises form the roof of the mesencephalon composed of two progenitor zones: The ventricular zone and the rhombic lip[27]. The rhombic lip gives rise to multiple glutamatergic derivatives including granule cells, whereas the ventricular zone gives rise to several GABAergic neurons, including Purkinje cells. These two cell types are crucial elements of early stages of cerebellar development, which can be grossly divided into two phases: Embryonic cellular migrations and postnatal organ expansion. Cell migration of granule cell precursors from the rhombic lip to the surface of the cerebellum occurs progressively from E13.5 to birth. Purkinje cells precursors behave differently: following their genesis within the ventricular zone (spanning days E10 to E12), they become rapidly post-mitotic before they begin migrating radially [27] (Figure 3 and 4).
Figure 3. Embryonic Cerebellar Development is Driven by Progenitor Cell Migration.
A schematic is shown of a sagittal view of the developing cerebellum during embryogenesis. At the indicated stages, cerebellar development involves primarily migration of immature cellular progenitors. There are two key zones from which the cells that will constitute the mature cerebellum originate, namely: the ventricular zone and the rhombic lip. Granule cell progenitors migrate out of the rhombic lip along the anterior-posterior axis from E13.5 to birth. The inward migration of Purkinje cell progenitors from the ventricular zone starts at E10.5 and finishes around E13.5.
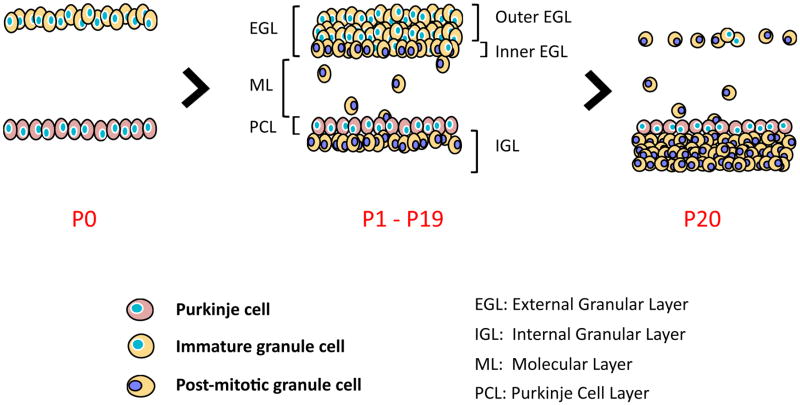
Figure 4. Postnatal cerebellum development involves the proliferation, migration and differentiation of granule cells.
(A) Following migration from the embryonic rhombic lip, granule precursor cells populate the external granular layer and engage in massive cellular expansion. (B) The initial proliferation stage of immature granule cells is closely followed by subsequent differentiation and inward migration of granule cells past the Purkinje cell layer (pink cells). Two sub-layers can then be distinguished: the proliferative outer layer (depicted with blue nuclei) and the differentiated inner layer (represented with purple nuclei). Post-mitotic granule cells migrate out of the external granular layer to invade and populate the internal granular layer (P1 to P19). (C) Around three weeks post-partum, all granule cells have left the external granular layer, completing the development of the cerebellum. The empty space between the external granular layer and the Purkinje cell layer is the molecular layer. It primarily contains the dendritic tree of Purkinje cells and axonal projections of different cell types of the cerebellar cortex: mature granule cells, Basket cells, Stellate cells, and Golgi cells. The cell bodies of Basket cells and Stellate cells are found in the molecular layer.
Around E18, cells from the external granular layer (EGL) start a cellular expansion primarily initiated by proliferating signals secreted by Purkinje cells (Figure 5) [27]. The latter produce the Sonic Hedgehog mitogen that is secreted outwardly into the external granular layer [28]. Following postnatal cellular expansion, the external granular layer can be subdivided into two layers: the outer layer that remains highly proliferative and the cells in the inner layer that engage cell cycle arrest and cellular differentiation. Differentiated granule cells then migrate inwards to form the internal granular layer. Cellular expansion in the external granular layer and cellular invasion of the internal granular layer are critical for the overall cerebellar foliation process [29]. Within the first two weeks post-partum, cellular expansion and migration continue. At this point, cells located in the internal granule layer have reached their mature state. By the third week post-partum, the cerebellum is now fully developed, at which point, the external granular layer is acellular (Figure 3 and 4).
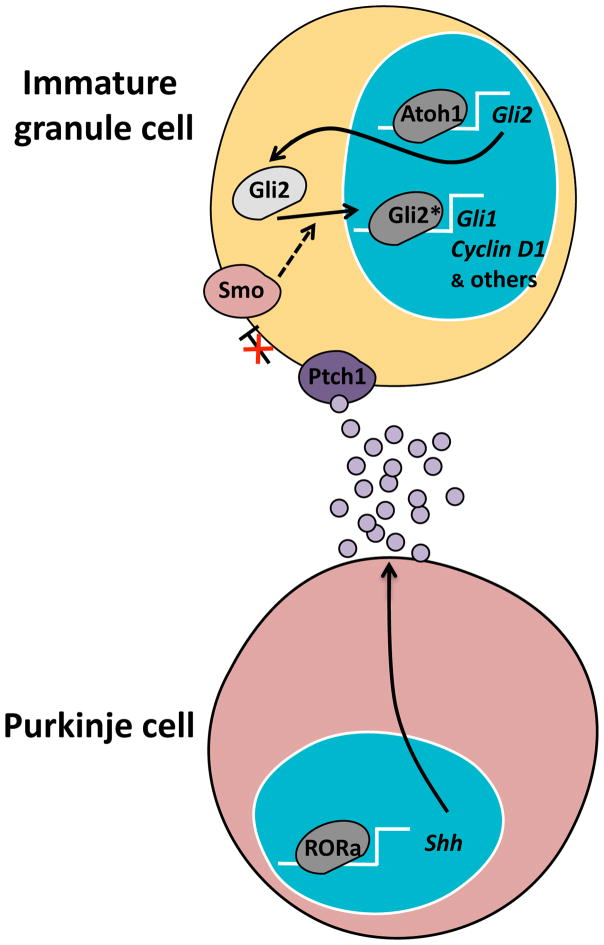
Figure 5. The Sonic Hedgehog (Shh) pathway is critical for cerebellar development.
During cerebellar development, Purkinje cells secrete Sonic Hedgehog (Shh) and other mitogens that stimulate the granule cells to proliferate. RORα is a transcription factor that upregulates Shh gene expression. Shh binds to the Patched receptor (Ptch1) on the surface of granule cells. This relieves Ptch1-mediated inhibition of Smoothened (Smo). Smo signaling leads to activation of Gli2 as a transcription factor and thus expression of genes encoding transcription factors (such as Gli1) and proteins needed for cell proliferation (such as Cyclin D1), among others. Atoh1 is a transcription factor expressed in immature granule cells that regulates basal gene expression of Gli2. This contributes to the sensitivity to proliferative signals coming from Purkinje cells.
The p53 Tumor Suppressor is Implicated in Cerebellar Development
Mdm2 is an E3 ubiquitin-ligase which binds specifically to p53 and targets it for proteasomal degradation [24]. Additionally, Mdm4 (also called Mdmx) can cooperate with Mdm2 to repress transcriptional activation by p53 (Figure 2) [25]. Upon detection of different kinds of stress signals, the activation of certain molecular cascades can trigger post-translational modifications stabilizing p53, which in turn can activate the transcription of its target genes [10]. As mentioned, although p53 null mice develop normally and progress through adulthood (despite an increased tendency for early tumorigenesis) [14], mouse models that alter one or more members of the p53 pathway have pointed to a role for p53 in development.
Alteration of p53 negative regulators
Mice null for Mdm2 are embryonic lethals, perishing after implantation of the embryo to the uterus wall (E4.5) [13]. Similarly, mice null for Mdm4 are also embryonic lethals but die later in gestation (E7.5) [30]. Incidentally, both mutations can be rescued by deletion of p53. These findings have demonstrated non-overlapping regulation of p53 by Mdm2 and Mdm4 but have also argued for the necessity of keeping p53 levels in check throughout development. This notion was further reinforced by studies that more finely modulated Mdm2, Mdm4, and p53 levels [31,32]. For instance, the cerebellar foliation defects observed in Mdm2+/− Mdm4+/− mice were abolished when just one allele of p53 was deleted. These defects were primarily due to increased levels of p53 protein accompanied by a higher apoptotic index. Additionally, mice null for Mdm2 in addition to harboring an arginine to proline mutation at codon 172 of p53 (apoptosis-deficient p53515C/515C) displayed reduced cellularity of the external granular layer at day P5 due to reduced proliferation of progenitor cells [33]. Similarly, hypoplasia and aberrant cerebellar foliation were also seen in mice with a hypomorphic Mdm2 allele (Mdm2puro). Taken together, these studies have indicated that the fine-tuning of the Mdm2-p53 network is critical during development.
Alterations of p53
A recent study in which the last 24 amino acids of p53 (p53ΔCTD) were deleted from p53 revealed an added layer of complexity by which p53 impacts cerebellum development in mice [34]. The p53 C-terminal domain shares features that are remarkably similar to those of histone tails; the C-terminus is highly basic and is subject to an extensive number of post-translational modifications [10]. This domain has been implicated in protein-protein interactions but the mechanisms by which expression of p53ΔCTD causes cerebellar defects remain to be deciphered. A similar mouse model lacking the last 31 amino acids of p53 (p53Δ31) has also resulted in a hypoplastic brain phenotype [35]. These results are consistent with the premise that p53 function during normal cerebellum development might be linked to its ability to interact with co-activators or co-repressors via its C-terminal domain.
p53 Regulates the Hedgehog Pathway in Mice
The Hedgehog pathway, through its best-studied ligand, Sonic Hedgehog (Shh), is critical for the regulation of vertebrate development and organogenesis. It is indeed one of the key drivers of cellular proliferation and organ patterning during post-natal cerebellar development [27,28]. Upon binding of the Shh ligand to its receptor, Patched 1 (Ptch1) on the receiving cell, the inhibition of the transmembrane protein Smoothened (Smo) by Ptch1 is disabled. This in turn allows the translocation of Smo to primary cilia [39–42] that will consequently further activate the pathway primarily through post-translational modifications and cleavage of effector Gli proteins, especially Gli2 (Figure 5) [41]. Elegant studies have identified additional partners of the Shh pathway at the membrane of the receiving cell that are either required for the activity of the pathway in multiple tissues, or are critical for specifying the outcomes of its subsequent activation, proliferation or patterning. Some Shh partners are co-receptors Gas1, Cdo and Boc [42] as well as certain proteoglycans [43].
Considering the influence of the Sonic Hedgehog pathway on cellular expansion, its different downstream targets, and its overall effect on cell cycle progression[41], it is evident that unbalanced regulation of this pathway may potently induce tumor formation. Numerous studies argue for a crosstalk between Sonic Hedgehog and p53 pathways. Indeed, the Shh mitogen produced by Purkinje cells in the perinatal cerebellum signals immature granule cells in the external granular layer for proliferation. Consequently, from birth to 21 days post-partum (P21) when the cerebellum has achieved its development, the organ size will have undergone a dramatic size increase (70, 71).
Evidence suggests that activated Sonic Hedgehog signaling can lead to the inhibition of p53 activity through Mdm2-dependent proteasomal degradation. This process requires post-translational modifications of Mdm2, especially phosphorylation at serine 166 that enhances its interaction with, and subsequent poly-ubiquitination of p53 [36–38]. Therefore, Sonic Hedgehog signaling may have a dual role: it could act as a driver of proliferation in the developing cerebellum and as an inhibitor of apoptosis and cell cycle exit. Concurrently, p53 is also able to down-regulate Sonic Hedgehog output through negative regulation of certain Gli proteins, including Gli1 and Gli2 [36,37,44,45]. However, even though several studies have reached similar conclusions regarding the interplay between effectors of the Sonic Hedgehog pathway and core members of the p53 pathway, the mechanisms by which such an interaction occurs still remain elusive. Taken together, these observations strengthen the notion of fine-tuned regulatory mechanisms occurring between antagonist signaling pathways during development.
Other studies have identified a potential indirect interaction between p53 and upstream members of the Sonic Hedgehog pathway. Studies on the retinoic acid receptor related orphan receptor (RORα) have revealed its function as a early factor controlling Purkinje cell differentiation [46]. Furthermore, RORα is directly involved in the transcriptional activation of Shh making it a determining factor of cerebellum development [47]. More recently, in vitro studies have identified an interesting interaction between p53 and RORα that seems to function in the context of DNA damage: RORα is not only a p53 target but also stabilizes the p53 tumor suppressor through inhibition of its proteasomal degradation. In turn, this enables p53 activity as a transcription factor [48,49]. Even though further analyses in vivo are required, multiple ways of regulation of cerebellum development by p53 could be envisioned. The tumor suppressor would be not only inhibiting proliferation in Shh-receiving cells – thus preventing uncontrolled cellular expansion – but might also act as an inducer of cell cycle arrest and apoptosis in RORα-expressing Purkinje cells following DNA damage. This could potentially block Shh secretion at the source.
A growing body of literature thus argues that activated signaling from the Sonic Hedgehog pathway can inhibit p53 activity in the cerebellum [36–38]. This suggests a fundamental balance between the anti-proliferative function of p53 and the induction of proliferation by Sonic Hedgehog signaling. However, the molecular details of this mechanism warrant further analysis and validation. The interplay between the Sonic Hedgehog and the p53 pathways may balance proliferation and cell death, in turn contributing to proper cerebellum development. It is quite conceivable that in cancers developing in the cerebellum, such as medulloblastomas, mutant p53 would be required for tumorigenesis. In fact, the majority of medulloblastoma cases harbor wild-type p53 proteins (Figure 1), possibly suggesting a novel role for the tumor suppressor in the formation of tumors arising in the cerebellum.
Medulloblastoma Subgroups
There are currently four molecularly characterized subtypes of medulloblastoma, namely: SHH, WNT, Group 3 and Group 4 (G4). The SHH subgroup, which is the best studied, finds its name from the Shh pathway shown to initiate tumorigenesis. Indeed, genetic analyses have shown that human tumors commonly display mutations or gene amplifications of particular members of the Sonic Hedgehog pathway, resulting in their specific constitutive activation [50,51]. For instance, the WNT subgroup is characterized by activation of the Wnt signaling pathway, in which two members (APC and CTNNB1) are often mutated in medulloblastoma [52]. This type has a much better prognosis than the SHH subgroup [53]. Group 3 comprises tumors with overexpression of c-Myc, not seen in the other subtypes. However, the definition of the inherent characteristics of Group 3 and Group 4 remains quite controversial and as yet, has been difficult to establish. Despite great efforts, the treatment of medulloblastomas (like other embryonic cancers) remains a serious challenge, further complicated by the histopathological and cytogenetic heterogeneities that are inherent to the disease [54].
Current Mouse Modeling of the Sonic Hedgehog and WNT Subgroups
It is currently thought that tumor cells of the SHH type of medulloblastoma originate from the granule cell progenitors in the external granular layer. Indeed the vast majority of these cells express markers of the proliferating external granular layer, the most common of which is ATOH1 (MATH1). More recently, it was demonstrated that Atoh1 protein expression was critical for the progression of this type of tumor [55], as this transcription factor potently regulates proliferation of granule cell progenitors in mice [56]. This likely occurs with Atoh1 keeping progenitors cells sensitive to Shh signaling through transcriptional activation of the Gli2 gene (Figure 5). Moreover, several groups have generated mouse models to key elements of the SHH subgroup of human medulloblastoma. Even though genetic analyses of human tumor samples have revealed mutations in PTCH1 and SUFU (and others), a full penetrance for medulloblastoma formation has not been generally achieved without additional deletion of other key genes such as p53 [57–61]. Of note, there have been exceptions for which fully penetrant inductions of medulloblastoma have been achieved with a constitutively active form of Smo, induced in either Atoh1-expressing cells, NeuroD2-expressing cells, or hGFAP-expressing cells. Nevertheless, SMO mutations have not been the most prevalent mutations in human forms of medulloblastoma [3–8]. By contrast, deletion of Patched in lineage-restricted progenitors or stem cells has been shown to induce fully penetrant medulloblastomas [62]. This strengthens the importance of neuronal lineage commitment for medulloblastoma formation. However, it also raises questions about current hypotheses regarding the driving factors of tumorigenesis, tumor progression, and the putative cellular origins of these types of medulloblastomas. Interestingly, Li and colleagues [63] identified another population of cells expressing the filament protein Nestin but not Atoh1. These cells seemed to originate from the ventricular zone as early as E14.5 and a subset of these was found to localize to the non-proliferative inner external granular layer. Besides having comparable sensitivity to Sonic Hedgehog signaling when compared with Atoh1-expressing cells in the external granular layer, Nestin-expressing progenitor cells were deficient in DNA repair, possibly explaining their increased tumorigenic potential. Consequently, one can imagine that some medulloblastomas from the SHH subgroup could originate from the Nestin-expressing progenitor cells or, that cerebellar tumorigenesis might involve the cumulative defects from multiple and distinct cell populations. This could explain, at least in part, the degree of heterogeneity described in medulloblastomas.
Gilbson and colleagues generated a mouse model recapitulating WNT subgroup medulloblastoma characteristics. This model showed the anatomical and molecular characteristics of the human WNT-subtype disease originated from the brainstem and led to the upregulation of markers such as BMP4, and DKK1[64], albeit under conditions of p53 deficiency. Even though the elucidation of the mechanisms leading to tumor formation has been substantive, a great deal of work in identifying effective treatments for medulloblastoma remains. Indeed, most current models of medulloblastoma are set in a p53 null background, which significantly increases the phenotype penetrance. However, p53 is not mutated in the majority of human tumors; the mutations range from 2.5% to 21%, depending on the report (Figure 1). Thus, it will be critical in future studies to reintegrate p53 status as seen in the human disease context in order to engineer better animal models. Hopefully, these experimental approaches will lead to more accurate predictions and consequently, a better potential for improved therapies.
Concluding Remarks
Despite the understanding gained from the use of existing mouse models of medulloblastomas, these generally rely on p53 deletion. These models contrast the frequency of p53 mutations in human tumors. Indeed, the majority of human medulloblastoma cases retain a wild-type form of the tumor suppressor. Even though these models do not fully recapitulate the genetic alterations observed in human medulloblastomas, they do nonetheless emphasize the role played by the tumor suppressor during the onset of the disease. They also highlight the potential timeframe during which aberrant cellular proliferation is likely to occur as a result of circumvented p53 responses. An increasing amount of evidence suggests that in the SHH medulloblastoma subgroup, a critical inhibition of p53 function occurs through the activation of its main repressor Mdm2 [36–38]. Incidentally, upregulation of the p53-inactivating oncogene WIP1 has also been reported in Group 3 and 4 medulloblastoma subtypes [65]. Collectively, these findings may help explain the certain lack of selective pressure that involves p53 loss or mutation in order to drive medulloblastoma tumorigenesis.
Other p53 deficient mouse models of medulloblastomas reveal how important the tumor suppressor is in regards to the genomic stability of the highly proliferative developing cerebellum. Mice simultaneously deficient for key effectors of the DNA repair pathway (Lig4, Xrcc4, or Brca2) and p53 have shown robust induction of medulloblastomas [66–69]. This fully penetrant phenotype would suggest that there is very likely a need for different DNA repair mechanisms in conjunction with p53 to prevent medulloblastoma formation. Given the inherent genomic instability of Nestin-expressing progenitor cells described by Li and colleagues [63], induction of DNA repair by p53 appears to be crucial during cerebellar development.
Although a multitude of questions remain, as emphasized in this short review (see Outstanding Questions), a deeper understating of cerebellar development in general, but also, of the contribution of p53 in this process, is likely to improve our ability to diagnose and treat different types of medulloblastomas, a serious clinical burden in pediatric medicine.
Outstanding Questions.
What role does p53 play during the genesis of medulloblastoma?
Since p53 is ubiquitously expressed, what is the basis of the observed tissue-specificities in mutant mice with differing alterations in the p53 pathway? How does this relate to development and cell differentiation?
What is the nature of the precise interplay between p53 and Sonic Hedgehog signaling?
Does p53 regulate the co-receptors involved in Shh signaling?
Why do the majority of medulloblastoma subtypes retain wild-type functional p53?
Is there a similar or redundant interplay between the p53 pathway and various oncogenic drivers of tumorigenesis in the various medulloblastoma subtypes?
Is p53 only relevant in granule cells or does it influence the development of other cell types in the cerebellum?
What is the significance of RORα as a p53 target in Purkinje cell signaling in the cerebellum?
Trends.
Mouse models of medulloblastoma depend on p53 ablation. However, most cases of the human disease are not associated with mutation(s) in p53. This raises the question of the role of p53 during the genesis of medulloblastomas.
Altering the dosage of the negative regulators of p53 (Mdm2 and Mdm4) in vivo has resulted in impaired cerebellar development in mice
p53 can down-regulate Hedgehog signaling output through the negative regulation of Gli proteins. This results in decreased proliferation of Sonic Hedgehog Shh-sensitive cells.
Activated Hedgehog signaling can lead to the inhibition of p53 activity through Mdm2-dependent proteasomal degradation. This in turn increases the activation of the Shh pathway and proliferation in Shh-sensitive cells.
Mouse models simultaneously deficient in key effectors of the DNA repair pathway (such as Lig4, Xrcc4, or Brca2) and p53 show early and strong induction of medulloblastoma with 100% penetrance. This reinforces the important role for DNA-damage repair mechanisms and possible cooperation with the p53 pathway during cerebellar development.
Acknowledgments
N.J.B and J.J.M. are supported by a research grant from the National Cancer Institute (R01 CA200256).
Glossary
- Hedgehog pathway
Crucial developmental pathway in mammals. Pathological activation has been implicated in the genesis of numerous human cancers including skin cancers (basal cell carcinoma) and the SHH subgroup of medulloblastomas.
- Medulloblastoma
The most common malignant pediatric brain tumors located in the cerebellum. Over 70% of Medulloblastoma are diagnosed before age 10 in patients. Prognoses are highly linked to the cancer subtype. These are usually good for SHH and WNT subtypes whereas they can be quite poor for group 3 and 4 subtypes.
- Cerebellum
organ playing an important role in motor control, coordination, motor learning and equilibrium. More recently, researchers have been pursuing a link between cerebellum functionality, cognitive development and autism.
- Ventricular zone
This progenitor zone is one of the two critical areas of the developing cerebellum. It primarily gives rise to inhibitory neurons such as Purkinje cells.
- Rhombic lip
This other progenitor zone of the developing cerebellum primarily giving rise to excitatory neurons such as granule cells.
- Granule cells
These excitatory neurons are among the smallest and most numerous cells of the brain. Their massive expansion accounts for the dramatic increase in size of the developing cerebellum from the late embryonic phases to the fully mature state. The SHH subtype of medulloblastoma is thought to be due to uncontrolled proliferation of these cells.
- Purkinje cells
In contrast to granule cells, Purkinje cells are some of the largest brain cells. Discovered in 1837, these inhibitory cells enable the fine regulation and coordination of motor movements.
- Mdm2−/− mice
Unviable and with embryonic lethality at the time of implantation. The phenotype is rescued by p53 deletion.
- Trp53−/− mice
Harbor apparent normal development but are subject to spontaneous tumors, primarily lymphomas and sarcomas in adults.
- Glutamatergic neurons
They use glutamate as a neurotransmitter; excitatory in the vertebrate nervous system.
- GABAergic neurons
They use γ-Aminobutyrate (GABA), a major inhibitory neurotransmitter in the mammalian central nervous system.
- Mdm2+/−; Mdm4+/− mice
Present bone marrow cell depletion and cerebellum hypoplasia. This phenotype can be rescued with monoallelic p53 deletion, further supporting the fine-tuning of p53 and its repressors during development.
- Trp53515C/515C mice
Express a mutant p53 protein, R172P. This is the mouse equivalent of R175P in humans. The mutant protein is deficient in inducing apoptosis but can still lead to partial cell cycle arrest, contributing to the maintenance of chromosome stability in the suppression of tumorigenesis.
- Mdm2puro mice
Harbor a puromycin cassette between exons 6 and 7 of Mdm2, thereby creating a hypomorphic Mdm2 allele. Exhibit aberrant foliation of the cerebellum due to increased granule cell progenitor death.
- Trp53ΔCTD/ΔCTD mice
express a truncated p53 lacking 24 amino acids in the C-terminus. DIsplay severe hematopoietic defects and reduced cellularity of the external granule layer of the cerebellum.
- Trp53Δ31/Δ31 mice
express a truncated p53 lacking 31 amino acids the C-terminus. Their primary phenotypes include aplastic anemia and pulmonary fibrosis.
- Nestin
type of intermediate filament expressed in certain neurons.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, et al. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 3.Jones DTW, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kool M, et al. Genome Sequencing of SHH Medulloblastoma Predicts Genotype-Related Response to Smoothened Inhibition. Cancer cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhukova N, et al. Subgroup-Specific Prognostic Implications of TP53 Mutation in Medulloblastoma. Journal of Clinical Oncology. 2013;31:2927–2935. doi: 10.1200/JCO.2012.48.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.RLS, et al. Infrequent p53 Gene Mutations in Medulloblastomas. Cancer research. 1991 [PubMed] [Google Scholar]
- 8.Adesina AM, et al. p53 gene mutation and mdm2 gene amplification are uncommon in medulloblastoma. Cancer research. 1994;54:5649–5651. [PubMed] [Google Scholar]
- 9.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wade M, et al. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nature reviews. Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones SN, et al. Rescue of embryonic lethality in mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 13.de Oca Luna RM, et al. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 14.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 15.Schmid P, et al. Expression of p53 during mouse embryogenesis. 1991;865:857–865. doi: 10.1242/dev.113.3.857. [DOI] [PubMed] [Google Scholar]
- 16.Choi J, Donehower LA. P53 in Embryonic Development: Maintaining a Fine Balance. Cellular and molecular life sciences : CMLS. 1999;55:38–47. doi: 10.1007/s000180050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eizenberg O, et al. p53 plays a regulatory role in differentiation and apoptosis of central nervous p53 Plays a Regulatory Role in Differentiation and Apoptosis of Central Nervous System-Associated Cells. 1996;16:5178–5185. doi: 10.1128/mcb.16.9.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira A, Kosik KS. Accelerated neuronal differentiation induced by p53 suppression. Journal of cell science. 1996;109:1509–1516. doi: 10.1242/jcs.109.6.1509. [DOI] [PubMed] [Google Scholar]
- 19.Molchadsky A, et al. P53 Plays a Role in Mesenchymal Differentiation Programs, in a Cell Fate Dependent Manner. PloS one. 2008;3:1–15. doi: 10.1371/journal.pone.0003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aloni-Grinstein R, et al. p53: the barrier to cancer stem cell formation. FEBS Lett. 2014;588:2580–2589. doi: 10.1016/j.febslet.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Jain AK, et al. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10:e1001268. doi: 10.1371/journal.pbio.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cicalese A, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siemens H, et al. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. 2013;4:1399–1415. doi: 10.18632/oncotarget.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. Genome-wide analysis of the p53 gene regulatory network in the developing mouse kidney. Physiol Genomics. 2013;45:948–964. doi: 10.1152/physiolgenomics.00113.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butts T, et al. Development of the cerebellum: simple steps to make a ‘little brain’. Development (Cambridge, England) 2014;141:4031–4041. doi: 10.1242/dev.106559. [DOI] [PubMed] [Google Scholar]
- 28.Hatten ME, Roussel MF. Development and cancer of the cerebellum. Trends in neurosciences. 2011;34:134–142. doi: 10.1016/j.tins.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez S, et al. Cellular and molecular basis of cerebellar development. Front Neuroanat. 2013;7:18. doi: 10.3389/fnana.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parant J, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nature genetics. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 31.Terzian T, et al. Haploinsufficiency of Mdm2 and Mdm4 in Tumorigenesis and Development. Molecular and cellular biology. 2007;27:5479–5485. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francoz S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu G, et al. The p53–Mdm2 network in progenitor cell expansion during mouse postnatal development. Journal of Pathology. 2007;213:360–368. doi: 10.1002/path.2238. [DOI] [PubMed] [Google Scholar]
- 34.Hamard PJ, et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes \& development. 2013;27:1868–1885. doi: 10.1101/gad.224386.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simeonova I, et al. Mutant mice lacking the p53 C-terminal domain model telomere syndromes. Cell reports. 2013;3:2046–2058. doi: 10.1016/j.celrep.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 36.Malek R, et al. The p53 Inhibitor MDM2 Facilitates Sonic Hedgehog-Mediated Tumorigenesis and Influences Cerebellar Foliation. PloS one. 2011;6:e17884. doi: 10.1371/journal.pone.0017884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Po A, et al. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. The EMBO journal. 2010;29:2646–2658. doi: 10.1038/emboj.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abe Y, et al. Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4838–4843. doi: 10.1073/pnas.0712216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohatgi R, et al. Patched1 Regulates Hedgehog Signaling at the Primary Cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 40.Pusapati GV, Rohatgi R. Location, location, and location: compartmentalization of Hedgehog signaling at primary cilia. The EMBO journal. 2014;33:1852–1854. doi: 10.15252/embj.201489294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrova R, Joyner AL. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development. 2014;141:3445–3457. doi: 10.1242/dev.083691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allen BL, et al. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Developmental cell. 2011;20:775–787. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan JA, et al. Proteoglycan interactions with Sonic Hedgehog specify mitogenic responses. Nature Publishing Group. 2009;12:409–417. doi: 10.1038/nn.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandner S. Nanog, Gli, and p53: a new network of stemness in development and cancer. 2010;29:2475–2476. doi: 10.1038/emboj.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stecca B, Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. The EMBO journal. 2009;28:663–676. doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen XR, et al. Mature Purkinje cells require the retinoic acid-related orphan receptor-α (RORα) to maintain climbing fiber mono-innervation and other adult characteristics. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:9546–9562. doi: 10.1523/JNEUROSCI.2977-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gold DA, et al. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–1131. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, et al. Regulation of p53 stability and apoptosis by a ROR agonist. PloS one. 2012;7:e34921. doi: 10.1371/journal.pone.0034921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H, et al. DNA damage-induced RORα is crucial for p53 stabilization and increased apoptosis. Molecular cell. 2011;44:797–810. doi: 10.1016/j.molcel.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 50.Slade I, et al. Heterogeneity of familial medulloblastoma and contribution of germline PTCH1 and SUFU mutations to sporadic medulloblastoma. Familial cancer. 2011;10:337–342. doi: 10.1007/s10689-010-9411-0. [DOI] [PubMed] [Google Scholar]
- 51.Northcott PA, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nature genetics. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Northcott PA, et al. Medulloblastomics: the end of the beginning. Nature reviews. Cancer. 2012;12:818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeSouza R-M, et al. Pediatric Medulloblastoma - Update on Molecular Classification Driving Targeted Therapies. Front Oncol. 2014;4:364–8. doi: 10.3389/fonc.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pomeroy SL, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 55.Flora A, et al. Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science (New York, NY) 2009;326:1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klisch TJ, et al. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodrich LV. Altered Neural Cell Fates and Medulloblastoma in Mouse patched Mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 58.Wetmore C, et al. Loss of p53 but not ARF Accelerates Medulloblastoma in Mice Heterozygous for patched. Cancer research. 2001 [PubMed] [Google Scholar]
- 59.Lee Y, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26:6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 60.Uziel T, et al. The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes \& development. 2005;19:2656–2667. doi: 10.1101/gad.1368605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hallahan AR, et al. The SmoA1 Mouse Model Reveals That Notch Signaling Is Critical for the Growth and Survival of Sonic Hedgehog-Induced Medulloblastomas. Cancer research. 2004 doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 62.Yang ZJ, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li P, et al. A population of Nestin-expressing progenitors in the cerebellum exhibits increased tumorigenicity. Nature neuroscience. 2013;16:1737–1744. doi: 10.1038/nn.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mendrysa SM, et al. p53 in the CNS: Perspectives on Development, Stem Cells, and Cancer. Genes \& cancer. 2011;2:431–442. doi: 10.1177/1947601911409736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buss MC, et al. The WIP1 oncogene promotes progression and invasion of aggressive medulloblastoma variants. Oncogene. 2015;34:1126–1140. doi: 10.1038/onc.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tong WM, et al. Null Mutation of DNA Strand Break-Binding Molecule Poly(ADP-ribose) Polymerase Causes Medulloblastomas in p53−/− Mice. American journal of pathology. 2003;162:343–352. doi: 10.1016/S0002-9440(10)63825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee Y, McKinnon PJ. DNA Ligase IV Suppresses Medulloblastoma Formation. Cancer research. 2002 [PubMed] [Google Scholar]
- 68.Yan CT, et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proceedings of the National Academy of Sciences. 2006;103:7378–7383. doi: 10.1073/pnas.0601938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frappart PO, et al. BRCA2 is required for neurogenesis and suppression of medulloblastoma. The EMBO journal. 2007;26:2732–2742. doi: 10.1038/sj.emboj.7601703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999 Jan;22(1):103–14. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 71.Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development. 2004 Nov;131(22):5581–90. doi: 10.1242/dev.01438. Epub 2004 Oct 20. [DOI] [PubMed] [Google Scholar]
- 72.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]