Summary
The p53 pro-apoptotic tumor suppressor is mutated or functionally altered in most cancers. In epithelial tumors induced by “high-risk” mucosal Human Papillomaviruses (hrm-HPVs), including human cervical carcinoma and a growing number of head-and-neck cancers 1, p53 is degraded by the viral oncoprotein E6 2. In this process, E6 binds to a short LxxLL consensus sequence within the cellular ubiquitin ligase E6AP 3. Subsequently, the E6/E6AP heterodimer recruits and degrades p53 4. Neither E6 nor E6AP are separately able to recruit p53 3,5, and the precise mode of assembly of E6, E6AP and p53 is unknown. Here, we solved the crystal structure of a ternary complex comprising full-length HPV16 E6, the LxxLL motif of E6AP and the core domain of p53. The LxxLL motif of E6AP renders the conformation of E6 competent for interaction with p53 by structuring a p53-binding cleft on E6. Mutagenesis of critical positions at the E6-p53 interface disrupts p53 degradation. The E6-binding site of p53 is distal from previously described DNA- and protein-binding surfaces of the core domain. This suggests that, in principle, E6 may avoid competition with cellular factors by targeting both free and bound p53 molecules. The E6/E6AP/p53 complex represents a prototype of viral hijacking of both the ubiquitin-mediated protein degradation pathway and the p53 tumor suppressor pathway. The present structure provides a framework for the design of inhibitory therapeutic strategies against HPV-mediated oncogenesis.
Papillomaviruses are small DNA viruses, which infect the mucosal and cutaneous epithelia of most vertebrate species. HPV16 is the most prevalent and best studied hrm-HPV, responsible for 50% of cervical carcinomas and for most HPV-positive head-and-neck cancers 1. The HPV oncoproteins E6 and E7 recognize numerous host proteins, in large part by hijacking cellular domain-motif interaction networks 6. In particular, most mucosal and cutaneous E6 proteins recognize cellular acidic leucine(L)-rich LxxLL motifs (reviewed in 7). In a recent structural study8, we have shown that LxxLL motifs bind to a conserved pocket of E6, which is contributed by the protein’s N- and C-terminal zinc-binding domains (E6N and E6C) and helix linker.
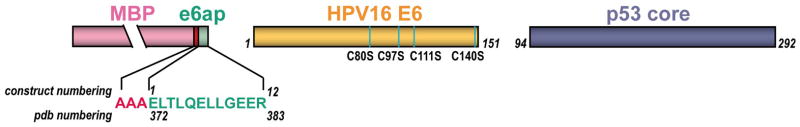
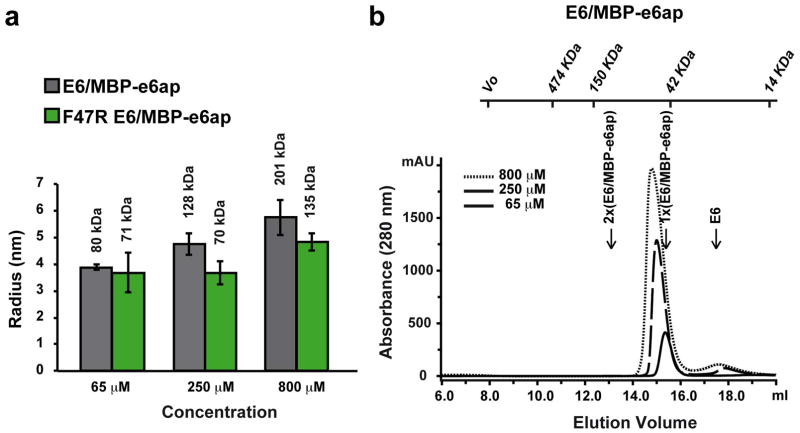
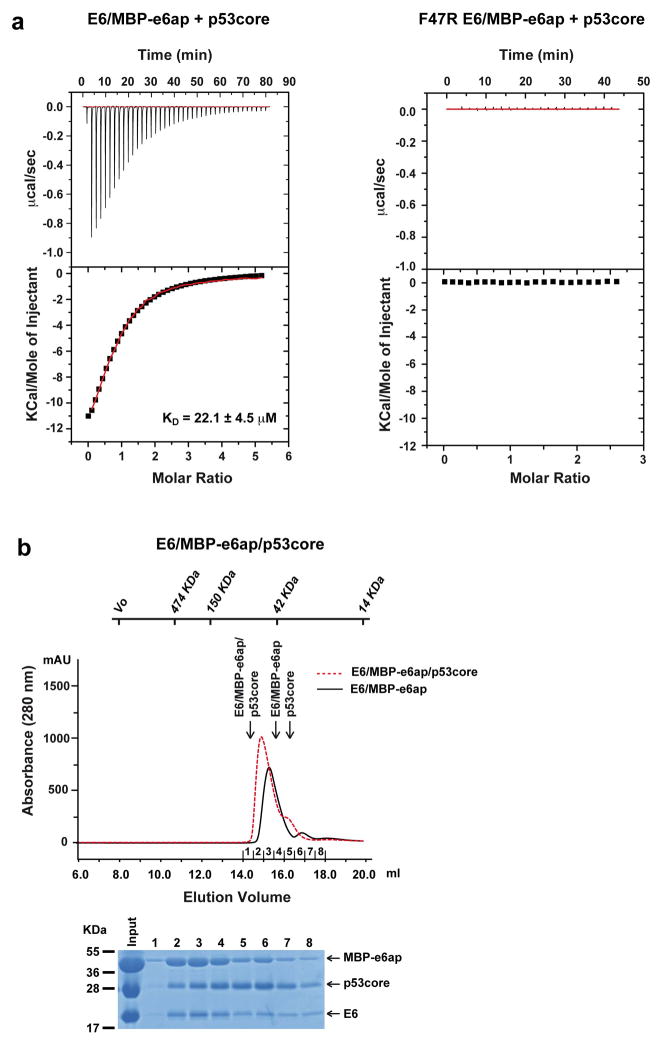
In E6-mediated degradation of p53, hrm-HPV E6 proteins interact with the LxxLL motif of E6AP, leading to recruitment and polyubiquitination of p53. The isolated LxxLL peptide of E6AP (named e6ap from here on) is sufficient to render E6 liable to interact with p53 5. Furthermore, several studies indicate that the “core” (DNA binding) domain of p53 is required for the interaction with E6/E6AP 9–11. We thus proceeded to reconstitute a minimal E6/E6AP/p53 ternary complex in vitro (Extended Data Fig. 1). The solubility enhanced HPV16 E6 4C/4S mutant (named E6 from here on), which degrades p53 with wild-type efficiency 12, was assembled with e6ap (sequence E1L2T3L4Q5E6L7L8G9E10E11R12) fused to a crystallization-prone mutant of the maltose binding protein (MBP) 8 (Extended Data Fig. 2). The resulting E6/MBP-e6ap heterodimer (named E6/e6ap from here on) was found to interact with the isolated p53 core domain (residues 94-292, named p53core from here on) by gel filtration chromatography and isothermal titration calorimetry (KD = 22 μM) (Extended Data Fig. 3 and Extended Data Table 1). In vivo, this affinity of p53 for E6/E6AP is likely to be enhanced by avidity effects, since p53 is tetrameric and E6AP can form trimers 13.
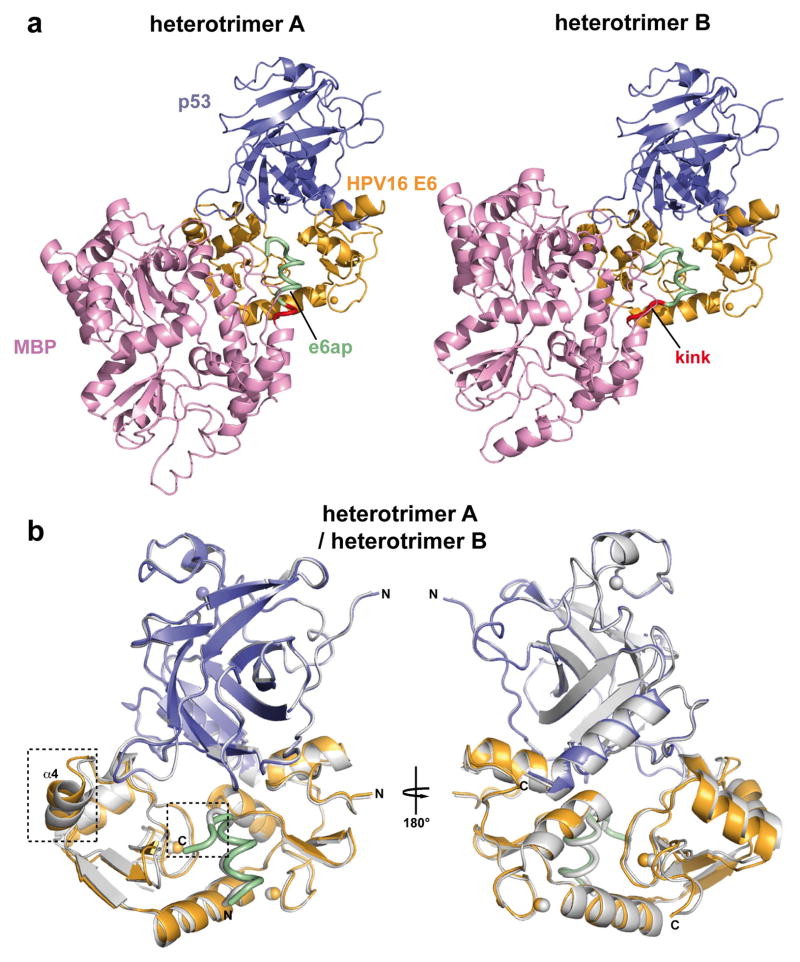
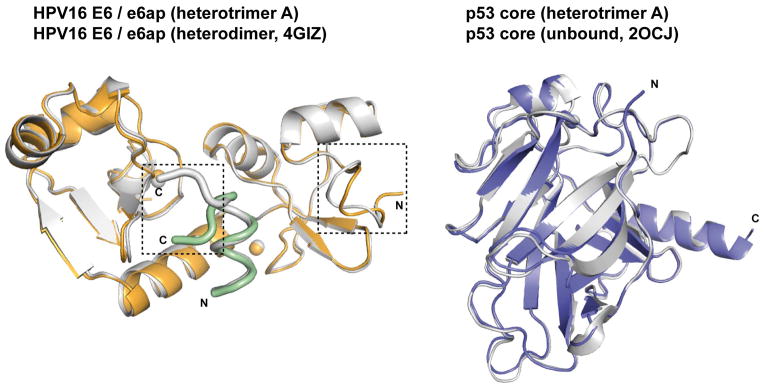
The E6/e6ap/p53core ternary complex raised several crystals diffracting up to 2.25 Å resolution using synchrotron radiation. This allowed structure determination by molecular replacement (Fig. 1a and Extended Data Table 2). The asymmetric unit of the crystal comprises two E6/e6ap/p53core heterotrimers, which contact each other mostly via MBP and display nearly identical structures except for the relative orientation of the MBP moieties (Extended Data Fig. 4). The structures of p53core and E6/e6ap observed in the heterotrimers are superimposable with previous structures of p53core and of E6/e6ap heterodimer, except for residues 1–8 of E6 and 10–12 of e6ap, which change conformation upon p53 binding (Extended Data Fig. 5). The similarities between the structures of the two heterotrimers in the crystal and previously solved structures of separate elements suggest that MBP does not significantly alter the overall conformation of the E6/e6ap/p53core complex.
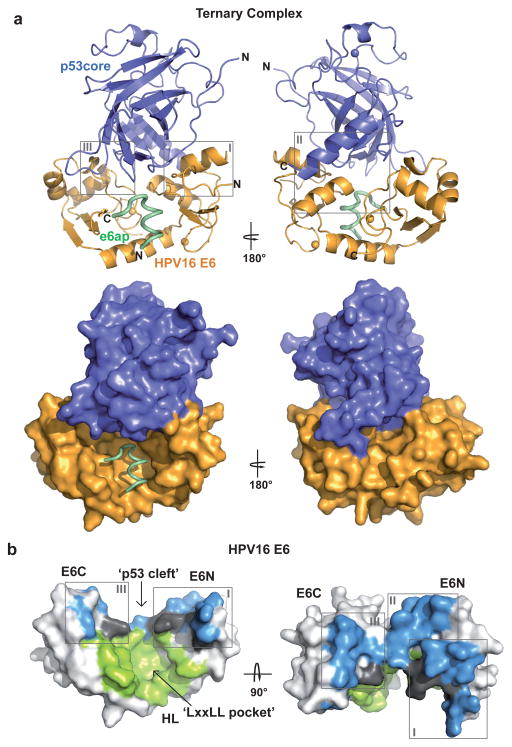
Figure 1. Structure of the HPV16 E6/e6ap/p53core ternary complex.
a, Ribbon (upper panel) and surface (lower panel) representations. Green: e6ap peptide; gold: HPV16 E6; blue: p53core. Spheres: zinc atoms. Boxes indicate sub-interfaces I–III (expanded in Fig. 2b). b, Surface representation of E6 colored for residues in atomic contact with p53 (light blue), e6ap (light green), and both p53 and e6ap (dark gray). E6N and E6C: N- and C-terminal zinc-binding domains; HL: helix linker. E6 molecules on the left side of panels a and b are in the same orientation.
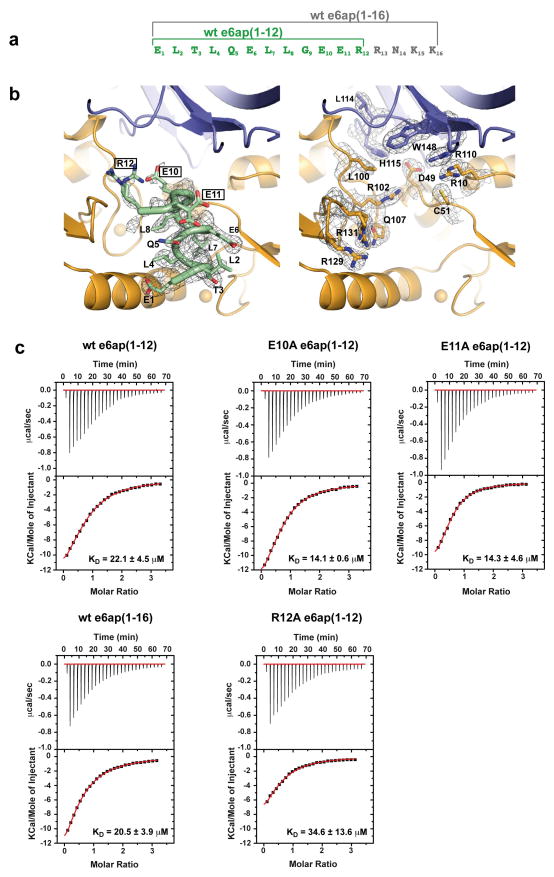
In each heterotrimer p53core binds to a cleft, which is formed by the E6N and E6C domains and held in place by contacts tethering the domains to the e6ap peptide (Fig. 1b and Fig 2a). The E6-p53 interface covers approximately 1200 Å2. The C-terminus of the e6ap peptide (residues 10–12, Extended Data Fig. 6a) also lies proximal to p53core (Extended Data Fig. 6b), but its structure is poorly defined, possibly due to an influence of the adjacent MBP tag. Nevertheless neither point mutations at residues 10–12 of e6ap nor extension of the e6ap peptide’s C-terminal boundary altered p53 binding affinity (Extended Data Fig. 6c and Extended Data Table 1), suggesting that the e6ap C-terminus does not contribute significant intermolecular contacts to p53.
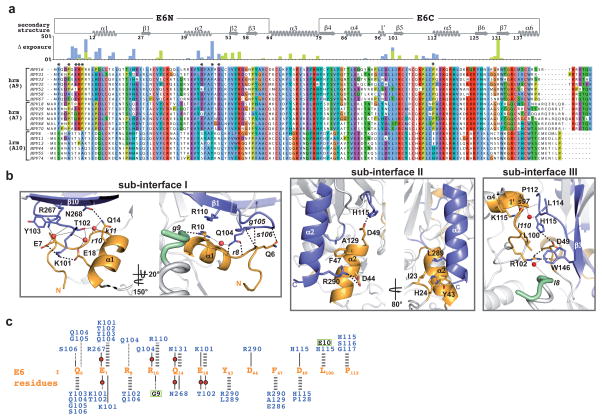
Figure 2. Intermolecular contacts at the E6-p53 interface.
a, Alignment of HPV E6 sequences from hrm- and lrm-HPV groups. Histograms: burial of residues at the interface with e6ap (light green) and p53 (light blue). *: positions conserved in hrm-HPV. b, Views of sub-interfaces I–III. Red spheres: water molecules; thick dashed lines: direct polar interactions; thin dotted lines: water-mediated interactions. Upper and lower case fonts: residues mediating polar interactions via side-chain and backbone, respectively. c, Key interaction networks. Gold fonts: E6 residues; blue fonts: p53 residues; green boxed black fonts: e6ap residues; thick gray dashed lines: non-polar interactions; black lines: polar interactions involving side-chain groups; black dotted lines: polar interactions involving backbone groups; red circles: water molecules.
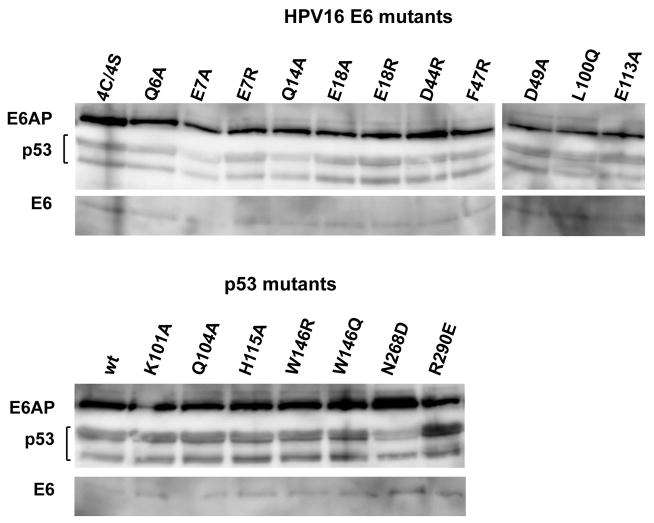
The E6-p53 interface can be divided into three sub-interfaces (Fig. 1a and Fig. 2b). Sub-interface I is dominated by polar interactions and brings together residues in the N-terminal arm and α1 helix of E6 and residues of the N-terminal arm, β1 and β10 strands of p53core (Fig. 2b–c). E6 residues Glu7 and Glu18 establish a bi-dentate salt bridge with Lys101 of p53 as well as water-mediated interactions with other p53 residues. In particular, Glu18 contributes to a network involving side-chains and backbone groups of Gln14, Arg10 and Lys11 of E6, and of Thr102 and Asn268 of p53. Consistently, E6 mutations E18A and E18R impair ternary complex assembly (Fig. 3a) and p53 degradation (Fig. 3b). Furthermore, Gln104 and Gly105 of p53 hydrogen bond to the backbone of E6 residues Arg8 and Gln6, respectively, thus altering the conformation of the N-terminal arm of E6 (residues 1–8) (Extended Data Fig. 5).
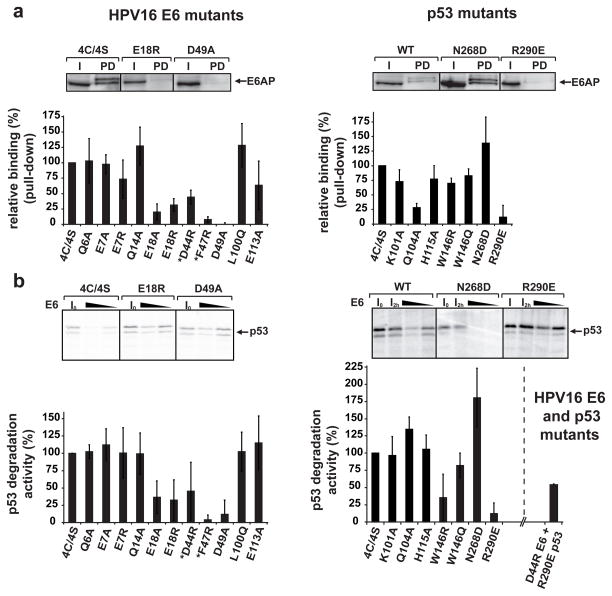
Figure 3. Ternary complex assembly and p53 degradation activities of E6 and p53 mutants.
a, Triple pull-downs. E6AP, E6 and biotinylated p53 were co-expressed in HEK 293T cells. E6AP bound to p53 was detected by Western blotting. All E6 mutations were introduced in the E6 4C/4S construct 12. *: previously investigated mutants 12. (Upper panel) Instances of binding efficiencies. I: input; PD: pull-down. (Lower panel) Percentage of E6AP bound to p53. See also Extended Data Fig. 8. b, p53 degradation assays. (Upper panel) Instances of degradation efficiencies showing p53 before and after incubation with 5 or 2.5 μl of E6 translation product. I0: p53 band at time=0; I2h : p53 band at time=2h without E6 (see Methods). (Lower panel) p53 degradation activities with 2.5 μl of E6 product.
The data (mean ± sd) were obtained from three independent experiments and normalized to 100% for the reference proteins (E6 4C/4S/wt p53). For gel source data see Supplementary Figure 1.
Sub-interface II mainly involves the α2 helix of the E6N domain and the α2 helix of p53core, interacting through hydrophobic, charged and polar contacts (Fig. 2b–c). E6 residue Phe47 intercalates between p53 residues Ala129 and Arg290. In turn, Arg290 establishes a salt bridge with Asp44 of E6. These key interactions explain the reported dominant-negative phenotype of the E6 F47R mutant, which is defective for p53 degradation, restores high p53 levels and drives senescence 14. Consistently, here we find that E6 F47R is defective for interaction with both p53core (Extended Data Fig. 3a) and full-length p53 (Fig. 3a). Besides Phe47, other E6 residues in sub-interface II (Ile23, His24 and Tyr43) provide hydrophobic contacts, whereas E6 residue Asp49 establishes polar interactions with His115 in the L1 loop of p53. Mutations disrupting the Asp44-Arg290 and Asp49-His115 interactions (D44R and D49A in E6 and the R290E in p53) impair both p53 ternary complex assembly (Fig. 3a) and p53 degradation (Fig. 3b). By contrast, creation of a swapped Glu290-Arg44 salt bridge, by combining D44R E6 and R290E p53 mutants, partially restores p53 degradation (Fig. 3b).
Finally, sub-interface III encompasses hydrophobic interactions between E6C residues Leu100 and Pro112 and p53core surface residues Leu114 and Trp146, as well as a number of water-mediated backbone-to-backbone contacts (Fig. 2b–c). Notably, Leu100 and Pro112 of E6C are proximal to conserved Arg102, which shapes the p53 binding cleft by mediating crucial interactions with the E6N domain and the e6ap peptide 8.
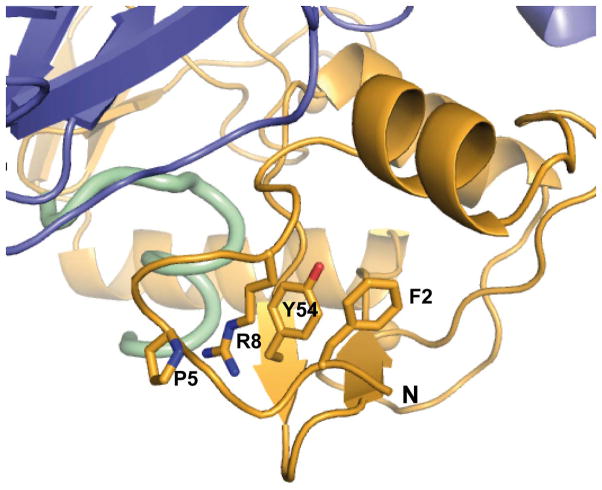
p53 degradation activity is a hallmark of hrm-HPV E6 proteins 15–17. The structure explains the critical contributions towards p53 degradation of particular residues, which are differentially conserved in high- versus low-risk mucosal (lrm) HPV E6. In sub-interface I, most conserved positions in hrm-HPV E6 proteins correspond to Phe2, Pro5, Glu7, Arg8 and Pro9 (Fig. 2a). While Glu7 mediates direct contacts to p53, Phe2, Pro5, Arg8 and Pro9 play an indirect role at the interface by shaping the conformation of the N-terminal region (Extended Data Fig. 7). Indeed, site-directed mutagenesis of Phe2, Pro5, Arg8 or Pro9 18–20 impairs ternary complex assembly and p53 degradation activities. In sub-interface II, most conserved positions in hrm-HPV correspond to Phe47, Asp44 and Asp49 (Fig. 2a), which are here found to mediate crucial contacts to p53. These residues were formerly found to be important for p53 degradation 14,18,21. Notably, Phe47 and Asp44 also participate in the dimerization of the E6N domain and subsequent in vitro self-association of the entire E6 protein 12. Therefore, E6 self-association and E6 binding to p53 are two distinct and competing processes mediated by partially overlapping interaction surfaces.
The LxxLL peptide of E6AP is absolutely required for E6 binding to p53, yet it does not contribute significant contacts to p53. Furthermore, in the absence of the LxxLL motif of E6AP, the few interactions connecting E6N, linker helix and E6C should be insufficient to maintain the overall E6 architecture observed in the E6/e6ap heterodimer structure (see 8 for further discussion). These observations indicate that the LxxLL motif structures the p53 binding cleft on E6, thereby rendering E6 competent for interaction with p53. Interestingly, we have shown that an in vitro selected peptide, targeting the LxxLL pocket of HPV16 E6, induces recruitment of p53 to E6 22, 23. Consequently, cellular proteins other than E6AP, which bind to the LxxLL pocket, might also promote the E6-p53 interaction.
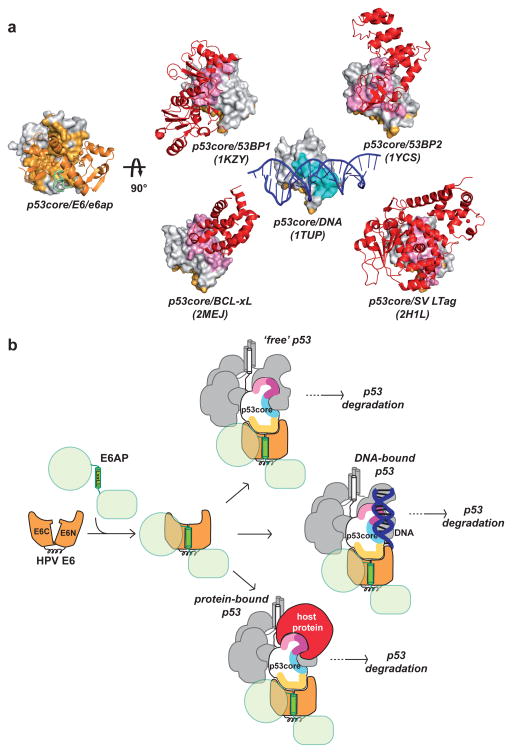
p53core is both a DNA binding domain and a protein-protein interaction hub. E6 interacts with the N-terminal arm of p53core and one of the edges of the β-sandwich. Previous mutagenesis studies had already suggested a role for these regions in E6 binding 9,11,24. The E6-binding interface on p53core is distal from both the DNA-binding region and protein-binding surfaces observed in all other solved complexes of p53core (Fig. 4a). Indeed, previously described protein-binding interfaces of p53core all overlap, albeit at different extents, with the DNA-binding region. In particular, the Large T antigen (LTag) of oncovirus SV40, which does not degrade p53, buries the entire DNA binding interface of p53core, thereby inhibiting p53 trans-activation activity 25. Consistent with these observations, two frequent cancer-associated mutations (R273C and R273L), affecting a prominent DNA-binding arginine residue of p53core, abolish binding to SV40 LTag, but do not alter the interaction with E6/E6AP 26. Therefore, E6 might avoid competition with cellular factors by targeting both ‘free’ p53 as well as p53 bound to DNA or other proteins (Fig. 4b). This, along with the irreversible character of the degradation process, renders E6 a potent inactivator of p53 functions.
Figure 4. p53core targeting by HPV E6.
a, Complexes of p53core (surface representation) with protein or DNA partners (ribbon representation). Left: p53core bound to HPV E6/e6ap. Right: p53core bound to 53BP2, 53BP1 and BCL-xL cellular proteins, to adenovirus SV LTag oncoprotein and to DNA. p53core binding interfaces: E6 (light orange), other proteins (pink) and DNA (cyan). b, Cartoon summarizing how the LxxLL motif shapes the p53 binding cleft on E6, which recognizes a distinct interface on p53core, enabling targeting of both free and bound p53 molecules. Tetrameric p53 is shown with one subunit colored for interfaces binding to HPV E6 (yellow), DNA (cyan and purple), and other host proteins (pink and purple).
Recent studies employing pro-apoptotic peptides 22 as well as small molecules 27, 28 directed against the LxxLL pocket have provided experimental evidence that this pocket is druggable. The p53-binding cleft observed in the present structure may represent a second potential binding site for drugs. Combinatorial strategies targeting both the LxxLL pocket and the p53-binding cleft could result in efficient disruption of the E6/E6AP/p53 complex.
Methods
Preparation of protein samples
DNA constructs
Residues 403 to 414 of human E6AP (peptide sequence E1L2T3L4Q5E6L7L8G9E10E11R12) were cloned via a three-alanine linker at the C-terminus of a mutant MBP used to promote crystallization 8. The point mutations introduced in MBP (D83A, K84A, K240A, E360A, K363A and D364A) have been previously described to increase the propensity of MBP to crystallize 29. The E6 4C/4S mutant of HPV16 12 (named E6 from here on) and the core (or DNA binding) domain of human p53 (residues 94 to 311) were cloned into the pETM-41 vector containing an N-terminal His6-MBP tag followed by a TEV cleavage site.
Protein expression and purification
MBP-e6ap, His6-MBP-tev-E6 and His6-MBP-tev-p53core constructs were over-expressed separately in E. coli BL21(DE3) cells at 15 °C for 18 hours. All constructs were purified separately by amylose affinity chromatography in buffer A (50 mM Tris pH 6.8, 400 mM NaCl, and 2 mM DTT). To remove soluble aggregates, all affinity purified samples were ultracentrifuged at 110,000 g in a swing SW41 rotor (Beckman) for 16 hours at 4 °C. Hence p53core and E6 samples were digested by TEV protease. In the case of p53core, after TEV digestion samples were additionally purified on an heparin column. The resulting MBP-e6ap, E6 and p53 samples were thus concentrated and loaded separately onto a Superdex 75 HiLoad 16/60 gel filtration column (GE Healthcare) equilibrated in buffer A. All purification buffers were filtered, degassed and saturated with argon.
Crystallization
The E6/e6ap/p53core complex was reconstituted by mixing MBP-e6ap and E6 and p53core samples in a 1:1:1 stoichiometric ratio in buffer B (50 mM Tris pH 6.8, 200 mM NaCl, 2 mM DTT, 5 mM maltose) and concentrated to 18 mg/ml prior to crystallization. Crystallization conditions were screened using commercially available kits (Qiagen, Hampton Research, Emerald Biosystems) by the sitting-drop vapor-diffusion method in 96-well MRC 2-drop plates (SWISSCI), employing a Mosquito robot (TTP Labtech). Initial crystals were obtained and used as seeds during further optimization steps.
After optimization, crystals (125 × 80 × 80 μm) grew in sitting drops made from 400 nl of protein solution at 18 mg/ml, 370 nl of reservoir solution containing 7.5 % polyethylene glycol (PEG) 20K, 50 mM MES (pH 6.5), and 30 nl of seeds. Drops were equilibrated against 50 μl of reservoir solution at 290 K. Crystals were sequentially transferred through two cryo solutions of reservoir solution supplemented with 15 % (v/v) and 25 % (v/v) of PEG 200, respectively. The crystals were flash-cooled and stored in liquid nitrogen.
Data collection and processing, and structure determination
X-ray diffraction data were collected on the X06DA beamline at the Synchrotron Swiss Light Source (SLS, Villigen, Switzerland). Data were acquired from a single cryo-cooled crystal (100 K) on a Pilatus-2M detector. 200° data were collected up to a resolution of 2.25 Å using 0.5° rotation and 0.8 sec exposure time with 20 % beam attenuation for each image. The data were indexed, processed and scaled using HKL2000 30. The crystals belong to the monoclinic space group P21 with unit cell parameters a=78.17Å, b=129.37Å, c=82.17Å and β=92.4°, with a refined crystal mosaicity of 0.28 – 0.34°. The asymmetric unit contains two copies of the E6/MBP-e6ap/p53core heterotrimer, with a corresponding Matthew’s coefficient of 2.48 Å3/Da and a solvent content of 50.34 %. The structure was solved by two sequential molecular replacements using Phaser 31 and the structures of MBP, E6/e6ap complex and p53core as templates (PDB IDs 4GIZ and 1TUP, respectively). Crystallographic refinement involved repeated cycles of conjugate-gradient energy minimization and temperature-factor refinement, and was performed using PHENIX 32 followed by iterative model building in Coot 33. All the disordered residues were fitted to the built electron density map, except from e6ap residue Arg12 in one of the two heterotrimers (corresponding to chain B residue Arg383 in the pdb file).
Data collection and refinement statistics are summarized in Extended Data Table 2. The quality of the refined models was assessed using MOLPROBITY 34. All molecular graphics figures were made using PyMOL 35. E6 residues at interface regions (maintext Fig. 1b and Fig 2a) were identified by the observation of an increase in solvent accessibility obtained upon removal of e6ap or p53core from the pdb file of the ternary complex. Positive “Δ exposure” values (maintext Fig. 2a) indicate E6 residues in atomic contact with or p53.
The refined model and the structure factor amplitudes have been deposited in the Protein Data Bank under the PDB code 4XR8.
DNA constructs for triple pull-down and p53 degradation assays
HPV16 E6 mutations were inserted in the background of the E6 4C/4S construct to prevent cross-linking of E6 molecules through intermolecular cysteine bridges. We have previously shown that E6 4C/4S and wt HPV16 E6 have indistinguishable p53 degradation activities in vitro 12. p53 mutations were inserted in the background of wild-type full-length human p53. All DNA constructs were verified by sequencing.
Triple pull-downs
DNA constructs encoding for full-length HPV16 E6 and p53, and a large fragment of E6AP (residues 291–875 of isoform II) were inserted into the final destination vectors by Gateway cloning (Invitrogen). HPV16 E6 and E6AP were inserted into the GPCA pSPICA-N2 and pSPICA-N1 vectors respectively. GPCA vectors pSPICA-N1 and pSPICA-N2 (both derived from the pCiNeo mammalian expression vector) allow expression of test proteins as fusions to the C-terminus of Gluc1 and Gluc2 complementary fragments of the Gaussia princeps luciferase, respectively 36. By contrast p53 constructs were inserted in the BioEase-DEST vector 36, which incorporates a 72-amino acid sequence from Klebsilla pneumonia that directs in vivo biotinylation.
Degradation assays
HPV16 E6 and full-length p53 proteins were cloned in the pXJ40 vector.
Triple pull-downs
HEK-293T cells were grown and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% FCS and 50 μg/ml of gentamycin at 37 °C with 5% CO2 and 95% humidity. Cells were seeded in 6-well plates at a concentration of 3×105 cells/well. After 24 h, cells were transfected using JetPEI® (Polyplus transfection) with 1 μg pSPICA-N2 plasmid expressing E6, 1 μg of pSPICA-N1 plasmid expressing E6AP protein and 1 μg of BioEase plasmid expressing p53. At 24 h post-transfection, cells were harvested and lysed by freeze-thawing in 100 μl of Renilla Lysis Buffer (Promega, E2820). Cellular lysates were cleared by centrifugation at 13 000 rpm in a microfuge for 15 minutes at 4 °C. Subsequently, 30 μl of pre-equilibrated Streptavidin Mag Sepharose beads (GE Healthcare) were incubated with 60 μl of cellular lysate supernatant for 2 hours at 4°C, thereby allowing capture of biotinylated p53. Streptavidin beads were washed 3 times with TNE buffer (50mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP40, 1mM EDTA and protease inhibitor) resuspended with 25 μl of protein loading buffer and loaded on a 15% SDS-PAGE gel. E6 and E6AP proteins were detected by Western blotting using rabbit anti-Gluc antibody (NEB, ref. E8023S), whereas p53 was detected using a mouse anti-p53 DO-1 antibody (Life Technologies, ref. 13–4000). The immunoreactive bands were visualized using WesternBright™ Sirius™ (Advansta) and LAS 4000 camera. Bands were quantified using ImageGauge software (Fujifilm). Error bars represent standard deviations from three independent experiments.
Cell lines
The HEK-293T cell line was kindly provided by Pierre Charneau (Institut Pasteur, Paris). These cells were authenticated and tested to be mycoplasma free.
p53 degradation assays
E6 and p53 proteins were in vitro translated in the presence of 35S-methionine using the TNT T7 coupled rabbit reticulocyte system. p53 degradation reactions were performed in 10 μl volumes by incubating 2 μl of p53 translation product with either 5 or 2.5 μl of E6 translation products at 28 °C for 2 hours according to previously described protocols 2. Reactions were resolved on a 15% SDS-PAGE gel. Gels were exposed to a PhosphorImager screen and scanned using a Typhoon FLA 9500 imaging system (GE Healthcare). Reactive p53 bands were quantified using ImageQuant TL software (GE Healthcare).
p53 degradation activity values reported in Fig. 3 of the main text were derived using the formula (I0−I)/I0, where I is the intensity of the p53 double band after incubation with E6 and I0 the p53 signal in the input lane. p53 degradation activity values were normalized to 100% for the reference proteins (E6 4C/4S/wt p53). For p53 mutant proteins (Fig. 3b of maintext), besides the I0 input control, the I2h control (corresponding to p53 at time=2h in the absence of E6) has been added to check for the intrinsic stability of p53 mutants. Error bars represent standard deviations from three independent experiments.
Extended Data
Extended Data Fig. 1.
Constructs of the minimal HPV16 E6/E6AP/p53 ternary complex. Green: E6AP residues 403–414 (e6ap 1–12) fused to the C-terminus of MBP (pink) via a AAA linker (red); gold: the HPV16 E6 4C/4S construct 12(named E6 from here on) comprising four cysteine to serine substitutions (cyan marks) that suppress aggregation mediated by disulfide cross-bridging; blue: p53 core domain (p53core).
Extended Data Fig. 2.
Assembly of the E6/MBP-e6ap heterodimer. a, Dynamic light scattering (DLS) analysis of E6/MBP-e6ap samples. Histograms report on the average particles’ hydrodynamic radii, whereas error bars indicate size polydispersity. Numbers above the histograms indicate molecular weight estimates assuming a spherical model. This analysis shows that binding to MBP-e6ap enhances the solubility of E6, which, in the unbound state, displays a solubility threshold of 50 μM. However, in the case of E6/MBP-e6ap samples (gray histograms), particle size increases with raising concentration. By contrast, introduction of the F47R mutation in E6 (green histograms) stabilizes particle size to values close to what is expected for a simple heterodimer (≈ 60 kDa). Therefore we conclude that, despite the increase in solubility, E6/MBP-e6ap still undergoes weak self-association via the E6N region hosting Phe47 (see 12 for further discussion). b, Gel filtration analysis of E6/MBP-e6ap samples. The elution volumes for molecular size markers are reported on top of the figure. The expected elution volumes of a simple E6/MBP-e6ap heterodimer (1x, 60 kDa) and of a dimer of heterodimers (2x, 120 kDa) are indicated. Note the relative small shift in the elution volumes of the different samples as compared to the differences in the hydrodynamic radii (panel a). This suggests that oligomers of the E6/MBP-e6ap heterodimer are rather weak and dissociate on the gel filtration column. See also Supplemental Methods section.
Extended Data Fig. 3.
Interaction of p53core with preformed E6/MBP-e6ap heterodimer. a, Isothermal titration calorimetry (ITC) experiments were performed by titrating increasing amounts of p53core into E6/MBP-e6ap heterodimer samples adjusted to a concentration of 45 μM, which limits heterodimer oligomerization. Note that the F47R mutation in E6 abolishes binding to p53core. See also Extended Data Table 1. b, (Upper panel) Comparison of gel filtration elution profiles of E6/MBP-e6ap/p53core (red dashed line) versus E6/MBP-e6ap samples (black line). Both samples were adjusted to a concentration of 250 μM before loading onto the gel filtration column. The expected elution volumes for p53core (24 kDa), monomeric E6/MBP-e6ap heterodimer (60 kDa) and E6/MBP-e6ap/p53core ternary complex (80 kDa) are indicated. (Lower panel) SDS-PAGE analysis of fractions comprising the elution peak of the ternary complex. Note the significant shift in the elution volumes of the main peak in the two chromatograms, indicating formation of a ternary complex. See also Supplemental Methods section.
Extended Data Fig. 4.
Comparison of the structures of the two E6/MBP-e6ap/p53core heterotrimers (heterotrimers A and B) observed in the asymmetric unit. Green: e6ap fused to the C-terminus of MBP (pink) via the AAA linker (red); gold: HPV16 E6; blue: p53core; spheres: zinc atoms. a, Different orientation of MBP in the two heterotrimers, which results from the different conformations of the two AAA-linkers. b, Superposition of the E6/e6ap/p53 regions of heterotrimers A (green/gold/blue) and B (gray). The backbone rms deviation for the E6/e6ap/p53 regions of the two heterotrimers was calculated by aligning backbone atoms of residues (i) 12–136 of HPV16 E6, (ii) 371–379 of e6ap and (iii) 109–191 of p53core and found to correspond to 0.9 Å. Regions displaying significant differences are boxed. These regions are the ill-defined C-terminal region of the e6ap peptide (see also Extended Data Fig. 6) and the a4 helix of E6, which is not involved in the E6-p53 interface.
Extended Data Fig. 5.
The structures of the E6/e6ap and p53core subunits of the ternary complex are superimposable with previously solved structures of the E6/e6ap heterodimer and p53core. (Left panel) Superposition of the previously solved E6/e6ap heterodimer (gray) 8 onto the E6/e6ap subunit of the ternary complex (heterotrimer A, gold/green) determined here. Dashed lines highlight regions of conformational change, namely the N-terminus of HPV16 E6 and the C-terminus of the e6ap peptide. (Right panel) Superposition of previously solved p53core in the unbound state (heterotrimer A, gray) 37 onto the p53core subunit of the ternary complex bound (blue).
Extended Data Fig. 6.
Contributions of the e6ap C-terminal region to ternary complex interface. a, Sequence of wild-type e6ap peptides used in the study. Green: wt e6ap(1–12) corresponding to the peptide used for crystallization of the ternary complex. Gray: wt e6ap(1–16) containing a four amino acid C-terminal extension. b, Electron density (2FO-FC) map (heterotrimer A) contoured at 1s level for e6ap (green, left panel) and for selected E6 (orange) and p53 (blue) interface residues in the proximity of the C-terminus of e6ap (right panel). Note the lack of electron density data for e6ap residues Glu10, Glu11 and Arg12. Two conformations for the side-chains Glu10 and Arg12 of e6ap and Arg131 of E6 are proposed in the model. c, ITC titration curves showing the interactions of p53core with pre-formed E6/e6ap complexes bearing either point mutations within e6ap C-terminus (E10A, E11A and R12A) or the wt e6ap(1–16) peptide construct. Note that neither the e6ap point mutations nor the e6ap C-terminal boundary affect the interaction between the E6/e6ap heterodimer and p53core. See also Extended Data Table 1.
Extended Data Fig. 7.
Interactions mediated by hrm-HPV conserved residues shaping the conformation of the N-terminal arm of HPV16 E6. Whereas Phe2 contributes to tethering of the N-terminal region to the core of the E6N domain, residues Pro5, Arg8 and Tyr54 are involved in a triple stacking interaction.
Extended Data Fig. 8.
Expression levels of HPV16 E6, E6AP and p53 proteins in for triple pull-down assays. HEK293T were transfected with 1 μg of pSPICA-N1 plasmid (expressing E6AP fused to the C-terminus of the Gaussia princeps luciferase Gluc1 fragment), 1 μg pSPICA-N2 plasmid (expressing the E6 fused to the C-terminus of the Gaussia princeps luciferase Gluc2 fragment) and 1 μg of BioEase plasmid (expressing N-terminally biotinylated p53). Cell lysates were resolved by SDS-PAGE electrophoresis and E6, E6AP and p53 fusion proteins detected by Western blotting. For gel source data, see Supplementary Figure 1.
Extended Data Table 1.
Thermodynamic parameters of p53core binding to preformed E6/e6ap complex.
| e6ap | a N | b cmin/cmax | KD(μM) | ΔH (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|---|
| wt c6ap(1–12) | 0.826±0.016 | 1.8/2.4 | 22.1±4.5 | −17.4±1.5 | −0.926±0.140 | −16.5 |
| wt e6ap(1–16) | 0.516±0.046 | 1.8/2.7 | 20.5±3.9 | −19.1±2.7 | −1.100±0.299 | −18.0 |
| E10A e6ap(1–12) | 0.827±0.080 | 3.1/3.3 | 14.1±0.6 | −17.1±0.5 | −0.878±0.051 | −16.2 |
| E11A e6ap(1–12) | 0.679±0.146 | 2.6/4.0 | 14.3±4.6 | −13.8±0.1 | −0.599±0.012 | −13.2 |
| R12A e6ap(1–12) | 0.719±0.111 | 1.0/2.2 | 34.6±13.6 | −13.9±2.7 | −0.651±0.250 | −13.2 |
All ITC experiments were performed at 25 °C.
N refers to the molar ratio of p53core : E6/e6ap complex.
c value is defined as: c=n[E6/e6ap]/KD.
Extended Data Table 2.
Data collection and refinement statistics.
| E6/e6ap/p53 | |
|---|---|
| Data collection | |
| Space group | P 21 |
| Cell dimensions | |
| a, b, c (A) | 78.15 129.41 82.26 |
| α, β, γ (°) | 90, 92.4, 90 |
| Resolution (Å) | 50-2.25 (2.33-2.25) (*) |
| Rsym or Rmerge | 6.0 (68.4) |
| I/σI | 17.6 (1.8) |
| Completeness (%) | 99.5 (99.6) |
| Redundancy | 3.2 (3.1) |
| Refinement | |
| Resolution (Å) | 50-2.25 |
| No. reflections | 77282 |
| Rwork/Rfree (%) | 19.4/24.6 (20.9/33.0) |
| No. atoms | 12034 |
| Protein | 11593 |
| Ligand/ion | 84 |
| Water | 354 |
| B-factors (Å2)(#) | |
| Protein | 50 |
| MBP-e6ap (chains A, B) | 50, 58 |
| p53 (chains C, D) | 43, 45 |
| E6 (chains F, H) | 49, 55 |
| Water | 47 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.37 |
| Ramachandran | |
| No. residues(&) | 1455 |
| Favored (%) | 95.94 |
| Allowed (%) | 3.99 |
| Outliers (%) | 0.07 |
Highest resolution shell is shown in parenthesis.
Values refer to occupancy-weighted average B-factors (OWAB)
The total number of residues in the chain cannot be analyzed: phi and psi angles cannot be analyzed for terminal residues, non-standard residues or incompletely modeled main chain residues.
Supplementary Material
Acknowledgments
This work received institutional support from CNRS, Université de Strasbourg, INSERM and Région Alsace. The work was supported by grants from ‘Ligue contre le Cancer’, NIH (grant R01CA134737 to SVP), ANR (ANR-13-JSV8-004-01), Instruct (ESFRI), the French Infrastructure for Integrated Structural Biology (FRISBI) and ‘Fondation pour La Recherche Medicale’ (fellowship to FXRF). The authors thank Pierre Poussin-Courmontagne (Structural Biology and Genomics platform, IGBMC, Strasbourg), Eric Ennifar (IBMC, Strasbourg), Vincent Olieric (SLS, Villigen) and Bruno Kieffer (IGBMC, Strasbourg) for advice.
Footnotes
Author contributions
DMZ, JP, AM, JRR, AF, ACS and KZ performed experiments; FXR, AP, DMZ and KZ performed structure determination; DMZ, FXR, JP, AM, AP and KZ analyzed the data; DMZ, JP and KZ prepared figures; KZ and GT wrote the manuscript together with comments from all authors; MM, SVP, AP, GT and KZ supervised the work.
Coordinates and structure factors have been deposited at the Protein Data Bank with accession code 4XR8.
The authors declare no competing financial interests.
The authors declare that the content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
References
- 1.Bosch FX, et al. Comprehensive control of human papillomavirus infections and related diseases. Vaccine. 2012;31(Suppl 6):G1–31. doi: 10.1016/j.vaccine.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 3.Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheffner M, Nuber U, Huibregtse J. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 5.Ansari T, Brimer N, Vande Pol SB. Peptide interactions stabilize and restructure human papillomavirus type 16 E6 to interact with p53. J Virol. 2012;86:11386–91. doi: 10.1128/JVI.01236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davey NE, Trave G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem Sci. 2011;36:159–69. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445:115–37. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanier K, et al. Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science. 2013;339:694–8. doi: 10.1126/science.1229934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu J, Rubin R, Yuan Z. A sequence element of p53 that determines its susceptibility to viral oncoprotein-targeted degradation. Oncogene. 2001;20:3519–3527. doi: 10.1038/sj.onc.1204454. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Coffino P. High-risk Human Papillomavirus E6 protein has two distinct binding sites within p53, of which one determines degradation. J Virol. 1996;70:4509–4516. doi: 10.1128/jvi.70.7.4509-4516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernard X, et al. Proteasomal degradation of p53 by human papillomavirus E6 oncoprotein relies on the structural integrity of p53 core domain. PLoS One. 2011;6:e25981. doi: 10.1371/journal.pone.0025981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanier K, et al. Solution structure analysis of the HPV16 E6 oncoprotein reveals a self-association mechanism required for E6-mediated degradation of p53. Structure. 2012;20:604–17. doi: 10.1016/j.str.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronchi VP, Klein JM, Edwards DJ, Haas AL. The active form of E6-associated protein (E6AP)/UBE3A ubiquitin ligase is an oligomer. J Biol Chem. 2014;289:1033–48. doi: 10.1074/jbc.M113.517805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ristriani T, Fournane S, Orfanoudakis G, Trave G, Masson M. A single-codon mutation converts HPV16 E6 oncoprotein into a potential tumor suppressor, which induces p53-dependent senescence of HPV-positive HeLa cervical cancer cells. Oncogene. 2009;28:762–72. doi: 10.1038/onc.2008.422. [DOI] [PubMed] [Google Scholar]
- 15.Fu L, et al. Degradation of p53 by human Alphapapillomavirus E6 proteins shows a stronger correlation with phylogeny than oncogenicity. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mesplede T, et al. p53 degradation activity, expression, and subcellular localization of E6 proteins from 29 human papillomavirus genotypes. J Virol. 2012;86:94–107. doi: 10.1128/JVI.00751-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiller T, Poppelreuther S, Stubenrauch F, Iftner T. Comparative analysis of 19 genital human papillomavirus types with regard to p53 degradation, immortalization, phylogeny, and epidemiologic risk classification. Cancer Epidemiol Biomarkers Prev. 2006;15:1262–7. doi: 10.1158/1055-9965.EPI-05-0778. [DOI] [PubMed] [Google Scholar]
- 18.Foster SA, Demers GW, Etscheid BG, Galloway DA. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J Virol. 1994;68:5698–5705. doi: 10.1128/jvi.68.9.5698-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper B, et al. Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology. 2003;306:87–99. doi: 10.1016/s0042-6822(02)00012-0. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, et al. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J Virol. 1999;73:7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakagawa S, et al. Mutational analysis of human papillomavirus type 16 E6 protein: transforming function for human cells and degradation of p53 in vitro. Virology. 1995;212:535–542. doi: 10.1006/viro.1995.1511. [DOI] [PubMed] [Google Scholar]
- 22.Zanier K, et al. The E6AP binding pocket of the HPV16 E6 oncoprotein provides a docking site for a small inhibitory peptide unrelated to E6AP, indicating druggability of E6. PLoS One. 2014;9:e112514. doi: 10.1371/journal.pone.0112514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stutz C, et al. Intracellular Analysis of the Interaction between the Human Papillomavirus Type 16 E6 Oncoprotein and Inhibitory Peptides. PLoS One. 2015;10:e0132339. doi: 10.1371/journal.pone.0132339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hengstermann A, Linares LK, Ciechanover A, Whitaker NJ, Scheffner M. Complete switch from Mdm2 to human papillomavirus E6-mediated degradation of p53 in cervical cancer cells. Proc Natl Acad Sci U S A. 2001;98:1218–23. doi: 10.1073/pnas.031470698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 2006;20:2373–82. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheffner M, Takahashi T, Huibregtse JM, Minna JD, Howley PM. Interaction of the human papillomavirus type 16 E6 oncoprotein with wild-type and mutant human p53 proteins. J Virol. 1992;66:5100–5. doi: 10.1128/jvi.66.8.5100-5105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherry JJ, et al. Structure based identification and characterization of flavonoids that disrupt human papillomavirus-16 E6 function. PLoS One. 2013;8:e84506. doi: 10.1371/journal.pone.0084506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malecka KA, et al. Identification and characterization of small molecule human papillomavirus E6 inhibitors. ACS Chem Biol. 2014;9:1603–12. doi: 10.1021/cb500229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon AF, Mueller GA, Zhong X, Pedersen LC. A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 2010;19:901–13. doi: 10.1002/pro.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: [Google Scholar]
- 36.Cassonnet P, et al. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat Methods. 2011;8:990–2. doi: 10.1038/nmeth.1773. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Rosengarth A, Luecke H. Structure of the human p53 core domain in the absence of DNA. Acta Crystallogr D Biol Crystallogr. 2007;63:276–81. doi: 10.1107/S0907444906048499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.