Abstract
A large number of genetic loci are associated with adult body mass index. However, the genetics of childhood body mass index are largely unknown. We performed a meta-analysis of genome-wide association studies of childhood body mass index, using sex- and age-adjusted standard deviation scores. We included 35 668 children from 20 studies in the discovery phase and 11 873 children from 13 studies in the replication phase. In total, 15 loci reached genome-wide significance (P-value < 5 × 10−8) in the joint discovery and replication analysis, of which 12 are previously identified loci in or close to ADCY3, GNPDA2, TMEM18, SEC16B, FAIM2, FTO, TFAP2B, TNNI3K, MC4R, GPR61, LMX1B and OLFM4 associated with adult body mass index or childhood obesity. We identified three novel loci: rs13253111 near ELP3, rs8092503 near RAB27B and rs13387838 near ADAM23. Per additional risk allele, body mass index increased 0.04 Standard Deviation Score (SDS) [Standard Error (SE) 0.007], 0.05 SDS (SE 0.008) and 0.14 SDS (SE 0.025), for rs13253111, rs8092503 and rs13387838, respectively. A genetic risk score combining all 15 SNPs showed that each additional average risk allele was associated with a 0.073 SDS (SE 0.011, P-value = 3.12 × 10−10) increase in childhood body mass index in a population of 1955 children. This risk score explained 2% of the variance in childhood body mass index. This study highlights the shared genetic background between childhood and adult body mass index and adds three novel loci. These loci likely represent age-related differences in strength of the associations with body mass index.
Introduction
Childhood obesity is an important public health problem with severe consequences, including an increased risk of premature death (1–5). Body mass index (BMI) has a strong genetic component with some reported heritability estimates being over 80% (6–8). Large genome-wide association studies (GWAS) have revealed many genetic loci associated with BMI or adiposity in adults (9–13). However, the genetic loci underlying BMI in children are less well known. The biological background of BMI may differ between children and adults. In addition, it may be that the relative contributions of the same genetic loci differ depending on age, for example due to different gene–environment interactions or body fat distributions (6,14,15). A limited number of loci have been identified to associate with dichotomous definitions of childhood obesity (16–18). Also, the roles of specific known adult loci for BMI, such as FTO and ADCY3, have been described in children (13,19). The age-specific effects are illustrated by longitudinal studies on the effects of the well-known adult BMI increasing risk allele of FTO with BMI throughout childhood (15). It has been reported that the adult BMI increasing risk allele is associated with lower BMI in infancy, an earlier adiposity rebound and a higher BMI from the age of 5 years onwards (14,15,20). To date, studies did not present a large GWAS meta-analysis on the full spectrum of childhood BMI (13,16–19).
To identify genetic loci influencing childhood BMI, we meta-analyzed 20 GWAS with a total of 35 668 children of European ancestry, combining data for around 2.5 million single-nucleotide polymorphisms (SNPs) imputed to the HapMap imputation panel. We used as outcome sex- and age-adjusted standard deviation scores at the oldest age between 2 and 10 years.
Results
Study characteristics are shown in Supplementary Material, Table S1. Childhood BMI was transformed into sex- and age-adjusted standard deviation scores (SDS) (LMS growth; Pan H, Cole TJ, 2012; http://www.healthforallchildren.co.uk).
Meta-analysis of genome-wide association studies
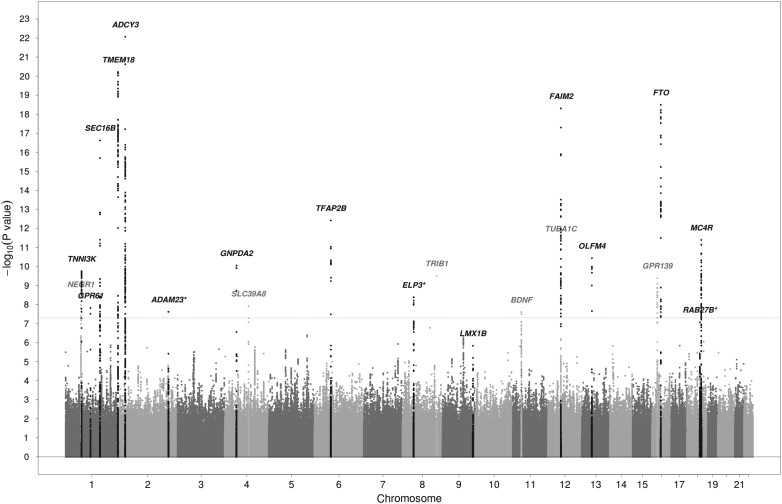
Inverse-variance weighted fixed-effects meta-analysis revealed 861 SNPs with genome-wide significant or suggestive P-values (<5 × 10−6). Two SNPs with high heterogeneity were not followed up (I2 values of 89.4 and 96.0), leaving 859 SNPs representing 43 loci. A locus was defined as a region of 500 kb to either side of the most significant SNP. The Manhattan and Quantile–Quantile plots of the discovery meta-analysis are shown in Figure 1 and Supplementary Material, Figure S1, respectively. The lambda for the discovery meta-analysis was 1.10. LD score regression analysis showed that this slight inflation was mainly due to polygenicity of the trait, rather than to population stratification, cryptic relatedness or other confounding factors (intercept 1.01). Individual study lambdas are shown in Supplementary Material, Table S2. All 43 loci were taken forward for replication in a sample of 11 873 children from 13 studies. Table 1 and Supplementary Material, Tables S3 and S4 show the results of the discovery, replication and joint analyses for the 43 genome-wide and suggestive loci.
Figure 1.
Manhattan plot of results of the discovery meta-analysis of 20 studies. Chromosomes are shown on the x-axis, the –log10 of the P-value on the y-axis. The gray dotted line represents the genome-wide significance cutoff of 5 × 10−8. Genes shown in black are the known loci that were significantly associated with childhood BMI in the joint discovery and replication analysis. Genes shown in gray were significant in the discovery, but not in the joint discovery and replication analysis. *indicates novel loci that were significantly associated with childhood BMI in the joint discovery and replication analysis. See also Table 1.
Table 1.
Results of the discovery, replication and joint analyses for 43 loci with P-values <5 × 10−6 in the discovery phase
| SNP | CHR | Position | Nearest gene | EA/Non-EA | EAFa | Betaa | SEa | P-value discovery | P-value replication | P-value joint |
|---|---|---|---|---|---|---|---|---|---|---|
| rs13130484b | 4 | 44870448 | GNPDA2 | T/C | 0.44 | 0.067 | 0.007 | 8.94 × 10−11 | 4.29 × 10−18 | 1.58 × 10−23 |
| rs11676272b | 2 | 24995042 | ADCY3 | G/A | 0.46 | 0.068 | 0.007 | 8.55 × 10−23 | 0.020 | 7.12 × 10−23 |
| rs4854349b | 2 | 637861 | TMEM18 | C/T | 0.83 | 0.090 | 0.009 | 6.00 × 10−21 | 0.005 | 5.41 × 10−22 |
| rs543874b | 1 | 176156103 | SEC16B | G/A | 0.20 | 0.077 | 0.009 | 2.38 × 10−17 | 8.77 × 10−4 | 2.20 × 10−19 |
| rs7132908b | 12 | 48549415 | FAIM2 | A/G | 0.39 | 0.066 | 0.008 | 4.99 × 10−19 | 0.043 | 1.57 × 10−18 |
| rs1421085b | 16 | 52358455 | FTO | C/T | 0.41 | 0.059 | 0.007 | 3.20 × 10−19 | 0.654 | 4.53 × 10−16 |
| rs12429545c | 13 | 53000207 | OLFM4 | A/G | 0.13 | 0.076 | 0.010 | 3.66 × 10−11 | 1.01 × 10−4 | 2.08 × 10−14 |
| rs987237b | 6 | 50911009 | TFAP2B | G/A | 0.19 | 0.062 | 0.009 | 3.81 × 10−13 | 0.224 | 1.80 × 10−12 |
| rs12041852b | 1 | 74776088 | TNNI3K | G/A | 0.46 | 0.046 | 0.007 | 1.77 × 10−10 | 0.142 | 2.28 × 10−10 |
| rs6567160b | 18 | 55980115 | MC4R | C/T | 0.23 | 0.050 | 0.008 | 4.06 × 10−12 | 0.996 | 1.21 × 10−9 |
| rs13253111 | 8 | 28117893 | ELP3 | A/G | 0.57 | 0.042 | 0.007 | 4.13 × 10−9 | 0.114 | 4.89 × 10−9 |
| rs8092503 | 18 | 50630485 | RAB27B | G/A | 0.27 | 0.045 | 0.008 | 8.55 × 10−8 | 0.034 | 8.17 × 10−9 |
| rs3829849b | 9 | 128430621 | LMX1B | T/C | 0.36 | 0.041 | 0.007 | 1.46 × 10−6 | 0.001 | 8.81 × 10−9 |
| rs13387838 | 2 | 206989692 | ADAM23 | A/G | 0.04 | 0.139 | 0.025 | 2.40 × 10−8 | 0.306 | 2.84 × 10−8 |
| rs7550711d | 1 | 109884409 | GPR61 | T/C | 0.04 | 0.105 | 0.019 | 1.50 × 10−8 | 0.401 | 4.52 × 10−8 |
| rs17309930b | 11 | 27705069 | BDNF | A/C | 0.21 | 0.045 | 0.009 | 2.47 × 10−8 | 0.540 | 1.41 × 10−7 |
| rs2590942b | 1 | 72657869 | NEGR1 | T/G | 0.82 | 0.047 | 0.009 | 3.88 × 10−9 | 0.966 | 1.91 × 10−7 |
| rs13107325b | 4 | 103407732 | SLC39A8 | T/C | 0.07 | 0.081 | 0.016 | 1.19 × 10−8 | 0.970 | 3.79 × 10−7 |
| rs10151686b | 14 | 29536217 | PRKD1 | A/G | 0.04 | 0.096 | 0.019 | 1.50 × 10−6 | 0.109 | 6.99 × 10−7 |
| rs25832 | 5 | 66211438 | LOC375449 | A/G | 0.71 | 0.039 | 0.008 | 2.41 × 10−6 | 0.177 | 1.62 × 10−6 |
| rs7869969 | 9 | 95257268 | FAM120A | G/A | 0.33 | 0.036 | 0.008 | 4.43 × 10−7 | 0.425 | 1.68 × 10−6 |
| rs11079830c | 17 | 44037629 | HOXB6 | A/G | 0.58 | 0.034 | 0.007 | 1.43 × 10−6 | 0.254 | 1.98 × 10−6 |
| rs4569924 | 5 | 153520218 | GALNT10 | T/C | 0.43 | 0.032 | 0.007 | 4.06 × 10−7 | 0.823 | 3.48 × 10−6 |
| rs8046312b | 16 | 19886835 | GPR139 | A/C | 0.81 | 0.042 | 0.009 | 4.06 × 10−10 | 0.185 | 3.97 × 10−6 |
| rs1838856 | 2 | 113822060 | PAX8 | A/C | 0.46 | 0.034 | 0.008 | 1.85 × 10−6 | 0.588 | 1.47 × 10−5 |
| rs633143 | 1 | 179716108 | CACNA1E | T/C | 0.14 | 0.044 | 0.011 | 3.40 × 10−6 | 0.648 | 2.44 × 10−5 |
| rs4923207 | 11 | 24713901 | LUZP2 | T/G | 0.79 | 0.039 | 0.010 | 1.60 × 10−6 | 0.834 | 3.52 × 10−5 |
| rs6971577 | 7 | 140350204 | MRPS33 | C/G | 0.78 | 0.036 | 0.009 | 1.17 × 10−6 | 0.690 | 6.80 × 10−5 |
| rs10866069 | 3 | 64366964 | ADAMTS9 | T/C | 0.17 | 0.041 | 0.011 | 3.05 × 10−6 | 0.687 | 8.43 × 10−5 |
| rs12457682 | 18 | 7216505 | LAMA1 | C/A | 0.23 | 0.035 | 0.009 | 3.81 × 10−6 | 0.942 | 9.01 × 10−5 |
| rs11165675b | 1 | 96812556 | PTBP2 | A/G | 0.27 | 0.031 | 0.008 | 2.93 × 10−6 | 0.520 | 1.01 × 10−4 |
| rs12096993 | 1 | 217931859 | SLC30A10 | T/C | 0.27 | 0.031 | 0.008 | 1.38 × 10−6 | 0.583 | 1.02 × 10−4 |
| rs760931 | 1 | 1637388 | CDC2L1 | C/G | 0.93 | 0.103 | 0.027 | 3.15 × 10−6 | 0.160 | 1.27 × 10−4 |
| rs2968990 | 4 | 131098524 | C4orf33 | C/T | 0.37 | 0.028 | 0.007 | 1.67 × 10−6 | 0.487 | 1.42 × 10−4 |
| rs1247117 | 10 | 120418792 | C10orf46 | G/A | 0.11 | 0.040 | 0.011 | 3.47 × 10−6 | 0.339 | 2.62 × 10−4 |
| rs6580706 | 12 | 47959818 | TUBA1C | C/G | 0.34 | 0.031 | 0.009 | 1.83 × 10−8 | 0.047 | 3.68 × 10−4 |
| rs8092620 | 18 | 41433991 | SLC14A2 | G/T | 0.48 | 0.024 | 0.007 | 3.31 × 10−6 | 0.199 | 7.32 × 10−4 |
| rs188584 | 3 | 62675007 | CADPS | C/A | 0.77 | 0.028 | 0.008 | 3.23 × 10−6 | 0.139 | 0.001 |
| rs4870949 | 8 | 126704776 | TRIB1 | T/C | 0.07 | 0.164 | 0.054 | 3.13 × 10−10 | 0.061 | 0.002 |
| rs1573972 | 4 | 171559399 | AADAT | C/T | 0.19 | 0.030 | 0.013 | 3.78 × 10−6 | 0.232 | 0.020 |
| rs214821 | 20 | 2258291 | TGM3 | T/C | 0.02 | 0.479 | 0.208 | 3.38 × 10−6 | 0.575 | 0.021 |
| rs8084077 | 18 | 49532928 | DCC | T/C | 0.73 | 0.014 | 0.008 | 3.47 × 10−6 | 2.72 × 10−5 | 0.062 |
| rs3845265 | 18 | 63690108 | DSEL | G/A | 0.71 | 0.013 | 0.008 | 8.86 × 10−7 | 1.44 × 10−6 | 0.087 |
CHR, chromosome; EA, effect allele; EAF, effect allele frequency; SE, standard error.
Bolded P-values indicate genome-wide significance in the joint analysis.
aFrom joint analysis.
bLocus previously reported in Ref. (11).
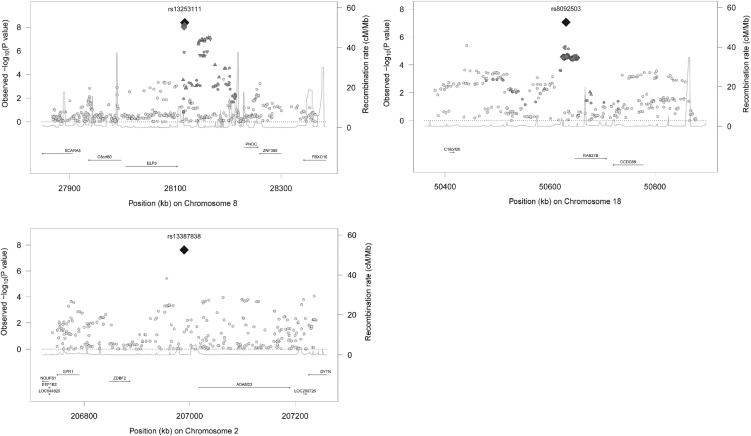
In total, 15 of these reached genome-wide significance in the joint analysis. Twelve out of these 15 had been reported previously for related phenotypes. SNPs in or close to ADCY3, GNPDA2, TMEM18, SEC16B, FAIM2, FTO, TFAP2B, TNNI3K, MC4R, GPR61, LMX1B and OLFM4 are associated with adult BMI or childhood obesity (11,13,16). We identified three novel loci: rs13253111 near ELP3, rs8092503 near RAB27B and rs13387838 near ADAM23. Per additional risk allele, BMI increased 0.04 Standard Deviation Score (SDS) [Standard Error (SE) 0.007], 0.05 SDS (SE 0.008) and 0.14 SDS (SE 0.025) for rs13253111, rs8092503 and rs13387838, respectively. Figure 2 and Supplementary Material, Figure S2 show the regional plots and the forest plots, respectively, for these loci.
Figure 2.
Regional plots of the three novel loci for childhood BMI. On the x-axis, the position of SNPs on the chromosome is shown. On the left y-axis is the –log10 of the P-values from the discovery analysis, on the right y-axis is the estimated recombination rate (from HapMap), shown by the light blue line in the figure. The named SNP is the most significant SNP in the locus from the discovery meta-analysis. The linkage disequilibrium of all SNPs with the most significant SNP is shown by the symbols, with dark gray diamonds indicating an R2 of ≥0.8, inversed dark gray triangles indicating an R2 of 0.6–0.8, dark gray triangles indicating an R2 of 0.4–0.6, dark gray circles indicating an R2 of 0.2–0.4 and light gray circles indicating an R2 of 0–0.2. Genes (from HapMap release 22) are plotted below the x-axis.
Genetic risk score
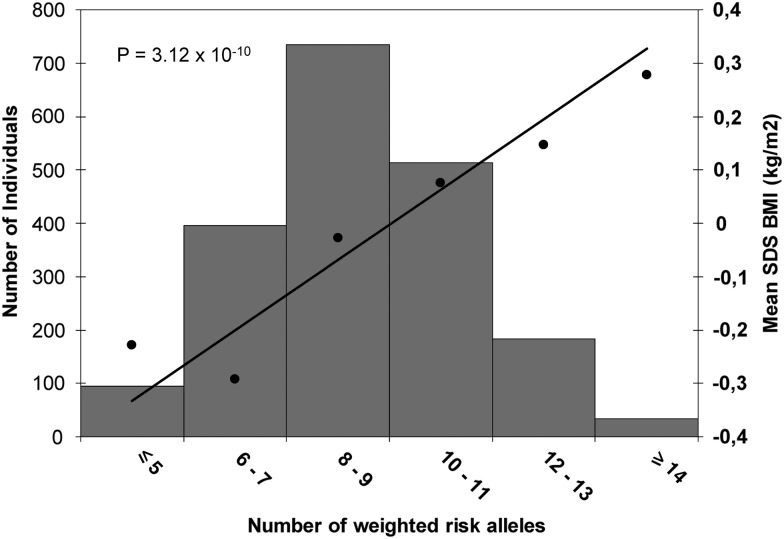
We combined the 15 identified genome-wide significant SNPs into a genetic risk score that summed the number of BMI-increasing alleles weighted by their betas from the discovery analysis and rescaled to a range of 0 to 30, which is the maximum number of risk alleles. The risk score was associated with childhood BMI (P-value = 3.12 × 10−10) in 1955 children from the PIAMA Study, one of our largest replication cohorts. For each additional average risk allele in the score, childhood BMI increased by 0.073 SDS (SE 0.011) (Fig. 3). This risk score explained 2.0% of the variance in childhood BMI.
Figure 3.
Association of the weighted risk score with BMI. Along the x-axis, categories of the weighted risk score are presented, the mean standard deviation score (SDS)-BMI per group is shown on the right y-axis, with the line representing the regression of the mean SDS–BMI values on the categories of the weighted risk score. The left y-axis represents the number of children in each risk score category, shown in the histogram. The P-value is derived from the analysis of the continuous risk score. Analysis was performed in the PIAMA Study (n = 1955).
Associations with adult body mass index and childhood obesity
The genetic correlation between childhood BMI and adult BMI was 0.73. A lookup of the 15 SNPs associated with childhood BMI in a recently published GWAS meta-analysis on adult BMI in >300 000 participants revealed that all SNPs showed evidence for association with adult BMI, with P-values of 0.005, 5.76 × 10−5 and 0.003 for the novel SNPs rs13253111, rs8092503 and rs13387838, respectively. Also, the direction of the effect estimates for all 15 SNPs was the same in children and adults (Supplementary Material, Table S5) (11). The 15 SNPs found in this study explained 0.94% of the variance in adult BMI in the GIANT consortium (11).
A reverse lookup in our dataset of the 97 known genome-wide significant loci previously reported to be associated with adult BMI showed that 22 out of the 97 loci were significantly associated with childhood BMI, using a Bonferroni-adjusted P-value cutoff of 5.2 × 10−4 for 97 SNPs. A total of 50 out of the 97 known adult BMI SNPs were nominally associated with childhood BMI (P-value <0.05). The direction of the effect estimates was the same in adults and children for 86 SNPs (P-value binomial sign test <1.0 × 10−4; Supplementary Material, Table S6).
We looked up the association of the three novel loci in a GWAS meta-analysis of childhood obesity. In this study, childhood obesity cases were defined as having a BMI ≥ 95th percentile, whereas childhood normal weight controls were defined as having a BMI < 50th percentile. This meta-analysis included 22 studies, of which 16 were also included in our current meta-analysis. All three SNPs were associated with childhood obesity (P-values 0.01, 0.005 and 6.0 × 10−4 for rs13253111, rs8092503 and rs13387838, respectively) (16).
Functional analysis
To explore functionality, we first analyzed whether the 15 identified SNPs affect messenger RNA expression (eQTLs). We analyzed eQTLs from peripheral blood samples from 5311 individuals, which revealed two cis-eQTLs [false discovery rate (FDR) P-value <0.05] for rs11676272, the top SNP in one of the previously identified loci (ADCY3). One of these eQTLs was for ADCY3, and one was for DNAJC27 (21). Also, we found a cis-eQTL for FAM125B for rs3829849, which is located in LMX1B (Supplementary Material, Table S7). eQTL analysis in adipose tissue, a more specific target tissue in relation to BMI, from 856 healthy female twins in the MuTHER resource in Genevar revealed two significant cis-eQTLs (distance to SNP < 1 Mb) for rs11676272, for transcripts of ADCY3 and POMC, with a Bonferroni-corrected P-value of <0.003 (22,23). The association of rs11676272 with expression of ADCY3 was also validated in a second eQTL analysis in a smaller set of 206 lymphoblastoid cell lines (24). We did not identify eQTLs related to our three novel loci.
Second, we performed functional analyses with the tool Data-Driven Expression Prioritized Integration for Complex Traits (DEPICT) using all SNPs with a P-value <1 × 10−5 in the discovery analysis (see Materials and Methods for details) (25). Gene prioritization analysis did not show prioritized genes, nor did the gene set enrichment analysis reveal evidence for enriched reconstituted gene sets and genes near the associated SNPs were not found to enrich for expression in a panel of 2009 tissue and cell types (FDR < 0.05; Supplementary Material, Tables S8a, b and c).
Discussion
In this GWAS meta-analysis of childhood BMI among >47 000 children, we identified 15 genome-wide significant loci, of which three loci, rs13253111 near ELP3, rs8092503 near RAB27B and rs13387838 near ADAM23, have not been associated with adiposity-related phenotypes before.
Large GWAS have revealed many genetic loci associated with BMI or adiposity in adults (9–13). A recent meta-analysis in up to 339 224 individuals identified 97 BMI-associated loci, explaining 2.7% of the adult BMI variation. Pathway analyses showed that the central nervous system may play a large role in obesity susceptibility. The number of identified loci associated with BMI or obesity in childhood is scarce. Of the total of 15 loci associated with childhood BMI in the current study, 12 have previously been associated with adiposity outcomes in adults or children. All 12 loci are known to be associated with adult BMI (11). Also, eight loci, including those in or near ADCY3 (annotated to the nearby gene POMC in the previous paper), TMEM18, SEC16B, FAIM2, FTO, TNNI3K, MC4R and OLFM4, have previously been associated with childhood obesity (16). All three novel loci were nominally associated with the more extreme outcome of childhood obesity in a largely overlapping population of child cohorts (16).
A recent meta-analysis of two studies showed that the known loci FTO, MC4R, ADCY3, OLFM4 are associated with BMI trajectories in childhood (26). Their findings also suggested that a locus annotated to FAM120AOS influences childhood BMI, which could not be replicated in the current study. The lead SNP in this locus, rs944990, had a P-value of 1.61 × 10−5 in the current analysis. These findings suggest that the overlap between the genetic background of childhood and adult BMI is relatively large, but not complete.
rs7550711 represents one of the 12 identified loci known to be associated with BMI or obesity in adults and children. rs7550711 is a proxy for rs17024258 and rs17024393 (R2 0.8 with both SNPs), which have previously been associated with adult obesity and BMI, respectively, and annotated with the GNAT2 gene. However, our proxy resides in GPR61, G protein-coupled receptor 61, the biology of which may be more relevant to BMI. Gpr61-deficient mice are obese and have hyperphagia, suggesting the role of Gpr61 in food intake regulation (27). Further studies, including expression studies in relevant human tissues, are needed to establish the causal genes underlying this association.
We identified three loci, rs13253111 near ELP3, rs8092503 near RAB27B and rs13387838 near ADAM23, which have not been associated with adiposity-related phenotypes before in adulthood or childhood. The nearest genes to the novel loci have varying functions. ELP3, Elongator Acetyltransferase Complex, subunit 3, has a potential role in the migration of cortical projection neurons and in paternal demethylation after fertilization in mice (28–30). RAB27B, RAS-associated protein RAB27B, encodes a membrane-bound protein with a role in secretory vesicle fusion and trafficking. It has been associated with pituitary hormone secretion, regulation of exocytosis of digestive enzyme containing granules from pancreatic acinar cells and with gastric acid secretion (31–33). Expression of ADAM23, A Disintegrin And Metalloproteinase Domain 23, may influence tumor progression and brain development (34,35). It has also been described to be expressed in mouse adipose tissue and to have a potential role in adipogenesis in vitro (36).
Two of our novel loci, rs13253111 near ELP3 and rs13387838 near ADAM23, are close to rs4319045 and rs972540, respectively. Both these SNPs were reported as subthreshold results in the GWAS meta-analysis on adult BMI (11). However, the linkage disequilibrium between the SNPs in both pairs is very low (R2 ≤ 0.1 for both) suggesting that these SNPs may represent different signals. It is important to note that, although both SNPs reached genome-wide significance in the joint discovery and replication analysis, the P-values in the replication stage were non-significant. This lack of significance may be due to the smaller sample size and lower power. Also, the joint P-values were slightly higher than the discovery P-values. Heterogeneity between the discovery and the replication stages was low to moderate, with I2 values of 61.1 and 27.8 for rs13253111 and rs13387838, respectively (P-values > 0.1 for both). These two signals need to be interpreted with some caution and further studies with larger sample sizes are needed to fully clarify the role of variants in these regions in the physiology of BMI.
Functional analysis showed cis-eQTLs for the lead SNPs in two of the known loci. rs11676272 was associated with eQTLs in ADCY3 and DNAJC27, also known as RBJ. Both these genes have been associated with adult BMI before and the association of rs11676272 with expression of ADCY3 has been previously described in childhood BMI (11,13,37). rs3829849 was associated with an eQTL in FAM125B, or MVB12B, multivesicular body subunit 12B. This gene encodes a component of ESCRT-I (endosomal sorting complex required for transport I), a plasma membrane complex with a role in vesicular trafficking was recently described to be associated with intra-ocular pressure (38). However, the LD of our SNPs with the peak markers for the DNAJC27 (R2 0.11) and the FAM125B (R2 0.03) transcripts was low. Our analysis using DEPICT did not show enriched gene sets. This may reflect the relatively limited sample size in our analysis. Further studies are needed to determine the potential functional impact of all SNPs associated with childhood BMI.
Using LD score regression analysis with our meta-analysis results and the results from the recently published GWAs meta-analysis on adult BMI as input, we found that the genetic correlation between childhood and adult BMI was high (11,39). The variance in adult BMI explained by the 15 SNPs identified in this study was lower than in children. The novel SNPs reported in this study may represent loci that specifically influence childhood BMI, but not adult BMI. An alternative explanation is that the effect sizes of these loci may be larger in children than in adults, which may explain the discovery in childhood studies but not in adult studies (11). The large overlap between childhood and adult BMI loci suggests that many of these loci may not represent childhood-specific effects, but rather involvement of the same loci with differential effect sizes at different ages. Age-specific effects of genetic variants associated with BMI in children have been described for the FTO locus (15). However, longitudinal studies with multiple measurements of BMI are needed to confirm and quantify such varying effects with age. In discussing the genetic overlap between childhood and adult BMI, it needs to be noted that, because of the differences in body proportions and body fat distribution, childhood BMI may be a different phenotype compared with adult BMI. Our outcome was the conventional measure of BMI calculated as weight/height2. Especially in early childhood, higher orders of magnitude for height may be more appropriate. Results from a previously published GWAS study on childhood BMI in two of the cohorts included in the current meta-analysis suggest that the results for SNPs close to ADCY3 are different when higher orders of magnitude for height are being used (37). Further studies are needed to identify loci related to more specific and directly assessed measures of adiposity and body fat distribution in young children.
In conclusion, we identified 15 loci associated with childhood BMI, of which three are novel. Our results highlight a considerable shared genetic background between childhood and adult BMI. The novel BMI-related loci may reflect childhood-specific genetic associations or differences in strength of associations between age groups.
Materials and Methods
Study populations
Characteristics of each discovery and replication study population can be found in Supplementary Material, Table S1 and Methods. The discovery analysis included 20 studies with an age range from 3 to 10 years: the Avon Longitudinal Study of Parents and Children (ALSPAC, 6887 children), the Children's Hospital of Philadelphia (CHOP, 2456 children), the Copenhagen Studies on Asthma in Childhood 2000 birth cohort (COPSAC2000, 309 children), the Danish National Birth Cohort (DNBC, 1020 children), the Generation R Study (GenerationR, 2226 children), the GOYA Study (GOYA, 199 children), the Helsinki Birth Cohort Study (HBCS, 1674 children), the INfancia y Medio Ambiente Project (INMA, 756 children), the Leipzig study (Leipzig, 555 children), the Lifestyle—Immune System—Allergy Study plus German Infant Study on the influence of Nutrition Intervention (LISA + GINI, 1147 children), the Manchester Asthma and Allergy Study (MAAS, 801 children), the Norwegian Mother and Child Cohort Study (MoBa, 126 children), the Northern Finland Birth Cohort 1966 (NFBC 1966, 3948 children), the Northern Finland Birth Cohort 1986 (NFBC 1986, 4000 children), the Netherlands Twin Register (NTR, 1810 children), the Physical Activity and Nutrition in Children Study (PANIC, 423 children), the Western Australian Pregnancy Cohort (Raine) Study (Raine, 1458 children), the Special Turku coronary Risk factor Intervention Project (STRIP, 569 children), the Young Finns Study (YFS, 1134 children), the British 1958 Birth Cohort Study, with two subcohorts that were entered into the meta-analysis separately (1958BC-T1DGC, 1974 children, and 1958BC-WTCCC2, 2196 children).
We included 13 replication studies. Eleven of these were cohort studies: 574 children from the Copenhagen Studies on Asthma in Childhood 2010 birth cohort (COPSAC2010), 676 additional children from the DNBC, 386 additional children from LISA + GINI, 3152 children from the TEDS Study, 1955 children from the Prevention and Incidence of Asthma and Mite Allergy birth cohort study (PIAMA), 1665 children from the BREATHE Study, 447 children from the Bone Mineral Density in Childhood Study (BMDCS), 200 children from the TEENs of Attica: Genes and Environment (TEENAGE) study, additional imputed data on 857 children from the Leipzig Study, 480 additional children from PANIC and additional imputed data for 569 children from STRIP. We also included two obesity case–control studies in the replication: the Danish Childhood Obesity Biobank (306 cases, 158 controls) and the French Young Study (304 cases, 144 controls). In the BREATHE Study, information was available about six SNPs only (rs8046312, rs12429545, rs13130484, rs3845265, rs543874, rs8084077).
All included children were of European ethnic origin. Sex- and age-adjusted standard deviation scores were created for BMI at the latest time point (oldest age, if multiple measurements existed) between 2 and 10 years using the same software across all studies (LMS growth; Pan H, Cole TJ, 2012; http://www.healthforallchildren.co.uk). Syndromic cases of obesity and children of non-European ethnic origin were excluded. In the case of twin pairs, only one twin was included, either randomly or based on genotyping or imputation quality.
Statistical approach
Cohort-specific genome-wide association analyses were first run in the discovery cohorts, using high-density Illumina or Affymetrix SNP arrays, followed by imputation to the HapMap CEU release 22 imputation panel. The MAAS study imputed to the combined 1000 Genomes (1000G) Pilot + HapMap 3 (release June 2010/Feb 2009) panel. Before imputation, studies applied study-specific quality filters on samples and SNP call rate, minor allele frequency and Hardy–Weinberg disequilibrium (see Supplementary Material, Table S1 for details). Leipzig (discovery sample), NFBC1986, STRIP (discovery sample) and PANIC (discovery sample) contributed unimputed data from the Metabochip. Linear regression models assuming an additive genetic model were run in each study, to assess the association of each SNP with SDS–BMI, adjusting for principal components if this was deemed needed in the individual studies. As SDS–BMI is age and sex specific, no further adjustments were made. Before the meta-analysis, we applied quality filters to each study, filtering out SNPs with a minor allele frequency below 1% and SNPs with poor imputation quality (MACH r2_hat ≤ 0.3, IMPUTE proper_info ≤0.4 or info ≤0.4). For studies contributing unimputed metabochip data to the discovery analysis, we excluded SNPs with a SNP call rate of <0.95 or with a Hardy–Weinberg Equilibrium P-value of ≤0.00001. We performed fixed-effects inverse-variance weighted meta-analysis of all discovery samples using Metal (40). Genomic control was applied to every study before the meta-analysis. Individual study lambdas ranged from 0.985 to 1.077 (Supplementary Material, Table S2). The lambda of the discovery meta-analysis was 1.10. After the meta-analysis, SNPs for which information was available in only one study were removed.
The final dataset consisted of 2 499 691 autosomal SNPs. The most significant SNP for each of 43 genome-wide significant or suggestive loci (P-value < 5 × 10−6) was taken forward for replication in 13 replication cohorts. A locus was defined as a region 500 kb to either side of the most significant SNP. All replication cohorts had in silico data available. One of them only had non-imputed data (BREATHE), two (TEENAGE and TEDS) had data imputed to HapMap release 22, one cohort (PANIC) used exome chip data and the other nine performed imputation to 1000G. The replication samples of the STRIP and Leipzig studies only contributed 20 and 21 imputed SNPs, respectively, as the unimputed SNPs were part of the discovery analysis. Fixed-effects inverse-variance meta-analysis was performed for these 43 SNPs combining the discovery samples and all replication samples, giving a joint analysis beta, standard error and P-value (Table 1 and Supplementary Material, Table S2).
Sensitivity analyses
Allele frequency differences between the discovery and the replication samples were small and stayed within a range of seven percentage points for all SNPs, except for rs1573972, which had a minor allele frequency of 9% in the discovery analysis and 28% in the replication analysis. This was likely due to the inclusion of one study (MAAS) that had imputed to the combined HapMap + 1000G panel, whereas all other studies with imputed data had imputed to HapMap. To increase homogeneity, we performed several sensitivity analyses. First, we reran the discovery meta-analysis excluding the MAAS study. This analysis did not materially change our findings, with one additional SNP (rs10055577) reaching the subthreshold level of significance (P-value = 1.10 × 10−6) and five SNPs (rs4870949, rs1838856, rs633143, rs10866069 and rs1573972) losing significance. None of these five SNPs had replicated in the primary analysis. Second, we reran the replication and joint meta-analysis including only those cohorts that imputed to 1000G. Results of this analysis were very similar to the primary analysis, with two additional replicated SNPs, rs17309930 near BDNF and rs13107325 in SLC39A8. Both of these are known loci for adult BMI (11,13). Third, we reran the replication including only the HapMap-imputed and unimputed studies (TEDS, TEENAGE and BREATHE). The results were very similar to those using all studies, with rs4870949 and rs2590942 now passing the significance threshold and rs8092503 and rs3829849 now just above it (results not shown). rs1573972 was not replicated in any of the analyses. As results of the third and fourth sensitivity analyses were very similar to those including all replication cohorts, we used the latter as our main analysis for reasons of power.
Genetic risk score and percentage of variance explained
A weighted risk score was computed as the sum of the number of SDS–BMI-increasing alleles (dosage) weighted by the effect sizes from the discovery meta-analysis. Then, the score was rescaled to range from zero to the maximum number of SDS–BMI-increasing alleles (30 alleles for 15 SNPs) and rounded to the nearest integer. The association of the risk score with SDS–BMI was assessed in one of the largest replication cohorts (PIAMA, N= 1955) by running a linear regression model. The variance in SDS–BMI explained by the risk score was estimated by the unadjusted R2 of this model. The percentage of variance in adult BMI explained by the 15 SNPs was calculated using the published data from the recently published large meta-analysis of GWAs studies on adult BMI (11). For each SNP, the variance explained was calculated as: 2 × (adult effect size2) × MAF × (1 − MAF), and these variances were then summed to give the total percentage of variance in adult BMI explained by the 15 SNPs (11,41).
LD score regression
LD score regression was used with the standard settings (39). Changing the minor allele frequency filter from 0 to 0.05 did not change the results. Therefore, we report the results of the unfiltered analysis only.
eQTL analysis
eQTL analysis was conducted using the most significant SNP from each of the 15 genome-wide significant loci from the joint analysis. There was no linkage disequilibrium between these SNPs. First, we assessed whether the top SNPs or their proxies, identified on the basis of R2 > 0.7, were associated with gene expression in whole-blood cells in a sample of 5311 individuals (21). Expression in this dataset was assessed using Illumina Whole-Genome Expression BeadChips (HumanHT-12). eQTLs were deemed cis when the distance between the SNP chromosomal position and the probe midpoint was <250 kb. eQTLs were mapped using Spearman's rank correlation, using imputation dosage values as genotypes. An FDR P-value of <0.05 was considered significant. Second, the 15 SNPs were introduced to the online eQTL database Genevar (www.sanger.ac.uk/resources/software/genevar) to explore their associations with expression transcripts of genes in proximity (<1 Mb distance) to the SNP in adipose tissue from 856 healthy female twins of the MuTHER resource (22,23). We used Bonferroni correction for the significance threshold (P-value <0.003).
Data-driven Expression Prioritized Integration for Complex Traits
DEPICT was run using SNPs with a P-value of <10−5 yielding 56 independent DEPICT loci comprising 100 genes (42). DEPICT was run using default settings, that is using 500 permutations for bias adjustment, 20 replications for FDR estimation, normalized expression data from 77 840 Affymetrix microarrays for gene set reconstitution [see Ref. (43) for details], 14 461 reconstituted gene sets for gene set enrichment analysis and testing 209 tissue/cell types assembled from 37 427 Affymetrix U133 Plus 2.0 Array samples for enrichment in tissue/cell type expression (42).
Supplementary Material
Supplementary material is available at HMG online.
Acknowledgements
ALSPAC Study
The MRC IEU is supported by the Medical Research Council and the University of Bristol (grant code MC_UU_12013/1–9). The authors are extremely grateful to all the families who took part in the ALSPAC study, the midwives for their help in recruiting them and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. ALSPAC GWAS data were generated by Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and LabCorp (Laboratory Corporation of America) supported by 23 and Me. The MRC IEU is supported by the Medical Research Council and the University of Bristol (grant code MC_UU_12013/1-9).
1958BC-T1DGC and 1958CB-WTCCC
DNA collection was funded by MRC grant G0000934 and cell-line creation by Wellcome Trust grant 068545/Z/02. This research used resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, National Institute of Child Health and Human Development and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of investigators who contributed to generation of the data is available from the Wellcome Trust Case-Control Consortium website. Funding for the project was provided by the Wellcome Trust under the award 076113. The work was supported by the Department of Health Policy Research Programme through the Public Health Research Consortium (PHRC). Information about the wider programme of the PHRC is available from http://phrc.lshtm.ac.uk. Great Ormond Street Hospital/University College London, Institute of Child Health and Oxford Biomedical Research Centre, University of Oxford receive a proportion of funding from the Department of Health's National Institute for Health Research (NIHR) (Biomedical Research Centres’ funding). This paper presents independent research and the views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. M.I.M. acknowledges support from the EU Framework 7 ENGAGE (HEALTH-F4-2007-201413) and The Wellcome Trust (098381 and 090367). M.I.M. is a Wellcome Trust Senior Investigator and a NIHR Senior Investigator.
BREATHE
The research leading to these results has received funding from the European Research Council under the ERC Grant Agreement number 268479—the BREATHE project. We are acknowledged with all the families participating into the study for their altruism and particularly to the schools Antoni Brusi, Baloo, Betània—Patmos, Centre d'estudis Montseny, Collegi Shalom, Costa i Llobera, El sagrer, Els Llorers, Escola Pia de Sarrià, Escola Pia Balmes, Escola concertada Ramon Llull, Escola Nostra Sra. de Lourdes, Escola Tècnica Professional del Clot, Ferran i Clua, Francesc Macià, Frederic Mistral, Infant Jesús, Joan Maragall, Jovellanos, La Llacuna del Poblenou, Lloret, Menéndez Pidal, Nuestra Señora del Rosario, Miralletes, Ramon Llull, Rius i Taulet, Pau Vila, Pere Vila, Pi d'en Xandri, Projecte, Prosperitat, Sant Ramon Nonat-Sagrat Cor, Santa Anna, Sant Gregori, Sagrat Cor Diputació, Tres Pins, Tomàs Moro, Torrent d'en Melis, Virolai.
BMDCS
The study was funded by R01 HD58886 and R01 HD076321 and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) contracts (N01-HD-1-3228, -3329, -3330, -3331, -3332, -3333) and the CTSA program Grant 8 UL1 TR000077.
CHOP
We thank the network of primary care clinicians, their patients and families for their contribution to this project and clinical research facilitated through the Pediatric Research Consortium (PeRC) at The Children's Hospital of Philadelphia. Rosetta Chiavacci, Elvira Dabaghyan, Hope Thomas, Kisha Harden, Andrew Hill, Kenya Fain, Crystal Johnson-Honesty, Cynthia Drummond, Shanell Harrison and Sarah Wildrick, Cecilia Kim, Edward Frackelton, George Otieno, Kelly Thomas, Cuiping Hou, Kelly Thomas and Maria L. Garris provided expert assistance with genotyping or data collection and management. We thank Smari Kristinsson, Larus Arni Hermannsson and Asbjörn Krisbjörnsson of Raförninn ehf for their extensive software design and contribution. This research was financially supported by an Institute Development Award from the Children's Hospital of Philadelphia, a Research Development Award from the Cotswold Foundation and NIH grant R01 HD056465.
COPSAC2000 and COPSAC2010
The authors gratefully express their gratitude to the children and families of the COPSAC cohort studies for all their support and commitment, and we acknowledge and appreciate the unique efforts of the COPSAC research team. COPSAC is funded by private and public research funds. The Lundbeck Foundation; the Pharmacy Foundation of 1991; Augustinus Foundation; the Danish Medical Research Council and The Danish Pediatric Asthma Centre provided the core support for the study. For full list, see copsac.com.
DNBC
Support for the Danish National Birth Cohort was obtained from the Danish National Research Foundation, the Danish Pharmacists' Fund, the Egmont Foundation, the March of Dimes Birth Defects Foundation, the Augustinus Foundation and the Health Fund of the Danish Health Insurance Societies. The generation of GWAS genotype data for the DNBC samples was carried out within the GENEVA consortium with funding provided through the NIH Genes, Environment and Health Initiative (GEI) (U01HG004423). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01HG004446). Genotyping was performed at Johns Hopkins University Center for Inherited Disease Research, with support from the NIH GEI (U01HG004438). B.F. is supported by an Oak Foundation Fellowship.
French Young
The authors thank the subjects and families who participated in this study. This work was supported by grants from the Agence Nationale de la Recherche, the Conseil Régional Nord-Pas de Calais as part of Fonds Européen de Développement Economique et Regional, Genome Quebec as part of Genome Canada and the UK MRC.
Generation R
The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam. We gratefully acknowledge the contribution of children and parents, general practitioners, hospitals, midwives and pharmacies in Rotterdam. The study protocol was approved by the Medical Ethical Committee of the Erasmus Medical Centre, Rotterdam. Written informed consent was obtained from all participants. The general design of Generation R Study is made possible by financial support from the Erasmus Medical Center, Rotterdam, the Erasmus University Rotterdam, the Netherlands Organization for Health Research and Development (ZonMw), the Netherlands Organisation for Scientific Research (NWO), the Ministry of Health, Welfare and Sport and the Ministry of Youth and Families. V.W.J. received an additional grant from the Netherlands Organization for Health Research and Development (VIDI 016.136.361) and a European Research Council Consolidator Grant (ERC-2014-CoG-648916). J.F.F. has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 633595. The generation and management of GWAS genotype data for the Generation R Study were done at the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. We thank Karol Estrada, Dr Tobias A. Knoch, Anis Abuseiris, Luc V. de Zeeuw and Rob de Graaf, for their help in creating GRIMP, BigGRID, MediGRID and Services@MediGRID/D-Grid, (funded by the German Bundesministerium fuer Forschung und Technology; grants 01 AK 803 A-H, 01 IG 07015 G) for access to their grid computing resources. We thank Mila Jhamai, Manoushka Ganesh, Pascal Arp, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating, managing and QC of the GWAS database. Also, we thank Karol Estrada for their support in creation and analysis of imputed data.
GOYA
This study was conducted as part of the activities of the Gene-diet Interactions in Obesity project (GENDINOB, www.gendinob.dk) and the MRC centre for Causal Analyses in Translational Epidemiology (MRC CAiTE). T.S.A. was supported by funding from GENDINOB project (www.gendinob.dk).
HBCS
The Helsinki Birth Cohort Study (HBCS/HBCS 1934-44) thanks Professor David Barker and Tom Forsen. Major financial support was received from the Academy of Finland (project grants 120315, 129287, 218029 and 267561). The DNA extraction, sample quality control, biobank up-keep and aliquotting were performed at the National Institute for Health and Welfare, Helsinki, Finland.
Danish Childhood Obesity Biobank
The Danish Childhood Obesity Biobank thanks all children, youths and their families for participation in the studies and thus providing opportunities to detect novel insights in regards to childhood obesity. This study was supported by grants from the Program Committee for Individuals, Disease and Society of the Danish Council for Strategic Research [grant numbers: 11-115917, TARGET (http://target.ku.dk) and 11-116714, BIOCHILD (http://biochild.ku.dk/)]. The Novo Nordisk Foundation Center for Basic Metabolic Research, an independent Research Center at the University of Copenhagen partially funded by an unrestricted donation from the Novo Nordisk Foundation. H.N.K. is supported by a EU-FP7 Marie Curie Actions—Career Integration Grant (CIG-293511).
INMA
This study was funded by grants from Instituto de Salud Carlos III (CB06/02/0041, G03/176, FIS PI041436, PI081151, PI041705, PI061756, PI091958 and PS09/00432, FIS-FEDER 03/1615, 04/1509, 04/1112, 04/1931, 05/1079, 05/1052, 06/1213, 07/0314, 09/02647, 11/01007, 11/02591, 11/02038, 13/1944, 13/2032 and CP11/0178), Spanish Ministry of Science and Innovation (SAF2008-00357), European Commission (ENGAGE project and grant agreement HEALTH-F4-2007-201413, HEALTH.2010.2.4.5-1, FP7-ENV-2011 cod 282957), Fundació La Marató de TV3, Generalitat de Catalunya-CIRIT 1999SGR 00241 and Conselleria de Sanitat Generalitat Valenciana. Part of the DNA extractions and genotyping was performed at the Spanish National Genotyping Centre (CEGEN-Barcelona). The authors are grateful to Silvia Fochs, Anna Sànchez, Maribel López, Nuria Pey, Muriel Ferrer, Amparo Quiles, Sandra Pérez, Gemma León, Elena Romero, Maria Andreu, Nati Galiana, Maria Dolores Climent, Amparo Cases and Cristina Capo for their assistance in contacting the families and administering the questionnaires. The authors would particularly like to thank all the participants for their generous collaboration. A full roster of the INMA Project Investigators can be found at http://www.proyectoinma.org/presentacion-inma/listado-investigadores/en_listado-investigadores.html.
Leipzig
The Leipzig Childhood Obesity cohort is supported by grants from Integrated Research and Treatment Centre (IFB) Adiposity Diseases FKZ: 01EO1001, from the German Research Foundation for the Clinical Research Center ‘Obesity Mechanisms’ CRC1052/1 C05 and the European Community's Seventh Framework Programme (FP7/2007-2013) project Beta-JUDO under grant agreement no 279153. We are grateful to all the patients and families for contributing to the study. We highly appreciate the support of the Obesity Team and Auxo Team of the Leipzig University Children's Hospital for management of the patients and to the Pediatric Research Center Lab Team for support with DNA banking.
LISA + GINI
The authors thank all families for participation in the studies; the obstetric units for allowing recruitment and the LISA and GINI study teams for excellent work. Generation of GWA data in the LISAplus and GINIplus studies in Munich were covered by Helmholtz Zentrum Munich, Helmholtz Centre for Environmental Research. Personal and financial support by the Munich Center of Health Sciences (MCHEALTH) as part of the Ludwig-Maximilians University Munich LMU innovative is gratefully acknowledged. In addition, this work was supported by the Kompetenznetz Adipositas (Competence Network Obesity) funded by the Federal Ministry of Education and Research (FKZ: 01GI1121A).
MAAS
We would like to thank the children and their parents for their continued support and enthusiasm. We greatly appreciate the commitment they have given to the project. We would also like to acknowledge the hard work and dedication of the study team (postdoctoral scientists, research fellows, nurses, physiologists, technicians and clerical staff).’ MAAS was supported by the Asthma UK Grants No 301 (1995–1998), No 362 (1998–2001), No 01/012 (2001–2004), No 04/014 (2004–2007) and The Moulton Charitable Foundation (2004-current); age 11 years clinical follow-up is funded by the Medical Research Council (MRC) Grant G0601361.
MoBa
This work was supported by grants from the Norwegian Research Council (FUGE 183220/S10, FRIMEDKLI-05 ES236011), Swedish Medical Society (SLS 2008-21198), Jane and Dan Olsson Foundations and Swedish government grants to researchers in the public health service (ALFGBG-2863, ALFGBG-11522) and the European Community's Seventh Framework Programme (FP7/2007-2013), ENGAGE Consortium, grant agreement HEALTH-F4-2007-201413. The Norwegian Mother and Child Cohort Study was also supported by the Norwegian Ministry of Health and the Ministry of Education and Research, NIH/NIEHS (contract no N01-ES-75558), NIH/NINDS (grant no.1 UO1 NS 047537-01 and grant no.2 UO1 NS 047537-06A1) and the Norwegian Research Council/FUGE (grant no. 151918/S10). We are grateful to all the participating families in Norway who take part in this ongoing cohort study. Researchers interested in using MoBa data must obtain approval from the Scientific Management Committee of MoBa and from the Regional Committee for Medical and Health Research Ethics for access to data and biological material. Researchers are required to follow the terms of an Assistance Agreement containing a number of clauses designed to ensure protection of privacy and compliance with relevant laws.
NFBC 1966 and 1986
NFBC1966 and 1986 has received financial support from, e.g. the Academy of Finland, University Hospital Oulu, Biocenter, University of Oulu, Sigrid Juselius Foundation, Finland, the European Commission (EURO-BLCS, Framework 5 award QLG1-CT-2000-01643, ENGAGE project and grant agreement HEALTH-F4-2007-201413, EurHEALTHAgeing (277849), European Regional Developmental Fund), NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), NIH/NIMH (5R01MH63706:02), USA, Stanley Foundation, USA, the Medical Research Council, UK and the Welcome Trust, UK. Our most sincere thanks go to all cohort members and their parents. The Section of Investigative Medicine, Imperial College London is funded by grants from the MRC, BBSRC, NIHR, an Integrative Mammalian Biology (IMB) Capacity Building Award, an FP7-HEALTH- 2009-241592 EuroCHIP grant and is supported by the NIHR Imperial Biomedical Research Centre Funding Scheme. A.I.F.B. is funded by the MRC, EPSRC and Diabetes UK.
NTR
We thank all participating twin families. N.T.R. is supported by ARRA RC2 2MH08995; the European Research Council (Genetics of Mental Illness, ERC-230374); Spinozapremie (NWO/SPI 56-464-14192) and Twin-family database for behavior genetics and genomics studies (NWO 480-04-004).
PANIC
We thank the children and their families who participated in The PANIC Study. The study protocol was approved by the Research Ethics Committee of the Hospital District of Northern Savo. All children and their parents gave their informed written consent. The PANIC Study has been financially supported by grants from the Ministry of Social Affairs and Health of Finland, the Ministry of Education and Culture of Finland, the University of Eastern Finland, the Finnish Innovation Fund Sitra, the Social Insurance Institution of Finland, the Finnish Cultural Foundation, the Juho Vainio Foundation, the Foundation for Paediatric Research, the Paulo Foundation, the Paavo Nurmi Foundation, the Diabetes Research Foundation, Kuopio University Hospital (EVO-funding number 5031343) and the Research Committee of the Kuopio University Hospital Catchment Area for the State Research Funding.
PIAMA
The PIAMA birth cohort study is a collaboration of the Institute for Risk Assessment Sciences, University Utrecht (B. Brunekreef), Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht (H.A. Smit), Centre for Prevention and Health Services Research, National Institute for Public Health and the Environment, Bilthoven (A.H. Wijga), Department of Pediatrics, Division of Respiratory Medicine, Erasmus MC -Sophia, Rotterdam (J.C. de Jongste), Pulmonology (D.S. Postma) and Pediatric Pulmonology and Pediatric Allergology (G.H.K.) of the University Medical Center Groningen and the Department of Immunopathology, Sanquin Research, Amsterdam (R.C. Aalberse), the Netherlands. The study team gratefully acknowledges the participants in the PIAMA birth cohort study, and all coworkers who helped conducting the medical examinations, field work and data management. The PIAMA study was funded by grants from the Dutch Asthma Foundation (grant 3.4.01.26, 3.2.06.022, 3.4.09.081 and 3.2.10.085CO), the ZON-MW Netherlands Organization for Health Research and Development (grant 912-03-031), the Stichting Astmabestrijding and the Ministry of the Environment. Genome-wide genotyping was funded by the European Commission as part of GABRIEL (A multidisciplinary study to identify the genetic and environmental causes of asthma in the European Community) contract number 018996 under the Integrated Program LSH-2004-1.2.5-1 Post genomic approaches to understand the molecular basis of asthma aiming at a preventive or therapeutic control and a Grant from BBMRI-NL (CP 29). G.H.K. received grants from the Dutch Lung Foundation and BBMRI-NL.
RAINE
The authors are grateful to the Raine Study participants and their families, and to the Raine Study research staff for cohort coordination and data collection. The authors gratefully acknowledge the NH&MRC for their long-term contribution to funding the study over the last 20 years and also the following Institutions for providing funding for Core Management of the Raine Study: the University of Western Australia (UWA), Raine Medical Research Foundation, UWA Faculty of Medicine, Dentistry and Health Sciences, Telethon Kids Institute and Women and Infants Research Foundation and Curtin University. The authors gratefully acknowledge the assistance of the Western Australian DNA Bank (National Health and Medical Research Council of Australia National Enabling Facility). This study was supported by the National Health and Medical Research Council of Australia (grant numbers 572613, 403981 and 003209) and the Canadian Institutes of Health Research (grant number MOP-82893).
STRIP
The STRIP study was financially supported by Academy of Finland (grants 206374, 251360 and 26080328); Juho Vainio Foundation; Finnish Cardiac Research Foundation; Finnish Cultural Foundation; Finnish Ministry of Education and Culture; Sigrid Juselius Foundation; Yrjö Jahnsson Foundation; C.G. Sundell Foundation; Special Governmental Grants for Health Sciences Research, Turku University Hospital; Foundation for Pediatric Research; and Turku University Foundation.
TEDS
We gratefully acknowledge the on-going contribution of the families in the Twins Early Development Study (TEDS). TEDS is supported by a program grant from the UK Medical Research Council (G0901245), with genotyping provided by the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z). R.P. is supported by a research professorship from the UK Medical Research Council (G19/2) and a European Research Council Advanced Investigator Award (295366).
TEENAGE
We would like to thank all study participants and their families as well as all volunteers for their contribution in this study. We thank the following staff from the Sample Management and Genotyping Facilities at the Wellcome Trust Sanger Institute for sample preparation, quality control and genotyping: Dave Jones, Doug Simpkin, Emma Gray, Hannah Blackburn, Sarah Edkins. The TEENAGE study has been co-financed by the European Union (European Social Fund—ESF) and Greek national funds through the Operational Program ‘Education and Lifelong Learning’ of the National Strategic Reference Framework (NSRF)—Research Funding Program: Heracleitus II. Investing in knowledge society through the European Social Fund. This work was funded by the Wellcome Trust (098051).
YFS
Young Finns Study was financially supported by the Academy of Finland [134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi)]; the Social Insurance Institution of Finland, Kuopio, Tampere; Turku University Hospital Medical Funds (grant 9M048 and 9N035 to Terho Lehtimäki); Juho Vainio Foundation; Paavo Nurmi Foundation; Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation; Tampere Tuberculosis Foundation; and Emil Aaltonen Foundation (to Terho Lehtimäki).
Further Acknowledgements
B.M. is supported by the National Natural Science Foundation of China (NO.61471078), and the China Postdoctoral Science Foundation (No.2014M551084). T.H.P. is supported by the Alfred Benzon Foundation.
Conflict of Interest statement. None declared.
Appendix: Members of the BMDCS
Heidi J. Kalkwarf, Division of General and Community Pediatrics, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA.
Joan M. Lappe, Division of Endocrinology, Department of Medicine, Creighton University, Omaha, NB, USA.
Vicente Gilsanz, Department of Radiology, Children's Hospital Los Angeles, Los Angeles, CA, USA.
Sharon E. Oberfield, Division of Pediatric Endocrinology, Diabetes, and Metabolism, Department of Pediatrics, Columbia University Medical Center, New York, NY, USA.
John A. Shepherd, Department of Radiology, Children's Hospital Los Angeles, Los Angeles, CA, USA.
Andrea Kelly, Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Babette S. Zemel, Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA; Division of Gastroenterology, Hepatology and Nutrition, The Children's Hospital of Philadelphia, Philadelphia, PA, USA.
Contributor Information
Collaborators: Bone Mineral Density in Childhood Study (BMDCS) Consortium, Heidi J. Kalkwarf, Joan M. Lappe, Vicente Gilsanz, Sharon E. Oberfield, John A. Shepherd, Andrea Kelly, and Babette S. Zemel
References
- 1.Baker J.L., Olsen L.W., Sorensen T.I. (2007) Childhood body-mass index and the risk of coronary heart disease in adulthood. N. Engl. J. Med., 357, 2329–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franks P.W., Hanson R.L., Knowler W.C., Sievers M.L., Bennett P.H., Looker H.C. (2010) Childhood obesity, other cardiovascular risk factors, and premature death. N. Engl. J. Med., 362, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng M., Fleming T., Robinson M., Thomson B., Graetz N., Margono C., Mullany E.C., Biryukov S., Abbafati C., Abera S.F. et al. (2014) Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet, 384, 766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reilly J.J., Methven E., McDowell Z.C., Hacking B., Alexander D., Stewart L., Kelnar C.J. (2003) Health consequences of obesity. Arch. Dis. Child., 88, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silventoinen K., Pietilainen K.H., Tynelius P., Sorensen T.I., Kaprio J., Rasmussen F. (2007) Genetic and environmental factors in relative weight from birth to age 18: the Swedish young male twins study. Int. J. Obes. (Lond), 31, 615–621. [DOI] [PubMed] [Google Scholar]
- 6.Elks C.E., den Hoed M., Zhao J.H., Sharp S.J., Wareham N.J., Loos R.J., Ong K.K. (2012) Variability in the heritability of body mass index: a systematic review and meta-regression. Front. Endocrinol. (Lausanne), 3, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maes H.H., Neale M.C., Eaves L.J. (1997) Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet., 27, 325–351. [DOI] [PubMed] [Google Scholar]
- 8.Silventoinen K., Rokholm B., Kaprio J., Sorensen T.I. (2010) The genetic and environmental influences on childhood obesity: a systematic review of twin and adoption studies. Int. J. Obes. (Lond), 34, 29–40. [DOI] [PubMed] [Google Scholar]
- 9.Berndt S.I., Gustafsson S., Magi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L., Croteau-Chonka D.C., Day F.R. et al. (2013) Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet., 45, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M.C., Speliotes E.K., Magi R. et al. (2010) Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet., 42, 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J. et al. (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature, 518, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Magi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E. et al. (2015) New genetic loci link adipose and insulin biology to body fat distribution. Nature, 518, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Magi R. et al. (2010) Association analyses of 249 796 individuals reveal 18 new loci associated with body mass index. Nat. Genet., 42, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haworth C.M., Carnell S., Meaburn E.L., Davis O.S., Plomin R., Wardle J. (2008) Increasing heritability of BMI and stronger associations with the FTO gene over childhood. Obesity (Silver Spring), 16, 2663–2668. [DOI] [PubMed] [Google Scholar]
- 15.Sovio U., Mook-Kanamori D.O., Warrington N.M., Lawrence R., Briollais L., Palmer C.N., Cecil J., Sandling J.K., Syvanen A.C., Kaakinen M. et al. (2011) Association between common variation at the FTO locus and changes in body mass index from infancy to late childhood: the complex nature of genetic association through growth and development. PLoS Genet., 7, e1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradfield J.P., Taal H.R., Timpson N.J., Scherag A., Lecoeur C., Warrington N.M., Hypponen E., Holst C., Valcarcel B., Thiering E. et al. (2012) A genome-wide association meta-analysis identifies new childhood obesity loci. Nat. Genet., 44, 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comuzzie A.G., Cole S.A., Laston S.L., Voruganti V.S., Haack K., Gibbs R.A., Butte N.F. (2012) Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One, 7, e51954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherag A., Dina C., Hinney A., Vatin V., Scherag S., Vogel C.I., Muller T.D., Grallert H., Wichmann H.E., Balkau B. et al. (2010) Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genet., 6, e1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Namjou B., Keddache M., Marsolo K., Wagner M., Lingren T., Cobb B., Perry C., Kennebeck S., Holm I.A., Li R. et al. (2013) EMR-linked GWAS study: investigation of variation landscape of loci for body mass index in children. Front. Genet., 4, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jess T., Zimmermann E., Kring S.I., Berentzen T., Holst C., Toubro S., Astrup A., Hansen T., Pedersen O., Sorensen T.I. (2008) Impact on weight dynamics and general growth of the common FTO rs9939609: a longitudinal Danish cohort study. Int. J. Obes. (Lond), 32, 1388–1394. [DOI] [PubMed] [Google Scholar]
- 21.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E. et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet., 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundberg E., Small K.S., Hedman A.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.P., Meduri E., Barrett A. et al. (2012) Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet., 44, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang T.P., Beazley C., Montgomery S.B., Dimas A.S., Gutierrez-Arcelus M., Stranger B.E., Deloukas P., Dermitzakis E.T. (2010) Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics, 26, 2474–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang L., Morar N., Dixon A.L., Lathrop G.M., Abecasis G.R., Moffatt M.F., Cookson W.O. (2013) A cross-platform analysis of 14 177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res., 23, 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geller F., Feenstra B., Carstensen L., Pers T.H., van Rooij I.A., Korberg I.B., Choudhry S., Karjalainen J.M., Schnack T.H., Hollegaard M.V. et al. (2014) Genome-wide association analyses identify variants in developmental genes associated with hypospadias. Nat. Genet., 46, 957–963. [DOI] [PubMed] [Google Scholar]
- 26.Warrington N.M., Howe L.D., Paternoster L., Kaakinen M., Herrala S., Huikari V., Wu Y.Y., Kemp J.P., Timpson N.J., St Pourcain B. et al. (2015) A genome-wide association study of body mass index across early life and childhood. Int. J. Epidemiol., 44, 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nambu H., Fukushima M., Hikichi H., Inoue T., Nagano N., Tahara Y., Nambu T., Ito J., Ogawa Y., Ozaki S. et al. (2011) Characterization of metabolic phenotypes of mice lacking GPR61, an orphan G-protein coupled receptor. Life Sci., 89, 765–772. [DOI] [PubMed] [Google Scholar]
- 28.Creppe C., Malinouskaya L., Volvert M.L., Gillard M., Close P., Malaise O., Laguesse S., Cornez I., Rahmouni S., Ormenese S. et al. (2009) Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell, 136, 551–564. [DOI] [PubMed] [Google Scholar]
- 29.Okada Y., Yamagata K., Hong K., Wakayama T., Zhang Y. (2010) A role for the elongator complex in zygotic paternal genome demethylation. Nature, 463, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson C.L., Lemmens R., Miskiewicz K., Broom W.J., Hansen V.K., van Vught P.W., Landers J.E., Sapp P., Van Den Bosch L., Knight J. et al. (2009) Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet., 18, 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Li C., Izumi T., Ernst S.A., Andrews P.C., Williams J.A. (2004) Rab27b localizes to zymogen granules and regulates pancreatic acinar exocytosis. Biochem. Biophys. Res. Commun., 323, 1157–1162. [DOI] [PubMed] [Google Scholar]
- 32.Suda J., Zhu L., Okamoto C.T., Karvar S. (2011) Rab27b localizes to the tubulovesicle membranes of gastric parietal cells and regulates acid secretion. Gastroenterology, 140, 868–878. [DOI] [PubMed] [Google Scholar]
- 33.Zhao S., Torii S., Yokota-Hashimoto H., Takeuchi T., Izumi T. (2002) Involvement of Rab27b in the regulated secretion of pituitary hormones. Endocrinology, 143, 1817–1824. [DOI] [PubMed] [Google Scholar]
- 34.Hu C., Lv H., Pan G., Cao H., Deng Z., Hu C., Wen J., Zhou J. (2011) The expression of ADAM23 and its correlation with promoter methylation in non-small-cell lung carcinoma. Int. J. Exp. Pathol., 92, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y.P., Deng K.J., Wang F., Zhang J., Huang X., Qiao S., Zhao S. (2004) Two novel isoforms of Adam23 expressed in the developmental process of mouse and human brains. Gene, 325, 171–178. [DOI] [PubMed] [Google Scholar]
- 36.Kim H.A., Park W.J., Jeong H.S., Lee H.E., Lee S.H., Kwon N.S., Baek K.J., Kim D.S., Yun H.Y. (2012) Leucine-rich glioma inactivated 3 regulates adipogenesis through ADAM23. Biochim. Biophys. Acta, 1821, 914–922. [DOI] [PubMed] [Google Scholar]
- 37.Stergiakouli E., Gaillard R., Tavare J.M., Balthasar N., Loos R.J., Taal H.R., Evans D.M., Rivadeneira F., St Pourcain B., Uitterlinden A.G. et al. (2014) Genome-wide association study of height-adjusted BMI in childhood identifies functional variant in ADCY3. Obesity (Silver Spring), 22, 2252–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nag A., Venturini C., Small K.S., Young T.L., Viswanathan A.C., Mackey D.A., Hysi P.G., Hammond C., International Glaucoma Genetics Consortium, (2014) A genome-wide association study of intra-ocular pressure suggests a novel association in the gene FAM125B in the TwinsUK cohort. Hum. Mol. Genet., 23, 3343–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium, (2015) LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet., 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park J.H., Wacholder S., Gail M.H., Peters U., Jacobs K.B., Chanock S.J., Chatterjee N. (2010) Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet., 42, 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pers T.H., Karjalainen J.M., Chan Y., Westra H.J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T. et al. (2015) Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun., 6, 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fehrmann R.S., Karjalainen J.M., Krajewska M., Westra H.J., Maloney D., Simeonov A., Pers T.H., Hirschhorn J.N., Jansen R.C., Schultes E.A. et al. (2015) Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet., 47, 115–125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.