Abstract
Sequences corresponding to a putative, novel rhabdovirus [designated Merida virus (MERDV)] were initially detected in a pool of Culex quinquefasciatus collected in the Yucatan Peninsula of Mexico. The entire genome was sequenced, revealing 11 798 nt and five major ORFs, which encode the nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G) and RNA-dependent RNA polymerase (L). The deduced amino acid sequences of the N, G and L proteins have no more than 24, 38 and 43 % identity, respectively, to the corresponding sequences of all other known rhabdoviruses, whereas those of the P and M proteins have no significant identity with any sequences in GenBank and their identity is only suggested based on their genome position. Using specific reverse transcription-PCR assays established from the genome sequence, 27 571 C. quinquefasciatus which had been sorted in 728 pools were screened to assess the prevalence of MERDV in nature and 25 pools were found positive. The minimal infection rate (calculated as the number of positive mosquito pools per 1000 mosquitoes tested) was 0.9, and similar for both females and males. Screening another 140 pools of 5484 mosquitoes belonging to four other genera identified positive pools of Ochlerotatus spp. mosquitoes, indicating that the host range is not restricted to C. quinquefasciatus. Attempts to isolate MERDV in C6/36 and Vero cells were unsuccessful. In summary, we provide evidence that a previously undescribed rhabdovirus occurs in mosquitoes in Mexico.
Introduction
The family Rhabdoviridae (order Mononegavirales) is composed of a large and versatile group of viruses that are ubiquitous in nature (Kuzmin et al., 2009). The family consists of 11 genera as well as several viruses that have not yet been assigned to a genus (http://www.ictvonline.org/virusTaxonomy.asp). Virions have a distinctive bullet or cone-shaped morphology or appear bacilliform. Rhabdoviruses have broad host ranges that include humans and other terrestrial mammals, birds, reptiles, fish, insects and plants (Hoffmann et al., 2005; Kuzmin et al., 2009; Mann & Dietzgen, 2014). Many rhabdoviruses are transmitted to vertebrate and plant hosts by insect vectors in which they replicate (Hogenhout et al., 2003; Ammar et al., 2009).
Rhabdoviruses have a negative-sense ssRNA genome of ∼11–16 kb. A universal feature of the rhabdovirus genome is the presence of at least five genes that code in 3′ → 5′ order for the structural proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G) and RNA-dependent RNA polymerase (L) (Fu, 2005; Walker et al., 2015). Each ORF is flanked by relatively conserved cis-acting transcription initiation and transcription termination/polyadenylation signals that regulate mRNA expression. Rhabdovirus genomes are often interspersed with smaller ORFs that encode accessory proteins, most of which are of unknown function (Walker et al., 2011). ORFs that encode accessory proteins can occur as alternative or overlapping ORFs within the major structural protein genes or as independent ORFs in the intergenic regions that separate the structural protein genes. Leader and trailer sequences are located at the 3′ and 5′ termini of the rhabdovirus genome, respectively. These sequences are non-coding, A/U-rich and usually 50–100 nt in length. The first 10–20 nt of the leader and trailer sequences commonly exhibit partial complementary, and function as promoter sequences required for the initiation of genome and anti-genome replication, respectively (Fu, 2005; Walker et al., 2015).
Many novel rhadboviruses have been discovered in recent years due to the advent of unbiased high-throughput sequencing (UHTS) (Quan et al., 2010; Ito et al., 2013; Kading et al., 2013; Tokarz et al., 2014; Binger et al., 2015; Sakai et al., 2015; Stremlau et al., 2015), including Bas-Congo virus which was associated with an outbreak of acute haemorrhagic fever in humans in the Democratic Republic of Congo (Grard et al., 2012). Here, we report the genomic organization and prevalence of an apparently novel rhabdovirus tentatively named Merida virus (MERDV) that was discovered by UHTS in mosquitoes in the Yucatan Peninsula of Mexico.
Results
Discovery of MERDV
UHTS of total RNA from a pool of Culex quinquefasciatus collected in Merida in the Yucatan Peninsula of Mexico generated a ∼11 kb sequence corresponding to a putative, novel rhabdovirus tentatively named MERDV. Several other novel virus-like sequences were also discovered and the data will be presented elsewhere. The MERDV genome terminal sequences were identified using a combination of 5′ and 3′ RACE and Sanger sequencing. Difficulties were encountered during the 3′ RACE because the reverse primer bound preferentially to an A-rich region located slightly upstream of the 3′ terminus. The 3′ end of the genome was eventually identified by taking advantage of the partial complementarity that exists between the 5′ and 3′ termini of the rhabdovirus genome. The 3′ end of the MERDV genome was amplified and sequenced using a reverse primer designed from the inverse complement of the 24 nt sequence at the distal end of the 5′ terminus. Therefore, our sequence may contain nucleotide errors in the 24 nt primer-binding region at the ultimate 3′ end of the genome.
Genomic organization
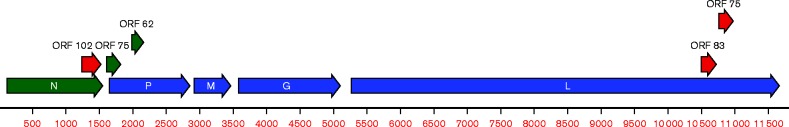
The MERDV genome consists of 11 798 nt (GenBank accession number KU194360) and its organization is consistent with that of the classical rhabdovirus genome: short leader and trailer sequences (68 and 74 nt, respectively) flank five structural protein genes in the order 3′-N–P–M–G–L-5′ (Fig. 1). The terminal nucleotides at the 5′ end of the MERDV genome are 5′-ACG-3′ and these same trinucleotides are located at the 5′ termini of other, related rhabdovirus genomes (Gubala et al., 2008, 2011; Kuwata et al., 2011; Zhu et al., 2011). The leader and trailer sequences are 59 and 58 % A/U-rich, respectively. Each ORF is separated by a non-coding region of 54–153 nt that contains transcription initiation and termination sequences identified as AACAU and CAUG[A]7, respectively, through sequence alignment of conserved nucleotides. The only exception to these consensus motifs is the CUUG[A]7 transcription termination sequence that regulates M mRNA expression (Table 1).
Fig. 1.
Coding capacity of MERDV sequence. Main ORFs, as well as minor ORFs unlikely to be expressed, are indicated along a scaled representation of the anti-genomic strand. The size of minor ORFs is indicated by their codon number: blue, frame 1; green, frame 2; red, frame 3.
Table 1. Non-coding sequences in the MERDV genome.
Solid underline indicates sequence overlap. Bold sequence indicates predicted transcription start/stop sites. The three nucleotides indicated by a dotted underline (AAC) and the AU of the start codon immediately downstream (not shown) are not predicted to serve as a transcription start site because they overlap the M coding sequence and are not in close proximity to any of the other coding sequences. Leader and trailer sequences are italicized.
| Gene | Non-coding sequences at the start of the gene | Non-coding sequences at the end of the gene | Intergenic region* |
|---|---|---|---|
| N | ACGAAAACAAAAAATCCCCACTCAACAGTCAGAATCCATGTTGTCGTTGGAGGTCTATGGAAATCCTT AACAUAACUAGUAUUAAUUAACUCUAAUAUUUGACACUUUUGGAUUUUCUGGAACGCCCGAC | GCCCUCACCUCGAGGACCCUCGGAUGCCCAGCAGGUUACAUGAAAAAAA | ACUCC |
| P | AACAUAACUAACUCGACCUCGGAAUCCGAUCAUUCACA | CAUGAAAAAAA | CUCC |
| M | AACAUCACUCACCUGAGACUCAUUCCCAGGUUAUUCUUGCC | AUCCCCCUUUAUAGACUUGGACCUUGUUAUUCCACAAUAAAGACAUAACAUAACUAGAAACU UGAAAAAAA | –† |
| G | AACAUAACUAGAAACUUGAAAAAAAGUUCCUCUGUGAAUUCCAGGUAGACGGGCCGAAAAAG | AGACCGUUAUGUAUCCCGCCUGUUCUUGGUUUGUCUGUGACCUGAAUUCAAUUUUGCCGUACUAUUGGAUAAUUCUUUUCCUCCUCUGAUUAUAUGUCUGUAAACUUUUAACAUGAAAAAAA | –† |
| L | AACAUGAAAAAAAUCAACAAAACUCAACGGGUAUCAUAUCAAAA | GACCAGAUCAAAGAGGGAAAAGAGACAGAAGAGAGAACAUGAAAAAAA CATGATCCCGATTCCTATAGTCTGATAAGGACCTCTAGGAGTATGCTTGTTGAGTGGGGATTTTTTGTTTTCGT |
Intergenic region located immediately downstream of the gene of interest that is neither translated nor transcribed to mRNA.
No intergenic region present due to gene overlap by 25 nt between the M and G genes and 13 nt between the G and L genes.
The genomic location and length of each predicted ORF is shown in Table 2. The ORF that encodes the N protein consists of 1437 nt. The predicted translation product is most closely related to the corresponding protein of Culex tritaeniorhynchus rhabdovirus (CTRV; 24 % identity and 45 % similarity) which was recently discovered in C. tritaeniorhynchus in Japan (Kuwata et al., 2011) and Yongjia tick virus 2 (also 24 % identity and 45 % similarity) from Haemaphysalis hystricis ticks in China (Li et al., 2015). The next two ORFs encode translation products that have no significant identity with any other sequences in GenBank. These two ORFs are assumed to encode the P and M proteins based on their positions in the genome. The ORF that encodes the G protein consists of 1530 nt and the predicted translation product is most closely related to the corresponding protein of CTRV (38 % identity and 58 % similarity). The next closest match is to a tandem rhabdovirus-like glycoprotein domain repeat sequence identified by the Aedes aegypti sequencing consortium (Nene et al., 2007). The largest ORF in the MERDV genome consists of 6411 nt and encodes the L protein. The predicted translation product is also most closely related to the corresponding region of CTRV (44 % identity and 65 % similarity). Minor ORFs of 102, 62, 83 and 75 codons overlap the N, P, L and L genes, respectively, but are not obviously accessible via ribosomal scanning (Fig. 1). The AUG of a fifth ORF (75 codons) that overlaps the 5′ end of the P ORF is apparently upstream of the P mRNA transcription start site, so is also unlikely to be accessible for translation.
Table 2. Predicted locations and lengths of the ORFs encoded by the MERDV genome.
| Protein | ORF genomic location | ORF length (nt) | Protein length (aa) | Protein mass (kDa) |
|---|---|---|---|---|
| N | 131–1567 | 1437 | 478 | 54.2 |
| P | 1660–2862 | 1203 | 400 | 43.8 |
| M | 2917–3477 | 564 | 187 | 21.0 |
| G | 3586–5112 | 1530 | 509 | 56.8 |
| L | 5266–11676 | 6411 | 2136 | 241.2 |
Predicted domains and post-translational modifications
The G proteins of rhabdoviruses have several common characteristics, including the presence of two to six potential N-linked glycosylation sites, 12 well-conserved cysteine residues, an N-terminal signal peptide, a transmembrane domain and a C-terminal short hydrophilic cytoplasmic domain (Coll, 1995; Walker & Kongsuwan, 1999). The G protein of MERDV is predicted to contain four potential N-linked glycosylation sites (one less than the G protein of CTRV) (Kuwata et al., 2011). All 12 conserved cysteines are present with two additional cysteines located at residues 12 and 485 (in CTRV one additional cysteine is present) (Kuwata et al., 2011). The G protein of MERDV is predicted to contain a signal peptide at residues 1–17, a hydrophobic transmembrane domain at residues 469–491 and a C-terminal hydrophilic cytoplasmic domain at residues 492–508, as common for rhabdoviral G proteins. Multiple protein kinase C (PKC) and tyrosine (TYR) phosphorylation sites are present in the N, P and M proteins of MERDV, consistent with many other rhabdoviruses including CTRV (Kuwata et al., 2011). Analysis with HHpred (Söding et al., 2005) revealed homology between the putative M protein of MERDV and Pfam family PF06326 (‘vesiculovirus matrix proteins’) indicating that it is indeed homologous to the M proteins of other rhabdoviruses.
Phylogenetic relationship to other rhabdoviruses
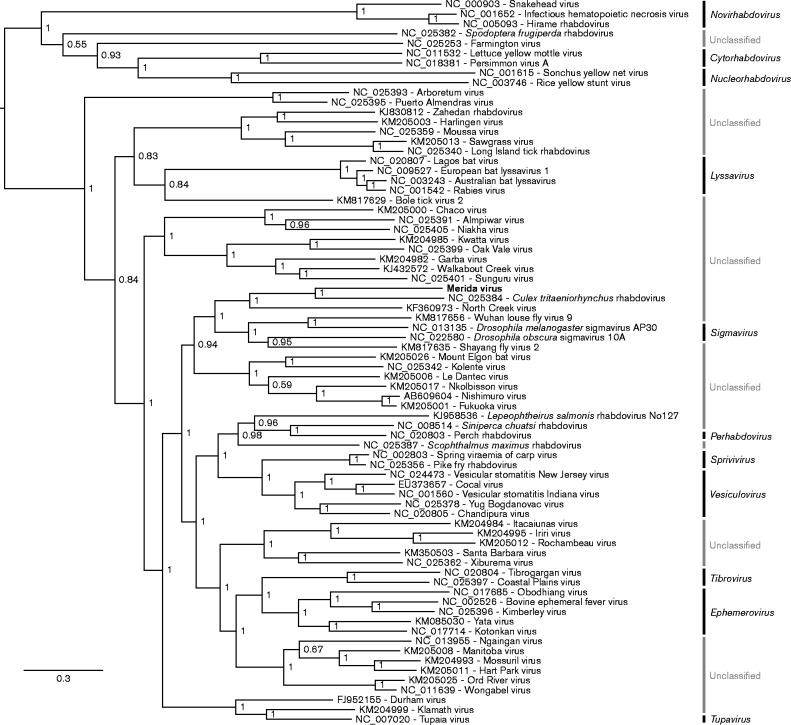
The L protein sequences from MERDV and 73 other rhabdovirus species were aligned using muscle (Edgar, 2004) and a phylogenetic tree was reconstructed using MrBayes (Ronquist et al., 2012) (Fig. 2). MERDV is most closely related to CTRV, consistent with the amino acid sequence alignments. More distantly, MERDV is related to North Creek virus which was identified in Culex sitiens in Australia (Coffey et al., 2014). Our analysis also indicated that MERDV cannot be assigned to one of the currently established rhabdovirus genera. Note that analyses of different genome regions (e.g. N) or of alignments with poorly aligning regions removed (e.g. with GBlocks; Castresana, 2000) provide different topologies in some of the deeper branches, but the clustering of MERDV with CTRV is consistent (data not shown).
Fig. 2.
Phylogenetic tree for MERDV and selected other rhabdovirus sequences. L protein amino acid sequences were aligned using muscle (Edgar, 2004). A maximum-likelihood phylogenetic tree was estimated using the Bayesian Markov chain Monte Carlo method implemented in MrBayes version 3.2.3 (Ronquist et al., 2012) sampling across the default set of fixed amino acid rate matrices with 10 million generations, discarding the first 25 % as burn-in. The original figure was produced using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The tree is midpoint-rooted and selected nodes are labelled with posterior probability values. Rhabdovirus genera, where defined, are labelled on the far right. GenBank accession numbers are indicated next to virus names. Bar indicates amino acid substitutions per site.
Prevalence in C. quinquefasciatus
A total of 27 571 C. quinquefasciatus, sorted into 728 pools of up to 50 individuals, were screened by MERDV-specific reverse transcription (RT)-PCR. Collections were made in Merida and Tixkokob in 2007–2008 using mosquito magnets, and in Merida in 2013 using Centers for Disease Control and Prevention (CDC) backpack-mounted aspirators. Mosquitoes obtained in 2007–2008 were tested according to gender, whereas those from 2013 were not. Overall, 256 pools (8038 mosquitoes) were composed of females, 195 pools (7196 mosquitoes) were composed of males and 277 pools (12 337 mosquitoes) were of mixed gender. Twenty-five mosquito pools were positive for MERDV RNA and the overall minimal infection rate (MIR; calculated as the number of positive mosquito pools per 1000 mosquitoes tested) was 0.9. The MIRs for female and male mosquitoes were similar (1.1 and 1.0, respectively). Evidence of MERDV infection was detected in mosquitoes collected in both study areas and during both time periods. See Tables 3 and 4.
Table 3. MIRs for MERDV in C. quinquefasciatus in the Yucatan Peninsula of Mexico, 2007–2008 and 2013.
| Study site | Date | No. mosquitoes tested | No. pools tested (positive) | MIR* |
|---|---|---|---|---|
| Tixkokob† | 2007–2008 | 9071 | 247 (2) | 0.2 |
| Merida† | 2007–2008 | 6163 | 204 (14) | 2.3 |
| Merida‡ | 2013 | 12 337 | 277 (9) | 0.7 |
| Total | 27 571 | 728 (25) | 0.9 |
Expressed as the number of positive mosquito pools per 1000 mosquitoes tested.
Mosquitoes were collected outdoors using mosquito magnets.
Mosquitoes were collected inside private residences using CDC backpack-mounted aspirators.
Table 4. Comparison of MIRs for MERDV in female and male C. quinquefasciatus in the Yucatan Peninsula of Mexico, 2007–2008.
| Study site | No. mosquitoes tested* | No. pools tested (positive) | MIR† | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Total | Female | Male | Total | Female | Male | Total | |
| Merida‡ | 3018 | 3145 | 6163 | 112 (7) | 92 (7) | 204 (14) | 2.3 | 2.2 | 2.3 |
| Tixkokob‡ | 5020 | 4051 | 9071 | 144 (2) | 103 (0) | 247 (2) | 0.4 | 0.0 | 0.2 |
| Total | 8038 | 7196 | 15 234 | 256 (9) | 195 (7) | 451 (16) | 1.1 | 1.0 | 1.1 |
C. quinquefasciatus collected in Merida in 2013 are not listed because males and females were not tested separately.
Expressed as the number of positive mosquito pools per 1000 mosquitoes tested.
Mosquitoes were collected outdoors using mosquito magnets.
Detection of MERDV sequence in other mosquito species
Another 5484 mosquitoes belonging to seven species were tested by RT-PCR using MERDV-specific primers RHAB-for and RHAB-rev as well as RHAB-121-for and RHAB-280-rev. Mosquito species tested were as follows: Aedes aegypti (n = 419), Anopheles albimanus (n = 727), Anopheles crucians (n = 691), Anopheles vestitipennis (n = 913), Ochlerotatus taeniorhynchus (n = 1000), Ochlerotatus trivittatus (n = 734) and Psorophora cyanescens (n = 1000) (Table 5). Collections were made using mosquito magnets at five study sites (Cozumel Island, Merida, Sian Ka'an, Tixkokob and Tzucacab) in 2007–2008. Mosquitoes had been sorted into 140 pools (20 pools per species) and all were female. MERDV RNA was detected in three pools of O. taeniorhynchus and in three pools of O. trivittatus using both primer pairs, whilst all other species were negative. The MERDV MIRs in O. taeniorhynchus and O. trivittatus were calculated as 3.0 and 4.1, respectively. All six PCR products generated using primers RHAB-121-for and RHAB-280-rev were analysed by Sanger sequencing. The resulting 114 nt sequences had at least 99.1 % nucleotide identity with the corresponding region of the MERDV genome sequence identified in C. quinquefasciatus (data not shown).
Table 5. MIRs for MERDV in selected Aedes, Anopheles, Ochlerotatus and Psorophora spp. mosquitoes.
| Species | No. mosquitoes tested* | No. pools tested (positive) | MIR† | Study site where positive pools were collected‡ |
|---|---|---|---|---|
| Aedes aegypti | 419 | 20 (0) | 0 | Tixkokob, Merida, Cozumel Island, Sian Ka'an and Tzucacab |
| Anopheles albimanus | 727 | 20 (0) | 0 | Tixkokob, Merida, Cozumel Island, Sian Ka'an and Tzucacab |
| Anopheles crucians | 691 | 20 (0) | 0 | Tixkokob, Merida, Cozumel Island, Sian Ka'an and Tzucacab |
| Anopheles vestitipennis | 913 | 20 (0) | 0 | Tixkokob, Merida, Cozumel Island, Sian Ka'an and Tzucacab |
| Ochlerotatus taeniorhynchus | 1000 | 20 (3) | 3.0 | Cozumel Island |
| Ochlerotatus trivittatus | 734 | 20 (3) | 4.1 | Cozumel Island, Merida and Tzucacab |
| Psorophora cyanescens | 1000 | 20 (0) | 0 | Tixkokob, Merida, Cozumel Island, Sian Ka'an and Tzucacab |
| Total | 5484 | 140 (6) |
All mosquitoes were female.
Expressed as the number of positive mosquito pools per 1000 mosquitoes tested.
Mosquitoes were collected using mosquito magnets at five study sites (Tixkokob, Merida, Cozumel Island, Sian Ka'an and Tzucacab) in 2007–2008.
Attempted virus isolations
An aliquot of every homogenate positive for MERDV RNA (25 for C. quinquefasciatus, three for O. taeniorhynchus and three for O. trivittatus) was tested by inoculation of C6/36 cells. Cytopathic effects were not observed in any cultures and a faint RT-PCR signal was only occasionally observed in supernatants or cell lysates harvested from the initially inoculated C6/36 cell monolayers; no RT-PCR signal was obtained after any of the second or third blind passages. Three positive homogenates from C. quinquefasciatus were also tested by virus isolation in Vero cells, but all were negative.
Dinucleotide usage preferences of MERDV
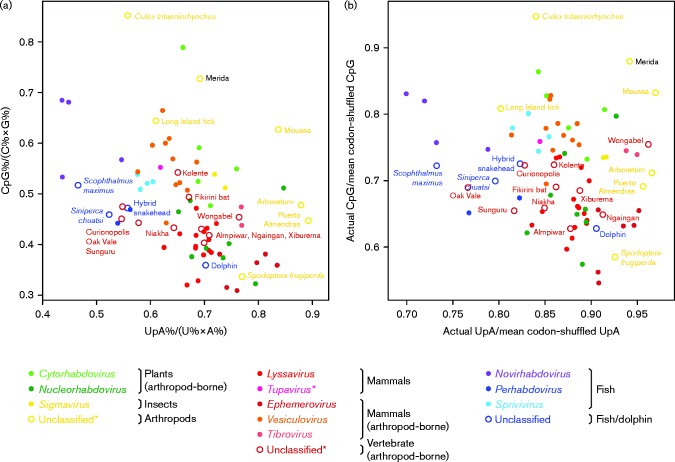
Vertebrate, invertebrate and plant virus hosts preferentially have certain codon and dinucleotide usage biases, e.g. vertebrate sequences display a strong under-representation of UpA and CpG, whilst insect sequences display a strong under-representation of UpA but not of CpG (Simmen, 2008). RNA virus sequences often have preferences that mimic those of their native hosts (Greenbaum et al., 2008; Atkinson et al., 2014; Tulloch et al., 2014). Thus, analysis of dinucleotide frequencies in virus genomes may be used to infer host taxa (Kapoor et al., 2010). In a comparison of UpA and CpG usage in the L protein ORF of 80 National Center for Biotechnology Information rhabdovirus RefSeqs and MERDV, CTRV had the least under-representation of CpG (observed/expected ratio close to unity) whilst MERDV ranked second or third depending on the randomization protocol utilized (Fig. 3), suggesting that MERDV, as well as CTRV, are not well adapted to vertebrate hosts.
Fig. 3.
Relative UpA and CpG frequencies in the L protein ORF of different rhabdovirus species. UpA and CpG frequencies were calculated in two different ways. (a) In each sequence, the numbers of UpA and CpG dinucleotides, and A, C, G and U mononucleotides, were counted. Dinucleotide frequencies, fXpY, were expressed relative to their expected frequencies, fX × fY, in the absence of selection. (b) To factor-out codon and amino acid usage, 1000 shuffled ORF sequences were generated for each virus sequence. In each shuffled sequence, the original amino acid sequence and the original total numbers of each of the 61 codons were maintained, but synonymous codons were randomly shuffled between the different sites where the corresponding amino acid is used in the original sequence. Next, the UpA and CpG frequencies in the original sequence were expressed relative to their mean frequencies in the codon-shuffled sequences. As codon usage is factored-out, the UpA and CpG relative frequencies tend to be less extreme in (b) compared with (a). Each point represents a single rhabdovirus sequence. Solid points correspond to species within defined genera, colour coded by genus (see key). Annotated open circles correspond to species that are currently unassigned at genus level, colour coded by host (or presumed host) taxa. Asterisks in the key indicate clades with uncertain host taxa: viruses in the unclassified ‘arthropod-infecting’ clades (yellow open circles) were isolated from arthropods but not from vertebrates; the sole representative of the genus Tupavirus was isolated from mammals but not from arthropods, although its phylogenetic position suggests that it may be arthropod-borne; the presence of viruses derived from vertebrates and viruses derived from arthropods in each of the unclassified ‘vertebrate (arthropod-borne)’ clades (brick-red open circles) suggests that all of these viruses are likely arboviruses. GenBank accession numbers of sequences used: NC_000855, NC_000903, NC_001542, NC_001560, NC_001615, NC_001652, NC_002251, NC_002526, NC_002803, NC_003243, NC_003746, NC_005093, NC_005974, NC_005975, NC_006429, NC_006942, NC_007020, NC_007642, NC_008514, NC_009527, NC_009528, NC_011532, NC_011542, NC_011639, NC_013135, NC_013955, NC_016136, NC_017685, NC_017714, NC_018381, NC_018629, NC_020803, NC_020804, NC_020805, NC_020806, NC_020807, NC_020808, NC_020809, NC_020810, NC_022580, NC_022581, NC_022755, NC_024473, NC_025251, NC_025253, NC_025255, NC_025340, NC_025341, NC_025342, NC_025353, NC_025354, NC_025356, NC_025358, NC_025359, NC_025362, NC_025364, NC_025365, NC_025371, NC_025376, NC_025377, NC_025378, NC_025382, NC_025384, NC_025385, NC_025387, NC_025389, NC_025391, NC_025392, NC_025393, NC_025394, NC_025395, NC_025396, NC_025397, NC_025398, NC_025399, NC_025400, NC_025401, NC_025405, NC_025406, NC_025408.
Discussion
The advent of UHTS has resulted in the discovery of many novel rhabdoviruses (Grard et al., 2012; Ito et al., 2013; Kading et al., 2013; Tokarz et al., 2014; Binger et al., 2015; Sakai et al., 2015; Stremlau et al., 2015), including several which were isolated from Anopheles, Culex, Ochlerotatus and Psorophora spp. mosquitoes (Quan et al., 2010; Coffey et al., 2014; Vasilakis et al., 2014). Here, we report the discovery of a putative, novel rhabdovirus in Culex and Ochlerotatus spp. mosquitoes from the Yucatan Peninsula of Mexico.
It is hypothesized that the majority of Rhabdoviruses are hosted by insects (Li et al., 2015), and numerous cyto-, nucleo- and dimarhabdoviruses are transmitted by arthropods to their plant or vertebrate hosts (Hogenhout et al., 2003; Bourhy et al., 2005). This includes the bite of haematophagous arthropods (Comer et al., 1990; Pérez De León et al., 2006), including Simulium vittatum blackflies, which can transmit vesicular stomatitis New Jersey virus to cattle under experimental conditions (Mead et al., 2009), and Phlebotomus argentipes sandflies, which efficiently transmitted Chandipura virus to laboratory mice (Mavale et al., 2007). It is currently unknown whether MERDV has the capacity to replicate in vertebrate hosts, but should its host range include vertebrates, it is unlikely that virus persistence in nature is dependent upon the bite of haematophagous arthropods. Male mosquitoes do not feed on blood, and thus a significant bias between male and female mosquito MIRs would be expected if that would be that case. Instead, the similar MIRs that we determined for MERDV are more compatible with vertical and venereal transmission. The occurrence of rhabdovirus transmission through these modes, in addition to horizontal transmission, has been demonstrated, for example, in Aedes aegypti mosquitoes for Chandipura virus (Mavale et al., 2005) and in phlebotomine sandflies for vesicular stomatitis Indiana virus (Tesh et al., 1972). A hallmark in the life cycle of insect-specific sigmaviruses is the exclusively vertical transmission through eggs and sperm (Longdon et al., 2011). Currently, no data are available to decide to what extent horizontal or vertical transmission contribute to the maintenance of MERDV in nature. Attempts to isolate MERDV by inoculation of Vero cells were unsuccessful, and together with the determined CpG and UpA dinucleotide usage biases, suggest that MERDV is likely not to infect vertebrates in nature. However, attempts to isolate MERDV in C6/36 cells also were unsuccessful thus far.
The inability to recover an isolate of MERDV in C6/36 cells is unexpected because this cell line supports the replication of a diverse range of mosquito-associated viruses, including several rhabdoviruses recently identified in Culex and Ochlerotatus spp. mosquitoes (Quan et al., 2010; Kuwata et al., 2011; Coffey et al., 2014; Vasilakis et al., 2014). However, it is not without precedent; Manitoba rhabdovirus from Culex tarsalis is reported to not propagate in C6/36 cells, although it does replicate in Vero, primary chick embryo and mouse neuroblastoma cells (Artsob et al., 1991). Shortcomings in sample handling and possible failures in the cold-chain during transport are unlikely for the failure to isolate MERDV because one-quarter of our mosquito homogenates induced virus-like cytopathic effect when inoculated onto C6/36 cells, indicating that other, undetermined viruses did successfully propagate. Another possibility is that MERDV does not actively replicate in mosquitoes. It cannot be excluded that some of the field-collected mosquitoes passively carried MERDV without the virus being capable of replicating in the mosquito, despite the detection over multiple years, several locations and specific species. Alternatively, we may have discovered another example of endogenous viral elements analogous to the rhabdovirus-like sequences described previously in various insect hosts (Nene et al., 2007; Katzourakis & Gifford, 2010; Li et al., 2015). However, such endogenous viral elements have thus far been reported to concern only partial sequences, at best covering one gene, but never what appears to be a complete, functional genome as we have found for MERDV.
Amino acid sequence alignments and phylogenetic analyses indicated CTRV as the closest known relative of MERDV. CTRV was isolated in C6/36 cells from C. tritaeniorhynchus in Japan (Kuwata et al., 2011), and later detected in Culex, Aedes, Anopheles and Armigeres spp. mosquitoes in China (Li et al., 2015; Shi et al., 2015). Recent studies have also shown that NIID-CTR cells, which were established from C. tritaeniorhynchus embryos, are persistently infected with CTRV (Gillich et al., 2015). CTRV establishes a non-cytolytic infection and, similar to sigmaviruses, employs vertical transmission. However, in contrast to sigmaviruses, CTRV replicates in the nucleus of the infected cell similar to nucleorhabdoviruses and it is the only known rhabdovirus that requires the cellular splicing machinery for its mRNA maturation. The coding region for the L protein of CTRV is interrupted by a 76 nt intron (Kuwata et al., 2011). Inspection of the MERDV sequence provided no evidence for the use of splicing similar to CTRV, suggesting that MERDV may not require a nuclear phase. Our proposed transcriptional signals match those confirmed for CTRV. Whilst conservation of the termination signal sequence is seen in comparison to other rhabdoviruses, including the Drosophila-specific sigmaviruses and Moussa virus, a potentially mosquito-specific rhabdovirus from Culex decans mosquitoes (Quan et al., 2010), the initiation signal sequence differs from the two other viruses. Additionally, whereas sigmaviruses and Moussa virus do not show overlap of genes, the G and L genes of CTRV and both the M/G and G/L genes of MERDV show overlap. Other rhabdoviruses also possess overlapping transcription termination and transcription initiation sequences in their genomes including two more recently discovered mosquito-associated rhabdoviruses: Malpais Spring virus and Oak Vale virus (Quan et al., 2011; Vasilakis et al., 2013). Indeed, the positioning of the initiation signal of the downstream gene in front of the termination signal of the preceding gene or the use of splicing are not unprecedented in mononegaviruses, e.g. in human metapneumovirus or in bornaviruses, where these mechanisms have been hypothesized to adjust transcription levels possibly in conjunction with persistent infection, or attenuate gene expression in addition to the 3′ → 5′ transcriptional gradient characteristic for mononegaviruses (Collins et al., 1987; Schneemann et al., 1994; Schneider et al., 1994).
The translated primary sequences of MERDV N, G and L ORFs show significant homology to the respective ORFs of other rhabdovirses. However, as also observed for CTRV, sigmaviruses and Moussa viruses, the P and M ORFs are far more diverse and do not exhibit homology to any other sequences in GenBank or to each other (except for the M proteins of sigmaviruses, which have a very distant relationship to the corresponding protein of Flanders virus). In addition, sigmaviruses contain an additional ORF (designated ORF X) between the P and G ORFs.
Although CTRV is the closest known relative of MERDV, these two viruses exhibit considerable sequence dissimilarity. The L proteins of MERDV and CTRV, which represent the most conserved genome regions of the rhabdoviruses, show an amino acid divergence of 56 %. This is reaching the divergence observed between rhabdoviruses belonging to different genera, which is commonly in a range of 47–83 % (Table S1, available in the online Supplementary Material). As species and genus demarcations for rhabdoviruses also include factors such as biological characteristics (e.g. host range) and serological cross-reactivity, additional work will be needed to accurately determine the taxonomic status of MERDV within the family Rhabdoviridae.
In summary, we provide evidence that a novel rhabdovirus occurs in mosquitoes in the Yucatan Peninsula of Mexico. This apparent virus, provisionally named MERDV, is most closely related to CTRV, although it shows considerable sequence and biological divergence. Our findings underscore the vast diversity of this virus family, highlight the power of next-generation sequence technology in the discovery of novel viruses and provide the basis for improved surveillance programmes to gain better insights into arbovirus evolution.
Methods
Mosquito collections
Mosquitoes were collected in five study areas in the Yucatan Peninsula of Mexico: Cozumel Island, Merida, Sian Ka'an, Tixkokob and Tzucacab. Descriptions of these study areas are provided elsewhere (Farfan-Ale et al., 2009, 2010). Collections were made in 2007 and 2008 using Mosquito magnets (all five study areas) and in 2013 using CDC backpack-mounted aspirators (Merida only). Mosquito magnets Pro-Liberty (American Biophysics) were baited with propane and octenol, and placed outdoors. Mosquito magnets were turned on between 16 : 00 and 18 : 00 and collection nets were replaced the following morning between 06 : 00 and 09 : 00. CDC back-pack mounted aspirators were used to collect resting mosquitoes inside private residences. Mosquitoes were transported alive to the Universidad Autonoma de Yucatan, frozen at − 80 °C, and identified on chill tables according to species and sex using morphological characteristics (Darsie, 1996). Mosquitoes were transported on dry ice from the Universidad Autonoma de Yucatan to Iowa State University by World Courier.
High-throughput sequencing
Mosquitoes were homogenized as described previously (Farfan-Ale et al., 2009) and total RNA was extracted using TRIzol (Invitrogen) following the manufacturer's instructions. Extracts were reverse transcribed using SuperScript III (Thermo Fisher) with random hexamers. The cDNA was RNase-H-treated prior to second-strand synthesis with Klenow fragment (NEB). The generated double-stranded cDNA was sheared to a mean fragment size of 200 bp using a Covaris focused-ultrasonicator E210 and the manufacturer's standard settings. Sheared products were purified (Agencourt Ampure DNA purification beads; Beckman Coulter) and libraries constructed. Sheared nucleic acid was end-repaired, dA-tailed, ligated to sequencing adapters (NEBNext modules; NEB), PCR-amplified (Phusion High-Fidelity DNA polymerase; NEB) and quantified by an Agilent Bioanalyzer for sequencing. Sequencing on the Illumina HiSeq 2500 platform (Illumina) resulted in a mean of 180 million reads per lane. Samples were de-multiplexed using Illumina software and FastQ files generated. Data were quality-filtered and trimmed (Slim-Filter), and de novo assembled using Dwight assembler at custom settings (Golovko et al., 2012). The generated contiguous sequences (contigs) and unique singleton reads were subjected to homology search using blastn and blastx against GenBank.
RT-PCR and Sanger sequencing
Total RNA was analysed by RT-PCR using MERDV-specific primers RHAB-for (5′-CAATCACATCGACTACTCTAAATGGA-3′) and RHAB-rev (5′-GATCAGACCTAGCTTGGCTGTTC-3′), which target a 490 nt region of the L protein gene, or RHAB-121-for (5′-AACGCCCGACATGACTACTATCG-3′) and RHAB-280-rev (5′-TTCCGTACCTCCCATATGAGTGG-3′), which target a 160 nt region of the N protein. cDNAs were generated using SuperScript III reverse transcriptase (Invitrogen), and PCRs were performed using Taq polymerase (Invitrogen) and the following cycling conditions: 94 °C for 3 min, then 35 cycles of 94 °C for 30 s, 56 °C for 45 s and 72 °C for 1 min followed by a final extension at 72 °C for 8 min. RT-PCR products were purified using the Purelink Gel Extraction kit (Invitrogen). Sanger sequencing was performed using a 3730 × 1 DNA sequencer (Applied Biosystems).
5′ and 3′ RACE
The extreme 5′ and 3′ ends of the MERDV genome were determine by 5′ and 3′ RACE, respectively. In the 5′ RACE reactions, total RNA was reversed transcribed using a MERDV-specific primer (5′-CTCAGAACGGAAGAGGTATACTG-3′). cDNAs were purified by ethanol precipitation and oligo(dC) tails were added to the 3′ ends using 15 U terminal deoxynucleotidyltransferase (Invitrogen) in 1 × tailing buffer (10 mM Tris/HCl, pH 8.4, 25 mM KCl, 1.5 mM MgCl2 and 0.02 mM dCTP). Tailing reactions were performed at 37 °C for 30 min and then terminated by heat inactivation (65 °C for 10 min). Oligo dC-tailed cDNAs were purified by ethanol precipitation, and then PCR-amplified using a consensus forward primer specific to the C-tailed termini (5′-GACATCGAAAGGGGGGGGGGG-3′) and a reverse primer specific to the MERDV cDNA sequence (5′-TTCCGTACCTCCCATATGAGTGG-3′). In the 3′ RACE reactions, polyadenylate [poly(A)] tails were added to the 3′ ends of the genomic RNA using 6 U poly(A) polymerase (Ambion) in 1 × reaction buffer (40 mM Tris/HCl, pH 8.0, 10 mM MgCl2, 2.5 mM MnCl2, 250 mM NaCl, 50 μg BSA ml− 1 and 1 mM ATP). Tailing reactions were performed at 37 °C for 1 h and terminated by heat inactivation (65 °C for 10 min). Poly(A)-tailed RNA was reverse transcribed using a poly(A) tail-specific primer (5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′). Complementary DNAs were PCR amplified using a forward primer specific to the MERDV cDNA sequence (5′-AAGAACATCGGGTATTGATCCGG-3′) and a reverse primer that matched the 5′ half of the poly(A)-specific RT primer (5′-GGCCACGCGTCGACTAGTAC-3′).
PCR products generated from the 5′ and 3′ RACE reactions were inserted into the pCR4-TOPO cloning vector (Invitrogen), and ligated plasmids were transformed into competent TOPO10 Escherichia coli cells (Invitrogen). Cells were grown on Luria–Bertani agar containing ampicillin (50 μg ml− 1) and kanamycin (50 μg ml− 1), and colonies were screened for inserts by PCR amplification. An aliquot of each PCR product was examined by 1 % agarose gel electrophoresis, and selected PCR products were purified by QIAquick spin column (Qiagen) and sequenced using a 3730x1 DNA sequencer.
Amino acid sequence alignments and prediction algorithms
The predicted amino acid sequences of MERDV were compared to all other sequences in GenBank by application of tblastn (Altschul et al., 1990). Per cent amino acid identities and similarities of select rhabdovirus protein sequences were calculated using clustal w2 (http://simgene.com/ClustalW). The following prediction algorithms were used for the amino acid sequence analysis: NetNGlyc 1.0 server (for the identification of potential N-linked glycosylation sites), SignalP 4.1 server (for the identification of potential signal peptides), tmhmm server 2.0 (for the identification of potential transmembrane domains and cytoplasmic domains) and NetPhos 2.0 Server (for the identification of potential PKC and TYR phosphorylation sites).
Virus isolation
An aliquot (200 μl) of each supernatant that tested positive for MERDV RNA was added to 2 ml Liebovitz's L15 medium (Invitrogen) supplemented with 2 % FBS, 2 mM l-glutamine, 100 U ml− 1 penicillin, 100 μg ml− 1 streptomycin and 2.5 μg fungizone ml− 1. Samples were filtered using a 0.22 μm filter and inoculated onto subconfluent monolayers of Aedes albopictus C6/36 cells in 75 cm2 flasks. Cells were incubated for at least 1 h at room temperature on an orbital shaker. Another 12 ml L15 maintenance medium was added to each flask and cells were incubated at 28 °C for 7 days. After two additional blind passages, supernatants were harvested and tested by RT-PCR for the presence of MERDV RNA.
Acknowledgements
The authors thank Valeria Bussetti for expert technical assistance. This study was supported by the National Institutes of Health (5R21AI067281, AI057158, 5R21AI067281 and AI088647), the US Department of Defense and an intramural grant from Iowa State University. A. E. F. is supported by a grant from the Wellcome Trust (106207).
Supplementary Data
Supplementary Data
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool J Mol Biol 215 403–410 10.1016/S0022-2836(05)80360-2 . [DOI] [PubMed] [Google Scholar]
- Ammar El-D., Tsai C. W., Whitfield A. E., Redinbaugh M. G., Hogenhout S. A. (2009). Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts Annu Rev Entomol 54 447–468 10.1146/annurev.ento.54.110807.090454 . [DOI] [PubMed] [Google Scholar]
- Artsob H., Doane F., Sekla L., Stackiw W., Brust R. (1991). Manitoba virus, a new rhabdovirus isolated from Culex tarsalis mosquitoes collected in Manitoba, Canada Can J Microbiol 37 329–332 10.1139/m91-053 . [DOI] [PubMed] [Google Scholar]
- Atkinson N. J., Witteveldt J., Evans D. J., Simmonds P. (2014). The influence of CpG and UpA dinucleotide frequencies on RNA virus replication and characterization of the innate cellular pathways underlying virus attenuation and enhanced replication Nucleic Acids Res 42 4527–4545 10.1093/nar/gku075 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binger T., Annan A., Drexler J. F., Müller M. A., Kallies R., Adankwah E., Wollny R., Kopp A., Heidemann H., other authors (2015). A novel rhabdovirus isolated from the straw-colored fruit bat Eidolon helvum, with signs of antibodies in swine and humans J Virol 89 4588–4597 10.1128/JVI.02932-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy H., Cowley J. A., Larrous F., Holmes E. C., Walker P. J. (2005). Phylogenetic relationships among rhabdoviruses inferred using the L polymerase gene J Gen Virol 86 2849–2858 10.1099/vir.0.81128-0 . [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis Mol Biol Evol 17 540–552 10.1093/oxfordjournals.molbev.a026334 . [DOI] [PubMed] [Google Scholar]
- Coffey L. L., Page B. L., Greninger A. L., Herring B. L., Russell R. C., Doggett S. L., Haniotis J., Wang C., Deng X., Delwart E. L. (2014). Enhanced arbovirus surveillance with deep sequencing: identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes Virology 448 146–158 10.1016/j.virol.2013.09.026 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll J. M. (1995). The glycoprotein G of rhabdoviruses Arch Virol 140 827–851 10.1007/BF01314961 . [DOI] [PubMed] [Google Scholar]
- Collins P. L., Olmsted R. A., Spriggs M. K., Johnson P. R., Buckler-White A. J. (1987). Gene overlap and site-specific attenuation of transcription of the viral polymerase L gene of human respiratory syncytial virus Proc Natl Acad Sci U S A 84 5134–5138 10.1073/pnas.84.15.5134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer J. A., Tesh R. B., Modi G. B., Corn J. L., Nettles V. F. (1990). Vesicular stomatitis virus, New Jersey serotype: replication in and transmission by Lutzomyia shannoni (Diptera: Psychodidae) Am J Trop Med Hyg 42 483–490 . [DOI] [PubMed] [Google Scholar]
- Darsie R. F., Jr. (1996). A survey and bibliography of the mosquito fauna of Mexico (Diptera: Culicidae) J Am Mosq Control Assoc 12 298–306 . [PubMed] [Google Scholar]
- Edgar R. C. (2004). muscle: a multiple sequence alignment method with reduced time and space complexity BMC Bioinformatics 5 113 10.1186/1471-2105-5-113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfan-Ale J. A., Loroño-Pino M. A., Garcia-Rejon J. E., Hovav E., Powers A. M., Lin M., Dorman K. S., Platt K. B., Bartholomay L. C., other authors (2009). Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico Am J Trop Med Hyg 80 85–95 . [PMC free article] [PubMed] [Google Scholar]
- Farfan-Ale J. A., Loroño-Pino M. A., Garcia-Rejon J. E., Soto V., Lin M., Staley M., Dorman K. S., Bartholomay L. C., Hovav E., Blitvich B. J. (2010). Detection of flaviviruses and orthobunyaviruses in mosquitoes in the Yucatan Peninsula of Mexico in 2008 Vector Borne Zoonotic Dis 10 777–783 10.1089/vbz.2009.0196 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. F. (2005). Genetic comparison of the rhabdoviruses from animals and plants Curr Top Microbiol Immunol 292 1–24 . [DOI] [PubMed] [Google Scholar]
- Gillich N., Kuwata R., Isawa H., Horie M. (2015). Persistent natural infection of a Culex tritaeniorhynchus cell line with a novel Culex tritaeniorhynchus rhabdovirus strain Microbiol Immunol 59 562–566 10.1111/1348-0421.12279 . [DOI] [PubMed] [Google Scholar]
- Golovko G., Khanipov K., Rojas M., Martinez-Alcántara A., Howard J. J., Ballesteros E., Gupta S., Widger W., Fofanov Y. (2012). Slim-filter: an interactive Windows-based application for Illumina genome analyzer data assessment and manipulation BMC Bioinformatics 13 166 10.1186/1471-2105-13-166 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grard G., Fair J. N., Lee D., Slikas E., Steffen I., Muyembe J. J., Sittler T., Veeraraghavan N., Ruby J. G., other authors (2012). A novel rhabdovirus associated with acute hemorrhagic fever in central Africa PLoS Pathog 8 e1002924 10.1371/journal.ppat.1002924 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum B. D., Levine A. J., Bhanot G., Rabadan R. (2008). Patterns of evolution and host gene mimicry in influenza and other RNA viruses PLoS Pathog 4 e1000079 10.1371/journal.ppat.1000079 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubala A. J., Proll D. F., Barnard R. T., Cowled C. J., Crameri S. G., Hyatt A. D., Boyle D. B. (2008). Genomic characterisation of Wongabel virus reveals novel genes within the Rhabdoviridae Virology 376 13–23 10.1016/j.virol.2008.03.004 . [DOI] [PubMed] [Google Scholar]
- Gubala A., Davis S., Weir R., Melville L., Cowled C., Boyle D. (2011). Tibrogargan and Coastal Plains rhabdoviruses: genomic characterization, evolution of novel genes and seroprevalence in Australian livestock J Gen Virol 92 2160–2170 10.1099/vir.0.026120-0 . [DOI] [PubMed] [Google Scholar]
- Hoffmann B., Beer M., Schütze H., Mettenleiter T. C. (2005). Fish rhabdoviruses: molecular epidemiology and evolution Curr Top Microbiol Immunol 292 81–117 . [DOI] [PubMed] [Google Scholar]
- Hogenhout S. A., Redinbaugh M. G., Ammar D. (2003). Plant and animal rhabdovirus host range: a bug's view Trends Microbiol 11 264–271 10.1016/S0966-842X(03)00120-3 . [DOI] [PubMed] [Google Scholar]
- Ito T., Suzaki K., Nakano M. (2013). Genetic characterization of novel putative rhabdovirus and dsRNA virus from Japanese persimmon J Gen Virol 94 1917–1921 10.1099/vir.0.054445-0 . [DOI] [PubMed] [Google Scholar]
- Kading R. C., Gilbert A. T., Mossel E. C., Crabtree M. B., Kuzmin I. V., Niezgoda M., Agwanda B., Markotter W., Weil M. R., other authors (2013). Isolation and molecular characterization of Fikirini rhabdovirus, a novel virus from a Kenyan bat J Gen Virol 94 2393–2398 10.1099/vir.0.053983-0 . [DOI] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Lipkin W. I., Zaidi S., Delwart E. (2010). Use of nucleotide composition analysis to infer hosts for three novel picorna-like viruses J Virol 84 10322–10328 10.1128/JVI.00601-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A., Gifford R. J. (2010). Endogenous viral elements in animal genomes PLoS Genet 6 e1001191 10.1371/journal.pgen.1001191 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata R., Isawa H., Hoshino K., Tsuda Y., Yanase T., Sasaki T., Kobayashi M., Sawabe K. (2011). RNA splicing in a new rhabdovirus from Culex mosquitoes J Virol 85 6185–6196 10.1128/JVI.00040-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin I. V., Novella I. S., Dietzgen R. G., Padhi A., Rupprecht C. E. (2009). The rhabdoviruses: biodiversity, phylogenetics, and evolution Infect Genet Evol 9 541–553 10.1016/j.meegid.2009.02.005 . [DOI] [PubMed] [Google Scholar]
- Li C. X., Shi M., Tian J. H., Lin X. D., Kang Y. J., Chen L. J., Qin X. C., Xu J., Holmes E. C., Zhang Y. Z. (2015). Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses eLife 4 e05378 10.7554/eLife.05378 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B., Wilfert L., Obbard D. J., Jiggins F. M. (2011). Rhabdoviruses in two species of Drosophila: vertical transmission and a recent sweep Genetics 188 141–150 10.1534/genetics.111.127696 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K. S., Dietzgen R. G. (2014). Plant rhabdoviruses: new insights and research needs in the interplay of negative-strand RNA viruses with plant and insect hosts Arch Virol 159 1889–1900 10.1007/s00705-014-2029-z . [DOI] [PubMed] [Google Scholar]
- Mavale M. S., Geevarghese G., Ghodke Y. S., Fulmali P. V., Singh A., Mishra A. C. (2005). Vertical and venereal transmission of Chandipura virus (Rhabdoviridae) by Aedes aegypti (Diptera: Culicidae) J Med Entomol 42 909–911 . [DOI] [PubMed] [Google Scholar]
- Mavale M. S., Fulmali P. V., Ghodke Y. S., Mishra A. C., Kanojia P., Geevarghese G. (2007). Experimental transmission of Chandipura virus by Phlebotomus argentipes (Diptera: Psychodidae) Am J Trop Med Hyg 76 307–309 . [PubMed] [Google Scholar]
- Mead D. G., Lovett K. R., Murphy M. D., Pauszek S. J., Smoliga G., Gray E. W., Noblet R., Overmyer J., Rodriguez L. L. (2009). Experimental transmission of vesicular stomatitis New Jersey virus from Simulium vittatum to cattle: clinical outcome is influenced by site of insect feeding J Med Entomol 46 866–872 10.1603/033.046.0419 . [DOI] [PubMed] [Google Scholar]
- Nene V., Wortman J. R., Lawson D., Haas B., Kodira C., Tu Z. J., Loftus B., Xi Z., Megy K., other authors (2007). Genome sequence of Aedes aegypti, a major arbovirus vector Science 316 1718–1723 10.1126/science.1138878 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez De León A. A., O'Toole D., Tabachnick W. J. (2006). Infection of guinea pigs with vesicular stomatitis New Jersey virus transmitted by Culicoides sonorensis (Diptera: Ceratopogonidae) J Med Entomol 43 568–573 . [DOI] [PubMed] [Google Scholar]
- Quan P. L., Junglen S., Tashmukhamedova A., Conlan S., Hutchison S. K., Kurth A., Ellerbrok H., Egholm M., Briese T., other authors (2010). Moussa virus: a new member of the Rhabdoviridae family isolated from Culex decens mosquitoes in Côte d'Ivoire Virus Res 147 17–24 10.1016/j.virusres.2009.09.013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan P. L., Williams D. T., Johansen C. A., Jain K., Petrosov A., Diviney S. M., Tashmukhamedova A., Hutchison S. K., Tesh R. B., other authors (2011). Genetic characterization of K13965, a strain of Oak Vale virus from Western Australia Virus Res 160 206–213 10.1016/j.virusres.2011.06.021 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., Larget B., Liu L., Suchard M. A., Huelsenbeck J. P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space Syst Biol 61 539–542 10.1093/sysbio/sys029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K., Hagiwara K., Omatsu T., Hamasaki C., Kuwata R., Shimoda H., Suzuki K., Endoh D., Nagata N., other authors (2015). Isolation and characterization of a novel Rhabdovirus from a wild boar (Sus scrofa) in Japan Vet Microbiol 179 197–203 10.1016/j.vetmic.2015.05.013 . [DOI] [PubMed] [Google Scholar]
- Schneemann A., Schneider P. A., Kim S., Lipkin W. I. (1994). Identification of signal sequences that control transcription of Borna disease virus, a nonsegmented, negative-strand RNA virus J Virol 68 6514–6522 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider P. A., Schneemann A., Lipkin W. I. (1994). RNA splicing in Borna disease virus, a nonsegmented, negative-strand RNA virus J Virol 68 5007–5012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Liu Y., Hu X., Xiong J., Zhang B., Yuan Z. (2015). A metagenomic survey of viral abundance and diversity in mosquitoes from Hubei province PLoS One 10 e0129845 10.1371/journal.pone.0129845 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen M. W. (2008). Genome-scale relationships between cytosine methylation and dinucleotide abundances in animals Genomics 92 33–40 10.1016/j.ygeno.2008.03.009 . [DOI] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A. N. (2005). The HHpred interactive server for protein homology detection and structure prediction Nucleic Acids Res 33 (Web Server), W244–W248 10.1093/nar/gki408 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M. H., Andersen K. G., Folarin O. A., Grove J. N., Odia I., Ehiane P. E., Omoniwa O., Omoregie O., Jiang P. P., other authors (2015). Discovery of novel rhabdoviruses in the blood of healthy individuals from West Africa PLoS Negl Trop Dis 9 e0003631 10.1371/journal.pntd.0003631 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh R. B., Chaniotis B. N., Johnson K. M. (1972). Vesicular stomatitis virus (Indiana serotype): transovarial transmission by phlebotomine sandflies Science 175 1477–1479 10.1126/science.175.4029.1477 . [DOI] [PubMed] [Google Scholar]
- Tokarz R., Sameroff S., Leon M. S., Jain K., Lipkin W. I. (2014). Genome characterization of Long Island tick rhabdovirus, a new virus identified in Amblyomma americanum ticks Virol J 11 26 10.1186/1743-422X-11-26 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch F., Atkinson N. J., Evans D. J., Ryan M. D., Simmonds P. (2014). RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies eLife 3 e04531 10.7554/eLife.04531 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N., Widen S., Travassos da Rosa A. P., Wood T. G., Walker P. J., Holmes E. C., Tesh R. B. (2013). Malpais spring virus is a new species in the genus vesiculovirus Virol J 10 69 10.1186/1743-422X-10-69 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N., Castro-Llanos F., Widen S. G., Aguilar P. V., Guzman H., Guevara C., Fernandez R., Auguste A. J., Wood T. G., other authors (2014). Arboretum and Puerto Almendras viruses: two novel rhabdoviruses isolated from mosquitoes in Peru J Gen Virol 95 787–792 10.1099/vir.0.058685-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P. J., Kongsuwan K. (1999). Deduced structural model for animal rhabdovirus glycoproteins J Gen Virol 80 1211–1220 10.1099/0022-1317-80-5-1211 . [DOI] [PubMed] [Google Scholar]
- Walker P. J., Dietzgen R. G., Joubert D. A., Blasdell K. R. (2011). Rhabdovirus accessory genes Virus Res 162 110–125 10.1016/j.virusres.2011.09.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P. J., Firth C., Widen S. G., Blasdell K. R., Guzman H., Wood T. G., Paradkar P. N., Holmes E. C., Tesh R. B., Vasilakis N. (2015). Evolution of genome size and complexity in the rhabdoviridae PLoS Pathog 11 e1004664 10.1371/journal.ppat.1004664 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R. L., Lei X. Y., Ke F., Yuan X. P., Zhang Q. Y. (2011). Genome of turbot rhabdovirus exhibits unusual non-coding regions and an additional ORF that could be expressed in fish cell Virus Res 155 495–505 10.1016/j.virusres.2010.12.006 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data