ABSTRACT
Gene duplication by retrotransposition, i.e., the reverse transcription of an mRNA and integration of the cDNA into the genome, is an important mechanism in evolution. Based on whole-genome bisulfite sequencing of monocyte DNA, we have investigated the methylation state of all CpG islands (CGIs) associated with a retrocopy (n = 1,319), their genomic environment, as well as the CGIs associated with the ancestral genes. Approximately 10% of retrocopies are associated with a CGI. Whereas almost all CGIs of the human genome are unmethylated, 68% of the CGIs associated with a retrocopy are methylated. In retrocopies resulting from multiple retrotranspositions of the same ancestral gene, the methylation state of the CGI often differs. There is a strong positive correlation between the methylation state of the CGI/retrocopy and their genomic environment, suggesting that the methylation state of the integration site determined the methylation state of the CGI/retrocopy, or that methylation of the retrocopy by a host defense mechanism has spread into the adjacent regions. Only a minor fraction of CGI/retrocopies (n = 195) has intermediate methylation levels. Among these, the previously reported CGI/retrocopy in intron 2 of the RB1 gene (PPP1R26P1) as well as the CGI associated with the retrocopy RPS2P32 identified in this study carry a maternal methylation imprint. In conclusion, these findings shed light on the evolutionary dynamics and constraints of DNA methylation.
KEYWORDS: CpG islands, DNA methylation, imprinting, RPS2P32, retrotransposition
Introduction
During evolution, new genes mainly arise from gene duplications.1 Two mechanisms for gene duplication are known: segmental duplication, which leads to a gene copy retaining the exon-intron-structure and nearby regulatory elements, and retrotransposition, which leads to a gene copy lacking introns and non-exonic regulatory elements. Whereas gene copies resulting from segmental duplication are usually found close to the ancestral gene, retrocopies are often located on another chromosome.1-3
For a long time, potential functions of retrocopies and their evolution into expressed (pseudo)genes were unknown.1,2,4 The genome-wide study of Vinckenbosch et al. analyzed the transcription of all human retrocopies and found more than 1,000 transcribed retrocopies.5 During evolution, more than 10% of these have developed into so called bona fide genes. Several mechanisms have been described regarding how retrocopies can develop into functional retrogenes, such as the use of a foreign promoter at the integration site or through natural selection at CpG-rich sequences or CpG islands.2,5 CpG islands (CGIs) are CpG-rich sequences that meet specific criteria regarding length and CG-content, and are mainly located at the 5′ end of a gene.6-8
Imprinted genes can also arise from retrotransposition, and this has been shown in several studies. In mouse, five imprinted genes (Mcts2, Nap1l5, U2af1-rs1, Inpp5f_v2, and Peg12) are known to have arisen from retrotransposition.9-11 This phenomenon can also be observed in the human genome for a number of genes, e.g., MAGEL2, MKRN3, NDN, and NPAPA1.9,12-14 In the case of RB1, a differentially methylated CpG island (CpG85), which is part of a retrocopy (PPP1R26P1) and located in intron 2, is responsible for imprinting of the human RB1 gene. Parent-of-origin-specific methylation of CpG85 has been confirmed by other studies.15-18 At present, nearly 100 imprinted human genes have been identified, and several studies have taken a genome-wide look at parent-of-origin-specific DNA methylation in order to identify novel imprinted genes.18,19
Retrocopies are instructive genomic elements for studying the evolutionary dynamics and constraints of DNA methylation. Based on a whole genome methylome data set with single base pair (bp) resolution, we have investigated the methylation status of all human CGIs associated with a retrocopy and of their genomic environment, which we defined as the region 1,000 bp up- and down-stream of a retrocopy. The aim of this study was to analyze the epigenetic fate of human retrocopies.
Results
Identification of CGIs overlapping a retrocopy
To analyze CGIs overlapping a retrocopy, we first investigated the location of 27,537 human CGIs with regard to 13,173 human retrocopies, including pseudogenes and expressed retrogenes (see Fig. 1 for a flow diagram of our study). Only 10% (1,319) of all human retrocopies overlap a CGI. Approximately 50% of these CGI/retrocopies are located in coding regions, while the other 50% are located between genes or in non-coding pseudogenes (RefSeq). Most of the 1,319 retrocopies are classified by the UCSC genome browser as expressed retrogenes (1,122; 85%) and only a small number as pseudogenes (197; 15%) without expression.
Figure 1.
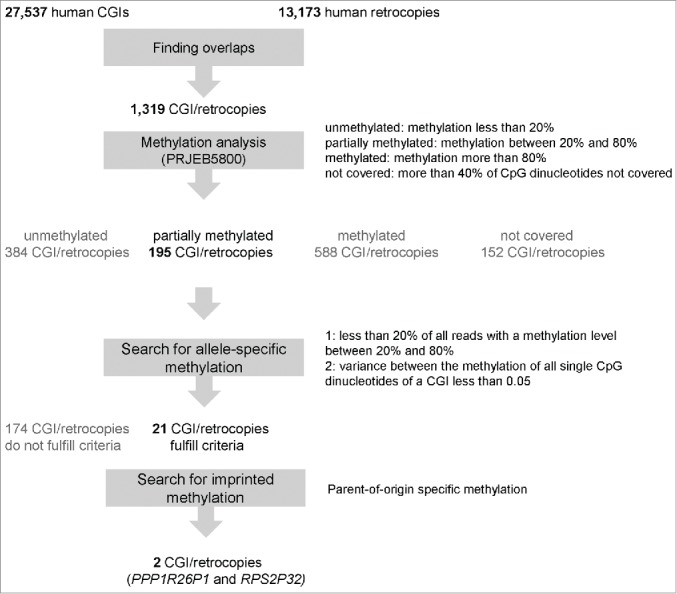
Flow diagram. This figure gives an overview of the analysis steps including the criteria for filtering.
Next, we analyzed whether the CGI/retrocopies were the result of a single or of multiple retrotransposition events. We found that 41% of the retrocopies are the result of a single event (Table S1). The other retrocopies are the result of two or more independent events.
Methylation analysis of human CGIs overlapping a retrocopy
The methylation level of all 1,319 CGIs overlapping a retrocopy was analyzed in a monocyte methylome data set obtained by whole-genome bisulfite sequencing (WGBS; ENA accession PRJEB5800). Fig. 2 visualizes the methylation level of all 1,319 CGIs, sorted by the number of independent transposition events. The majority of CGIs are either fully methylated or fully unmethylated, but the methylation level of different CGI/retrocopies derived from the same ancestral gene often differs.
Figure 2.
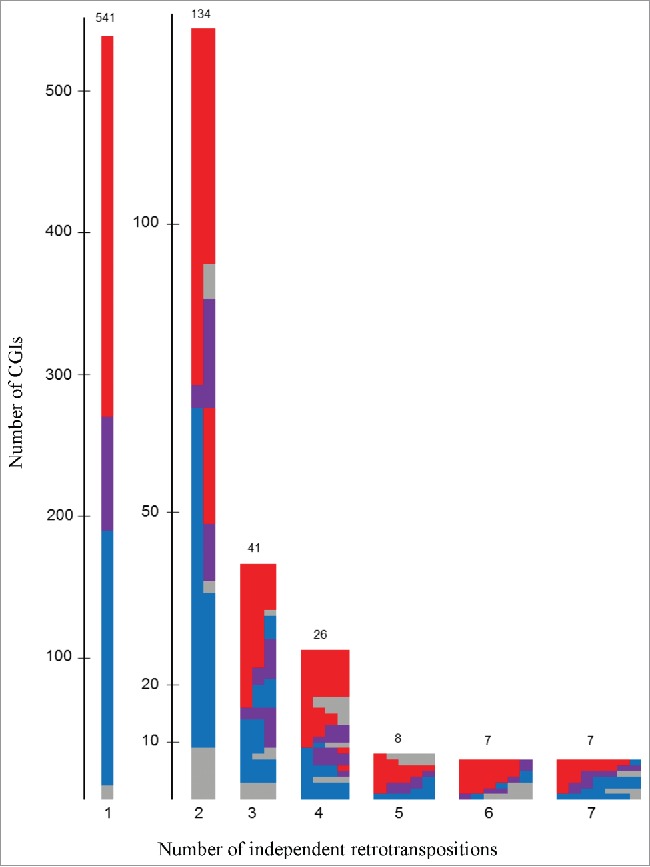
Methylation status of 1,319 CGIs overlapping a retrogene in monocytes. This figure shows the methylation (m) status of single CGIs overlapping a retrogene sorted by the number of independent retrotranspositions. Red: methylated CGIs (m > 80%); blue: unmethylated CGIs (m < 20%); violet: methylation between 20% and 80% and gray: CGIs with more than 40% of single CpG dinucleotides not covered and excluded for further analysis. The numbers can be found in the supplemental material, Table S1.
Because of low coverage (more than 40% of CpG dinucleotides were not covered; Fig. 2 colored in gray), 152 CGIs were excluded from further analysis. The methylation level of the remaining 1,167 CGIs/retrocopies is divided into 384 (32%) unmethylated CGIs (methylation less than 20%), 588 (50%) methylated CGIs (methylation more than 80%), and 195 (17%) partially methylated CGIs (methylation level between 20% and 80%). We did not find any methylation differences between intergenic/intragenic CGIs/retrocopies and expressed/not-expressed retrogenes (Table S2-S6).
Moreover, we compared the methylation level of all 1,167 CGI/retrocopies for which we have high quality methylation data to the methylation level of the surrounding region. For this, the methylation level of the region 1,000 bp upstream and downstream of the retrocopy was analyzed. First, we calculated the Pearson correlation coefficient (r = 0.78), which indicates a very strong positive relationship between the methylation level of the CGI and the surrounding region. Further we analyzed the data by k-means cluster analysis (k = 4; Fig. 3). Approximately 52% of the CGIs overlapping a retrocopy are methylated and so is the surrounding region of the retrocopy (red cluster; 603 elements, 52%). Unmethylated CGIs (32%) can be found in unmethylated regions (blue cluster; 204 elements, 17%) and methylated regions (dark gray cluster; 136 elements, 12%), with a slight bias for unmethylated regions. Only a small number of CGIs (12%) have a methylation level between 20% and 80%. These CGIs are found in regions containing a methylation level between 40% and 100%.
Figure 3.
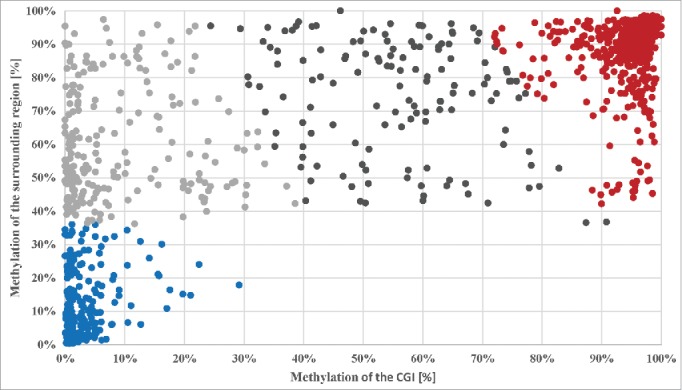
Cluster analysis of the methylation level of the 1,167 CGI dependent on the methylation status of the surrounding region (± 1,000 bp) of the retrogene using k-means. The vertical axis gives the percentage of the methylation level of the surrounding region of the retrogene and the horizontal axis the percentage of the methylation level of the CGI. The cluster analysis resulted in four cluster (k = 4); Cluster 1: dark gray, 136 elements (12%), centroid (56%/74%); Cluster 2: red, 603 elements (52%), centroid (95%/89%); Cluster 3: blue, 204 elements (17%), centroid (4%/13%); Cluster 4: light gray, 224 elements (19%), centroid (9%/60%).
In addition, we analyzed if the presence of a DNA repeat close to the retrocopy is linked to the methylation status of the CGI. In most cases (839; 72%) both the 1,000 bp upstream region and the 1,000 bp downstream region overlapped with at least one repeat. The absence of a repeat on both sides (47), in the upstream region only (137) or in the downstream region only (144) did not change the clustering of the CGIs (Figs. S1-S4).
Comparative methylation analysis of ancestral CGIs and retrocopy-associated CGIs
Nearly all ancestral genes (96%) contain at least one CGI. We compared the sequence identity of the CGIs overlapping a retrocopy with the ancestral CGIs to distinguish between retrotransposed CGIs and newly formed CGIs. Only 372/1167 CGIs (32%) showed an identity over 60% to the ancestral CGI, suggesting that most of the CGIs developed during evolution. In 310 of 372 cases, the ancestral CGI was unmethylated, whereas 262/372 of the retrotransposed copies were methylated, suggesting a gain of methylation.
Search for allele-specific methylation
For identification of allele-specific methylation, we analyzed the 195 partially methylated CGIs in more detail. Allele-specific methylation should be reflected by the presence of mainly unmethylated and mainly methylated single sequence reads in the WGBS data set and only a small number of reads with a methylation level between 20% and 80% (less than 20% of all reads). As described in Rademacher et al. 2014, we checked the methylation of each single CpG dinucleotide by calculating the variance (variance equal or less than 0.05) to exclude CGIs having a high methylation in one part und a low degree of methylation in another part. Based on the two criteria, we found 21 CGIs (Table 1; Figs. S5-S21 and Table S11) including the four human/non-murine intronic CGIs reported previously by Rademacher et al.20
Table 1.
Read analysis and CpG methylation with intermediate methylation levels.
| Reads unmethylated(< 20% meth.) |
Reads methylated(≥ 80% meth.) |
Reads partially methylated (≥ 20% and < 80% meth) |
CpG methylation |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CGI_ID | Genomic coordinates | Ancestral gene | Mean meth. [%] | Mean cov. | Number reads | Mean mapping quality of all reads*** | [#] | [%] | [#] | [%] | [#] | [%] | VAR | SD |
| 2391_1_hg19 | chr1:240656253-240656720 | GREM1 | 56% | 12 | 83 | 60 | 29 | 35% | 44 | 53% | 10 | 12% | 0.04 | 0.2 |
| 6141_1_hg19 | chr4:144833114-144833512 | SAV1 | 49% | 18 | 111 | 57 | 67 | 60% | 39 | 35% | 5 | 5% | 0.04 | 0.2 |
| 9224_1_hg19** | chr7:16890768-16891087 | ARHGAP20 | 58% | 13 | 62 | 59 | 24 | 39% | 27 | 44% | 11 | 18% | 0.06 | 0.23 |
| 9261_1_hg19 | chr7:23530434-23530690 | RPS2 | 72% | 15 | 59 | 60 | 16 | 27% | 40 | 68% | 3 | 5% | 0.01 | 0.11 |
| 9377_1_hg19 | chr7:36010997-36011407 | PPP1R14B | 39% | 7 | 48 | 60 | 25 | 52% | 15 | 31% | 8 | 17% | 0.04 | 0.21 |
| 9473_1_hg19 | chr7:52341468-52342266 | CCDC115 | 79% | 12 | 108 | 56 | 19 | 18% | 74 | 69% | 15 | 14% | 0.03 | 0.17 |
| 13085_1_hg19 | chr10:66813635-66814061 | NEK4 | 76% | 8 | 58 | 49 | 9 | 16% | 40 | 69% | 9 | 16% | 0.02 | 0.16 |
| 13250_1_hg19 | chr10:91596974-91597792 | MARK2 | 22% | 10 | 109 | 58 | 83 | 76% | 23 | 21% | 3 | 3% | 0.01 | 0.07 |
| 14414_1_hg19* | chr11:62138621-62138873 | ASRGL | 55% | 10 | 40 | 58 | 17 | 43% | 21 | 53% | 2 | 5% | 0.02 | 0.13 |
| 15224_1_hg19* | chr12:3947922-3948620 | PARP11 | 42% | 11 | 108 | 49 | 53 | 49% | 45 | 42% | 10 | 9% | 0.05 | 0.20 |
| 15400_1_hg19 | chr12:31405184-31405545 | RPL13AP5 | 69% | 16 | 97 | 53 | 26 | 27% | 61 | 63% | 10 | 10% | 0.02 | 0.15 |
| 15512_1_hg19 | chr12:49782965-49783193 | FGFR1OP2 | 54% | 18 | 59 | 60 | 22 | 37% | 30 | 51% | 7 | 12% | 0.02 | 0.15 |
| 16448_1_hg19 | chr13:21893156-21893605 | GRK6 | 79% | 13 | 88 | 58 | 9 | 10% | 67 | 76% | 12 | 14% | 0.02 | 0.12 |
| 16458_1_hg19 | chr13:23412207-23412623 | IPMK | 31% | 11 | 66 | 59 | 41 | 62% | 17 | 26% | 8 | 12% | 0.02 | 0.15 |
| 16634_1_hg19* | chr13:48892635-48893857 | PPP1R26 | 75% | 13 | 193 | 60 | 69 | 36% | 116 | 60% | 8 | 4% | 0.02 | 0.13 |
| 17031_1_hg19 | chr14:21191657-21191860 | XPO6 | 42% | 7 | 30 | 60 | 18 | 60% | 11 | 37% | 1 | 3% | 0.02 | 0.16 |
| 19100_1_hg19* | chr16:15083366-15084045 | KIAA2013 | 75% | 11 | 104 | 44 | 22 | 21% | 67 | 64% | 15 | 14% | 0.02 | 0.16 |
| 20403_1_hg19 | chr17:15686218-15686474 | IL6ST | 20% | 13 | 49 | 57 | 41 | 84% | 7 | 14% | 2 | 4% | 0.01 | 0.10 |
| 23548_1_hg19 | chr19:21860792-21861016 | MTDH | 59% | 14 | 52 | 59 | 14 | 27% | 27 | 52% | 11 | 21%** | 0.03 | 0.17 |
| 24982_1_hg19 | chr20:30135076-30135292 | MCTS1 | 55% | 20 | 64 | 60 | 25 | 39% | 37 | 58% | 2 | 3% | 0.01 | 0.10 |
| 26859_1_hg19 | chrX:37026348-37026706 | FAM47A | 77% | 8 | 44 | 46 | 6 | 14% | 34 | 77% | 4 | 9% | 0.02 | 0.15 |
This table shows the results of the read methylation and CpG methylation analysis of 17 CGIs overlapping a retrogene and containing an intermediate methylation level and four already published CGIs by Rademacher et al. (same dataset; indicated by *).20 In addition to CGI_ID, genome coordinates, ancestral gene, mean methylation, mean coverage the number of reads is shown. The reads are divided into three classes, unmethylated (< 20% methylation), methylated (≥ 80% methylation) and partially methylated (≥ 20% and < 80% methylation). For each class, the total number of reads and the percentage is shown. The last column shows the results of the CpG methylation analyses; variance (VAR) and standard derivation (SD) over all single CpGs were calculated. More information of the CGIs, retrocopies, and ancestral genes can be found in the supplemental material (Table S11).
Published by Rademacher et al.20
Fulfill only one criteria (out of two) described by Rademacher et al.20
Mean of the phred quality score of all analyzed reads. The score quantifies the probability that a read is misplaced (for further information see supplement material).
In addition to monocytes, we analyzed the newly discovered 17 CGIs/retrocopies in an additional methylome data set (Rademacher et al; EGA accession: EGAS000001000719), 16 already published methylome data sets by Ziller et al. and two oocyte methylome data sets published by Okae et al. (Table S8).20-22 Out of these 17 CGIs/retrocopies, seven CGIs (2391_1_hg19, 9224_1_hg19, 9261_1_hg19, 9377_1_hg19, 17031_1_hg19, 23548_1_hg19 and 24982_1_hg19) showed an intermediate methylation level in nearly all tissues except for human sperm DNA, where all CGIs are typically unmethylated. No methylation in nearly all tissues was found in three CGIs (13250_1_hg19, 16458_1_hg19, and 20403_1_hg19), six CGIs are fully methylated in at least three tissues, and one CGI (6141_1_hg19) is not methylated in fetal tissues. Without exception, all analyzed CGIs show a methylation level lower than 20% in human sperm DNA. In oocytes, 13 out of 17 CGIs/retrocopies showed no methylation and the other four CGIs/retrocopies were fully methylated in these cells. Single reads were not available for these data sets.
Deep bisulfite amplicon sequencing of 17 CGIs overlapping a retrocopy
We performed deep bisulfite amplicon sequencing on monocyte DNA samples from unrelated donors to find out whether the intermediate methylation levels of the newly discovered 17 CGIs overlapping a retrocopy resulted from allele-specific methylation. We did not find heterozygous samples (we analyzed 21 unrelated monocyte DNA samples) for a single-nucleotide polymorphism for five CGIs out of 17. For the remaining 12 CGIs we were able to establish amplicons for deep bisulfite sequencing. Table 2 shows the results of this analysis. We found two heterozygous samples for 11 CGIs, except for CGI 16458_1_hg19, where we just found only one heterozygous sample out of 21. In contrast to the in silico methylation analysis, two CGIs (13250_1_hg19 and 20403_1_hg19) appeared to be fully unmethylated after deep bisulfite amplicon sequencing. Five CGIs showed more than 10% methylation differences between both alleles in two samples. For two CGIs (6141_1_hg19 and 9473_1_hg19), one allele was always more methylated than the other allele in both samples. This could be an indication of a cis-acting SNP. The remaining three CGIs (9261_1_hg19, 15400_1_hg19, and 23548_1_hg19) show also more than 10% methylation differences between both alleles, but the methylation status of the single alleles differ between both samples. These results could hint to genomic imprinting. CGI 9261_1_hg19 shows a methylation difference greater than 40% between both alleles and this CGI is methylated in oocytes and unmethylated in sperm. Because of these results, we decided to analyze 9261_1_hg19 in more detail.
Table 2.
Results of deep bisulfite amplicon sequencing on monocyte DNA samples.
| Allele a |
Allele b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CGI_ID | Sample | Mean meth. [%] | Analyzed CpGs | Number reads | Base | Meth.[%] | Number reads | Base | Meth.[%] | Number reads |
| 2391_1_hg19 | R3 | 59% | 13 | 714 | G | 60% | 213 | A | 58% | 478 |
| R9 | 57% | 13 | 607 | G | 60% | 135 | A | 55% | 458 | |
| 6141_1_hg19 | R7 | 28% | 41 | 1452 | A | 47% | 393 | T | 20% | 1057 |
| R13 | 23% | 41 | 1127 | A | 39% | 366 | T | 16% | 759 | |
| 9224_1_hg19 | R6 | 32% | 13 | 2179 | G | 8% | 960 | A | 51% | 1150 |
| R7 | 23% | 13 | 2800 | G | 26% | 1226 | A | 21% | 1484 | |
| 9261_1_hg19 | R5 | 64% | 16 | 897 | G | 87% | 536 | A | 27% | 326 |
| R13 | 47% | 16 | 1002 | G | 23% | 428 | A | 66% | 542 | |
| *9377_1_hg19 | − | − | − | − | − | − | − | − | − | − |
| 9473_1_hg19 | R8 | 77% | 18 | 2491 | A | 86% | 1393 | G | 66% | 929 |
| R10 | 79% | 18 | 2542 | A | 86% | 1369 | G | 70% | 1017 | |
| **13085_1_hg19 | − | − | − | − | − | − | − | − | − | − |
| 13250_1_hg19 | R3 | 3% | 19 | 1801 | G | 4% | 677 | A | 2% | 934 |
| R5 | 4% | 19 | 3060 | G | 6% | 1338 | A | 2% | 1499 | |
| 15400_1_hg19 | R4 | 70% | 29 | 1840 | G | 62% | 1012 | A | 81% | 810 |
| R14 | 78% | 29 | 1482 | G | 82% | 1063 | A | 66% | 411 | |
| *15512_1_hg19 | − | − | − | − | − | − | − | − | − | − |
| 16448_1_hg19 | R3 | 61% | 31 | 516 | A | 81% | 240 | G | 44% | 272 |
| R5 | 94% | 31 | 982 | A | 94% | 406 | G | 94% | 566 | |
| 16458_1_hg19 | P2 | 22% | 27 | 984 | A | 15% | 302 | G | 25% | 509 |
| 17031_1_hg19 | R4 | 29% | 18 | 4187 | G | 19% | 2038 | T | 40% | 2124 |
| R14 | 17% | 18 | 4814 | G | 12% | 2313 | T | 21% | 2469 | |
| 20403_1_hg19 | R1 | 3% | 32 | 3397 | T | 2% | 1503 | G | 3% | 1889 |
| R8 | 3% | 32 | 2255 | T | 3% | 1116 | G | 3% | 1135 | |
| 23548_1_hg19 | R2 | 25% | 22 | 3633 | G | 38% | 1910 | T | 11% | 1700 |
| R15 | 25% | 22 | 3438 | G | 19% | 1569 | T | 30% | 1851 | |
| *24982_1_hg19 | − | − | − | − | − | − | − | − | − | − |
| *26859_1_hg19 | − | − | − | − | − | − | − | − | − | − |
This table summarizes the results of the deep bisulfite amplicon sequencing on monocyte DNA samples. In addition to the CGI_ID, the in-house sample number, the mean methylation level, the number of analyzed CpG dinucleotides and the total number of reads are shown. For each separated allele (allele a and allele b) base, methylation level and number of reads are specified.
non-informative sample;
no Amplicon design possible.
Parent-of-origin of CGI 9261_1_hg19 methylation
To determine whether allelic methylation of CGI 9261_1_hg19 is parent-of-origin-specific, we analyzed blood of three parent-child trios informative for SNP rs10228640 (I-III; Fig. 4A). The methylation level of CGI 9261_1_hg19 is between 49% and 60% in all three children (Fig. 4B). Similar to the results in monocytes, one allele is always more methylated than the other allele (more than 55% methylation differences between both alleles). In all three analyzed trios, the more methylated allele was maternally inherited (Fig. 4B: I maternally methylated G-allele; II and III maternally methylated A-allele) and the less methylated allele was paternally inherited (Fig. 4B: I paternally less methylated A-allele; II and III maternally less methylated G-allele). This result excludes a cis-acting genetic effect on DNA methylation and points to imprinted DNA methylation.
Figure 4.
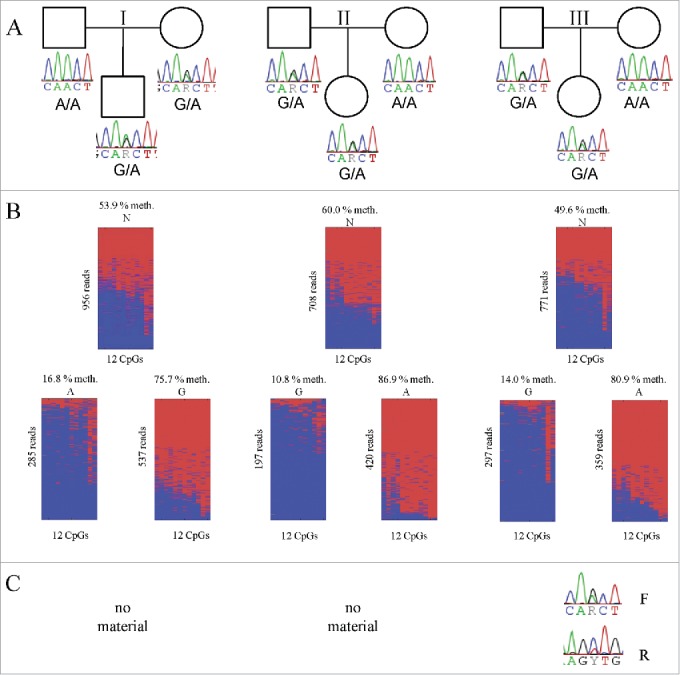
DNA methylation analysis and expression analysis of CGI 9261_1_hg19 in blood. (A). Pedigrees of the four trios (I-IV) investigated including the genomic genotype of all family members. (B). Single molecular methylation analysis of the CGI 9261_1_hg19 in heterozygous individuals (children of trios shown in A). The amplicon covers 16 out of 21 CpGs of the CGI. The upper part of the figure shows all amplicon reads, whereas the lower part shows the sequence reads sorted by SNP allele. Lines represent sequence reads, columns CpGs. Mean methylation level, SNP allele and number of reads is given. Blue unmethylated; red methylated. Part (C) shows the results of the expression analysis. For one individual (trio II) no RNA was available.
Next, we analyzed the methylation of CGI 9261_1_hg19 in six patients with a known multilocus imprinting defect (MLID) (Fig. 5). Four of the MLID patients (MLID 3, MLID 4, MLID 5, and MLID 6) showed a hypomethylation (methylation level less than 20%) in the analyzed amplicon of CGI 9261_1_hg19. The remaining two patients (MLID 1 and MLID 2) showed a methylation level about 50% as the four controls (NC 1, NC 2, NC 3 and NC 4).
Figure 5.
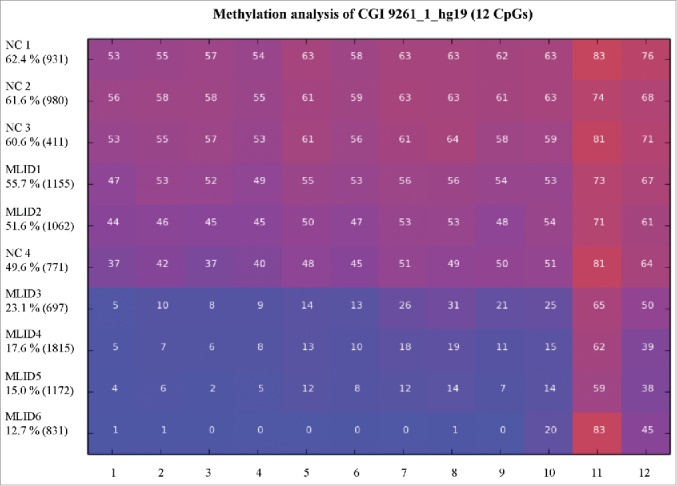
Comparative methylation analysis of CGI 9261_1_hg19 in blood. This figure shows the results of the methylation analyses by deep bisulfite sequencing of six individuals with a confirmed MLID (MLID 1–6) and four controls (NC 1–4). Each line represents one analyzed individual and each column an analyzed CpG dinucleotide. Mean methylation and number of reads for each individual are given. The color indicates the average methylation level of each CpG dinucleotide from blue unmethylated to red methylated. For clinical classification of patients with a confirmed MLID see Material.
Expression analysis
For determining if RPS2P32 is monoallelically or biallelically expressed, we performed strand-specific expression analyses of RNA from blood of child III (other RNA samples were not available). As shown in Fig. 4C, RPS2P32 is expressed from both alleles in both directions. By quantitative primer extension analyses (Table S12) we found an equal expression of the forward strand (ratio = 1.18) and a slight skewing (ratio = 1.78) of the reverse strand in favor of the paternal (unmethylated) allele.
Because the CGI does not appear to regulate the expression of RPS2P32, we inspected the neighboring genes (Fig. 6). About 15 kb upstream is the TRA2A (ENSG00000164548) gene, which encodes a sequence-specific RNA-binding protein involved in the regulation of pre-mRNA splicing. Circa 20 kb downstream is the IGF2BP3 gene, which encodes the insulin-like growth factor 2 mRNA binding protein 3 (ENSG00000136231).
Figure 6.

Genomic position of RPS2P32 on chromosome 7. This figure visualizes the genomic position of RPS2P32 in the human genome. The structure of the gene and the location of CGI 9261_1_hg19 are shown in (A). In (B) the neighboring genes (IGF2BP3 and TRA2A) including CGIs are represented. Gray: RefSeq genes including transcription direction (white arrow); green: CpG islands.
Owing to the lack of a common expressed SNP in TRA2A, we could not analyze this gene. Since IGF2BP3 is poorly expressed in blood, we analyzed DNA and RNA from placentas of different trimesters of pregnancy. As shown in Fig. S22, we detected biallelic expression in all informative samples.
Discussion
Based on whole-genome bisulfite sequencing of human monocyte DNA, which provided DNA methylation data at single base pair resolution, we have investigated the methylation status of all CGIs associated with a retrocopy and their genomic environment as well as the CGIs associated with the ancestral genes. Our major findings are as follows:
In contrast to the majority of human genes, only 10% of retrocopies are associated with a CGI.
Whereas almost all CGIs are unmethylated, 68% of the CGIs associated with a retrocopy are methylated.
In retrocopies resulting from multiple retrotranspositions of the same ancestral gene, the methylation state of the CGI often differs.
There is a strong positive correlation between the methylation state of the CGI/retrocopy and their genomic environment.
In addition to the previously reported CGI/retrocopy in intron 2 of the RB1 gene (CpG85), we have identified one other retrocopy (RPS2P32) associated CGI with a maternal methylation imprint.
Most of the CGIs in the human genome span the promoter/exon 1 region of a gene and are unmethylated. Apparently, the binding of transcription factors to these regions prevents their methylation.23 Lack of methylation has led to retention of the cytosine residues during evolution, which is in contrast to other regions of the human genome where deamination of methylcytosine has led to C->T transitions. Since only exons, but not promoter regions are retrotransposed, retrocopies lack a CGI or have a shorter CGI. Furthermore, exonic CGI fragments may have deteriorated after retrotransposition. On the other hand, other CGIs may have newly evolved after retrotransposition. All of this explains why only 10% of retrocopies are associated with a CGI.
As mentioned above, the binding of transcription factors to CGIs appears to prevent their methylation.23 This is the case for most of the human genes. In contrast, retrocopies are non-functional or expressed at low levels only. This may explain why only one third of the retrocopy associated CGIs are unmethylated. Furthermore, retrotransposed sequences may be subject to host defense mechanisms that methylate invading DNA. Unmethylated CGIs may have escaped this mechanism, or they have gained a function after retrotransposition.
Methylation differences between CGI/retrocopies derived from the same ancestral gene (Fig. 2) demonstrate that the methylation status is not or not only determined by the retrotransposed DNA sequence. As discussed below, a major determinant appears to be the genomic environment.
In fact, there is a strong correlation between the methylation state of the genomic environment and the CGI/retrocopy. We did not detect any link between methylation and the expression status of the retrocopies or the presence of repeats in their immediate vicinity. The presence of methylated CGIs in a methylated environment only (red cluster in Fig. 3) suggests that methylation at the integration site has a strong effect on the methylation status of a CGI/retrocopy.24 With regard to imprinted CGIs, there are several examples showing that a retrogene can acquire the epigenetic state of their integration site: NDN, which is derived from the X chromosome, and NPAP1, which is derived from chromosome 7.14,25,26 Both genes jumped into the imprinted chromosome 15q11q13 domain and came under the control of a cis-acting imprinting center. NDN is in the list of 1,319 CGIs associated with a retrocopy in this study, but it did not pass the threshold for differential methylation, because more than 40% of the WGBS reads had mixed methylation, which is a rather unusual finding for an imprinted gene. This may be due to the very large size of the CGI (1 kb). NPAP1 and some other retrogenes, such as PEG10, are not in the list of 1,319 CGIs, because they are not indicated as retrocopies in the UCSC data.27 The origin of NPAP1 has only recently been described by Neumann et al.14
However, it is also possible that methylation of the retrocopy by the host defense mechanism may have spread into adjacent regions.28 In this case, the correlation between the methylation state of the genomic environment and the CGI/retrocopy were due to de novo methylation after retrotransposition. Therefore, we do not know whether the presence of methylated CGIs in methylated regions and unmethylated CGIs in unmethylated regions indicates that retrocopies can integrate in methylated and unmethylated regions with equal probability.
Interestingly, CGI/retrocopies with intermediate levels of methylation are exclusively found in regions with > 40% methylation. In a pure cell population like monocytes, intermediate levels of methylation can be due to several situations: cellular heterogeneity, patchy methylation of both alleles, a gradient of methylation along the CGI, allele-specific methylation due to cis-acting genetic variation, and imprinted methylation. Among the 195 CGI/retrocopies with intermediate levels of methylation, we identified only 2 CGIs with a methylation imprint: The previously reported CpG85, which is associated with PPP1R26P1 and located within the RB1 gene, and the CGI associated with the RPS2P32 retrocopy described here. In both cases, the retrocopy jumped into a non-imprinted region and acquired maternal methylation. The genomic environment of PPP1R26P1 has a methylation level of 79%. CpG85 harbors an alternative start exon, which is spliced onto exon 3 of the RB1 gene. This alternative RB1 transcript is transcribed from the paternal allele only. The integration of this retrocopy led to RB1 imprinting as its expression is skewed in favor of the maternal allele.16,17
RPS2P32 on chromosome 7 is derived from the gene RPS2 on chromosome 16 (Fig. 6). The genomic environment shows a degree of methylation of 90%. The ancestral gene also contains a CGI, but this is unmethylated. As demonstrated by oocyte-specific methylation, maternal methylation in blood cells, hypermethylation in maternal uniparental disomy 7 (matUPD729), and loss of methylation in 4 of 6 patients with MLID, the RPS2P32 associated CGI is maternally imprinted. However, we found biallelic expression of RPS2P32 in blood, although there may be a slight skewing of allelic expression of the reverse strand (Fig. 4C). Also, Hannula-Jouppi et al. did not find a significantly different expression between matUPD7 and controls.29 At first sight, this excludes RPS2P32 as a novel imprinted gene, but it is possible that it is monoallelically expressed in other tissues.
As we could not detect monoallelic expression for RPS2P32, we looked at the neighboring genes TRA2A and IGF2BP3. IGF2BP3 encodes a protein that regulates the stability and translation of the mRNA of IGF2, an imprinted gene involved in development and growth.30-32 Thus, IGF2BP3 is growth promoting and thus is an excellent candidate for an imprinted gene. However, in blood the gene is expressed from both alleles, as has already been shown by Monk et al.33 Li et al. showed that IGF2BP3 is highly expressed in placenta, where the expression is strongest in the 1st trimester and decreases in the 3rd trimester of pregnancy.34 Moreover, they detected that IGF2BP3 expression is decreased in pre-eclamptic placentas, which is accompanied by intrauterine growth retardation. Thus, we also investigated the expression in 1st, 2nd, and 3rd trimester placenta samples. Our analyses revealed that in all analyzed placenta samples IGF2BP3 is biallelically expressed (Fig. S22). Still, as mentioned above for RPS2P32, it is possible that imprinted IGF2BP3 expression is restricted to certain tissues or cell types. A good example for this is the UBE3A gene, which is biallelically expressed in almost all tissues except for brain, where it is transcribed from the maternal allele only.35 Its transcription is controlled by an antisense-transcript that in turn is regulated by an ICR.36-39 This ICR is paternally unmethylated and maternally methylated in all tissues.40
In conclusion, our study has shed light on the evolutionary dynamics and constraints of DNA methylation and identified a novel human locus carrying a genomic imprint.
Materials and methods
Material
Genomic DNA for the four informative trios was isolated from peripheral blood using the FlexiGene DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer's manual. RNA from peripheral blood was extracted with PAXgene blood Kit (PreAnalytiX, Hombrechtikon, Switzerland) following the manufacturer's instructions.
DNA and RNA from human placenta were ordered from the Baby Bio Bank (University College London and Imperial College London, London, United Kingdom) and from the Institute of Molecular Biology / Department of Gynecology and Obstetrics, University Hospital Essen, Germany. Blood and placenta samples were obtained after informed consent.
MLID patients have been recruited through the BMBF consortium ‘Diseases caused by imprinting defects’ (01GM1513) and are described by Bens, S., Kolarova, J., Beygo, J., Buiting, K., Caliebe, A., Eggermann, T., Gillessen-Kaesbach, G., Prawitt, D., Thiele-Schmitz, S., Begemann, M., Enklaar, T., Gutwein, J., Haake, A., Paul, U., Richter, J., Soellner, J., Vater, I., Monk, D., Horsthemke, B., Ammerpohl, O. and Siebert, R. (submitted). Three MLID patients (MLID1, MLID3, and MLID5) have a Silver-Russell syndrome-like phenotype; two patients (MLID2 and MLID6) have a Beckwith-Wiedemann syndrome-like phenotype and one patient (MLID4) has a pseudohypoparathyrodism type 1b-like phenotype.
Data collection
The following tracks for humans (CRCh37/hg19) were downloaded from the UCSC Genome Browser for chromosomes 1 to 22 and chromosome X: CpG islands (information and sequences; n = 27,537; last accessed 04.09.2014), retrocopies (information; n = 13,173; last accessed 11.11.2014), RefSeq genes (information; n = 49,451; last accessed 16.12.2014), and all repeats (Repeat Masker; n = 5,189,085; last accessed 16.12.2015).41
The monocyte methylome data (methylome 1: ENA accession PRJEB5800; methylome 2: EGA accession EGAS000001000719) were already published by Rademacher et al.20 In addition, 16 methylomes of different tissues published by Ziller et al. were downloaded from the gene expression omnibus (GSE46644) and two oocyte methylomes published by Okae et al. were downloaded from the Japanese Genotype-phenotype archive under the Accession number JGA S00000000006.21,22
Data analysis
All human CGIs were serially numbered with a unique ID as described by Rademacher and colleagues.20 By comparing the position of CGIs and retrocopies using the Perl programming language (http://www.perl.org/), all CGIs overlapping a retrocopy were determined. To divide CGIs/retrocopies in intergenic and intragenic once, we compared the positions of all CGIs/retrocopies to the positions of RefSeq genes. For sequence comparison between CGIs overlapping a retrocopy and CGIs of the ancestral gene blast2seq (blast two sequences) is used.42
Whole genome bisulfite sequencing and whole genome methylation analysis
‘Whole Genome Bisulfite Sequencing’ and ‘Whole Genome Methylation Analysis’ were performed as previously described in Rademacher et al.20
In addition to the methylation status of single CGIs, we also calculated the average methylation level of 1000 bp upstream to the retrocopy start and 1000 bp downstream to the retrocopy end.
Statistical analysis
To statistically determine a correlation between the methylation level of a CGI/retrogene and the surrounding region of a retrogene we used the Pearson correlation coefficient. The cluster analysis of these methylation data was done by a k-means cluster analysis (k = 4) using the algorithm of Hartigan and Wong.43 For both statistically analyses the statistical software R was used (https://www.r-project.org/; cor(stats); kmeans(stats)).
Genotyping
Primers for genotyping of the 17 CGIs overlapping a retrogene are listed in Table S8 of the supplemental material. For 14 out of 17 CGIs (indicated in Table S8 with a ‘no’ in row ‘betain’), each 25 μl reaction contained 100 ng of genomic DNA, 200 μM of each dNTP (dATP, dCTP, dTTP, and dGTP), 0.4 μM of each primer, 1.5 mM MgCl2, 1 x Green GoTaq Reaction Buffer and 1.25 units GoTaq G2 DNA Polymerase (Promega, Fitchburg, USA). The PCR conditions for these 17 CGIs were as follows (for Tm = X see supplemental material, Table S8): 95°C for 2 min; 35 cycles of 95°C for 30 s, X°C for 30 s and 72°C for 45 s; and 72°C for 5 min.
For the remaining three CGIs, each 25 μl reaction contained 130 ng of genomic DNA, 80 μM of each dNTP (dATP, dCTP, and dTTP), 32 μM of dGTP, 48 μM of 7-deaza-2′-deoxy-guanosine-5′-triphosphate (Roche, Basel, Switzerland), 0.4 μM of each primer, 1.5 mM MgCl2, 0.5 mM betaine (USB Corporation, Cleveland, OH, USA), 1 x Green GoTaq Reaction Buffer and 5 units GoTaq G2 DNA Polymerase (Promega, Fitchburg, USA). The PCR conditions were as follows (for Tm = X see supplemental material, Table S8): 95°C for 2 min; 45 cycles of 96°C for 30 s, X°C for 30 s and 72°C for 45 s; and 72°C for 7 min. The PCR products were sequenced using Big Dye Terminators (BigDye Terminator v1.1 Cycle Sequencing Kit; Applied Biosystems, Foster City, CA) on an ABI 3130xl Genetic Analyzer. For analyses, Sequencing Analysis software (Applied Biosystems) and Geneious (Biomatters, Auckland, New Zealand) were used.
Deep bisulfite amplicon sequencing
Genomic DNA was bisulfite treated using the EZ DNA Methylation-Gold Kit (Zymo Research Europe, Freiburg, Germany) according to the manufacturer's protocol. The following steps including generation of the bisulfite amplicon libraries, sample preparation and sequencing on the Roche 454 GS Junior system were realized as previously described.44 Primer sequences and annealing temperatures can be found in the supplement (supplemental material, Table S9). The Python-based amplikyzer software was used to analyze the bisulfite sequencing data generated by the Roche 454 GS junior.45
Expression analysis
All RNA samples were treated with RQ1 DNase (Promega, Fitchburg, USA) to remove residual traces of genomic DNA. For the RPS2P32 locus a strand-specific reverse transcriptase (RT) for each strand was performed using SuperScript™ III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. For each informative sample 2 μg RNA from PAX-blood (PreAnalytiX, Hombrechtikon, Switzerland) was reverse transcribed with strand-specific primers (9261_1_hg19_ F and 9261_1_hg19_R; see supplemental material Table S10). For the PCR each 50 μl reaction contained 12 μl of the RT reaction, 0.2 mM of dNTPs, 0.4 μM of each primer, 1.5 mM MgCl2, 1x Green GoTaq Reaction Buffer and 2.5 U GoTaq G2 DNA Polymerase (Promega, Fitchburg, USA). The PCR conditions for RPS2P32 were as follows: 95°C for 2 min; 40 cycles of 95°C for 30 s, 63°C for 30 s and 72°C for 45 s; and 72°C for 5 min. Since both DNA and RNA products have the same size a PCR for the ß-actin gene was carried out to rule out DNA contamination and to verify the integrity of the RNA samples.46
For the IGF2BP3 locus the reverse transcription was performed using the GeneAmp RNA PCR Kit (Applied Biosystems, Foster City, CA, USA). For each informative sample 2 μg total RNA from PAX-blood or rather placenta (1st, 2nd, 3rd trimester of pregnancy) was reverse transcribed using random hexamers (reaction volume 50 µl).16 The subsequent touch down PCR (reaction volume 50 μl) contained 16 μl of the RT reaction, 0.2 mM dNTPs, 0.4 μM of each primer, 1.5 mM MgCl2, 1x Green GoTaq Reaction Buffer and 5 U GoTaq G2 DNA Polymerase (Promega, Fitchburg, USA). PCR conditions were as follows: 95°C for 2 min; 14 cycles of 95°C for 30 s, 68°C for 30 s and 72°C for 30 s (annealing temperature was gradually reduced for 0.5°C/cycle); 45 cycles of 95°C for 30 s, 62°C for 30 s and 72°C for 30 s; and 72°C for 5 min.
The PCR products for both loci were gel-purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Fitchburg, USA). Sequencing and sequence analysis were performed as described above.
Primer extension analysis
For measuring the allelic ratios of heterozygous mRNA (after conversion to cDNA) and genomic DNA (as reference), single nucleotide primer extension assays were performed. For this, we used the ABI Prism SNaPshot ddNTP Primer Extension Kit (Applied Biosystems, Foster City, CA, USA). Equal amounts of amplicons of cDNA and genomic DNA were subjected to capillary gel-electrophoresis on ABI 3130 (Applied Biosystems, Foster City, CA, USA). Gene Mapper 4.0 software was used to analyze the electropherograms. Allelic cDNA ratios were normalized using the allelic DNA ratios. Primers for SNaPshot can be found in Table S10.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Dr. M. Schmidt, Duisburg, for providing the placenta samples; Prof. T. Eggermann, Aachen, for providing MLID patients; Dr J. Beygo, Essen, for helpful discussions; S. Kaya and C. Mertel, Essen, for expert technical assistance and the Bundesministerium für Bildung und Forschung for financial support (01GM1513).
Financial disclosure
The funder has no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res 2010; 20:1313-26; PMID:20651121; http://dx.doi.org/ 10.1101/gr.101386.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet 2009; 10:19-31; PMID:19030023; http://dx.doi.org/ 10.1038/nrg2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res 2003; 13:2541-58; PMID:14656962; http://dx.doi.org/ 10.1101/gr.1429003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mighell AJ, Smith NR, Robinson PA, Markham AF. Vertebrate pseudogenes. FEBS Lett 2000; 468:109-14; PMID:10692568; http://dx.doi.org/ 10.1016/S0014-5793(00)01199-6 [DOI] [PubMed] [Google Scholar]
- 5.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A 2006; 103:3220-5; PMID:16492757; http://dx.doi.org/ 10.1073/pnas.0511307103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A 2002; 99:3740-5; PMID:11891299; http://dx.doi.org/ 10.1073/pnas.052410099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol 1987; 196:261-82; PMID:3656447; http://dx.doi.org/ 10.1016/0022-2836(87)90689-9 [DOI] [PubMed] [Google Scholar]
- 8.Illingworth RS, Bird AP. CpG islands–'a rough guide'. FEBS Lett 2009; 583:1713-20; PMID:19376112; http://dx.doi.org/ 10.1016/j.febslet.2009.04.012 [DOI] [PubMed] [Google Scholar]
- 9.Cowley M, Oakey RJ. Retrotransposition and genomic imprinting. Brief Funct Genomics 2010; 9:340-6; PMID:20591835; http://dx.doi.org/ 10.1093/bfgp/elq015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet 2007; 3:e20; PMID:17291163; http://dx.doi.org/ 10.1371/journal.pgen.0030020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCole RB, Loughran NB, Chahal M, Fernandes LP, Roberts RG, Fraternali F, O'Connell MJ, Oakey RJ. A case-by-case evolutionary analysis of four imprinted retrogenes. Evolution 2011; 65:1413-27; PMID:21166792; http://dx.doi.org/ 10.1111/j.1558-5646.2010.01213.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanber D, Giltay J, Wieczorek D, Zogel C, Hochstenbach R, Caliebe A, Kuechler A, Horsthemke B, Buiting K. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet 2009; 17:582-90; PMID:19066619; http://dx.doi.org/ 10.1038/ejhg.2008.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang A, Skaar DA, Li Y, Huang D, Price TM, Murphy SK, Jirtle RL. Novel retrotransposed imprinted locus identified at human 6p25. Nucleic Acids Res 2011; 39:5388-400; PMID:21421564; http://dx.doi.org/ 10.1093/nar/gkr108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann LC, Feiner N, Meyer A, Buiting K, Horsthemke B. The imprinted NPAP1 gene in the Prader-Willi syndrome region belongs to a POM121-related family of retrogenes. Genome Biol Evol 2014; 6:344-51; PMID:24482533; http://dx.doi.org/ 10.1093/gbe/evu019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steenpass L, Kanber D, Hiber M, Buiting K, Horsthemke B, Lohmann D. Human PPP1R26P1 functions as cis-repressive element in mouse Rb1. PLoS One 2013; 8:e74159; PMID:24019952; http://dx.doi.org/ 10.1371/journal.pone.0074159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanber D, Berulava T, Ammerpohl O, Mitter D, Richter J, Siebert R, Horsthemke B, Lohmann D, Buiting K. The human retinoblastoma gene is imprinted. PLoS Genet 2009; 5:e1000790; PMID:20041224; http://dx.doi.org/ 10.1371/journal.pgen.1000790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanber D, Buiting K, Roos C, Gromoll J, Kaya S, Horsthemke B, Lohmann D. The origin of the RB1 imprint. PLoS One 2013; 8:e81502; PMID:24282601; http://dx.doi.org/ 10.1371/journal.pone.0081502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakabayashi K, Trujillo AM, Tayama C, Camprubi C, Yoshida W, Lapunzina P, Sanchez A, Soejima H, Aburatani H, Nagae G, et al.. Methylation screening of reciprocal genome-wide UPDs identifies novel human-specific imprinted genes. Hum Mol Genet 2011; 20:3188-97; PMID:21593219; http://dx.doi.org/ 10.1093/hmg/ddr224 [DOI] [PubMed] [Google Scholar]
- 19.Court F, Tayama C, Romanelli V, Martin-Trujillo A, Iglesias-Platas I, Okamura K, Sugahara N, Simon C, Moore H, Harness JV, et al.. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res 2014; 24:554-69; PMID:24402520; http://dx.doi.org/ 10.1101/gr.164913.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rademacher K, Schroder C, Kanber D, Klein-Hitpass L, Wallner S, Zeschnigk M, Horsthemke B. Evolutionary origin and methylation status of human intronic CpG islands that are not present in mouse. Genome Biol Evol 2014; 6:1579-88; PMID:24923327; http://dx.doi.org/ 10.1093/gbe/evu125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al.. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013; 500:477-81; PMID:23925113; http://dx.doi.org/ 10.1038/nature12433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okae H, Chiba H, Hiura H, Hamada H, Sato A, Utsunomiya T, Kikuchi H, Yoshida H, Tanaka A, Suyama M, et al.. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet 2014; 10:e1004868; PMID:25501653; http://dx.doi.org/ 10.1371/journal.pgen.1004868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al.. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011; 480:490-5; PMID:22170606; http://dx.doi.org/ 10.1038/nature10716 [DOI] [PubMed] [Google Scholar]
- 24.Gunzburg WH, Groner B. The chromosomal integration site determines the tissue-specific methylation of mouse mammary tumour virus proviral genes. EMBO J 1984; 3:1129-35; PMID:6329738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapkins RW, Hore T, Smithwick M, Ager E, Pask AJ, Renfree MB, Kohn M, Hameister H, Nicholls RD, Deakin JE, et al.. Recent assembly of an imprinted domain from non-imprinted components. PLoS Genet 2006; 2:e182; PMID:17069464; http://dx.doi.org/ 10.1371/journal.pgen.0020182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai JH, Locke DP, Ohta T, Greally JM, Nicholls RD. Retrotransposed genes such as Frat3 in the mouse Chromosome 7C Prader-Willi syndrome region acquire the imprinted status of their insertion site. Mamm Genome 2001; 12:813-21; PMID:11845283; http://dx.doi.org/ 10.1007/s00335-001-2083-1 [DOI] [PubMed] [Google Scholar]
- 27.Suzuki S, Ono R, Narita T, Pask AJ, Shaw G, Wang C, Kohda T, Alsop AE, Marshall Graves JA, Kohara Y, et al.. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet 2007; 3:e55; PMID:17432937; http://dx.doi.org/ 10.1371/journal.pgen.0030055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature 1985; 315:594-7; PMID:2989695; http://dx.doi.org/ 10.1038/315594a0 [DOI] [PubMed] [Google Scholar]
- 29.Hannula-Jouppi K, Muurinen M, Lipsanen-Nyman M, Reinius LE, Ezer S, Greco D, Kere J. Differentially methylated regions in maternal and paternal uniparental disomy for chromosome 7. Epigenetics 2014; 9:351-65; PMID:24247273; http://dx.doi.org/ 10.4161/epi.27160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao B, Hu Y, Herrick DJ, Brewer G. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J Biol Chem 2005; 280:18517-24; PMID:15753088; http://dx.doi.org/ 10.1074/jbc.M500270200 [DOI] [PubMed] [Google Scholar]
- 31.Liao B, Hu Y, Brewer G. RNA-binding protein insulin-like growth factor mRNA-binding protein 3 (IMP-3) promotes cell survival via insulin-like growth factor II signaling after ionizing radiation. J Biol Chem 2011; 286:31145-52; PMID:21757716; http://dx.doi.org/ 10.1074/jbc.M111.263913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suvasini R, Shruti B, Thota B, Shinde SV, Friedmann-Morvinski D, Nawaz Z, Prasanna KV, Thennarasu K, Hegde AS, Arivazhagan A, et al.. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J Biol Chem 2011; 286:25882-90; PMID:21613208; http://dx.doi.org/ 10.1074/jbc.M110.178012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monk D, Bentley L, Beechey C, Hitchins M, Peters J, Preece MA, Stanier P, Moore GE. Characterisation of the growth regulating gene IMP3, a candidate for Silver-Russell syndrome. J Med Genet 2002; 39:575-81; PMID:12161597; http://dx.doi.org/ 10.1136/jmg.39.8.575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Liu D, Chang W, Lu X, Wang YL, Wang H, Zhu C, Lin HY, Zhang Y, Zhou J, et al.. Role of IGF2BP3 in trophoblast cell invasion and migration. Cell Death Dis 2014; 5:e1025; PMID:24457969; http://dx.doi.org/ 10.1038/cddis.2013.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet 1997; 17:14-5; PMID:9288088; http://dx.doi.org/ 10.1038/ng0997-14 [DOI] [PubMed] [Google Scholar]
- 36.Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics 2001; 73:316-22; PMID:11350123; http://dx.doi.org/ 10.1006/geno.2001.6543 [DOI] [PubMed] [Google Scholar]
- 37.Landers M, Calciano MA, Colosi D, Glatt-Deeley H, Wagstaff J, Lalande M. Maternal disruption of Ube3a leads to increased expression of Ube3a-ATS in trans. Nucleic Acids Res 2005; 33:3976-84; PMID:16027444; http://dx.doi.org/ 10.1093/nar/gki705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng L, Person RE, Beaudet AL. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum Mol Genet 2012; 21:3001-12; PMID:22493002; http://dx.doi.org/ 10.1093/hmg/dds130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol Cell 2008; 100:149-66; PMID:18271756; http://dx.doi.org/ 10.1042/BC20070126 [DOI] [PubMed] [Google Scholar]
- 40.Horsthemke B, Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am J Med Genet A 2008; 146A:2041-52; PMID:18627066; http://dx.doi.org/ 10.1002/ajmg.a.32364 [DOI] [PubMed] [Google Scholar]
- 41.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 2004; 32:D493-6; PMID:14681465; http://dx.doi.org/ 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol 2000; 7:203-14; PMID:10890397; http://dx.doi.org/ 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]
- 43.Hartigan JA, Wong MA. Algorithm AS 136: A K-Means Clustering Algorithm. J Royal Statistical Society Series C (Applied Statistics) 1979; 28:100-8; http://dx.doi.org/ 10.2307/2346830 [DOI] [Google Scholar]
- 44.Beygo J, Ammerpohl O, Gritzan D, Heitmann M, Rademacher K, Richter J, Caliebe A, Siebert R, Horsthemke B, Buiting K. Deep bisulfite sequencing of aberrantly methylated loci in a patient with multiple methylation defects. PLoS One 2013; 8:e76953; PMID:24130816; http://dx.doi.org/ 10.1371/journal.pone.0076953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahmann S, Beygo J, Kanber D, Martin M, Horsthemke B, Buiting K. Amplikyzer: Automated methylation analysis of amplicons from bisulfite flowgram sequencing. PeerJ PrePrints 2013; 1:e122v2; http://dx.doi.org/ 10.7717/peerj.122 [DOI] [Google Scholar]
- 46.Buiting K, Barnicoat A, Lich C, Pembrey M, Malcolm S, Horsthemke B. Disruption of the bipartite imprinting center in a family with Angelman syndrome. Am J Hum Genet 2001; 68:1290-4; PMID:11283796; http://dx.doi.org/ 10.1086/320120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


