Abstract
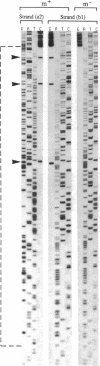
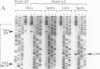
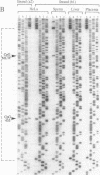
The modulation of DNA-protein interactions by methylation of protein-binding sites in DNA and the occurrence in genomic imprinting, X chromosome inactivation, and fragile X syndrome of different methylation patterns in DNA of different chromosomal origin have underlined the need to establish methylation patterns in individual strands of particular genomic sequences. We report a genomic sequencing method that provides positive identification of 5-methylcytosine residues and yields strand-specific sequences of individual molecules in genomic DNA. The method utilizes bisulfite-induced modification of genomic DNA, under conditions whereby cytosine is converted to uracil, but 5-methylcytosine remains nonreactive. The sequence under investigation is then amplified by PCR with two sets of strand-specific primers to yield a pair of fragments, one from each strand, in which all uracil and thymine residues have been amplified as thymine and only 5-methylcytosine residues have been amplified as cytosine. The PCR products can be sequenced directly to provide a strand-specific average sequence for the population of molecules or can be cloned and sequenced to provide methylation maps of single DNA molecules. We tested the method by defining the methylation status within single DNA strands of two closely spaced CpG dinucleotides in the promoter of the human kininogen gene. During the analysis, we encountered in sperm DNA an unusual methylation pattern, which suggests that the high methylation level of single-copy sequences in sperm may be locally modulated by binding of protein factors in germ-line cells.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Boyes J., Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990 Aug 10;62(3):503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Bell M. V., Hirst M. C., Nakahori Y., MacKinnon R. N., Roche A., Flint T. J., Jacobs P. A., Tommerup N., Tranebjaerg L., Froster-Iskenius U. Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell. 1991 Feb 22;64(4):861–866. doi: 10.1016/0092-8674(91)90514-y. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Chaillet J. R., Vogt T. F., Beier D. R., Leder P. Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell. 1991 Jul 12;66(1):77–83. doi: 10.1016/0092-8674(91)90140-t. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comb M., Goodman H. M. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990 Jul 11;18(13):3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi H., Magewu A. N., Gonzales F., Jones P. A. Changes in the allelic methylation patterns of c-H-ras-1, insulin and retinoblastoma genes in human development. Dev Suppl. 1990:115–123. [PubMed] [Google Scholar]
- Gregor P. D., Sawadogo M., Roeder R. G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990 Oct;4(10):1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- Hayatsu H., Wataya Y., Kai K., Iida S. Reaction of sodium bisulfite with uracil, cytosine, and their derivatives. Biochemistry. 1970 Jul 7;9(14):2858–2865. doi: 10.1021/bi00816a016. [DOI] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989 May;3(5):612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
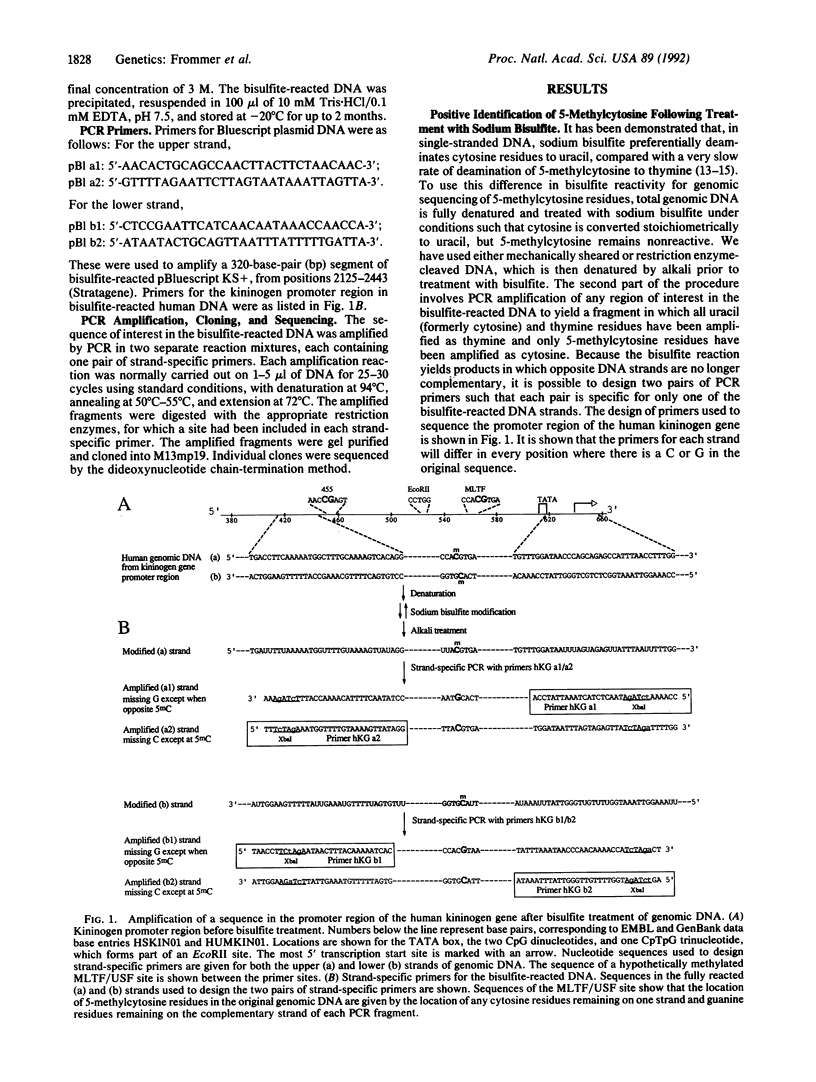
- Kitamura N., Kitagawa H., Fukushima D., Takagaki Y., Miyata T., Nakanishi S. Structural organization of the human kininogen gene and a model for its evolution. J Biol Chem. 1985 Jul 15;260(14):8610–8617. [PubMed] [Google Scholar]
- Kochanek S., Toth M., Dehmel A., Renz D., Doerfler W. Interindividual concordance of methylation profiles in human genes for tumor necrosis factors alpha and beta. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8830–8834. doi: 10.1073/pnas.87.22.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovesdi I., Reichel R., Nevins J. R. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M. Changes in DNA methylation during mouse embryonic development in relation to X-chromosome activity and imprinting. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):299–312. doi: 10.1098/rstb.1990.0013. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Saluz H. P., Jiricny J., Jost J. P. Genomic sequencing reveals a positive correlation between the kinetics of strand-specific DNA demethylation of the overlapping estradiol/glucocorticoid-receptor binding sites and the rate of avian vitellogenin mRNA synthesis. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluz H., Jost J. P. A simple high-resolution procedure to study DNA methylation and in vivo DNA-protein interactions on a single-copy gene level in higher eukaryotes. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2602–2606. doi: 10.1073/pnas.86.8.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Shapiro R., Braverman B., Louis J. B., Servis R. E. Nucleic acid reactivity and conformation. II. Reaction of cytosine and uracil with sodium bisulfite. J Biol Chem. 1973 Jun 10;248(11):4060–4064. [PubMed] [Google Scholar]
- Stein R., Sciaky-Gallili N., Razin A., Cedar H. Pattern of methylation of two genes coding for housekeeping functions. Proc Natl Acad Sci U S A. 1983 May;80(9):2422–2426. doi: 10.1073/pnas.80.9.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M., Müller U., Doerfler W. Establishment of de novo DNA methylation patterns. Transcription factor binding and deoxycytidine methylation at CpG and non-CpG sequences in an integrated adenovirus promoter. J Mol Biol. 1990 Aug 5;214(3):673–683. doi: 10.1016/0022-2836(90)90285-T. [DOI] [PubMed] [Google Scholar]
- Wang R. Y., Gehrke C. W., Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980 Oct 24;8(20):4777–4790. doi: 10.1093/nar/8.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F., Molloy P. L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988 Sep;2(9):1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- Woodcock D. M., Crowther P. J., Diver W. P. The majority of methylated deoxycytidines in human DNA are not in the CpG dinucleotide. Biochem Biophys Res Commun. 1987 Jun 15;145(2):888–894. doi: 10.1016/0006-291x(87)91048-5. [DOI] [PubMed] [Google Scholar]