A histone methyltransferase negatively regulates a set of genes coordinating seed size and lipid biosynthesis pathways in developing Arabidopsis embryos.
Abstract
CURLY LEAF (CLF), a histone methyltransferase of Polycomb Repressive Complex 2 (PRC2) for trimethylation of histone H3 Lys 27 (H3K27me3), has been thought as a negative regulator controlling mainly postgermination growth in Arabidopsis (Arabidopsis thaliana). Approximately 14% to 29% of genic regions are decorated by H3K27me3 in the Arabidopsis genome; however, transcriptional repression activities of PRC2 on a majority of these regions remain unclear. Here, by analysis of transcriptome profiles, we found that approximately 11.6% genes in the Arabidopsis genome were repressed by CLF in various organs. Unexpectedly, approximately 54% of these genes were preferentially repressed in siliques. Further analyses of 118 transcriptome datasets uncovered a group of genes that was preferentially expressed and repressed by CLF in embryos at the mature-green stage. This observation suggests that CLF mediates a large-scale H3K27me3 programming/reprogramming event during embryonic development. Plants of clf-28 produced bigger and heavier seeds with higher oil content, larger oil bodies, and altered long-chain fatty acid composition compared with wild type. Around 46% of CLF-repressed genes were associated with H3K27me3 marks; moreover, we verified histone modification and transcriptional repression by CLF on regulatory genes. Our results suggest that CLF silences specific gene expression modules. Genes operating within a module have various molecular functions, but they cooperate to regulate a similar physiological function during embryo development.
Epigenetic chromatin repression mediated by trimethylation of histone H3 Lys 27 (H3K27) is an evolutionarily conserved mechanism with important regulatory roles in higher eukaryotes. Trimethylation (H3K27me3) of H3K27 is catalyzed by Polycomb Repressive Complex 2 (PRC2) via its Enhancer of Zeste homolog 2 (EZH2) and EZH1 subunits. On the other hand, H3K27me3 repressive marks can be erased by histone demethylases at specific developmental stages (Cao and Zhang, 2004; Schubert et al., 2006; Agger et al., 2008; Hennig and Derkacheva, 2009; Ingouff et al., 2010). The histone methyltransferase activity of EZH2 is dependent on its assembly with several other PRC2 subunits such as ESC, SUZ12, and p55/ RbAp48 (Cao and Zhang, 2004; Yoo and Hennighausen, 2012). The Arabidopsis (Arabidopsis thaliana) genome encodes 3 EZH2 homologs, including CURLY LEAF (CLF), SWINGER (SWN), and MEDEA (MEA); 1 ESC protein (FERTILIZATION-INDEPENDENT ENDOSPERM; FIE); and more than 30 putative demethylases of the jmjC and lsd1 (Chanvivattana et al., 2004; Hennig and Derkacheva, 2009). Genetics and genomic studies have identified CLF as the major H3K27 methyltransferase, with SWN sharing partially redundant function. Furthermore, there is evidence that the maternal allele of MEA is specifically expressed during early seed development, FIE is a core component of PRC2, and Early Flowering 6 (ELF6) and its homolog REF6 are functional demethylases (Chanvivattana et al., 2004; Schultes et al., 2005; Spillane et al., 2007; Lafos et al., 2011; Lu et al., 2011; Holec and Berger, 2012; Deng et al., 2013; Crevillén et al., 2014).
In animal cells, PRC2 is required to maintain normal embryogenesis and prevent inappropriate cell differentiation (Hansen et al., 2008). Ectopic expression of EZH2 in humans can result in several proliferative and neoplastic disorders, including carcinoma, sarcoma, lymphoma, leukemia, and myeloproliferative disorder (Fiskus et al., 2009; Ernst et al., 2010; Xu et al., 2013). Different from PRC2 in animal cells, plant PRC2 shows the ability to promote cell differentiation (Aichinger et al., 2009). In Arabidopsis, transcriptional repression by PRC2 regulates genes specifying leaf morphology, vegetative to reproductive phase transition, cell pluripotency, and floral organogenesis (Schubert et al., 2006; Spillane et al., 2007; Hennig and Derkacheva, 2009; Lafos et al., 2011; Lu et al., 2011; Zheng and Chen, 2011; He et al., 2012).
Being heritable during cell division, H3K27me3 marks can be transmitted to the next developmental stage (Hansen et al., 2008; Ingouff et al., 2010). In animal cells, genomic evidence indicates that 85% of H3K27me3 marks in embryonic stem cells can be transmitted to hemopoietic cells during differentiation (Young et al., 2011). Maintenance of repressive marks is necessary for normal embryogenesis and determination of cell identity (Hansen et al., 2008). In Arabidopsis, H3K27me3 modifications are preferentially enriched on genomic regions with putative transcription activities. Around 15 to 28% genes are with associated H3K27me3 marks in seedlings (Zhang et al., 2007; Lafos et al., 2011). Recent studies showed that the majority (91% and 89%, respectively) of H3K27me3 marks are transmitted to the next developmental stage during meristem-to-leaf and leaf-to-callus transition (Lafos et al., 2011; He et al., 2012). However, expression levels of the majority of these genes, even for many with high levels of H3K27me3 modifications, have not been shown to be derepressed in clf and swn mutants at the same developmental stages (Farrona et al., 2011). It has been reported that a large-scale replacement and reprogramming of histone H3 occurred in the zygotic nucleus during fertilization in animals and plants (Ooi et al., 2006; Ingouff et al., 2010; Santenard et al., 2010). A recent study showed that H3K27me3 modifications on Flowering Locus C (FLC) were erased by ELF6 histone demethylase during Arabidopsis seed development (Crevillén et al., 2014). Double mutant of clf and swn showed somatic embryo phenotype, suggesting that PRC2-regulated genes may be involved in embryonic development (Chanvivattana et al., 2004). A high CLF expression level in embryos also suggests that CLF may play roles in postfertilization development (Spillane et al., 2007). Based on these observations, we hypothesized that the majority of H3K27me3 marks detected in seedlings may have been retained at a maintenance status. Thus, large-scale PRC2-mediated transcriptional repression may be a tissue-specific event and may occur at an early developmental stage(s) after fertilization and prior to seed germination. Furthermore, given the high complexity of the epigenetic gene-silencing network, eukaryotic organisms may evolve gene regulatory modules in which a large number of genes with various molecular functions within pathways can be deliberately and coordinately repressed to modulate biological traits for evolutionary adaptation.
Here, we found that CLF-mediated repression is highly tissue-specific on a genome-wide basis. CLF-repressed genes (CRGs) were preferentially expressed and repressed by CLF in several embryonic tissues at the mature-green stage. Plants of clf showed phenotypes of cell size and lipid biosynthesis during embryonic development. We also detected transcriptional repression and H3K27me3 modifications on several regulatory genes controlling these morphological traits. Combining RNA-sequencing (RNA-seq) results, H3K27me3 datasets, and genetic evidence, we proposed a rheostat module to explain coordination of PRC2-mediated transcriptional silencing of genes in multiple pathways to govern expression of biological traits.
RESULTS
CLF Represses a Large Number of Genes in Embryos at the Mature-Green Stage
Recently, a clf loss-of-function allele (clf-28) was isolated, which shows comparable PRC2-deficient phenotypes but is capable of producing a moderate amount of seeds (Farrona et al., 2011; Deng et al., 2013). We sequenced 24 cDNA libraries obtained from roots, shoots, inflorescences, and siliques of Arabidopsis wild-type and clf-28 samples using strand-specific RNA-seq protocols with three biological replicates (Supplemental Data Set S1). On average, around 83% of the sequences could be aligned to the Arabidopsis TAIR10 genome using TopHat (Trapnell et al., 2009). Using transcriptome assembly and differential expression analysis (fold-change ≥2, DESeq P-value < 0.01, false discovery rate < 0.1; Liu et al., 2012), we identified 3,744 significantly up-regulated genes in clf-28 plants (Fig. 1A; Supplemental Data Set S2), which collectively account for approximately 11.6% of 32,243 annotated genes in the Arabidopsis TAIR10 genome. The majority (3,729, 99.6%) of these genes were preferentially repressed by CLF in specific organs, and 2,038 (54%) were derepressed in clf-28 siliques (Supplemental Data Set S2). The expression level of the CRGs was significantly lower than that of other genes in wild-type siliques (P-value of Mann-Whitney-Wilcoxon test < 1.2e−5) but became significantly higher than that of other genes in clf-28 siliques (P-value < 2.2e−16), indicating a genome-wide transcriptional repression by CLF in siliques (Fig. 1D). FLC and AGAMOUS (AG) are 2 extensively investigated loci decorated with H3K27me3 marks (Bowman et al., 1989; Schubert et al., 2006; Crevillén et al., 2014). On the FLC locus, H3K27me3 modifications are programed/reprogrammed during embryonic development and can be further enhanced during the vernalization process (Heo and Sung, 2011; Crevillén et al., 2014), whereas H3k27me3-mediated chromatin repression on AG locus mainly occurs after germination (Saleh et al., 2007). In our dataset, genes derepressed by CLF in siliques showed a similar regulation pattern like that of FLC rather than that of AG (Supplemental Fig. S1).
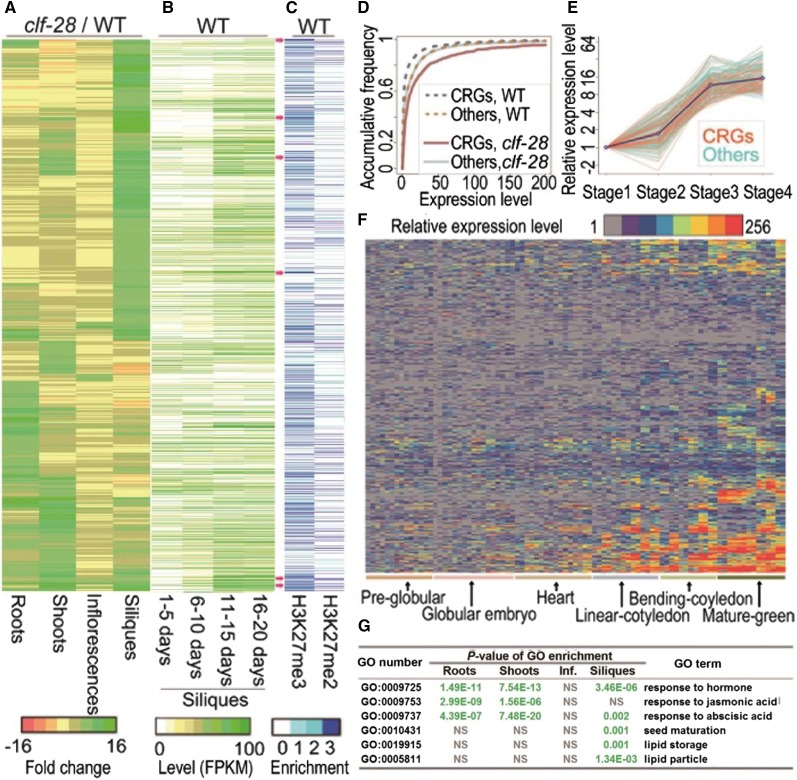
Figure 1.
Expression levels and histone modifications of CRGs. A, Expression changes (clf-28/wild type) of CRGs in four organs. Ratios of fragments mapped of assembled transcripts changes were used for clustering. Genes in A-C were shown in the same cluster order. B, Expression levels of CRGs in siliques at four different stages. C, Histone modifications on the genomic regions of CRGs in seedlings. Red arrows indicate some CRGs with high levels of H3K27me3 modifications. D, Distribution of expression of CRGs and other genes in siliques of wild type and clf-28. E, Relative expression levels of a gene cluster showing differential expression pattern at four different silique stages. Blue line shows average relative expression levels. F, Relative expression levels of CRGs in 42 embryonic compartments at six stages of seed development (Zuber et al., 2010). Samples were as described in Supplemental Data Set 1. Average expression levels in globular embryo were used as the reference for comparison (P-value of Empirical-Bayes t test < 0.01, fold-change cutoff > 8). (G) Representative GO enrichments of CRGs in four organs. “NS” means nonsignificant.
To further investigate developmental specificities of CRGs during postfertilization, we profiled transcriptome at four different stages of silique development using RNA-seq (Supplemental Fig. S2). We identified 5,608 stage-specific genes (Supplemental Data Set S3). Further analysis showed that the CRGs were preferentially expressed in 11- to 20-d-old maturing siliques (Fig. 1B; Supplemental Fig. S3), within which embryos were at approximately the mature-green stage. Fig. 1E shows a representative cluster of genes preferentially expressed in developing siliques, in which 93 CRGs in siliques were over-represented (41%, P-value of hypergeometric-test enrichment, P-value-H < 1.9 e−13).
To extend our analysis to the tissue-/cell-type level, we compared our results with 87 published microarray datasets profiling 42 tissues and/or cell types isolated by laser capture microdissection at six different stages of embryo development (Le et al., 2010; Zuber et al., 2010; Belmonte et al., 2013) and reanalyzed the datasets according to the Arabidopsis TAIR10 annotation. CRGs were preferentially expressed at the late stages of embryo development, especially in several tissue/cell types at the mature-green stage, such as embryo proper, peripheral endosperm, chalazal endosperm, chalazal seed coat, and seed coat, followed by the bending-cotyledon and the linear-cotyledon stages (Fig. 1F; Supplemental Data Set S1). On the other hand, CLF was highly expressed in the embryo proper at the preglobular stage and the globular embryo stage, and its expression decreased along with embryo development (Supplemental Fig. S1). This is consistent with previous study that CLF is detectable at early stage(s) of embryo development (Spillane et al., 2007). The developmental stage-specific expression pattern of CRGs was also similar to that of FLC with preferentially expression at the mature-green stage. By contrast, AG is expressed at the preglobular and globular embryo stages (Supplemental Fig. S1). Our result is consistent with a recent study that shows that reprogramming of H3K27me3 on FLC mainly occurs at the mature-green stage of embryo development (Crevillén et al., 2014).
Next, we isolated developing embryos at the mature-green stage from wild-type and clf-28 siliques and profiled transcriptome of these samples using strand-specific RNA-seq experiments with two biological replicates (Supplemental Data Sets S1 and S2). Of the 2,038 CRGs in siliques, expression levels of 328 CRGs were also significantly up-regulated in clf-28 embryos at the mature-green stage (Supplemental Fig. S4). Note that these genes include many important regulatory genes for late embryo development (Supplemental Fig. S4). We further verified expression levels of the stage-specific marker genes in clf-28 and wild-type embryos by quantitative RT-PCR (qRT-PCR), confirming that the RNA libraries were derived from embryos at the mature-green stage (Supplemental Fig. S4).
CLF Regulates Gene Sets Specifying Seed Size and Lipid Biosynthesis during Postfertilization Development
Using Gene Ontology (GO) analysis (Zheng and Wang, 2008; Zhang et al., 2010), we uncovered distinctive biological functions of CRGs in siliques. Consistent with previous studies (Kim et al., 2010; Bouyer et al., 2011), CRGs repressed in roots and/or shoots were mostly involved in stress response and hormone signaling pathways (Fig. 1G; Supplemental Data Set S4), whereas newly identified CRGs that were repressed in siliques preferentially encode proteins associated with lipid particles, lipid storage, and seed maturation, in addition to hormonal responses. To complement function enrichment analysis, we also found that several derepressed transcription factor (TF) genes in clf-28 siliques were involved in regulating lipid biosynthesis and controlling organ size and/or embryonic development (Supplemental Data Set S5). Although, these CLF-repressed TFs and downstream targets belong to various pathways, genetic evidence showed that they have related biological functions in regulating seed size. Taken together, we found that although CRGs repressed in siliques are involved in diversified pathways, they congregate as gene sets to execute similar biological functions. To simplify CRG networks, we defined a set of genes controlled by the same regulator and cooperating to affect a similar biological process or physiological function as a molecular module. Our results indicate CLF may regulate molecular modules specifying seed size and lipid biosynthesis during postfertilization development.
Morphological Phenotypes Confirm Biological Functions of CLF-Regulated Molecular Modules
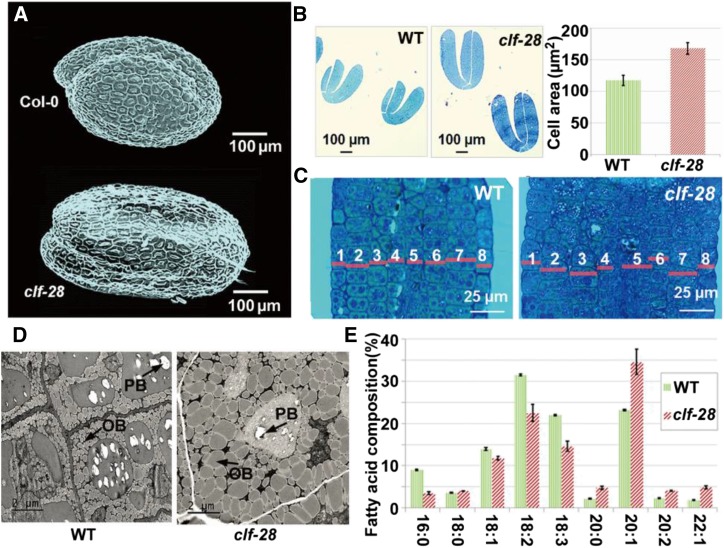
Next, we investigated morphological changes of clf-28. Consistent with previous studies (Hennig and Derkacheva, 2009; Lafos et al., 2011), clf-28 plants showed multiple developmental defects in vegetative and reproductive organs such as reduced biomass, fewer and shorter siliques, and reduced seed number (Supplemental Fig. S5). This notwithstanding, clf-28 seeds were larger than wild-type seeds (Fig. 2A), and the weight of clf-28 seeds increased to 175% that of wild-type seeds (Supplemental Fig. S6). Paraffin section showed that the cell size of clf-28 seeds increased to 140% that of wild type (Fig. 2B). Electron microscopic examination of seed sections showed that clf-28 seeds contained similar number of cells as wild-type seeds (Fig. 2C). Since clf-28 seeds have the same number of cell layers as wild-type seeds, the increased seed size of clf-28 can be largely accounted for by the cell size increase. The cell size increase in clf-28 seeds verifies the regulation by CLF on seed size gene module.
Figure 2.
CLF regulates seed development. A‑C, Seed cell size of wild type and clf-28 determined by microscopy and paraffin section. D, Paraffin section (10-μm × 10-μm area) of wild-type and clf-28 seed cells. OBs and protein bodies (PBs) are indicated by arrows. E, Fatty acid composition of wild-type and clf-28 seeds.
Consistent with the existence of CLF-regulated molecular module controlling lipid biosynthesis (Fig. 1H), clf-28 seed oil content increased to 146% that of wild type, and the total oil yield per 100 seeds in clf-28 seeds reached approximately 255% that of wild-type seeds (Supplemental Fig. S6, A–C). Oil bodies (OBs) are major oil storage organelles in plant seeds (Siloto et al., 2006).The sizes and shapes of OBs are determined by a balance between the amount of storage oil and the abundance of oleosin family proteins (OLEs) covering OBs (Siloto et al., 2006). We found OBs in clf-28 seeds were larger than those in wild type (Fig. 2D; Supplemental Fig. S7), reflecting altered levels of storage oil and OLEs. Using gas chromatography, we found altered fatty acid composition and increased eicosenoic acid (C20:1) levels in clf-28 seeds (Fig. 2E; Supplemental Fig. S6D). Our results showed that CLF regulates embryonic lipid biosynthesis not only in terms of accumulation levels but also in the fatty acid composition.
To further confirm the function of CLF, we isolated another clf mutant allele, salk_088542, which carries a T-DNA insertion in the 13th intron of CLF. The T-DNA insertion may possibly perturb transcription and intron splicing or trigger nonsense-mediated mRNA decay of the aberrant mRNA produced. Mutant plants of salk_088542 displayed similar phenotypes as clf-28 plants, such as curved leaves, early flowering, and bigger seeds compared to wild type (Supplemental Fig. S8). Consistent with this phenotype, RT-qPCR experiments verified that AG and FT were highly expressed in 2-week-old salk_088542 seedlings. Expression levels of FUS3, ABI3, WRINKLED 1 (WRI1), and genes controlling cell size and biosynthesis pathways were elevated in developing siliques of salk_088542 compared to wild type (Supplemental Fig. S8).
H3K27me3 Marks on CRGs Are Modified by PRC2
The high heritability of H3K27me3 transmission through development stages suggested that a proportion of H3K27me3 marks modified during postfertilization may possibly be maintained in seedlings (Hansen et al., 2008; Lafos et al., 2011; He et al., 2012). We determined H3K27me3 enrichment in the Arabidopsis genome by reanalysis of Chromatin-Immunoprecipitation-on-chip (ChIP-chip) datasets derived from seedling samples (Supplemental Data Set S1; Oh et al., 2008; Bouyer et al., 2011; He et al., 2012). The genomic regions encoding 1,723 (46%) of the 3,744 CRGs showed a significant enrichment (P-value-H < 1.5e−148) of H3K27me3 epigenetic marks (Fig. 1C; Supplemental Data Set S2). Of these CRGs with H3K27me3 marks, 935 (54%) also showed a tissue-preferential derepression in clf-28 siliques (Fig. 1A).
Being the only Arabidopsis homolog of the ESC protein serving as a core component of PRC2, FIE can interact with CLF (Katz et al., 2004; Deng et al., 2013). By reanalysis of public ChIP-chip datasets based on a custom array (Bouyer et al., 2011), we found that the majority (approximately 98%) of H3K27me3 modifications on CRGs were abolished in fie seedlings (Supplemental Fig. S9A), which is consistent with a previous study. Moreover, by analysis of FIE binding sites on genomic DNA (Deng et al., 2013), we found that FIE was significantly associated with approximately 15% of these genes (P-value-H < 1.0e−6; Fig. 3A). These lines of evidence confirmed that the H3K27me3 marks on CRGs were modified by FIE/PRC2 and suggested that those CRGs with H3K27me3 marks are more likely to be CLF direct targets. Hereafter, these genes are designated as putative CLF-targeted genes (pCTGs). In addition, we verified expression levels of several pCTGs using qRT-PCR and their H3K37me3 levels by locus-specific PCR on ChIP-derived DNA-materials (ChIP-PCR; Supplemental Fig. S10).
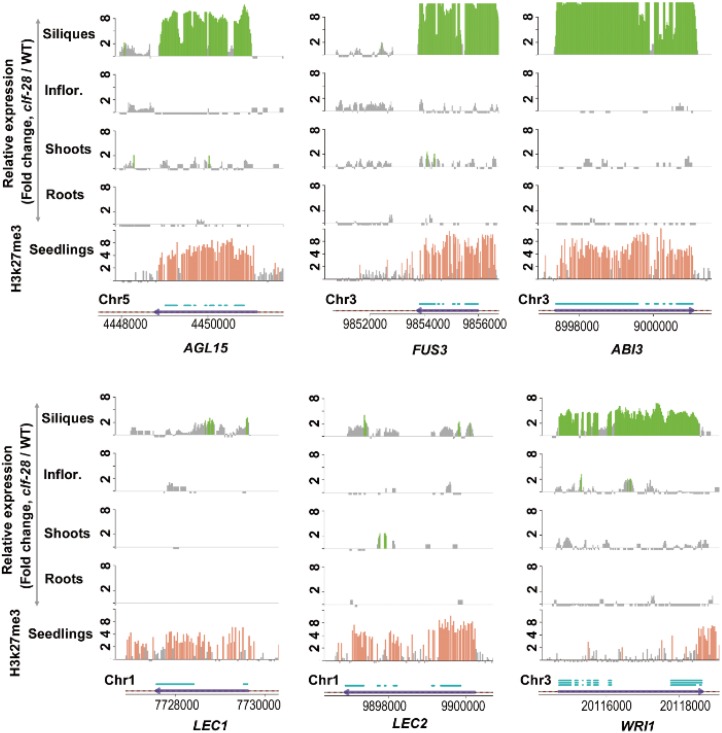
Figure 3.
CLF regulates transcription factors controlling lipid biosynthesis. Relative expression levels of six transcription factor genes regulating lipid biosynthesis were profiled by RNA-seq in four organs and H3K27me3 modification levels on these gene loci in seedlings. The genomic positions (10-bp sliding windows) with signal -fold changes >2-fold between clf-28 and wild type were presented using bars for gene expression levels or H3K27me3 modification levels. Gray bars present those with gene expression or histone modification changes <2-fold. Segments present the genomic positions of exons. Inflor., inflorescences.
CLF Coordinates Key Regulators within Molecular Modules
In Arabidopsis, organ size is controlled by more than 50 regulatory genes (Breuninger and Lenhard, 2010; Petricka et al., 2012). Based on genetic evidence (Breuninger and Lenhard, 2010), genes controlling organ size can be classified into three subgroups: genes regulating cell number, genes regulating cell size, and other regulatory genes (Supplemental Data Set S6). In clf-28, we found that 12 organ-size genes were derepressed in siliques, and most of these genes regulate cell size but not cell number (P-value-H < 1.1e−4; Supplemental Data Set S6). Among them, AIL5, AIL6, ARGOs, and CYP78A7 carrying H3K27me3 marks are pCTGs (Supplemental Fig. S9). By analysis of public microarray datasets derived from tissue/cell types samples of embryos (Le et al., 2010; Zuber et al., 2010; Belmonte et al., 2013), we found these five organ-size genes were preferentially expressed at the mature-green, bent-cotyledon, and/or the linear-cotyledon stages (Supplemental Fig. S11). Experimental verification by qRT-PCR confirmed their derepression in clf-28 siliques (Supplemental Fig. S12). Our results uncovered the role of CLF in the epigenetic silencing of key regulators in a molecular module controlling cell size in embryos.
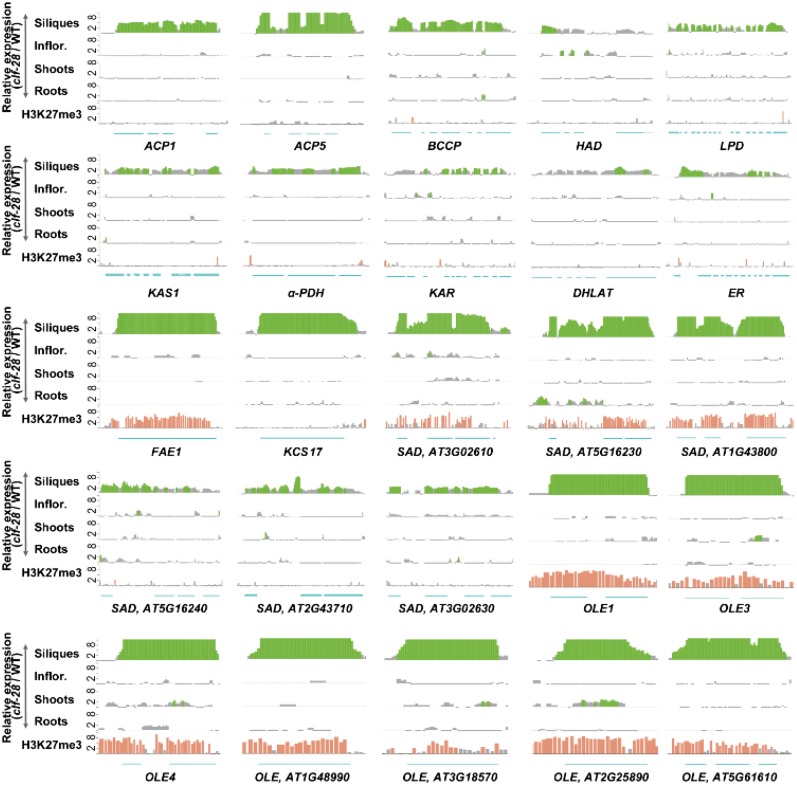
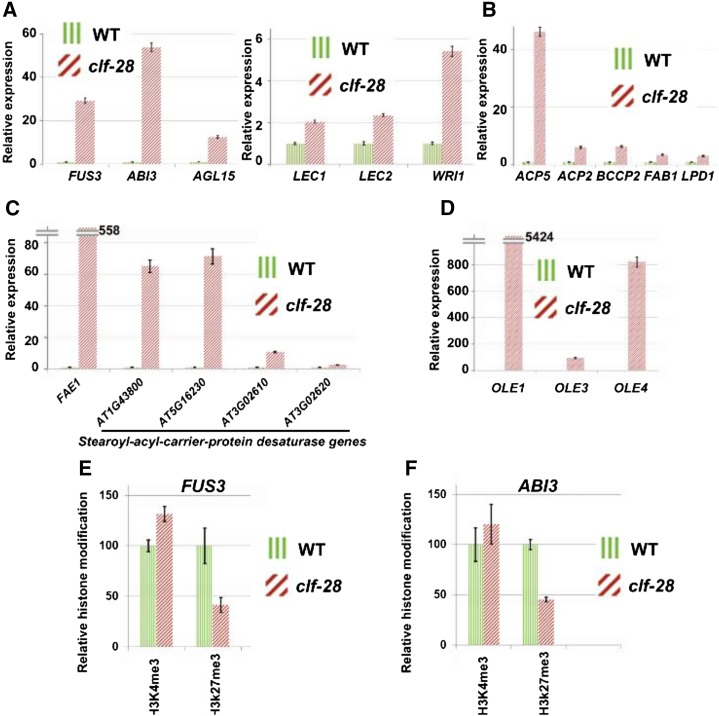
In Arabidopsis, the WRI1 transcription factor, which promotes lipid biosynthesis (Santos-Mendoza et al., 2008), is regulated by several upstream TFs, including AGAMOUS-Like 15 (AGL15), FUSCA 3 (FUS3), ABA INSENSITIVE 3 (ABI3), LEAFY COTYLEDON 1 (LEC1), and LEAFY COTYLEDON 2 (LEC2; Zheng et al., 2009). We found that AGL15, FUS3, and ABI3 were derepressed in clf-28 siliques (Fig. 3). Their genomic regions were covered by H3K27me3 modifications in wild-type seedlings (Supplemental Fig. S9). Microarray analysis showed that AGL15, FUC3, and ABI3 were preferentially expressed at the mature-green stage, whereas LEC1 and LEC2 were at the heart and the linear-cotyledon stages, respectively (Supplemental Fig. S13). For WRI1 downstream genes, 10 genes with catalytic activities in seven fatty acid biosynthesis reactions were also derepressed in clf-28 siliques, but none was associated with H3K27me3, suggesting indirect regulations by CLF (Fig. 4; Supplemental Fig. S14). In plants, synthesis of very-long-chain fatty acids such as eicosenoic acid is catalyzed by enlongases. Four of the 21 Arabidopsis enlongases have the capacity to extend the chain length of fatty acids from C18 to C20 and FATTY ACID ELONGATION 1 (FAE1) has the highest catalytic activity (Trenkamp et al., 2004). Two of the four enlongases were up-regulated in clf-28 siliques, and the most de-repressed one was FAE1, which was associated with H3K27me3 marks (Fig. 4). Stearoyl-acyl-carrier-protein desaturase (SAD) catalyzes fatty acid desaturation. The Arabidopsis genome encodes six SADs, and all of them showed increased expression levels in clf-28 siliques and three of them carried H3K27me3 marks (Fig. 4). In vascular plants, OLEs are required to stabilize oil bodies (Siloto et al., 2006). All of the 13 OLEs in the Arabidopsis genome were found derepressed in clf-28 siliques, and 11 of them were associated with H3K27me3 modifications (Fig. 4; Supplemental Data Set S6. Finally, we confirmed derepression of the TF genes, enlongase genes, SADs, and OLEs in clf-28 siliques using qRT-PCR and reduced levels of H3K27me3 modifications on genomic regions of FUS3 and ABI3 in clf-28 seedlings using ChIP-PCR (Fig. 5). Our results support the view that CLF silences several key regulators within the molecular module specifying lipid biosynthesis.
Figure 4.
Derepressed expression level of lipid biosynthesis genes in clf-28. Relative expression levels of 10 fatty acid biosynthesis genes, 2 elongase genes, 6 SAD, and 7 OLE genes profiled by RNA-seq in four different organs and H3K27me3 modification levels on these gene loci in seedlings. The genomic positions with signal -fold changes >2-fold between clf-28 and wild type were presented using bars for gene expression levels or H3K27me3 modification levels. Gray bars present those with gene expression or histone modification changes <2-fold. Segments present the genomic positions of exons. Inflor., inflorescences.
Figure 5.
Experimental verfication of lipid regulatory genes by qRT-PCR and ChIP. Experimental verification of expression levels of six transcription factor genes regulating lipid biosynthesis (A), five genes encoding catalytic enzymes in fatty acid synthesis pathway (B), FEA1 and four SAD genes (C), and three OLE genes (D) by RT-qPCR in siliques. Error bars give ses (n = 3). (E and F) Histone modification levels on FUS3 and ABI3 detected by ChIP-PCR in seedlings. Error bars give standard deviations (n = 3).
DISCUSSION
H3K27me3 Modifications and Transcriptional Repression
In contrast to animal cells where H3K27me3 marks are often deposited on large genomic regions, H3K27me3-enriched regions in Arabidopsis are relatively short and are mainly associated with genomic regions with transcription activities (Zhang et al., 2007). Around 4,400 to 7,900 annotated genes in Arabidopsis are associated with H3K27me3 marks, and these are expressed at a relatively low level on a genome-wide basis (Zhang et al., 2007; Lafos et al., 2011). On several genomic regions such as AG and FLOWERING LOCUS T, H3K27me3 modification and associated transcriptional repression occurred at the same developmental stage in seedlings and inflorescences (Goodrich et al., 1997; Zhang et al., 2007; Farrona et al., 2011; Lafos et al., 2011). Our transcriptome profiling results of roots, shoots, and inflorescences of clf-28 are consistent with these studies. However, for the majority of H3K27me3-modified genes, genome-wide study has not detected transcriptional derepression in seedlings of PRC2-deficient mutants (Farrona et al., 2011). These observations raised the issue whether the majority of the thousands of H3K27me3-modified regions detected in seedlings have any transcriptional repression functions.
Our study showed that PRC2-mediated transcriptional repression is highly tissue specific. Derepression of CRGs occurred mainly in siliques and developing seeds of clf-28, providing evidence that H3K27me3 marks are functional in transcriptional repression in Arabidopsis. A recent study showed that ELF6 erased repressive H3K27me3 marks on FLC locus to reactivate its transcription mainly in mature green embryos (Crevillén et al., 2014). Our experiments identified a large number of genes repressed by CLF with a tissue-specific regulation pattern like that of FLC. This result suggests that the deliberate reprograming/programing of H3K27me3 mark by ELF6/CLF on FLC in mature-green embryos may be not an isolated case, but this mechanism may also apply to a large number of genes on a genome-wide basis. Our result also suggests that the dynamics of H3K27me3 mark on many genomic loci may only be detected in specific tissues and at specific developmental stages
CLF and PICKLE in Lipid Biosynthesis Regulation
It has long been known that a CHD3/4 chromatin remodeling factor, PICKLE (PKL), is required for regulating lipid biosynthesis genes in plants (Eshed et al., 1999; Ogas et al., 1999). Many PKL downstream genes, including LEC1, LEC2, FUS3, and ABI3, also carry H3K27me3 marks (Aichinger et al., 2009, 2011; Zhang et al., 2012). PKL and some of its regulated genes are highly expressed in siliques, and deficiency in any of these genes showed phenotypes in seeds, suggesting their regulatory role in embryonic development (Dean Rider et al., 2003). Most of the previous studies used seedling, root, leaf, and inflorescence samples rather than siliques and seeds to investigate the mechanism of PKL and CLF regulation. Here, we found that CLF-regulated transcriptional repression on lipid biosynthesis genes is a tissue-specific event and that the regulation mainly occurs in embryos. In view of our results, it will be interesting to further investigate epigenetic repression mechanism of PKL in embryonic samples.
Coordination of CRGs in Regulating Multiple Pathways
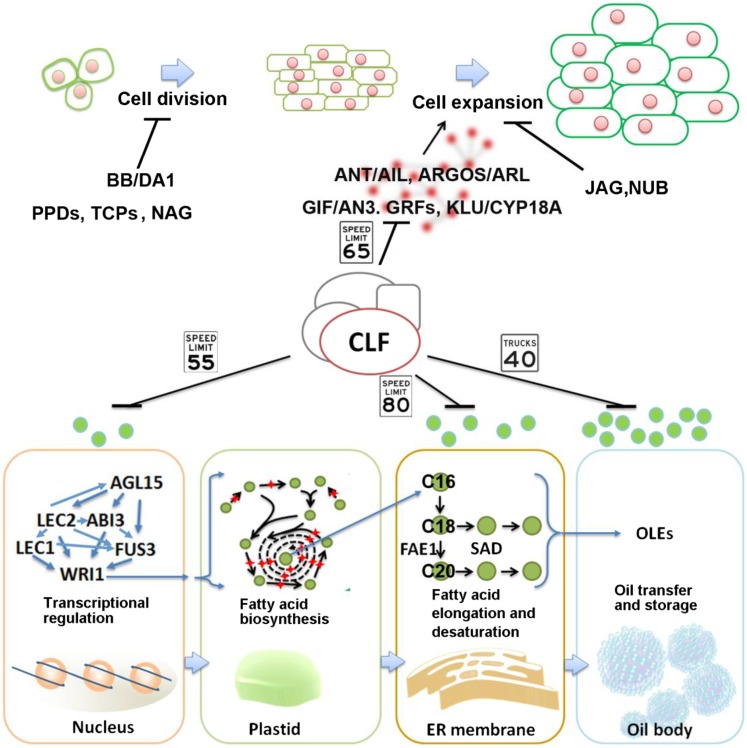
We uncovered several transcriptional silencing events mediated by CLF during postfertilization and embryo development. CLF can silence a molecular module comprising AILs, ARGOS, GRFs, and CYP78A7, thereby regulating downstream genes to control cell size in seeds. CLF also functions as a negative regulator of ABI3, FUS3, AGL15, FEA1, SADs, and OLEs to control a molecular module governing seed lipid biosynthesis and storage. Previous studies indicated that these genes have dosage-effect functions (Siloto et al., 2006; Santos-Mendoza et al., 2008; Breuninger and Lenhard, 2010; Petricka et al., 2012). According to this view, the epigenetic regulation on these genes mediated by CLF may deliberately adjust expression of developmental traits for evolutionary adaptation. In brief, our results suggested CLF-mediated tissue-specific transcriptional gene silencing may act as a rheostat to coordinate the activities of molecular modules governing multiple pathways (Fig. 6).
Figure 6.
CLF regulates molecular module controlling cell size and lipid biosynthesis. A cartoon showing CLF-regulated molecular modules.
Regulation on Embryo Development by CLF May Be a Programmed Process
Our morphological and molecular results showed that CRG sets control cell size and lipid biosynthesis at the mature-green stage of embryos. At this stage of embryo development, the cell number and cell identities have largely been determined (Belmonte et al., 2013). However, transcriptome profiling results showed that CLF was highly expressed at earlier stages of embryo development during which cells in embryos are still undergoing division and differentiation (Belmonte et al., 2013). These observations suggested that on many genomic regions H3K27me3 marks may be decorated during cell differentiation prior to transcriptional repression that presumably occurs during cell expansion. Therefore, CLF-mediated epigenetic regulation during embryo development may be a programmed process in which cell size control and lipid biosynthesis at the latter developmental stage(s) are predetermined at earlier developmental stage(s).
MATERIALS AND METHODS
Plant Materials
Seeds clf-28 in ecotype Columbia (Col-0) background were provided by the Arabidopsis Biological Resource Center under accession number SALK_139371 (Alonso et al., 2003). The T-DNA insertion on the fourth exon of CLF disrupted its open reading frame (Doyle and Amasino, 2009; Lafos et al., 2011). Col-0 (wild-type, wild-type) and clf-28 plants of Arabidopsis (Arabidopsis thaliana) were grown under long-day conditions (22°C, 16-/8-h photoperiod cycles). Shoots including leaves and roots were collected from 2-week-old plants grown on Murashige and Skoog plates. Inflorescences and whole siliques were collected from 5-week-old plants grown on soil. Siliques were also collected at four different stages, and developing embryos at the third stage were isolated (Supplemental Fig. S2). All samples were harvested and frozen immediately in liquid nitrogen. Total RNAs extracted using Trizol reagent (Invitrogen) was treated with TURBO DNase I (Ambion) for 30 min and purified using RNeasy Plant Mini Kit (QIAGEN).
RNA-seq Experiments
Strand-specific or regular RNA sequence libraries were prepared with TruSeq RNA sample Prep V2 kit according to the manufacturer’s instructions (Illumina). The quality and size of cDNA libraries for sequencing were checked using Agilent 2200 TapeStation system (Agilent, Santa Clara, CA). RNA libraries were sequenced using HiSEquation 2000 sequencing system (Illumine) with 100-cycle single-end sequencing protocol at the Genomic Center of Rockefeller University (Liu et al., 2012). Samples of the four different silique stages were sequenced using the Illumina TruSeq paired-end protocol (Supplemental Data Set S1).
Analysis of RNA-seq Datasets
The Arabidopsis genome sequence (TAIR10) and annotation files were downloaded from the Arabidopsis Information Resource (Swarbreck et al., 2008). After quality control analysis of raw sequencing datasets, RNA-seq sequences were aligned to the Arabidopsis genome using TopHat with TAIR10 annotation as the reference (Trapnell et al., 2009). The mapped sequences of each sample were assembled by Cufflinks (Trapnell et al., 2010). Fragments per kilobase of exon per million fragments mapped of assembled transcripts were calculated and normalized using Cuffcompare and Cuffdiff with global normalization parameters (Trapnell et al., 2010). A custom PERL script was used to analyze Cuffdiff results.
We used plant long noncoding RNA database (PLncDB) to browse the high through-put datasets (Jin et al., 2013). RNA-seq or ChIP-seq sequences were aligned to the TAIR10 genome using TopHat, SAMtools, BEDTools, and a custom PERL script (Li et al., 2009; Quinlan and Hall, 2010). Converter programs for files with Bed, bedGraph, WIG, and BigWiG formats were downloaded from UCSC Genome Bioinformatics Site (Dreszer et al., 2012). BigWig-formatted files were integrated into PLncDB. Construction methods of PLncDB were described in our previous study (Jin et al., 2013).
Differential Expression Analysis
For Affymetrix ATH1 genome array datasets, we updated the probe information to TAIR10 based on the NetAffx Annotation release 34. The normalized signals of CRGs in samples were compared to the average signals derived from the two samples of globular embryo using Empirical Bayes t test. The CRGs with P-value of Empirical Bayes t test < 0.01 and fold-change in at least one sample > 8 were considered as significant induction. Hierarchical clusters of CRGs expression levels in 87 samples were analyzed using R and “Hclust” package with “complete” parameter (Cameron et al., 2012).
For analysis of RNA-seq datasets, we used Tophat, SAMtools, Cufflinks, Cuffcompare, HTseq-count, and DESeq2 to perform differential expression analysis for strand-specific RNA-seq datasets (Li et al., 2009; Trapnell et al., 2009; Anders and Huber, 2010; Trapnell et al., 2010; Chandramohan et al., 2013). Read numbers on each gene were aligned and calculated using Tophat, SAMtools, and HTseq-count based on a GTF-formatted file produced by Cufflinks and Cuffcompare (Li et al., 2009; Trapnell et al., 2009; Trapnell et al., 2010; Chandramohan et al., 2013). Then, we applied DESeq2 to normalize expression levels and perform differential expression analysis based on the negative binomial distribution (Anders and Huber, 2010). Genes with normalized expression fold-change >2, significance P-value < 0.01, and Benjamini-Hochberg adjusted P-value/false discovery rate < 0.1 were considered as differentially expressed genes.
Analysis of ChIP-chip and ChIP-seq Datasets
The signal intensity files of ChIP-chip and the fastaq-formatted sequences of ChIP-seq datasets were downloaded from public databases (Supplemental Data Set 1; Oh et al., 2008; Bouyer et al., 2011; Deng et al., 2013). For analysis of Affymetrix tiling array datasets, we updated probe information to TAIR10 using the annotation files provided by Affymetrix. Signal intensities with replicates were normalized using Quantile method (Bolstad et al., 2003). Statistical significance of each H3K27me3 peak on chromosomes and statistical significance values were scanned based on hidden Markov model using CisGenome and TileMap (Ji et al., 2008). For analysis of NimbleGen tiling array datasets, we selected the best and unique match for each probe on the Arabidopsis TAIR10 genome using BLASTn. Then, we scanned for modification peaks by the method used in the original paper (Bouyer et al., 2011). For analysis of ChIP-seq datasets, we aligned the fastaq-formatted datasets to the Arabidopsis genome TAIR10 using bowtie2 (Langmead and Salzberg, 2012). The protein binding sites on genome were identified using MACS (Zhang et al., 2008).
GO and Pathway Analysis
GO enrichment analysis was performed using GOEAST based on hypergeometric test, where all the TAIR10 annotated genes were considered as the entire population and significantly up-regulated ones in clf-28 siliques as sampling points (Zheng and Wang, 2008; Zhang et al., 2010). Pathway annotation was downloaded from AraCyc pathway database and the Arabidopsis Acyl-Lipid Metabolism Web site (Mueller et al., 2003; Li-Beisson et al., 2010). Transcription factor annotation datasets were downloaded from AGRIS and DATF databases (Guo et al., 2005; Yilmaz et al., 2011).
Light Microscopy
Seed samples were excised from mature Arabidopsis seeds and fixed overnight in 2.5% glutaraldehyde in 0.1 m phosphate buffer, pH 7.2. After rinsing three times in 0.1 m phosphate buffer for 15 min, seed samples were then postfixed in 1% (w/v) aqueous OsO4 for 1 h. Tissues were dehydrated in an ethanol series and embedded in Spurr’s resin. Semithin sections (500 nm) were stained in 0.1% toluidine blue and photographed with a Zeiss Axioplan2 microscope (Carl Zeiss). Cell number in a specific region of seed was counted under a microscopy. The cell size in each sample was obtained using ImageJ (Hartig, 2013). We used five seed sections of Col-0 and clf-28 to calculate the mean and se.
Transmission Electron Microscope
Seed samples were fixed overnight in 2.5% glutaraldehyde solution in 0.1 m phosphate buffer (pH 7.2) at 4°C, washed three times in 0.1 m phosphate buffer, and postfixed in 1% osmium tetroxide in 0.1 m phosphate buffer for 1 h at 4°C. The samples were then gradually dehydrated with ethanol, embedded in Spurr’s resin, and sectioned on the Lecia Ultracut UCT ultramicrotome equipped with diamond knives. Ultrathin sections were double stained with uranyl acetate and lead citrate and observed under a JEOL JEM-1230 (JEOL) electron microscope at 120 kV.
Scanning Electron Microscope
Following freezing in liquid N2, seeds were fixed with a tape inside a sample chamber. Images were collected using a scanning electron microscope (JSM-6360LV, JEOL).
Gas Chromatography
Total lipid was extracted and transmethylated from 100 dry Arabidopsis seeds as previously described (Li et al., 2006). Fatty acid methyl esters were generated and separated by gas chromatography and detected using Agilent 7890 GC with 5975C MS (Agilent). The results were presented as means ± standard deviations.
RT-PCR on ChIP-Derived DNA Materials
We performed chromatin preparation and immunoprecipitation using the method described in our previous study (Jang et al., 2011). Seedling samples were fixed in 1% formaldehyde for 10 min. The reaction was terminated by incubation for 5 min in a vacuum. Seedlings were rinsed three times and frozen in liquid nitrogen. Chromatin was sheared to approximately 500-bp fragments by sonication (Diagenode Bioruptor UCD-200). We used anti-H3K4me3 (Active Motif; 39,159) and anti-H3K27me3 (Millipore; 07-449) antibodies to perform immunoprecipitation. Immune complexes were eluted from the protein A or G agarose/salmon sperm DNA beads (Millipore) and reverse cross-linked by incubation for 6 h at 65°C. The samples were then treated with proteinase K for 1 h. DNA was extracted using the QIAquick PCR purification kit (Qiagen). qRT-PCR reactions were performed using SYBR Premix Ex Taq (TaKaRa) in the CFX96 real-time system (Bio-Rad) and ACTIN7 was used as a control. Error bars in each graph gave sd (n = 3). The primers are listed in Supplemental Data Set S7.
qRT-PCR
A total of 1 to 2 µg of DNase I (RNeasy plant mini kit) treated RNA was reverse transcribed using SuperScript III (Invitrogen) and oligo(dT) primer. cDNA was analyzed by quantitative PCR using SYBR Premix Ex Taq (Takara) in a Biorad CFX96 real-time PCR system. All qRT-PCR reactions were performed in triplicates for each cDNA sample with an annealing temperature of 60°C and a total of 40 cycles of amplification. The primers are listed in Supplemental Data Set S7.
Accession Numbers
Detailed information of RNA-seq experiment and the fastaq-formatted sequence datasets are available on the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE61545, GSE55866, and GSE68080.
Supplemental Data
The following supplemental materials are available.
Supplemental Fig. S1. Expression specificities of FLC, AG, and CLF in embryos.
Supplemental Fig. S2. Siliques at four different developmental stages used for RNA-seq experiments.
Supplemental Fig. S3. Gene clusters with enriched CRGs are preferentially expressed at the mature-green stage.
Supplemental Fig. S4. Regulation of CRGs by CLF in embryos at the mature-green stage.
Supplemental Fig. S5. Developmental defects in vegetative and reproductive organs of clf-28.
Supplemental Fig. S6. Seed weight and fatty acid content in wild-type and clf-28 seeds.
Supplemental Fig. S7. Oil bodies in wild-type and clf-28 seeds.
Supplemental Fig. S8. Phenotype and qRT-PCR verification in salk_088542 plants.
Supplemental Fig. S9. H3K27me3 marks on pCTGs.
Supplemental Fig. S10. Experimental verification of pCTGs by qRT-PCR and ChIP.
Supplemental Fig. S11. Expression specificities of the genes controlling cell size in embryos.
Supplemental Fig. S12 Experimental verification of CRGs controlling cell size by qRT-PCR.
Supplemental Fig. S13 Expression specificities of the genes controlling fatty acid biosynthesis in embryos.
Supplemental Fig. S14. CLF regulates several reactions in fatty acid synthesis pathway.
Supplemental Datasets 1-7.
Supplementary Material
Acknowledgments
We thank C. Zhao for technical support and J. Goodrich and X-J. Wang for discussion.
Glossary
- ChIP-chip
Chromatin-Immunoprecipitation-on-chip
- ChIP-PCR
PCR on ChIP-derived DNA-materials
- Col-0
ecotype Columbia
- CRG
CLF-repressed gene
- GO
Gene Ontology
- OB
oil body
- OLE
oleosin family protein
- pCTG
putative CLF-targeted gene
- PLncDB
plant long noncoding RNA database
- qRT-PCR
quantitative RT-PCR
- RNA-seq
RNA-sequencing
- SAD
stearoyl-acyl-carrier-protein desaturase
Footnotes
Articles can be viewed without a subscription.
References
- Agger K, Christensen J, Cloos PA, Helin K (2008) The emerging functions of histone demethylases. Curr Opin Genet Dev 18: 159–168 [DOI] [PubMed] [Google Scholar]
- Aichinger E, Villar CB, Di Mambro R, Sabatini S, Köhler C (2011) The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23: 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E, Villar CB, Farrona S, Reyes JC, Hennig L, Köhler C (2009) CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, Goodrich J, Renou JP, Grini PE, Colot V, et al. (2011) Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 7: e1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuninger H, Lenhard M (2010) Control of tissue and organ growth in plants. Curr Top Dev Biol 91: 185–220 [DOI] [PubMed] [Google Scholar]
- Cameron DA, Middleton FA, Chenn A, Olson EC (2012) Hierarchical clustering of gene expression patterns in the Eomes + lineage of excitatory neurons during early neocortical development. BMC Neurosci 13: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y (2004) The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14: 155–164 [DOI] [PubMed] [Google Scholar]
- Chandramohan R, Wu PY, Phan JH, Wang MD (2013) Benchmarking RNA-Seq quantification tools. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society Annual Conference 2013: 647–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C (2014) Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean Rider S Jr, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Buzas DM, Ying H, Robertson M, Taylor J, Peacock WJ, Dennis ES, Helliwell C (2013) Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. BMC Genomics 14: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Amasino RM (2009) A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol 151: 1688–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. (2012) The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res 40: D918–D923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. (2010) Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42: 722–726 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209 [DOI] [PubMed] [Google Scholar]
- Farrona S, Thorpe FL, Engelhorn J, Adrian J, Dong X, Sarid-Krebs L, Goodrich J, Turck F (2011) Tissue-specific expression of FLOWERING LOCUS T in Arabidopsis is maintained independently of polycomb group protein repression. Plant Cell 23: 3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, Ustun C, Rao R, Fernandez P, Chen J, et al. (2009) Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood 114: 2733–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J (2005) DATF: a database of Arabidopsis transcription factors. Bioinformatics 21: 2568–2569 [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K (2008) A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300 [DOI] [PubMed] [Google Scholar]
- Hartig SM (2013) Basic image analysis and manipulation in ImageJ. In Frederick M. Ausubel, ed Current Protocols in Molecular Biology, John Wiley and Sons, Inc., New Jersey, the United States of America Chap 14, p 15 [DOI] [PubMed] [Google Scholar]
- He C, Chen X, Huang H, Xu L (2012) Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet 8: e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M (2009) Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Holec S, Berger F (2012) Polycomb group complexes mediate developmental transitions in plants. Plant Physiol 158: 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Rademacher S, Holec S, Soljić L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F (2010) Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20: 2137–2143 [DOI] [PubMed] [Google Scholar]
- Jang IC, Chung PJ, Hemmes H, Jung C, Chua NH (2011) Rapid and reversible light-mediated chromatin modifications of Arabidopsis phytochrome A locus. Plant Cell 23: 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Liu J, Wang H, Wong L, Chua NH (2013) PLncDB: plant long non-coding RNA database. Bioinformatics 29: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A, Oliva M, Mosquna A, Hakim O, Ohad N (2004) FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Kim SY, Zhu T, Sung ZR (2010) Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiol 152: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7: e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2010) Acyl-lipid metabolism. Arabidopsis Book 8: e0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43: 715–719 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Park S, van Nocker S (2008) Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet 4: e1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Priess JR, Henikoff S (2006) Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genet 2: e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Al-Abdallat A, Ndamukong I, Alvarez-Venegas R, Avramova Z (2007) The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res 35: 6290–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME (2010) Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol 12: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes EA, Spasic A, Mohanty U, Bartel DP (2005) Compact and ordered collapse of randomly generated RNA sequences. Nat Struct Mol Biol 12: 1130–1136 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane C, Schmid KJ, Laoueillé-Duprat S, Pien S, Escobar-Restrepo JM, Baroux C, Gagliardini V, Page DR, Wolfe KH, Grossniklaus U (2007) Positive Darwinian selection at the imprinted MEDEA locus in plants. Nature 448: 349–352 [DOI] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36: D1009–D1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenkamp S, Martin W, Tietjen K (2004) Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc Natl Acad Sci USA 101: 11903–11908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Abourbih S, Sircar K, Kassouf W, Mansure JJ, Aprikian A, Tanguay S, Brimo F (2013) Enhancer of zeste homolog 2 expression is associated with metastasis and adverse clinical outcome in clear cell renal cell carcinoma: a comparative study and review of the literature. Arch Pathol Lab Med 137: 1326–1336 [DOI] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E (2011) AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo KH, Hennighausen L (2012) EZH2 methyltransferase and H3K27 methylation in breast cancer. Int J Biol Sci 8: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Willson TA, Wakefield MJ, Trounson E, Hilton DJ, Blewitt ME, Oshlack A, Majewski IJ (2011) ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res 39: 7415–7427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bishop B, Ringenberg W, Muir WM, Ogas J (2012) The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol 159: 418–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE (2007) Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu J, Jia C, Li T, Wu R, Wang J, Chen Y, Zou X, Chen R, Wang XJ, Zhu D (2010) Systematic identification and evolutionary features of rhesus monkey small nucleolar RNAs. BMC Genomics 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Chen X (2011) Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr Opin Plant Biol 14: 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ (2008) GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 36: W358-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber H, Davidian JC, Aubert G, Aimé D, Belghazi M, Lugan R, Heintz D, Wirtz M, Hell R, Thompson R, et al. (2010) The seed composition of Arabidopsis mutants for the group 3 sulfate transporters indicates a role in sulfate translocation within developing seeds. Plant Physiol 154: 913–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.