Sucrose promotes early leaf growth by stimulating cell proliferation and repressing chloroplast transcription through the induction of GPT2 expression.
Abstract
Leaves are the plant’s powerhouses, providing energy for all organs through sugar production during photosynthesis. However, sugars serve not only as a metabolic energy source for sink tissues but also as signaling molecules, affecting gene expression through conserved signaling pathways to regulate plant growth and development. Here, we describe an in vitro experimental assay, allowing one to alter the sucrose (Suc) availability during early Arabidopsis (Arabidopsis thaliana) leaf development, with the aim to identify the affected cellular and molecular processes. The transfer of seedlings to Suc-containing medium showed a profound effect on leaf growth by stimulating cell proliferation and postponing the transition to cell expansion. Furthermore, rapidly after transfer to Suc, mesophyll cells contained fewer and smaller plastids, which are irregular in shape and contain fewer starch granules compared with control mesophyll cells. Short-term transcriptional responses after transfer to Suc revealed the repression of well-known sugar-responsive genes and multiple genes encoded by the plastid, on the one hand, and up-regulation of a GLUCOSE-6-PHOSPHATE TRANSPORTER (GPT2), on the other hand. Mutant gpt2 seedlings showed no stimulation of cell proliferation and no repression of chloroplast-encoded transcripts when transferred to Suc, suggesting that GPT2 plays a critical role in the Suc-mediated effects on early leaf growth. Our findings, therefore, suggest that induction of GPT2 expression by Suc increases the import of glucose-6-phosphate into the plastids that would repress chloroplast-encoded transcripts, restricting chloroplast differentiation. Retrograde signaling from the plastids would then delay the transition to cell expansion and stimulate cell proliferation.
The energy needed for plant growth and development is produced by photosynthesis in leaves, capturing and converting light into chemical energy, which is stored in sugars and transported to all other plant organs to meet their energy demands.
Arabidopsis (Arabidopsis thaliana) leaves arise from the shoot apical meristem as leaf primordia, which initially grow exclusively by cell proliferation. Subsequently, cell proliferation ceases at the tip of the leaf and, gradually, the cells start to expand in a tip-to-base direction (Donnelly et al., 1999; Andriankaja et al., 2012). After a few days, this cell cycle arrest front abruptly disappears at the base of the leaf, and further leaf growth is driven by cell expansion (Kazama et al., 2010; Andriankaja et al., 2012) and the asymmetric division of meristemoids (i.e. precursors of stomata in the epidermis; Geisler et al., 2000; Kazama et al., 2010; Gonzalez et al., 2012).
Leaves that actively perform photosynthesis, so-called source leaves, produce their own energy and carbon sources for growth and development. In contrast, plant tissues that are unable to photosynthesize, such as roots, flowers, and young growing leaves, depend on these source leaves for carbon supply to grow (Turgeon, 1989). The primary end products of photosynthesis are triose phosphates, which are rearranged into glucose-6-phosphate (G6P) and used for the formation of starch as storage molecules or transported into the cytosol to form Suc. In the source leaves, Suc can be metabolized to its hexose products (i.e. Glc and Fru), it can be stored in the vacuole, or it can be transported through the phloem to the sink tissues (Lemoine et al., 2013). Suc is either imported directly in the sink cells via active Suc transporters located at the plasma membrane (Kühn and Grof, 2010) or via plasmodesmata or it is first cleaved to its hexose products by cell wall invertases in the apoplast (Sturm, 1999; Ruan et al., 2010). In the sink cells, hexoses are imported in the plastids for starch biosynthesis or for the oxidative pentose phosphate pathway. The main Glc transporters in photosynthetically inactive sink cells are the plastid-located G6P transporters (i.e. GPT1 and GPT2), which import G6P in exchange for phosphates (Kammerer et al., 1998). GPT1 is expressed throughout plant development (Niewiadomski et al., 2005). However, GPT2 expression is limited to certain tissues, such as senescing leaves, and is induced under different conditions, such as during relief of seed dormancy (Finch-Savage et al., 2007), during acclimatization to high light (Dyson et al., 2015), during Glc-induced senescence (Pourtau et al., 2006), in starch-free mutants (Kunz et al., 2010; Heinrichs et al., 2012), and when sugar levels increase (Price et al., 2004; Gonzali et al., 2006; Müller et al., 2007; Osuna et al., 2007).
Chloroplasts are the central organelles performing photosynthesis and producing sugars. Photosynthetically active chloroplasts are derived from proplastids present in the meristematic cells (Sakamoto et al., 2009; Charuvi et al., 2012). Functional chloroplasts contain about 3,000 different proteins mainly involved in photosynthesis, transcription, and translation, of which most are encoded by the nuclear genome. However, plastids also have their own DNA, the so-called plastome, consisting of 133 genes in Arabidopsis, of which 87 encode proteins with different functions, such as photosynthetic and ribosomal proteins (Sato et al., 1999; Wicke et al., 2011). Genes of the plastome are transcribed by two different RNA polymerases: a nucleus-encoded polymerase and a plastid-encoded polymerase (PEP; Shiina et al., 2005; Liere et al., 2011). PEP consists of the plastome-encoded core subunits rpoA, rpoB, rpoC1, and rpoC2 and one of the six nucleus-encoded σ factors that define promoter specificity (Lerbs-Mache, 2011). Besides this core PEP complex, some noncore subunits have been identified to exhibit additional transcriptional functions, the so-called polymerase-associated proteins (Steiner et al., 2011). During leaf development, both the nucleus-encoded polymerase and PEP actively transcribe their specific target genes (Zoschke et al., 2007), of which most are organized in operons and transcribed into polycistronic mRNA from a single promoter, such as their bacterial ancestors (Wicke et al., 2011).
Obviously, the nucleus and chloroplasts have to exchange information to regulate photosynthesis as a function of environmental conditions. To date, different signals have been described to be involved in this chloroplast-to-nucleus, or retrograde, signaling (for review, see Kleine and Leister, 2013). The best-known retrograde signals are the intermediates of tetrapyrrole synthesis (i.e. the precursors of chlorophyll), which have been identified through the analysis of genomes uncoupled mutants, in which nuclear photosynthesis-related gene expression is maintained when chloroplast differentiation is perturbed by norflurazon (NF) treatment (Susek et al., 1993; Terry and Smith, 2013). Furthermore, reactive oxygen species, the redox state of the plastoquinone pool of the chloroplasts and of redox components such as glutathione and ascorbate (Oelze et al., 2012; Pfalz et al., 2012; Shapiguzov et al., 2012), as well as different hormone signals and the plastid gene expression itself (Tiller and Bock, 2014) have been reported to exert signals from chloroplasts to regulate nuclear gene expression. Additionally, sugars can act as signals in retrograde and other signaling pathways, integrating environmental and developmental changes during plant growth (Häusler et al., 2014; Smeekens and Hellmann, 2014). For example, sugars can modulate nuclear gene expression, especially the repression of nucleus-encoded photosynthesis genes, such as CHLOROPHYLL A/B BINDING PROTEIN and the small subunit of Rubisco, to control the feedback regulation of photosynthesis (Krapp et al., 1993). However, our knowledge of sugar-regulated transcripts comes from studies using a wide variety of plant organs and tissues, developmental stages, and treatments with different sugars and growth conditions. Furthermore, in most studies, sugars are applied to cell suspension cultures, detached leaves, or liquid cultures (Price et al., 2004; Li et al., 2006; Müller et al., 2007; Osuna et al., 2007; Usadel et al., 2008; Kunz et al., 2014), highlighting the need for more targeted experimental designs to study the effect of Suc on organ growth such as leaves. In addition, most sugar-feeding experiments make use of high, nonphysiological Glc or Suc concentrations (Price et al., 2004; Gonzali et al., 2006; Li et al., 2006; Müller et al., 2007; Heinrichs et al., 2012).
During leaf development, it has been observed that the transition from cell proliferation to cell expansion occurs simultaneously with the onset of photosynthesis (Andriankaja et al., 2012). Furthermore, an up-regulation of transcripts encoding proteins involved in tetrapyrrole synthesis has been observed in leaves just before the start of the transition to cell expansion. These findings suggest a role for differentiation of the photosynthetic machinery and associated retrograde signaling in the transition to cell expansion. In contrast, in a recent study using the crumpled leaf mutant deficient in chloroplast development, it has been demonstrated that impaired chloroplast differentiation affects cell proliferation and induces an early onset of cell differentiation (Hudik et al., 2014). Young proliferating leaves first depend on the supply of sugars, produced by photosynthetically active source leaves, to grow. The reduced photosynthetic activity of source leaves or reduced sugar availability triggers young, proliferating leaves to produce their own sugars and energy for further growth (Li et al., 2006). To do so, chloroplasts need to differentiate to start photosynthesis, producing sugars and other retrograde signals, which could trigger the transition to cell expansion. Hence, it is obvious that a cross talk exists between sugars, chloroplasts, and leaf growth, but which process (i.e. cell proliferation or expansion) is affected by sugars and how sugars are sensed during leaf growth still need to be investigated in detail.
Here, we exogenously supplied Suc during the growth of Arabidopsis seedlings and found that Suc increases final leaf size by promoting cell proliferation and postponing the transition to cell expansion. Furthermore, transcriptome and microscopic analyses of the growing leaves revealed a central role for chloroplast differentiation and GPT2 during the Suc-induced promotion of leaf growth. The transfer of seedlings to Suc resulted in reduced plastome transcription and smaller chloroplasts per mesophyll cell, which were irregular in shape and less differentiated compared with control seedlings. Also, in gpt2 mutant seedlings, cell proliferation was not stimulated and chloroplast transcription was not repressed upon transfer to Suc.
RESULTS
Development of an Experimental Setup to Analyze the Influence of Exogenously Supplied Suc on Final Leaf Size
To investigate in detail how sugars regulate early leaf growth, we designed an experimental assay in which the sugar status is changed at a specific developmental stage during the growth of Arabidopsis seedlings and in which the impact of this change on leaf growth can easily be monitored.
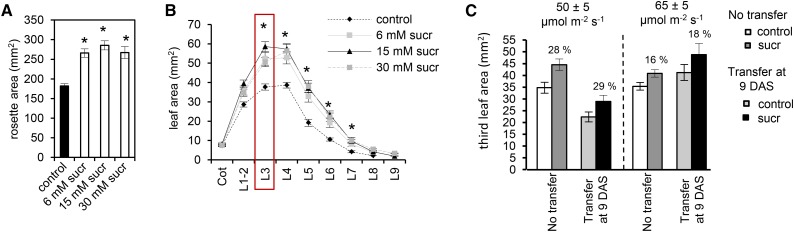
We first tested the effect of three different Suc and Glc concentrations (6, 15, and 30 mm) and found none of the Glc concentrations to reproducibly increase the third leaf size at 21 d after stratification (DAS; Supplemental Fig. S1). On the other hand, when plants were germinated and grown under a 16-h-day/8-h-night cycle on Suc-containing medium, a clear effect could be measured at 21 DAS on both rosette and individual leaf areas (Fig. 1, A and B). The three concentrations tested (i.e. 6, 15, and 30 mm) resulted in significant average increases in rosette area (40%, 56%, and 44%, respectively; P < 0.05) compared with plants grown on medium without Suc (Fig. 1A). The measurements of individual leaf area showed that all leaves (with the exception of the cotyledons and the two first leaves) of the plants grown on Suc-containing medium were larger (P < 0.05) compared with control plants (Fig. 1B). Because a concentration of 15 mm Suc resulted in the largest significant increase in size both of the whole rosette and of the third true leaf (P < 0.0001), it was retained for further characterization at the cellular level.
Figure 1.
Rosette and individual leaf area increase upon Suc treatment. A and B, Plants were germinated on different Suc (sucr) concentrations (0 [control], 6, 15, or 30 mm), and the average rosette area (A) and average individual leaf area (B) were measured at 21 DAS. Cot, Cotyledons; Lx, leaf position x in the order of appearance on the rosette. Values are means of three biological repeats with their se. Rosette and leaf area were measured for six to 10 plants in each repeat. C, Third leaf area of plants germinated on 15 mm Suc or germinated on Suc-free (control) medium (No transfer) and transferred at 9 DAS to 15 mm Suc or control medium, measured at 21 DAS at a light intensity of approximately 65 µmol m−2 s−1 or at a lower light intensity of 50 µmol m−2 s−1. Values are means of three biological repeats with their se. Leaf area was measured for an average of 18 leaves in each repeat. Asterisks indicate adjusted P < 0.05 for log-transformed values in A and B, mixed models (see Supplemental Methods S1).
We also tested two different light intensities to optimize the effect of the supplemented Suc on the final leaf size, because different light intensities may change the photosynthetic capacity and, consequently, the endogenous sugar production in the leaf. At a light intensity of 65 ± 5 µmol m−2 s−1, germination on medium containing 15 mm Suc resulted at 21 DAS in an average increase in the third leaf size of 16% as compared with control plants germinated on medium without Suc (Fig. 1C). However, when plants were grown at a lower light intensity (50 ± 5 µmol m−2 s−1), 15 mm Suc resulted in an average 28% increase of the third leaf size (Fig. 1C). Additionally, to study the short-term effects of Suc during early leaf growth, seedlings were first grown on a mesh (see “Materials and Methods”) covering a sugar-free medium and subsequently transferred to medium supplemented with 15 mm Suc. Transfer was done at 9 DAS, the time point at which the third leaf is fully proliferating (Andriankaja et al., 2012), demonstrating a similar average increase of the third leaf area at 21 DAS of 18% and 29% at the two light intensities (65 ± 5 and 50 ± 5 µmol m−2 s−1, respectively) compared with the control plants transferred to medium without Suc (Fig. 1C). Statistical analysis revealed a significant average increase in the third leaf area upon Suc supplementation independent of the use of meshes or different light intensities (P < 0.05).
In conclusion, we developed an experimental assay in which the transfer of plants grown at a light intensity of 50 ± 5 µmol m−2 s−1 at 9 DAS to a growth medium with 15 mm Suc reproducibly increases the size of the third leaf at 21 DAS. This setup, using the meshes, was used in all the following experiments to study the underlying cellular and molecular mechanisms by which Suc affects plant growth.
Suc Positively Affects Leaf Growth by Promoting Cell Proliferation
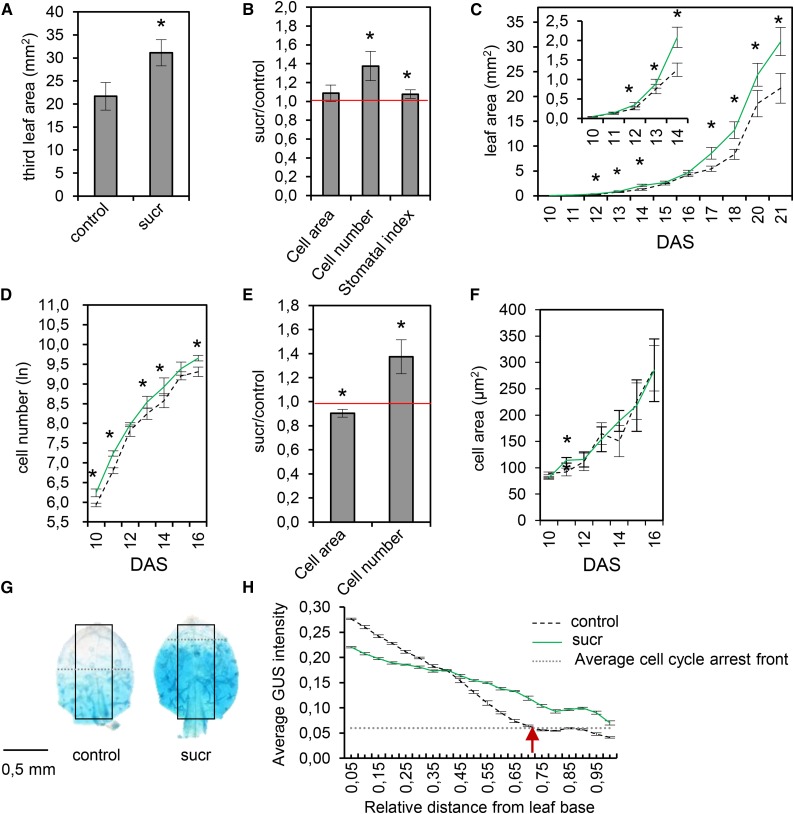
To identify the cellular process involved in the Suc-induced enlarged leaf size, the pavement cell number, cell size, and stomatal index were measured at 21 DAS, 12 d after the transfer of seedlings to medium with or without 15 mm Suc (at 50 ± 5 µmol m−2 s−1). Transfer of plants to Suc resulted in a significant average increase of the third leaf area of 47% (P < 0.05; Fig. 2A) due to a significantly higher total pavement cell number (37%; P < 0.05), whereas the cell size remained unchanged (P = 0.11; Fig. 2B). Also, the stomatal index (i.e. the fraction of guard cells in the total population of epidermal cells) was slightly but significantly increased compared with the control (7%; P < 0.05; Fig. 2B). Thus, Suc increases the final third leaf size mainly by promoting cell proliferation.
Figure 2.
Cellular changes upon transfer to Suc. Seedlings were first grown on medium without Suc (sucr) and, at 9 DAS, transferred to medium supplemented with or without 15 mm Suc. A, Third leaf area at 21 DAS. B, Ratio of the pavement cell number, cell area, and stomatal index of the third leaf of plants transferred to 15 mm Suc relative to the control (0 mm Suc) at 21 DAS. C, Leaf area from 10 to 21 DAS. The inset shows a closeup of 10 to 14 DAS. D, Cell number from 10 to 16 DAS. E, Ratio of the pavement cell area and number of the third leaf of seedlings 24 h after transfer to 15 mm Suc relative to the control (0 mm Suc). F, Cell area from 10 to 16 DAS. G and H, GUS-stained third leaves at 13 DAS of pCYCB1;1::CYCB1;1-D-box:GUS seedlings transferred to control or Suc-containing medium (G) and the GUS intensity plot of these leaves (H). GUS staining was in a defined region from the base to the tip of each leaf, as indicated by the black rectangles in G. The dotted lines indicate the cell cycle arrest front. Above this front is the division zone and below the expansion zone. The red arrow in H indicates the position of the average cell cycle arrest front of control leaves. Values in A to C are means of three biological repeats with their se. Leaf area was measured for five to 20 leaves in each repeat. Cellular data are from five leaves in each repeat. Values in D to F are means of four to five leaves with their se. Values in H are means of two biological repeats with their se. GUS intensity was measured for eight to 10 leaves in each repeat. Asterisks indicate adjusted P < 0.05 for log-transformed values in A to F, mixed models (see Supplemental Methods S1).
To analyze the effect of Suc on cell proliferation in more detail, a time-course experiment was performed by harvesting the third leaf daily after transfer, from 10 DAS until 21 DAS, and measuring its leaf area. At 12 DAS (3 d after transfer), the third leaf size of Suc-transferred plants was significantly larger than that of control plants, with an average increase of 39% (Fig. 2C; P < 0.05). Additionally, the relative leaf growth rate of Suc-grown plants was slightly, but not significantly, higher, whereas later during development, the growth rates remained unchanged compared with control plants (Supplemental Fig. S2). Because Suc significantly increased the third leaf area within 3 d after transfer, cellular measurements were performed on early time points during leaf growth (10–16 DAS; Fig. 2, D–F). The total pavement cell number was increased significantly by 35% (P = 0.02; Fig. 2E) already 24 h (10 DAS) after transfer (Fig. 2D), whereas the cell area did not change (P = 0.32; Fig. 2F). The total pavement cell number remained higher until 16 DAS (Fig. 2D). No consistent changes in the average cell size were found between control and Suc-transferred plants at early time points during leaf growth (10–16 DAS; Fig. 2F).
To further analyze the effect of the transfer to Suc on cell proliferation and the transition to cell expansion, the pCYCB1;1::CYCB1;1-D-box:GUS (Eloy et al., 2012) reporter line, which allows visualizing actively dividing cells, was used. After 9 d of growth without Suc, pCYCB1;1::CYCB1;1-D-box:GUS seedlings were transferred to control and Suc-supplemented medium and grown for an additional 4 d until 13 DAS. Subsequently, the third leaf was harvested and stained with 5-bromo-4-chloro-3-indolyl-β-glucuronide (X-Gluc). The GUS intensity was measured in a defined region from the base to the tip of each leaf, as indicated in Figure 2G. At 13 DAS, control leaves showed a cell cycle arrest front positioned closer to the leaf base (Fig. 2H, dotted line) compared with Suc-transferred leaves. Hence, a large number of third leaf cells of plants grown for 4 d on Suc were still proliferating, whereas cell expansion was initiated in most cells of the control leaves at 13 DAS.
Taken together, the above results indicate that the addition of Suc to the medium promotes early leaf growth by stimulating cell proliferation. Although the positive effect of Suc on leaf size was only clear after 3 d of growth on Suc, a significant underlying effect on the cell number was observed already after 24 h.
Short-Term Effects of Suc on the Transcriptome
To gain more insight into the molecular mechanisms driving leaf growth upon exogenous Suc application, short-term transcriptional responses were analyzed using RNA sequencing (RNA-seq). Because Suc solely affects cell proliferation, we microdissected the third leaf very early during development (at an average size of 0.04 mm2) and extracted RNA to be used for RNA-seq. Nine-day-old plants were transferred to medium with or without Suc for 3 and 24 h. Only 19 and 69 genes were found to be differentially expressed 3 and 24 h after transfer, respectively (log2 fold change [Log2FC] > 0.58 and false discovery rate [FDR] < 0.05; Fig. 3A; Supplemental Table S1).
Figure 3.
Suc-induced transcriptional responses in growing leaves. A, Overlap between differentially expressed genes in the third leaf, microdissected at 3 and 24 h, of seedlings transferred to 15 mm Suc or control medium, and Log2FC at 3 and 24 h of the six common genes. Data are from an RNA-seq analysis. B, Pie chart of differentially expressed transcripts 24 h after transfer to 15 mm Suc. C, MapMan representations of enriched genes differentially repressed 24 h after transfer to 15 mm Suc in the third leaf.
At 3 h, only three of the 19 differentially expressed genes were found to be induced: AT3G49110 and AT5G58390, encoding peroxidase proteins, and GPT2, which was the highest up-regulated gene, with a Log2FC of 2.04. Six of the 16 repressed genes at 3 h (DIN6, SEN1, AT2G05540, DRM2, BT2, and AT3G15630) remained repressed 24 h after transfer (Fig. 3A). Two of these genes, SEN1 and DIN6, belong to the so-called dark-induced class of genes (Fujiki et al., 2000), both well known as sugar starvation markers, and showed highly reduced transcript levels at 3 h with Log2FC of −2.34 and −3.49, respectively (Fig. 3A). The remaining four genes, which were repressed at both 3 and 24 h, encode a protein with telomerase activity (BT2), a Gly-rich protein (AT2G05540), a dormancy/auxin-associated protein (DRM2), and a protein with unknown function (AT3G15630). The other 10 genes, which were down-regulated at 3 h but regained normal expression levels at 24 h, encode three hydrolase superfamily proteins (AT2G32150, AT2G39400, and AT1G04280), an unknown protein (AT1G68440), an aluminum-induced protein (AT3G15450), an oxidative stress protein (OXS3 [AT5G56550]), another Gly-rich protein (AT2G05380) as well as another dormancy-associated protein (DRM1), a chloroplast-targeted DnaJ protein J8 (AT1G80920), and PV42a (AT1G15330). PV42a was the second highest repressed gene at 3 h and encodes a protein belonging to the class of γ-subunits of the plant-specific SUCROSE NONFERMENTING1 (SNF1)-RELATED PROTEIN KINASE1 (SnRK1) complexes, which are central metabolic sensors activated when environmental stress conditions deplete carbon and energy supply and which are known to link the sugar status with organ growth (Gissot et al., 2006; Baena-González et al., 2007; Fang et al., 2011).
The transcript levels of 69 genes were significantly changed 24 h after transfer to Suc, of which the vast majority (66) showed decreased expression compared with the control. The three up-regulated genes encode a cytochrome P450 protein, CYP710A2 (AT2G34490), with C22-sterol desaturase activity, a pentatricopeptide repeat superfamily protein (AT5G06400), and a stearoyl acyl-carrier-protein desaturase family protein (AT1G43800). Surprisingly, of the 66 (six also at 3 h and 60 only at 24 h) repressed genes, 30 were encoded by the nuclear genome, while 29 were located on the chloroplast DNA and seven on mitochondrial DNA (Fig. 3B). A list of the Suc-repressed, chloroplast-encoded transcripts at 3 and 24 h is shown in Table I. From the genes encoded by the nuclear genome, several have been reported to be induced by sugar starvation and were found here to be repressed by short-term Suc treatment (e.g. genes encoding anhydrases, oxygenases, and hydrolases as well as DIN genes; Fujiki et al., 2000; Lee et al., 2007). Suc-repressed genes located on mitochondrial DNA encode subunits of the electron transport chain complexes and mitochondrial ribosomal proteins. The repressed genes located on chloroplast DNA represented a mixture of genes belonging to different operons and coding for photosynthesis-related proteins involved in the light reactions, such as PSI and PSII proteins (psa and psb), proteins that are part of the cytochrome b6f complex (pet), and subunits of NADPH dehydrogenase and ATP synthase (atp; Fig. 3C). Furthermore, genes encoding proteins involved in PSI and PSII assembly and stability (ycf), chloroplast ribosomal proteins (rps), a maturase involved in intron splicing (matK), an acetyl-CoA carboxylase subunit (accD), the large subunit of Rubisco (rbcL), and one gene encoding the β-subunit of PEP (rpoC2) were found to be down-regulated. These transcriptional changes were confirmed by quantitative reverse transcription (qRT)-PCR analysis for a set of selected chloroplast genes (Supplemental Fig. S3). Furthermore, because not all genes encoded by the plastome were found to be significantly differentially expressed by Suc at 24 h, the effect of Suc on the complete plastome was studied by a gene set enrichment analysis, in which all chloroplast-encoded transcripts were analyzed together as a single gene set. We found that the chloroplast gene set was significantly down-regulated compared with all other genes (21,481 genes; P = 0) 24 h after transfer to Suc. The repression of plastome expression upon transfer to Suc can result from either a reduced plastome copy number or fewer chloroplasts per cell. However, quantitative PCR on total cellular DNA demonstrated no differences in chloroplast DNA copy number in the third leaf 3 and 24 h after transfer to Suc-supplemented or control medium (Supplemental Fig. S4).
Table I. Log2FC and corresponding P values for the 29 Suc-repressed chloroplast-encoded transcripts 3 and 24 h after transfer to Suc.
| Gene Identifier | Name | Description | 3 h |
24 h |
||
|---|---|---|---|---|---|---|
| Log2FC | P | Log2FC | P | |||
| ATCG01090 | ndhI | Subunit of the chloroplast NAD(P)H dehydrogenase complex | −0.54 | 0.47 | −3.57 | 2.42E-04 |
| ATCG00360 | ycf3 | Protein required for PSI assembly and stability | 0.10 | 0.72 | −3.38 | 3.42E-05 |
| ATCG00020 | psbA | PSII reaction center protein A | −0.14 | 0.93 | −3.37 | 2.28E-09 |
| ATCG00730 | petD | Subunit IV of the cytochrome b6/f complex | −0.18 | 0.94 | −3.27 | 3.06E-07 |
| ATCG00340 | psaB | D1 subunit of PSI and PSII reaction centers | −0.09 | 0.81 | −3.14 | 1.85E-08 |
| ATCG00280 | psbC | CP43 subunit of the PSII reaction center | −0.16 | 0.96 | −3.12 | 2.67E-08 |
| ATCG00520 | ycf4 | Protein required for PSI assembly and stability | −0.23 | 1.00 | −3.07 | 8.37E-06 |
| ATCG00720 | petB | Cytochrome b6 subunit of the cytochrome b6f complex | −0.23 | 0.89 | −3.03 | 1.38E-07 |
| ATCG01100 | ndhA | NADH dehydrogenase ND1 | −0.09 | 0.89 | −3.02 | 4.18E-05 |
| ATCG00490 | rbcL | Large subunit of Rubisco | −0.18 | 0.95 | −2.99 | 5.49E-08 |
| ATCG00350 | psaA | Protein comprising the reaction center for PSI along with the psaB protein | −0.13 | 0.92 | −2.93 | 1.09E-07 |
| ATCG00540 | petA | Cytochrome f apoprotein | −0.31 | 0.65 | −2.89 | 9.87E-07 |
| ATCG00650 | rps18 | Chloroplast ribosomal protein S18 | −0.50 | 0.38 | −2.76 | 1.89E-04 |
| ATCG01040 | ycf5 | Hypothetical protein | −0.49 | 0.31 | −2.76 | 3.72E-04 |
| ATCG00140 | atpH | ATPase III subunit | 0.10 | 0.77 | −2.68 | 2.23E-04 |
| ATCG00040 | matK | Maturase located in the trnK intron in the chloroplast genome | −0.32 | 0.68 | −2.55 | 8.28E-08 |
| ATCG00160 | rps2 | Chloroplast ribosomal protein S2 | −0.39 | 0.65 | −2.55 | 2.03E-04 |
| ATCG00270 | psbD | PSII reaction center protein D | −0.05 | 0.74 | −2.54 | 4.03E-06 |
| ATCG00680 | psbB | PSII reaction center protein B | −0.11 | 0.87 | −2.52 | 5.41E-07 |
| ATCG00330 | rps14 | Chloroplast ribosomal protein S14 | 0.20 | 0.55 | −2.48 | 5.54E-05 |
| ATCG00420 | ndhJ | NADH dehydrogenase subunit J | −0.31 | 0.63 | −2.46 | 6.86E-05 |
| ATCG00500 | accD | Carboxyltransferase β-subunit of the acetyl-CoA carboxylase complex | −0.70 | 0.18 | −2.32 | 3.90E-04 |
| ATCG01050 | ndhD | Subunit of a NAD(P)H dehydrogenase complex | −0.11 | 0.90 | −2.26 | 8.12E-05 |
| ATCG00380 | rps4 | Chloroplast ribosomal protein S4 | −0.07 | 1.00 | −2.19 | 2.25E-04 |
| ATCG00120 | atpA | ATP synthase subunit α | −0.20 | 0.92 | −2.13 | 5.70E-06 |
| ATCG01110 | ndhH | 49-kD plastid NAD(P)H dehydrogenase subunit H protein | −0.52 | 0.28 | −2.08 | 3.45E-04 |
| ATCG00170 | rpoC2 | DNA-directed RNA polymerase β′-subunit 2 | −0.16 | 0.98 | −1.87 | 8.94E-05 |
| ATCG01130 | ycf1.2 | Hypothetical protein | −0.35 | 0.60 | −1.81 | 2.03E-05 |
| ATCG00150 | atpI | Subunit of ATPase complex CF0 | −0.36 | 0.59 | −1.78 | 3.43E-04 |
In conclusion, transfer to Suc resulted in the repression of plastome transcription, while plastome copy number was not affected, which suggests reprogramming of chloroplasts upon transfer to Suc.
Blocking Chloroplast Differentiation Also Induces Cell Proliferation
To investigate if changes in chloroplast differentiation affect the Suc-induced cellular processes, seedlings were treated with NF, an herbicide that inhibits phytoene desaturase by competition with the cofactors. Phytoene desaturase is involved in carotenoid biosynthesis, and treatment of plants with NF inhibits chloroplast development (Koussevitzky et al., 2007).
Seedlings were transferred at 9 DAS to Murashige and Skoog (MS) medium with or without Suc and medium with or without Suc supplemented with 5 µm NF. Three days after transfer to NF with or without Suc (12 DAS), smaller seedlings with bleached leaves could be observed, indicating a clear effect on chloroplast differentiation (Fig. 4A). At 10 DAS, or 24 h after transfer, the third leaf area was increased significantly with Suc by 42% (P < 0.05; Fig. 4B) and by 16%, although not significantly, when transferred to Suc-containing medium supplemented with NF (P = 0.65; Fig. 4B). To study the underlying cellular processes induced by Suc, the relative increases in cell size and pavement cell number on Suc-containing medium supplemented with NF or not were determined. Suc did not result in an altered cell size, and the addition of NF did not change this, whereas the pavement cell number was increased significantly with Suc by 32% (P < 0.05) and by 34% by the addition of NF, albeit not significantly (P = 0.1), compared with control seedlings (Fig. 4C). Furthermore, cell size was decreased significantly when NF was added to the medium, and this reduction was equal between leaves of seedlings grown on Suc and without Suc in the medium (P < 0.05; Fig. 4D, left). Remarkably, third leaves of seedlings grown without Suc, but with NF, had a similar average increase in total pavement cell number (27%; P = 0.10) as seedlings grown with Suc and without NF (34%; P < 0.05) compared with control seedlings on MS medium (Fig. 4D, right). Moreover, the combination of both Suc and NF resulted in a significant increase in pavement cell number of 62% (P < 0.05) compared with control leaves, which was equal to the sum of the effects of NF and Suc separately (61%; Fig. 4D, right). Therefore, we hypothesize that NF and Suc act additively on cell number.
Figure 4.
Cellular effects of Suc with or without NF. Seedlings were transferred at 9 DAS to normal MS medium with Suc (MS+S), MS medium without Suc (MS-S), and MS+S supplemented with 5 µm NF (MS+S+NF). A, Images of seedlings 3 d after transfer to MS+S or MS+S+NF. B, Third leaf area 24 h after transfer to MS+S or MS+S+NF. C, Relative increase of cell area and cell number of the third leaf 24 h after transfer to MS+S or MS+S+NF. D, Cell area and cell number of the third leaf of seedlings transferred to MS+S or MS+S+NF. Values are means of five biological repeats with their se. Leaf area was measured for four to 15 leaves in each repeat. Cellular data are from three to five leaves in each repeat. Asterisks indicate adjusted P < 0.05 for log-transformed values, mixed models (see Supplemental Methods S1).
Transfer to Suc Results in Fewer, Smaller, and Less Differentiated Chloroplasts
Because Suc rapidly represses chloroplast-encoded transcripts, on the one hand, and cell proliferation can be stimulated by blocking chloroplast differentiation by NF, on the other hand, we set out to study the effect of Suc on chloroplast number and morphology by transmission electron microscopy.
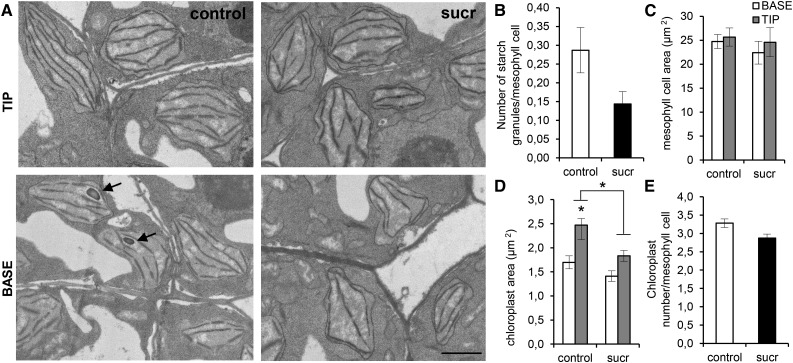
Leaves were harvested 24 h (10 DAS) after transfer to Suc-containing or control medium, and subsequently, transverse sections were made to examine differences in chloroplast thylakoid structure, chloroplast size, and chloroplast number. For each leaf, 17 to 87 mesophyll cells of the tip and the base of the leaf, representing the cell expansion and proliferation regions, respectively, were analyzed. A clear difference in thylakoid structure and organization, as well as chloroplast shape, could be observed between leaves of control and Suc-transferred seedlings (Fig. 5). Generally, in control leaves, chloroplasts seem to be more differentiated compared with leaves of seedlings transferred to Suc. Some chloroplasts already start to form starch granules; they generally have more thylakoid membranes and start to form the typical lens shapes of mature chloroplasts (Fig. 5A). In contrast, the chloroplasts of Suc-transferred seedlings are generally more irregular in shape, and less starch formation could be observed. To quantify the difference in starch formation, starch grains were counted in transverse sections of mesophyll cells of the control and Suc-transferred leaves. In general, leaves of Suc-transferred seedlings contained on average 0.14 starch granules per mesophyll cell, whereas control leaves had 0.29 starch granules (Fig. 5B). Similarly, increased starch accumulation was seen by Lugol’s staining at 12 DAS in control leaves compared with leaves of Suc-treated seedlings (Supplemental Fig. S5). Mesophyll cell area, chloroplast number, and chloroplast size were measured and analyzed statistically, taking into account the differences between the tip and the base of the leaf. The average mesophyll cell area did not differ significantly between leaves of control and Suc-transferred seedlings (Fig. 5C). Chloroplasts were significantly larger at the tip compared with the base of the leaves in control seedlings (44%; P < 0.05; Fig. 5D). In Suc-transferred leaves, the chloroplast areas were not significantly different between the tip and the base (P = 0.45). Additionally, chloroplasts were significantly larger in the tip of control leaves compared with the tip of Suc-transferred leaves (50%; P < 0.05; Fig. 5D). Furthermore, leaves of Suc-transferred seedlings had fewer chloroplasts in transverse sections, although not significantly (P = 0.12; Fig. 5E).
Figure 5.
Differences in chloroplast morphology, number, and size in the tip and base of Suc-treated and control leaves. A, Transmission electron micrographs of the tip and base of the 10-d-old third leaf 24 h after transfer to control or 15 mm Suc (sucr)-supplemented medium. Arrows point to starch granules. Bar = 1 µm. B, Average number of starch granules per mesophyll cell counted in approximately 60 cells of two leaves of control and Suc-transferred seedlings. C and D, Average mesophyll cell area (C) and average chloroplast size (D) in the tip and base of the third leaf of control and Suc-treated seedlings. E, Chloroplast number of the third leaf of control and Suc-treated seedlings. Values are means of two independent leaves with their se. Chloroplast data are from 17 to 87 mesophyll cells in the tip and the base of each leaf, respectively. Asterisks indicate adjusted P < 0.05 for log-transformed values in D, mixed models (see Supplemental Methods S1).
In conclusion, transfer of seedlings to Suc resulted in fewer, smaller, and less differentiated chloroplasts with limited formation of thylakoid membranes and fewer starch granules.
Role of GPT2 in Suc-Induced Stimulation of Cell Proliferation
The above-described results demonstrate a clear negative effect of Suc on plastome transcription as well as chloroplast development, resulting in the stimulation of cell proliferation. Remarkably, one of the three nucleus-encoded genes that was induced by Suc 3 h after transfer encodes GPT2. Recently, a central role of GPT2 in seedling development was described; gpt2 seedlings lacking GPT2 expression exhibit a delayed establishment and greening of the cotyledons (Dyson et al., 2014).
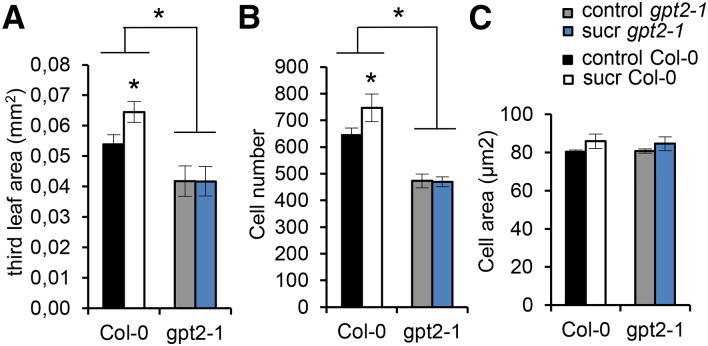
To explore whether GPT2 also has a pivotal role in the Suc-induced stimulation of cell proliferation, we subjected gpt2-1 mutant seedlings to the experimental Suc assay. gpt2-1 seedlings were grown together with their corresponding wild-type seedlings on control medium for 9 d, after which they were transferred to control or Suc-supplemented medium for 24 h, after which leaf area, cell size, and pavement cell number were determined. Generally, third leaves of gpt2-1 seedlings were significantly smaller than wild-type leaves at 10 DAS (P < 0.05; Fig. 6A), due to a significant decrease in pavement cell number (P < 0.05; Fig. 6B). Suc significantly increased third leaf size by 20% in wild-type seedlings, also due to a significantly increased total pavement cell number (P < 0.05). Remarkably, gpt2-1 seedlings showed a completely insensitive cell proliferation response to the transfer to Suc (P = 0.81; Fig. 6B) and no change of third leaf size (Fig. 6A). Cell sizes remained unchanged between both wild-type and gpt2-1 leaves, independent of the transfer to Suc-containing or control medium (P = 0.44; Fig. 6C).
Figure 6.
gpt2 mutant seedlings show an insensitive cell proliferation response to the transfer to Suc. gpt2-1 mutant seedlings were grown together with their corresponding wild-type Columbia (Col-0) on medium without Suc for 9 d and, subsequently, transferred to medium supplemented with 15 mm Suc (sucr) or without Suc. At 10 DAS, 24 h after transfer, the third leaf area (A), cell number (B), and cell area (C) were determined and compared. Values are means of three biological repeats with their se. Leaf area was measured for nine to 40 leaves in each repeat. Cellular data are from three to 14 leaves in each repeat. Asterisks indicate adjusted P < 0.05 for log-transformed values, mixed models (see Supplemental Methods S1).
In conclusion, GPT2 has an essential role in the short-term stimulation of cell proliferation by Suc. Seedlings without functional GPT2 have fewer cells and completely abolish the expected cellular response to Suc, leading to growth promotion.
GPT2 Is Required for the Suc-Mediated Repression of Plastome Transcription
In the absence of GPT2, no stimulation of cell proliferation by Suc could be observed. Subsequently, to investigate whether GPT2 expression is also required for the downstream Suc-induced transcriptional responses, a comparative analysis was done between our transcriptomics data set and the published microarray data set of Dyson et al. (2015). In the latter study, transcriptome analysis was performed on mature leaves of the gpt2.2 mutant and Wassilewskija-4 wild-type plants (Dyson et al., 2015). The data set of Dyson et al. (2015) was first filtered using the same criteria as the transcriptome analysis described here (i.e. Log2FC > 0.58 and P < 0.05). Consequently, comparison between the differentially expressed genes in the gpt2.2 mutant and the 66 Suc-repressed genes 24 h after transfer to Suc revealed a significant overlap of 20 genes (P = 8.37E-8, χ2 test; Supplemental Fig. S6A). Remarkably, 19 of the 20 overlapping genes were chloroplast-encoded transcripts, representing a mixture of genes coding for different photosynthesis-related proteins, and only one mitochondria-encoded transcript, rpl16 (Supplemental Fig. S6B). Furthermore, 18 of the 20 transcripts demonstrated an opposite effect on gene expression, namely, up-regulated in the gpt2.2 mutant compared with the wild type and down-regulated upon transfer to Suc. The two transcripts that did not show this opposite effect were the mitochondria-encoded transcript, rpl16, and the chloroplast-encoded transcript, rps14.
Consequently, this significant overlap between Suc-repressed and gpt2.2 up-regulated chloroplast-encoded transcripts, as well as the insensitivity of the gpt2-1 mutant to stimulate cell proliferation upon transfer to Suc, prompted us to test the expression of several Suc-responsive genes with qRT-PCR in microdissected third leaves of wild-type and gpt2-1 mutant seedlings 24 h after transfer to control or Suc-supplemented medium. The expression levels of 10 chloroplast-encoded genes (psbA, petD, psaA, psaB, ycf3, ndhI, atpH, rps18, rbcL, and rpoC2) were determined and compared between Suc-transferred and control leaves in the wild type and the gpt2-1 mutant separately (Supplemental Fig. S6C). These 10 chloroplast-encoded genes were selected based on the above-described transcriptome analysis and also were used for the confirmation of the Suc-responsive repression (Supplemental Fig. S3). Notwithstanding that no significant difference for each gene could be detected (P > 0.05), a clear contrasting trend in relative expression levels was observed between wild-type and gpt2-1 seedlings. All chloroplast transcripts were expressed at lower levels upon transfer to Suc in wild-type leaves, whereas eight of 10 of these transcripts were up-regulated in gpt2-1 mutant leaves (Supplemental Fig. S6C).
Taken together, these results demonstrate that chloroplast DNA transcription is affected in the gpt2 mutant but that this transcriptional response is opposite to that of wild-type plants transferred to Suc, suggesting a central role for GPT2 in mediating the Suc-induced repression of chloroplast DNA transcription.
DISCUSSION
The aim of this study was to identify the underlying cellular and transcriptional mechanisms of sugars in regulating early leaf growth in Arabidopsis. For this, we developed an experimental setup in which the sugar status was changed during the proliferating phase of the third leaf, which normally depends on other photosynthetically active leaves for carbon and energy supply. This is in contrast to the two first leaves, which probably mainly depend on carbon provided by the cotyledons for their growth. We showed that Suc had a pronounced effect on the leaf pavement cell proliferation phase. This observation is in agreement with previously described roles of Suc in cell cycle regulation. In higher plants, the cell cycle is controlled by cyclin-dependent kinases and their interacting cyclins (CYCs), which in turn respond to developmental and environmental signals (Inzé and De Veylder, 2006; Komaki and Sugimoto, 2012). A link between Suc and the cell cycle was first demonstrated through the use of Suc to synchronize Arabidopsis cell suspension cultures (Goetz and Roitsch, 1999). Suc starvation induces a reversible arrest in the G1 or G0 phase of the cell cycle, and after resupplementing Suc to the growth medium, the cell cultures are synchronized. Suc mainly regulates the expression of D-type CYCs involved in the G1-to-S phase progression (Riou-Khamlichi et al., 2000). However, none of these major cell cycle regulators were differentially expressed in our transcriptome analysis 3 or 24 h after transfer of seedlings to Suc. Hence, these findings suggest that Suc has no impact on the transcription of the cell cycle machinery during leaf growth stimulation. Nonetheless, at 13 DAS, we clearly observed a difference in cell proliferation between control and Suc-transferred plants through staining of the pCYCB1;1::CYCB1;1-D-box:GUS reporter line. Suc-treated leaves demonstrated a cell cycle arrest front closer to the tip, which suggests an increase in cell proliferation. This effect on cell proliferation could result from (post)translational regulation. Suc starvation-induced translational control of cell division and cell growth has already been described in Arabidopsis cell cultures (Nicolaï et al., 2006; Rahmani et al., 2009). Several transcripts involved in protein synthesis, cell cycle, and growth were less abundant in polysomal RNA compared with their total RNA and, thus, translationally repressed by Suc starvation.
It is well known that sugars trigger conserved signaling systems regulating plant growth and development (Smeekens et al., 2010; Lastdrager et al., 2014). One of these major regulators is the conserved SnRK1s in plants, SNF1 in yeast, and AMP-activated kinase in animals, which are heterotrimeric Ser/Thr kinases consisting of a catalytic α-subunit and two regulatory β- and γ-subunits (Polge and Thomas, 2007; Ghillebert et al., 2011; Hardie et al., 2012). These proteins act as metabolic sensors activated when environmental stress conditions deplete carbon and energy supply (Baena-González et al., 2007; Baena-González and Sheen, 2008). Interestingly, PV42a (AT1G15330), one of the cystathionine-β-synthase domain-containing proteins that belong to the γ-type subunits of SnRK1, was significantly repressed 3 h after the transfer of seedlings to Suc. Trehalose-6-phosphate (T6P) and G6P are known to inhibit SnRK1 activity (Toroser et al., 2000; Paul et al., 2008; Zhang et al., 2009; Nunes et al., 2013) and are tightly correlated with the cellular Suc levels (Lunn et al., 2006). T6P is synthesized from G6P and UDP-Glc by TREHALOSE-6-PHOSPHATE SYNTHASE1 (TPS1; Gómez et al., 2010), demonstrating a close link between G6P and T6P levels. G6P allosterically activates Suc phosphate synthase (Huber and Huber, 1996), whereas inorganic phosphate inhibits this enzyme, by which Suc synthesis is buffered upon increased Suc levels. T6P has been described to act as a Suc signal, via the inhibition of SnRK1, in the regulation of many different aspects of plant development, such as growth, flowering, and senescence (for review, see Lunn et al., 2014). Arabidopsis plants lacking TPS1 are embryo lethal, due to an embryonic developmental arrest between the transition from cell proliferation to cell expansion (Eastmond et al., 2002). Plants overexpressing TPS1, and, thus, with a higher T6P content and lower G6P levels, have small dark green leaves, whereas plants overexpressing trehalose-phosphate hydrolase result in large pale green leaves due to low T6P and high G6P levels (Schluepmann et al., 2003). These phenotypes are in line with our results in which higher Suc and, thus, G6P/T6P levels affect chloroplast differentiation.
The effect of sugars during early leaf growth has not yet been investigated, and understanding of the molecular processes that regulate growth requires the microdissection of these growing tissues. The use of whole young seedlings for transcriptome experiments mainly reveals differential gene expression in expanding tissues (Skirycz et al., 2010). To elucidate the Suc-regulated transcriptional responses during the growth of a young leaf, we developed a setup integrating plant developmental timing. Surprisingly, only 19 and 69 genes were differentially expressed in the developing leaf at 3 and 24 h, respectively, after transfer of the seedlings to Suc-containing medium, whereas previously published transcriptomics data sets generally resulted in approximately hundreds of sugar-induced, differentially expressed transcripts (Price et al., 2004; Gonzali et al., 2006; Müller et al., 2007; Osuna et al., 2007; Usadel et al., 2008). This difference in the abundance of differentially expressed genes might be explained by the differences in the harvested samples (microdissected proliferating leaves used here compared with whole seedlings or mature plants in other studies) as well as differences in sugar concentrations. Nevertheless, similar sugar-responsive genes were found in our transcriptional analysis of microdissected third leaves. A significant repression of specific plastid-encoded genes has already been reported in response to the sugar treatment of whole seedlings grown in liquid cultures or whole rosettes (Price et al., 2004; Gonzali et al., 2006; Osuna et al., 2007). However, we found that the complete plastome transcriptome was significantly repressed without a change in plastome copy number per cell. At 3 h after exposure to Suc, most of these chloroplast-encoded transcripts were already down-regulated, albeit not significantly (FDR > 0.05). These findings clearly demonstrate an effect of Suc on plastome expression. Almost all proteins that are present in the chloroplasts are encoded by the nuclear genome. These proteins assemble in large complexes, such as the photosynthetic systems PSI and PSII, around core protein components encoded by the plastid (Jarvis and López-Juez, 2013). Hence, repressing chloroplast transcription can disturb the establishment of these important complexes and, consequently, might impair further chloroplast differentiation. Indeed, transfer of seedlings to Suc-supplemented medium resulted in significant differences in chloroplast morphology. Generally, the chloroplasts were smaller and showed fewer differentiated thylakoid membranes and starch granules compared with control leaves. Besides that, chloroplasts were significantly larger in the tip compared with the base of control leaves. It has been shown that, before cells start to expand at the leaf tip, transcripts involved in photosynthesis and retrograde signaling are up-regulated, which suggests a profound role of chloroplast differentiation in controlling the onset of cell expansion (Andriankaja et al., 2012). Moreover, leaves treated with NF, a chemical inhibitor of chloroplast differentiation, show a delay in the onset of cell expansion (Andriankaja et al., 2012). In concert, we found that leaves transferred to medium without Suc but with NF show a similar increase in total pavement cell number to seedlings transferred to medium with Suc. This phenotype was even more enhanced when both Suc and NF were present, suggesting that both molecules would act additively to stimulate cell proliferation. NF acts directly on the chloroplasts itself, blocking chloroplast differentiation at an early stage, whereas transfer to Suc affects plastome expression, which probably leads to fewer differentiated chloroplasts. Consequently, less retrograde signals are sent to stimulate the onset of cell expansion. Leaves of seedlings transferred to Suc contained fewer chloroplasts in transverse sections compared with control leaves, showing that Suc treatment not only results in smaller chloroplasts with reduced differentiation but also negatively affects chloroplast division.
Besides the effects of Suc on chloroplast-encoded transcripts and chloroplast morphology, Suc induced the expression of GPT2. GPT2, together with its homolog GPT1, acts as a plastid phosphate antiporter involved in the transport of G6Ps between the cytosol and plastids in exchange for inorganic phosphate (Knappe et al., 2003). Mutants lacking GPT1 have been described to be embryo lethal (Andriotis et al., 2010), whereas a disruption of GPT2 does not result in obvious growth defects in the final growth stages (Niewiadomski et al., 2005). However, recently, several studies identified GPT2 as an important regulator in seedling development and during acclimation to high light (Athanasiou et al., 2010; Dyson et al., 2014). In addition, a microarray analysis revealed higher transcript levels for photosynthesis-related and chloroplast-encoded genes in gpt2 mutants (Dyson et al., 2015). A significant overlap showing opposite gene expression profiles was found between this microarray analysis and the Suc-repressed chloroplast-encoded transcripts 24 h after transfer, and almost all selected chloroplast-encoded genes were not repressed upon transfer to Suc in the gpt2 mutant background. Furthermore, no increase in pavement cell number by Suc could be observed in gpt2 mutant seedlings. These observations further indicate the involvement of GPT2 in the Suc-induced promotion of cell proliferation as well as in mediating the Suc-induced repression of the chloroplast-encoded transcripts. Suc could directly induce the transcription of GPT2, by which possibly more G6P, which is correlated with cellular Suc levels (Lunn et al., 2006), is imported in the plastid. Alternatively, GPT2 expression also could be regulated by Glc, which is cleaved from Suc and which is further converted in G6P. The metabolic regulation of GPT2 transcription has been reported in different studies. Microarray analysis of the pho3 mutant, impaired in the SUC2 gene encoding a Suc transporter for phloem loading of Suc, revealed a remarkable up-regulation of both GPT1 and GPT2 expression, but the causative metabolic signal was not investigated in detail (Lloyd and Zakhleniuk, 2004). Another study analyzing GPT2 expression in dark-grown Arabidopsis seedlings treated for 6 h with a broad range of different sugar concentrations (0–200 mm) showed that mainly Suc induces GPT2 expression, whereas Glc treatment only affects its transcription moderately (Gonzali et al., 2006). Our results also show that low concentrations of Glc do not stimulate leaf growth, probably because Glc is not transported through the phloem (Liu et al., 2012). Whether the intracellular conversion of Suc to G6P in the sink tissue is needed to regulate GPT2 transcription remains elusive. Further experiments using a different kind of experimental setup in which sugars can be supplied directly to the sink tissue, for example using plant cell cultures, would be interesting to identify the causative metabolic signal.
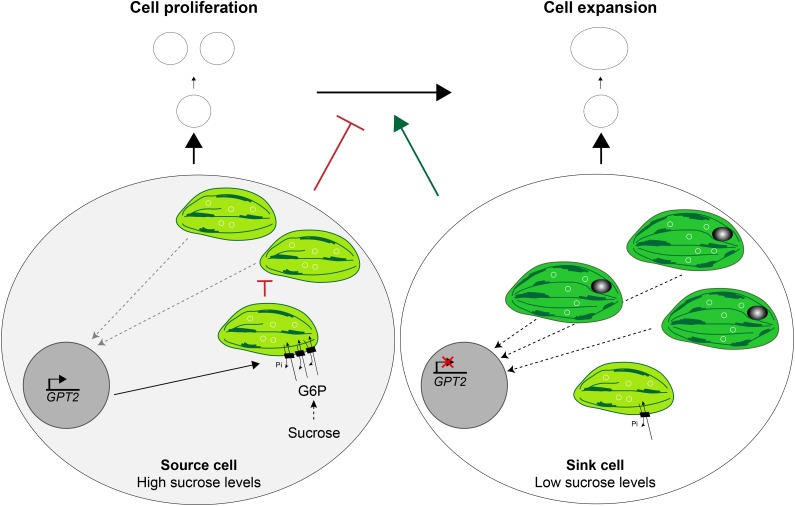
Taken together, GPT2 imports G6P in the chloroplast, which signals the Suc status of the cytosol to the chloroplasts to adjust their development and, thus, to regulate photosynthesis according to the carbon demand of the growing leaf. We hypothesize that, during early leaf development, exogenously applied Suc or Suc produced by source leaves delays chloroplast differentiation in sink leaves, by which cell expansion is postponed and cell proliferation is stimulated (Fig. 7). Conversely, when Suc levels are limiting, a faster transition from Suc-requiring sink tissue to Suc-producing photosynthetically active source tissue will ensure sufficient energy supply and proper plant development. In the sink cells, Suc will be cleaved to Fru and Glc, which will result in higher levels of cytosolic G6P, which then can be transported into the chloroplasts by the Suc-induced plastid transporter GPT2. Furthermore, higher Suc levels result in the repression of chloroplast transcription, leading to a stop in chloroplast differentiation, which might be due to higher levels of G6P inside the chloroplast stroma. Consequently, less retrograde signals because of fewer differentiated chloroplasts could delay the transition to cell expansion, making it possible to use sugars for further stimulation of cell proliferation until the leaf becomes too large and needs autonomous sugar production to sustain growth.
Figure 7.
Model of the central role of chloroplasts in the Suc-induced stimulation of cell proliferation. Low Suc levels in sink cells trigger a rapid formation of photosynthetically active chloroplasts. Consequently, these chloroplasts sent retrograde signals to the nucleus to start the transition to cell expansion. However, higher Suc levels in source cells (left) result in higher levels of G6Ps that are transported into the chloroplasts by the Suc-induced plastid transporter GPT2 (black rectangles). In addition, high Suc levels repress chloroplast transcription (plastome represented by white circles), causing the chloroplasts to stop differentiating and dividing. Consequently, less retrograde signals are sent to the nucleus, which postpones the onset of the transition to cell expansion and stimulates cell proliferation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All experiments were performed on Arabidopsis (Arabidopsis thaliana) ecotype Columbia. Seedlings were grown in vitro on one-half-strength MS medium (Murashige and Skoog, 1962) without Suc for 9 d (9 DAS) under a 16-h-day (50 µmol m−2 s−1) and 8-h-night regime, unless specified differently. Plates were overlaid with nylon mesh of 20-µm pore size. At 9 DAS, seedlings were transferred to plates containing control medium without Suc or medium supplemented with different concentrations of Suc and Glc (6, 15, and 30 mm). In the NF experiments, seedlings were transferred at 9 DAS to one-half-strength MS medium with or without 15 mm Suc supplemented with 5 µm NF. Homozygous seeds of the gpt2-1 mutant, a T-DNA insertion GABI-kat line in the Columbia background (GK-454H06-018837), were a kind gift of Dr. Giles Johnson (University of Manchester; Dyson et al., 2014).
Growth Analysis
For the leaf area analysis, leaves were cleared in 100% (v/v) ethanol, mounted in lactic acid on microscope slides, and photographed. Leaf areas were measured with ImageJ software (http://rsb.info.nih.gov//ij/). Abaxial epidermal cells of the leaves were drawn with a DMLB microscope (Leica) fitted with a drawing tube and a differential interference contrast objective. Drawings were scanned and analyzed using automated image-analysis algorithms (Andriankaja et al., 2012). Subsequently, drawings were used to measure average pavement cell area, from which the total pavement cell number was calculated. The stomatal index was defined as the percentage of stomata compared with all cells.
GUS Staining and Analysis
Seedlings of two biological repeats were harvested at 13 DAS, 4 d after transfer to control or 15 mm Suc-containing medium, incubated in heptane for 10 min, and subsequently left to dry for 5 min. Then, they were submersed in X-Gluc buffer [100 mm Tris-HCl, 50 mm NaCl buffer, pH 7, 2 mm K3Fe(CN)6, and 4 mm X-Gluc], vacuum infiltrated for 10 min, and incubated at 37°C overnight. Seedlings were cleared in 100% (v/v) ethanol and then kept in 90% (v/v) lactic acid. The third leaf was microdissected, mounted on slides, and photographed with a light microscope. Leaf length and GUS staining were measured with ImageJ software (http://rsb.info.nih.gov/ij/). The position of the cell cycle arrest front along the length of each leaf was calculated based on the color intensities between stained (proliferation zone) and nonstained (expanding zone) regions according to the method described by Vercruyssen et al. (2014). The average cell cycle arrest front was determined by taking the average grayscale intensities of the expansion zones of the control leaves.
RNA Extraction and Expression Analysis by qRT-PCR
Seedlings were harvested in liquid nitrogen, put in RNAlater ice solution, and incubated at −20°C for at least 1 week, after which the third leaf was microdissected using a binocular light microscope. Leaves were frozen in liquid nitrogen, and RNA was extracted using Trizol (Invitrogen) and the RNeasy Plant Mini Kit (Qiagen). DNase treatment was done on columns with RNase-free DNase I (Promega). The iScript complementary DNA (cDNA) synthesis kit (Bio-Rad) was used to prepare cDNA from 200 ng of RNA, and qRT-PCR was done on the LightCycler 480 with SYBR Green I Master (Roche) according to the manufacturer’s instructions. Normalization was done against the average of three housekeeping genes, AT1G13320, AT2G32170, and AT2G28390. Primer sequences are listed in Supplemental Table S2.
RNA-Seq Analysis
Library preparation was done using the TruSeq RNA Sample Preparation Kit version 2 (Illumina). Briefly, poly(A)-containing mRNA molecules were reverse transcribed, double-stranded cDNA was generated, and adapters were ligated. After quality control using the 2100 Bioanalyzer (Agilent), clusters were generated through amplification using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina), followed by sequencing on an Illumina HiSeq2000 with the TruSeq SBS Kit v3-HS (Illumina). Sequencing was performed in paired-end mode with a read length of 100 nucleotides. The quality of the raw data was verified with FastQC version 0.9.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Next, quality filtering was performed using the FASTX-Toolkit version 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/). Reads where globally filtered; for at least 75% of the reads, the quality exceeded Q20 and 3′ trimming was performed to remove bases with a quality below Q10, ensuring a remaining minimum length of 90 nucleotides. Repairing was performed using a custom Perl script. Reads were subsequently mapped to the Arabidopsis reference genome (The Arabidopsis Information Resource 10) using GSNAP version 2012-07-20 (Wu and Nacu, 2010), allowing a maximum of five mismatches. These steps were performed through Galaxy (Goecks et al., 2010). The concordantly paired reads that uniquely mapped to the genome were used for quantification on the gene level with HTSeq-count from the HTSeq.py python package (Anders et al., 2015). The analysis was performed with the R (version 3.1.2) software package edgeR (Robinson et al., 2010). Trimmed mean of M values normalization (Robinson and Oshlack, 2010) was applied using the calcNormFactors function. Differentially expressed genes were analyzed with the exact binomial test. FDR adjustments of the P values were done with the method described by Benjamini and Hochberg (1995).
Gene Set Enrichment Analysis
Gene set enrichment analysis was performed with the R package Piano (Väremo et al., 2013) based on P values and Log2FC. Three methods were compared: Fisher’s combined probability test, Stouffer’s method, and the tail strength method. Permutation-based null distributions were calculated by permuting the genes 1,000 times. P values were adjusted with the FDR method (Benjamini and Hochberg, 1995).
Transmission Electron Microscopy
Leaves were immersed in a fixative solution of 2.5% (v/v) glutaraldehyde and 4% (v/v) formaldehyde in 0.1 m sodium cacodylate buffer, placed in a vacuum oven for 30 min, and then left rotating for 3 h at room temperature. This solution was later replaced with fresh fixative, and samples were left rotating overnight at 4°C. After washing, samples were postfixed in 1% (w/v) OsO4 with K3Fe(CN)6 in 0.1 m sodium cacodylate buffer, pH 7.2. Samples were dehydrated through a graded ethanol series, including a bulk staining with 2% (v/v) uranyl acetate at the 50% (v/v) ethanol step, followed by embedding in Spurr’s resin. In order to have a larger overview of the phenotype, semithin sections were first cut at 0.5 µm and stained with Toluidine Blue. Ultrathin sections of a gold interference color were cut using an ultramicrotome (Leica EM UC6), followed by poststaining with uranyl acetate and lead citrate in a Leica EM AC20, and collected on Formvar-coated copper slot grids. Two leaves of control and three leaves of Suc-treated seedlings were viewed with a JEM 1010 transmission electron microscope (JEOL), operating at 80 kV, using Image Plate Technology from Ditabis. For each leaf, 17 to 68 mesophyll cells of the leaf tip and 21 to 87 mesophyll cells of the base of the leaf, representing expanding and proliferating cells, respectively, were analyzed.
Next-generation sequence data from this article were deposited in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-4262.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Glc treatment did not result in an increase in final leaf size.
Supplemental Figure S2. Relative leaf growth rate.
Supplemental Figure S3. Repression of chloroplast transcripts by Suc.
Supplemental Figure S4. Plastome copy numbers per cell of the third leaf upon Suc transfer.
Supplemental Figure S5. Increased starch accumulation in control leaves.
Supplemental Figure S6. Transcriptional responses in the gpt2 mutant.
Supplemental Table S1. Differentially expressed genes 3 and 24 h after transfer to Suc.
Supplemental Table S2. qRT-PCR primer sequences of selected chloroplast-encoded transcripts.
Supplemental Methods S1. Plastome copy number determination and statistical analysis of growth experiments and chloroplast measurements.
Supplementary Material
Acknowledgments
We thank all colleagues of the Systems Biology of Yield research group for many fruitful discussions, Frederik Coppens for helping with the RNA-seq analysis, and Annick Bleys for help in preparing the article.
Glossary
- G6P
glucose-6-phosphate
- PEP
plastid-encoded polymerase
- NF
norflurazon
- DAS
days after stratification
- RNA-seq
RNA sequencing
- FDR
false discovery rate
- Log2FC
log2 fold change
- qRT
quantitative reverse transcription
- MS
Murashige and Skoog
- T6P
trehalose-6-phosphate
- X-Gluc
5-bromo-4-chloro-3-indolyl-β-glucuronide
- cDNA
complementary DNA
Footnotes
This work was supported by the Interuniversity Attraction Poles Program (grant no. IUAP P7/29 MARS) initiated by the Belgian Science Policy Office, by Ghent University (Bijzonder Onderzoeksfonds Methusalem Project grant no. BOF08/01M00408), and by the Research Foundation-Flanders (postdoctoral fellowship to S.D. and FWO grant no. G046512N).
Articles can be viewed without a subscription.
References
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GTS, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Andriotis VME, Pike MJ, Bunnewell S, Hills MJ, Smith AM (2010) The plastidial glucose-6-phosphate/phosphate antiporter GPT1 is essential for morphogenesis in Arabidopsis embryos. Plant J 64: 128–139 [DOI] [PubMed] [Google Scholar]
- Athanasiou K, Dyson BC, Webster RE, Johnson GN (2010) Dynamic acclimation of photosynthesis increases plant fitness in changing environments. Plant Physiol 152: 366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J (2008) Convergent energy and stress signaling. Trends Plant Sci 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57: 289–300 [Google Scholar]
- Charuvi D, Kiss V, Nevo R, Shimoni E, Adam Z, Reich Z (2012) Gain and loss of photosynthetic membranes during plastid differentiation in the shoot apex of Arabidopsis. Plant Cell 24: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Dyson BC, Allwood JW, Feil R, Xu Y, Miller M, Bowsher CG, Goodacre R, Lunn JE, Johnson GN (2015) Acclimation of metabolism to light in Arabidopsis thaliana: the glucose 6-phosphate/phosphate translocator GPT2 directs metabolic acclimation. Plant Cell Environ 38: 1404–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson BC, Webster RE, Johnson GN (2014) GPT2: a glucose 6-phosphate/phosphate translocator with a novel role in the regulation of sugar signalling during seedling development. Ann Bot (Lond) 113: 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJ, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JD, Smeekens SC, Graham IA (2002) Trehalose-6-phosphate synthase 1 which catalyses the first step in trehalose synthesis is essential for Arabidopsis embryo maturation. Plant J 29: 225–235 [DOI] [PubMed] [Google Scholar]
- Eloy NB, Gonzalez N, Van Leene J, Maleux K, Vanhaeren H, De Milde L, Dhondt S, Vercruysse L, Witters E, Mercier R, et al. (2012) SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. Proc Natl Acad Sci USA 109: 13853–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Hou X, Lee LYC, Liu L, Yan X, Yu H (2011) AtPV42a and AtPV42b redundantly regulate reproductive development in Arabidopsis thaliana. PLoS ONE 6: e19033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Cadman CS, Toorop PE, Lynn JR, Hilhorst HW (2007) Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J 51: 60–78 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Ito M, Nishida I, Watanabe A (2000) Multiple signaling pathways in gene expression during sugar starvation: pharmacological analysis of din gene expression in suspension-cultured cells of Arabidopsis. Plant Physiol 124: 1139–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghillebert R, Swinnen E, Wen J, Vandesteene L, Ramon M, Norga K, Rolland F, Winderickx J (2011) The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS J 278: 3978–3990 [DOI] [PubMed] [Google Scholar]
- Gissot L, Polge C, Jossier M, Girin T, Bouly JP, Kreis M, Thomas M (2006) AKINβγ contributes to SnRK1 heterotrimeric complexes and interacts with two proteins implicated in plant pathogen resistance through its KIS/GBD sequence. Plant Physiol 142: 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Roitsch T (1999) The different pH optima and substrate specificities of extracellular and vacuolar invertases from plants are determined by a single amino-acid substitution. Plant J 20: 707–711 [DOI] [PubMed] [Google Scholar]
- Gómez LD, Gilday A, Feil R, Lunn JE, Graham IA (2010) AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J 64: 1–13 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci 17: 332–340 [DOI] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Solfanelli C, Novi G, Alpi A, Perata P (2006) Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J Plant Res 119: 115–123 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA (2012) AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler RE, Heinrichs L, Schmitz J, Flügge UI (2014) How sugars might coordinate chloroplast and nuclear gene expression during acclimation to high light intensities. Mol Plant 7: 1121–1137 [DOI] [PubMed] [Google Scholar]
- Heinrichs L, Schmitz J, Flügge UI, Häusler RE (2012) The mysterious rescue of adg1-1/tpt-2—an Arabidopsis thaliana double mutant impaired in acclimation to high light—by exogenously supplied sugars. Front Plant Sci 3: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Huber JL (1996) Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 431–444 [DOI] [PubMed] [Google Scholar]
- Hudik E, Yoshioka Y, Domenichini S, Bourge M, Soubigout-Taconnat L, Mazubert C, Yi D, Bujaldon S, Hayashi H, De Veylder L, et al. (2014) Chloroplast dysfunction causes multiple defects in cell cycle progression in the Arabidopsis crumpled leaf mutant. Plant Physiol 166: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Kammerer B, Fischer K, Hilpert B, Schubert S, Gutensohn M, Weber A, Flügge UI (1998) Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter. Plant Cell 10: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama T, Ichihashi Y, Murata S, Tsukaya H (2010) The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol 51: 1046–1054 [DOI] [PubMed] [Google Scholar]
- Kleine T, Leister D (2013) Retrograde signals galore. Front Plant Sci 4: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S, Flügge UI, Fischer K (2003) Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol 131: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S, Sugimoto K (2012) Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiol 53: 953–964 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Krapp A, Hofmann B, Schäfer C, Stitt M (1993) Regulation of the expression of rbcS and other photosynthetic genes by carbohydrates: a mechanism for the ‘sink regulation’ of photosynthesis? Plant J 3: 817–828 [Google Scholar]
- Kühn C, Grof CPL (2010) Sucrose transporters of higher plants. Curr Opin Plant Biol 13: 288–298 [DOI] [PubMed] [Google Scholar]
- Kunz HH, Häusler RE, Fettke J, Herbst K, Niewiadomski P, Gierth M, Bell K, Steup M, Flügge UI, Schneider A (2010) The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol (Stuttg) (Suppl 1) 12: 115–128 [DOI] [PubMed] [Google Scholar]
- Kunz S, Pesquet E, Kleczkowski LA (2014) Functional dissection of sugar signals affecting gene expression in Arabidopsis thaliana. PLoS ONE 9: e100312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastdrager J, Hanson J, Smeekens S (2014) Sugar signals and the control of plant growth and development. J Exp Bot 65: 799–807 [DOI] [PubMed] [Google Scholar]
- Lee EJ, Matsumura Y, Soga K, Hoson T, Koizumi N (2007) Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol 48: 405–413 [DOI] [PubMed] [Google Scholar]
- Lemoine R, La Camera S, Atanassova R, Dédaldéchamp F, Allario T, Pourtau N, Bonnemain JL, Laloi M, Coutos-Thévenot P, Maurousset L, et al. (2013) Source-to-sink transport of sugar and regulation by environmental factors. Front Plant Sci 4: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2011) Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Mol Biol 76: 235–249 [DOI] [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW (2006) Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res 16: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T (2011) The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Liu DD, Chao WM, Turgeon R (2012) Transport of sucrose, not hexose, in the phloem. J Exp Bot 63: 4315–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J Exp Bot 55: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M (2014) Trehalose metabolism in plants. Plant J 79: 544–567 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JH, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143: 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nicolaï M, Roncato MA, Canoy AS, Rouquié D, Sarda X, Freyssinet G, Robaglia C (2006) Large-scale analysis of mRNA translation states during sucrose starvation in Arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiol 141: 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C, Primavesi LF, Patel MK, Martinez-Barajas E, Powers SJ, Sagar R, Fevereiro PS, Davis BG, Paul MJ (2013) Inhibition of SnRK1 by metabolites: tissue-dependent effects and cooperative inhibition by glucose 1-phosphate in combination with trehalose 6-phosphate. Plant Physiol Biochem 63: 89–98 [DOI] [PubMed] [Google Scholar]
- Oelze ML, Vogel MO, Alsharafa K, Kahmann U, Viehhauser A, Maurino VG, Dietz KJ (2012) Efficient acclimation of the chloroplast antioxidant defence of Arabidopsis thaliana leaves in response to a 10- or 100-fold light increment and the possible involvement of retrograde signals. J Exp Bot 63: 1297–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Bläsing OE, Höhne M, Günter M, Kamlage B, Trethewey R, Scheible WR, et al. (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49: 463–491 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Primavesi LF, Jhurreea D, Zhang Y (2008) Trehalose metabolism and signaling. Annu Rev Plant Biol 59: 417–441 [DOI] [PubMed] [Google Scholar]
- Pfalz J, Liebers M, Hirth M, Grübler B, Holtzegel U, Schröter Y, Dietzel L, Pfannschmidt T (2012) Environmental control of plant nuclear gene expression by chloroplast redox signals. Front Plant Sci 3: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–28 [DOI] [PubMed] [Google Scholar]
- Pourtau N, Jennings R, Pelzer E, Pallas J, Wingler A (2006) Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 224: 556–568 [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smeekens S, Hanson J (2009) Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol 150: 1356–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JMS, Murray JAH (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 20: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Jin Y, Yang YJ, Li GJ, Boyer JS (2010) Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol Plant 3: 942–955 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Uno Y, Zhang Q, Miura E, Kato Y, Sodmergen (2009) Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant Cell Physiol 50: 2069–2083 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res 6: 283–290 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A, Vainonen JP, Wrzaczek M, Kangasjärvi J (2012) ROS-talk: how the apoplast, the chloroplast, and the nucleus get the message through. Front Plant Sci 3: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS (2005) Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int Rev Cytol 244: 1–68 [DOI] [PubMed] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Hellmann HA (2014) Sugar sensing and signaling in plants. Front Plant Sci 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13: 274–279 [DOI] [PubMed] [Google Scholar]
- Steiner S, Schröter Y, Pfalz J, Pfannschmidt T (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A. (1999) Invertases: primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol 121: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Terry MJ, Smith AG (2013) A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toroser D, Plaut Z, Huber SC (2000) Regulation of a plant SNF1-related protein kinase by glucose-6-phosphate. Plant Physiol 123: 403–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R. (1989) The sink-source transition in leaves. Annu Rev Plant Physiol Plant Mol Biol 40: 119–138 [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väremo L, Nielsen J, Nookaew I (2013) Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res 41: 4378–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, Han SK, Jégu T, Archacki R, Van Leene J, Andriankaja M, et al. (2014) ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26: 210–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76: 273–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RAC, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50: 710–722 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.