The STEROL 4α-METHYL OXIDASE2 family proteins SMO2-1 and SMO2-2 function partially through effects on auxin accumulation, auxin response, and PIN1 expression to regulate embryogenesis in Arabidopsis.
Abstract
Sterols are essential molecules for multiple biological processes, including embryogenesis, cell elongation, and endocytosis. The plant sterol biosynthetic pathway is unique in the involvement of two distinct sterol 4α-methyl oxidase (SMO) families, SMO1 and SMO2, which contain three and two isoforms, respectively, and are involved in sequential removal of the two methyl groups at C-4. In this study, we characterized the biological functions of members of the SMO2 gene family. SMO2-1 was strongly expressed in most tissues during Arabidopsis (Arabidopsis thaliana) development, whereas SMO2-2 showed a more specific expression pattern. Although single smo2 mutants displayed no obvious phenotype, the smo2-1 smo2-2 double mutant was embryonic lethal, and the smo2-1 smo2-2/+ mutant was dwarf, whereas the smo2-1/+ smo2-2 mutant exhibited a moderate phenotype. The phenotypes of the smo2 mutants resembled those of auxin-defective mutants. Indeed, the expression of DR5rev:GFP, an auxin-responsive reporter, was reduced and abnormal in smo2-1 smo2-2 embryos. Furthermore, the expression and subcellular localization of the PIN1 auxin efflux facilitator also were altered. Consistent with these observations, either the exogenous application of auxin or endogenous auxin overproduction (YUCCA9 overexpression) partially rescued the smo2-1 smo2-2 embryonic lethality. Surprisingly, the dwarf phenotype of smo2-1 smo2-2/+ was completely rescued by YUCCA9 overexpression. Gas chromatography-mass spectrometry analysis revealed a substantial accumulation of 4α-methylsterols, substrates of SMO2, in smo2 heterozygous double mutants. Together, our data suggest that SMO2s are important for correct sterol composition and function partially through effects on auxin accumulation, auxin response, and PIN1 expression to regulate Arabidopsis embryogenesis and postembryonic development.
Sterols are isoprenoid-derived molecules that have diverse and essential functions in eukaryotes. Sterols are structural membrane components required for proper membrane permeability, fluidity, and membrane protein trafficking/localization. Silencing of the SQUALENE SYNTHASE1 gene compromised the resistance of tobacco (Nicotiana tabacum) to bacterial pathogens because of increased membrane leakage (Wang et al., 2012). Disturbed membrane sterol composition in the Arabidopsis (Arabidopsis thaliana) cyclopropylsterol isomerase1-1 (cpi1-1) mutant affected the polar localization of the PIN2 auxin efflux protein and the cell plate distribution of KNOLLE syntaxin by inhibiting the endocytosis of these proteins (Men et al., 2008; Boutté et al., 2010). Moreover, sterol-enriched plasma membrane microdomains (lipid rafts) have been proposed to act as signaling platforms (Mongrand et al., 2004, 2010; Martin et al., 2005).
Plant sterols are precursors for the biosynthesis of brassinosteroids (BRs), a group of phytohormones that are essential for plant growth and development. However, increasing evidence indicates that plant sterols per se can act as signaling molecules. Plant sterol biosynthetic mutants, such as smt1/cph, cpi1-1, cyp51A2, fackel (fk), and hyd1, could not be rescued by BR application (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002; Willemsen et al., 2003; Kim et al., 2005; Men et al., 2008). Sterols were found to have intimate interactions with plant hormones, such as auxin and ethylene. Polar auxin transport (PAT) or auxin response was hindered in sterol biosynthetic mutants (Willemsen et al., 2003; Men et al., 2008; Carland et al., 2010; Pullen et al., 2010). Inhibition of ethylene biosynthesis or signaling partially rescued fk, hyd1, and cyp51A2 mutants (Kim et al., 2010; Pullen et al., 2010). Sterols also can regulate the generation of reactive oxygen species, which play important roles in plant growth and cell death (Posé et al., 2009; Kim et al., 2010). A specific plant sterol biosynthetic intermediate (SBI) was speculated to have signaling functions. Sterols, such as sitosterol and stigmasterol, and SBIs, such as obtusifoliol, can regulate the expression of genes involved in cell expansion and cell division (He et al., 2003; O’Brien et al., 2005). An SBI, 4-carboxy-4-methyl-24-methylenecycloartanol (CMMC), which is derived from the first C-4 demethylation reaction, can inhibit PAT and can cause PAT-related phenotypes when applied to wild-type Arabidopsis plants (Mialoundama et al., 2013).
Plant sterol biosynthesis differs from that in animals and fungi in the removal of the two methyl groups at the C-4 position (Benveniste, 2004; Rahier, 2011). Each C-4 demethylation reaction is performed with the sequential participation of three enzymes, a sterol 4α-methyl oxidase (SMO), a 4α-carboxysterol-C-3-dehydrogenase/C-4-decarboxylase (CSD), and a sterone ketoreductase, which are tethered into a complex by the scaffold protein ERG28 (Mo et al., 2002; Rahier, 2011). In animals and fungi, the two C-4 demethylation reactions occur during consecutive rounds of action and are catalyzed by the same enzymes (Mo et al., 2002; Rahier, 2011). However, in higher plants, the two C-4 demethylation reactions are interrupted by several other steps (Supplemental Fig. S1; Rahier, 2011), and two distinct families of SMO enzymes (SMO1 and SMO2, containing three and two members, respectively) are involved in the first and second C-4 demethylation steps (Darnet et al., 2001; Darnet and Rahier, 2004). Members of the SMO1 and SMO2 protein families share 32% and 29% sequence identity, respectively, with the yeast (Saccharomyces cerevisiae) homolog ERG25, but only SMO2 family proteins can complement the yeast erg25 mutant (Darnet et al., 2001; Darnet and Rahier, 2004). Virus-induced gene silencing (VIGS) in tobacco showed that, after the silencing of SMO1, 4,4-dimethyl-9β,19-cyclopropylsterols were accumulated, whereas after the silencing of SMO2, 4α-methyl-Δ7-sterols were accumulated (Darnet and Rahier, 2004; Rahier, 2011). These results suggest that the two SMO family proteins perform specific C-4 demethylation, with SMO1 participating in the first C-4α-methyl group removal and SMO2 participating in the second C-4 methyl group removal (Rahier, 2011). In yeast, ERG25 was demonstrated to be essential because the yeast erg25 mutant was lethal (Bard et al., 1996). In humans, SMO deficiency was reported to cause growth delays during infancy (He et al., 2011). However, mutants have not been characterized in either plant SMO gene family, and the roles of SMOs in plant growth and development remain unknown.
In this work, we investigated the developmental roles of SMO2 family genes in Arabidopsis by analyzing their knockout mutants. Single null smo2-1 and smo2-2 mutants showed no obvious phenotype, but the smo2-1 smo2-2 double mutant was embryonic lethal. Moreover, the heterozygous smo2-1 smo2-2/+ double mutant displayed a dwarf phenotype, whereas the smo2-1/+ smo2-2 mutant was relatively normal. Our data indicate that the embryogenesis defects of the smo2-1 smo2-2 double mutant are caused by perturbed PAT and auxin accumulation. We further show that auxin application and crossing with YUCCA9 overexpression (YUC9 OE) plants partially rescued the smo2-1 smo2-2 embryonic lethality. Moreover, the dwarf phenotype of the heterozygous smo2-1 smo2-2/+ double mutant was fully rescued by YUC9 OE. Our results show that both smo2-1 smo2-2/+ and smo2-1/+ smo2-2 mutants accumulate 4α-methylsterols that are substrates of SMO2. We conclude that SMO2s are essential for Arabidopsis embryogenesis and postembryonic development, and they function partially through auxin accumulation, auxin response, and PIN1 expression by modulating sterol composition.
RESULTS
SMO2-1 and SMO2-2 Have Different Expression Patterns, and Their Encoded Proteins Localize to the Endoplasmic Reticulum
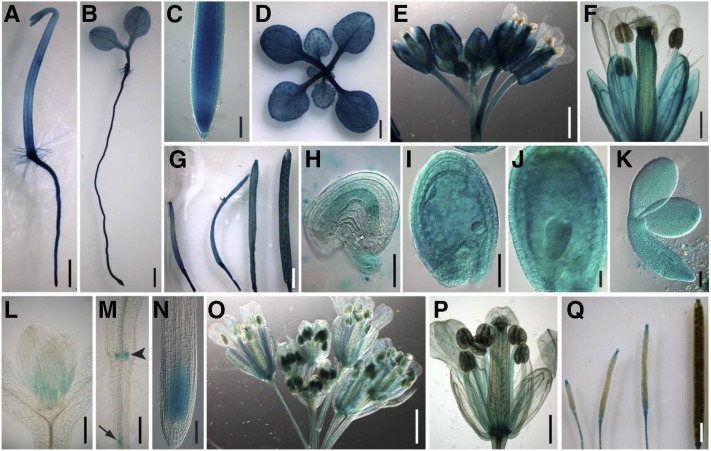
An alignment of the SMO2-1 and SMO2-2 proteins showed that they are highly similar, sharing 91% amino acid sequence identity (Supplemental Fig. S2A). The predicted topology of SMO2-1 and SMO2-2 also shares high similarity (Supplemental Fig. S2, B and C). To determine whether the expression patterns of the SMO2-1 and SMO2-2 genes are different, we expressed the GUS reporter gene under the control of SMO2-1 and SMO2-2 genomic cis-regulatory sequences located upstream and downstream of their open reading frames (ProSMO2-1:GUS and ProSMO2-2:GUS). Strong GUS staining was observed in most tissues throughout the ProSMO2-1:GUS transgenic plants (Fig. 1, A–K). In roots, ProSMO2-1:GUS expression was detected in all tissues at an exceptionally high level (Fig. 1, A–C). In leaves, the GUS signal was strong in vascular tissues and trichomes (Fig. 1D). In flowers, SMO2-1 was highly expressed in sepals, stamen filaments, pollen grains, and pistils, but it was barely detectable in petals (Fig. 1, E and F). Siliques also were strongly stained at all stages of development (Fig. 1G). ProSMO2-1:GUS was highly expressed in both embryos and endosperms, and a high expression level was maintained throughout embryonic development (Fig. 1, H–K). By contrast, SMO2-2 showed a restricted and weaker expression pattern. In shoots, ProSMO2-2:GUS was expressed only in the apical meristem (Fig. 1L). In roots, GUS activity was detected only in adventitious root primordia, lateral root primordia, and the root tip meristem (Fig. 1, M and N). In flowers, SMO2-2 was expressed in vascular tissues of sepals and petals as well as in stamen filaments and pollen (Fig. 1, O and P). In young siliques, SMO2-2 expression was restricted to the style and silique funiculus; in mature siliques, GUS activity was restricted to the abscission zone (Fig. 1Q).
Figure 1.
Tissue-specific expression of ProSMO2-1:GUS and ProSMO2-2:GUS. A to K, Expression of ProSMO2-1:GUS, showing seedlings grown on Murashige and Skoog (MS) medium in the dark (A) and in light (B); root tip (C); rosette of a 2-week-old plant (D); inflorescence (E) and flower (F); siliques at different developmental stages (G); and ovule, seeds, and embryos (H–K). L to Q, Expression of ProSMO2-2:GUS showing the shoot apical meristem (L), adventitious root primordia (arrowhead) and lateral root primordium (arrow; M), and root tip (N) of 5-d-old seedlings; inflorescence (O) and flower (P); and siliques at different developmental stages (Q). Bars = 1 mm (A, B, D, E, G, O, and Q), 100 μm (C, K, and N), 0.5 mm (F, L, M, and P), and 50 μm (H–J).
Because we did not detect ProSMO2-2:GUS expression in developing seeds, we used quantitative reverse transcription (RT)-PCR to determine whether SMO2-2 is expressed during seed development. Consistent with the seed expression of ProSMO2-1:GUS, SMO2-1 transcript levels were high at all seed developmental stages and were highest in seeds containing embryos at torpedo stage (Supplemental Fig. S3A). By contrast, SMO2-2 was expressed at very low levels in Arabidopsis seeds at the early developmental stages; then, its expression was increased in the late stages. SMO2-2 expression also peaked in seeds containing embryos at the torpedo stage (Supplemental Fig. S3A). We also analyzed the expression patterns of SMO2 genes in Arabidopsis embryos using publicly available transcriptome databases generated by the Raju Datla laboratory (Xiang et al., 2011). The expression patterns of these genes in embryos were very similar to the results we obtained in seeds, except that the SMO2-2 expression levels in early-stage embryos were not very low (compare Supplemental Fig. S3, B and A). Together, these data indicate that both SMO2 genes are expressed in embryos, with SMO2-2 expressed at lower levels than SMO2-1.
To address the subcellular localization of SMO2 proteins, we generated a translational fusion between the SMO2 genomic sequences and the EGFP gene, and the construct was driven by the native SMO2 genomic cis-regulatory sequence (the same sequence used in the transcriptional fusion with GUS). These ProSMO2:SMO2-EGFP constructs were transformed into wild-type plants. We analyzed T2 progeny from two different transgenic lines for each construct by confocal laser microscopy and found a similar reticulate pattern in these lines. To confirm that SMO2-EGFP localizes to the endoplasmic reticulum (ER), we performed immunolocalization on the roots of these transgenic plants using an antibody against BiP (an ER-intrinsic chaperone, used as an ER marker; Men et al., 2008). The results showed that the SMO2-EGFP signal (Supplemental Fig. S4; shown in green) overlapped with the immunoprocessed BiP signal (shown in red), suggesting that SMO2 proteins are localized to the ER.
The smo2-1 smo2-2 Double Mutant Was Not Recovered, and the smo2-1 smo2-2/+ Mutant Showed a Dwarf Phenotype
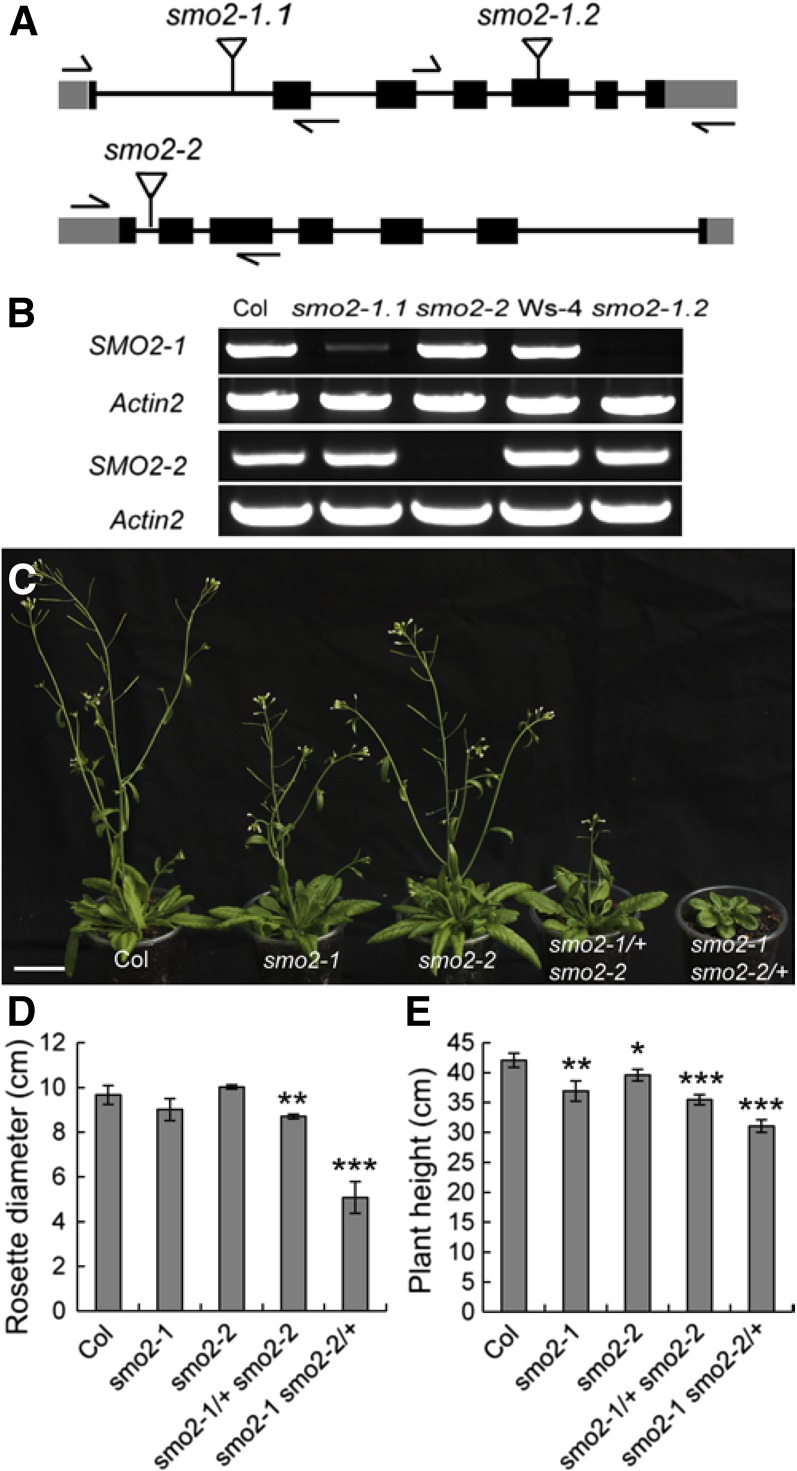
To study the biological functions of the SMO2 genes, transfer DNA (T-DNA) insertion alleles were isolated for SMO2-1 within the first intron (SALK_105017, smo2-1.1) and within the fifth exon (FLAG_522B02, smo2-1.2) and for SMO2-2 within the first intron (SALK_030719, smo2-2; Fig. 2A). RT-PCR analyses revealed that no transcripts were detectable in the homozygous smo2-1.1, smo2-1.2, and smo2-2 mutants (Fig. 2B), indicating that these mutants were complete knockouts. We did not observe abnormal phenotypes for these single mutants except that their hypocotyls and root lengths were slightly shorter than in the wild type (Supplemental Fig. S5, A–E). These results implied that the SMO2-1 and SMO2-2 genes are functionally redundant. Therefore, we crossed smo2-1 and smo2-2 plants to generate double mutants. Both smo2-1.1 and smo2-1.2 mutants were crossed with smo2-2. However, no homozygous smo2-1 smo2-2 double mutant plants were recovered; only plants homozygous for one allele and heterozygous for the other (smo2-1/+ smo2-2 and smo2-1 smo2-2/+) were obtained.
Figure 2.
Expression and phenotype analysis of T-DNA insertion mutants for the SMO2-1 and SMO2-2 genes. A, Schematic diagrams of T-DNA insertion sites in smo2-1 and smo2-2 mutants. Black boxes indicate exons; gray boxes indicate 5′ and 3′ untranslated regions; lines indicate introns; and flags indicate T-DNA insertion sites. Arrows indicate the positions of gene-specific primers used for PCR verification of insertions. B, RT-PCR analysis of the expression levels of SMO2-1 and SMO2-2 genes in the wild-type Columbia-0 (Col) and mutants. C, Phenotypes of 5-week-old wild-type and smo2 single and double mutant plants. Bar = 3 cm. D, Rosette diameter of 4-week-old plants. E, Height of mature (9-week-old) plants. The data were derived from three experiments and are presented as means ± sd (n = 10 for each experiment; *, P < 0.05; **, P < 0.01; and ***, P < 0.001 by Student’s t test).
Compared with the wild type, smo2-1 smo2-2/+ seedlings had slightly shorter roots (Supplemental Fig. S5F). The smo2-1 smo2-2/+ plants had dwarf and late-flowering phenotypes, with small, round, dark green leaves (Fig. 2, C–E). These phenotypes were similar to those of sterol biosynthetic mutants, such as dwarf7 (dwf7)/ste1, dwf5, and dwf1/diminuto (dim; Klahre et al., 1998; Choe et al., 1999a, 1999b, 2000). However, unlike the dwf7/ste1, dwf5, and dwf1/dim mutants, the short-root phenotype of the smo2-1 smo2-2/+ mutant could not be rescued by BR (Supplemental Fig. S5G). Moreover, the smo2-1 smo2-2/+ mutant exhibited a high degree of phenotypic variability (Supplemental Fig. S5H). Unlike the smo2-1 smo2-2/+ mutant, smo2-1/+ smo2-2 showed only a moderate phenotype, with slightly smaller rosettes, shorter plant height, and later flowering than the wild type (Fig. 2, C–E).
The smo2-1 smo2-2 Double Mutant Is Arrested Early during Embryogenesis
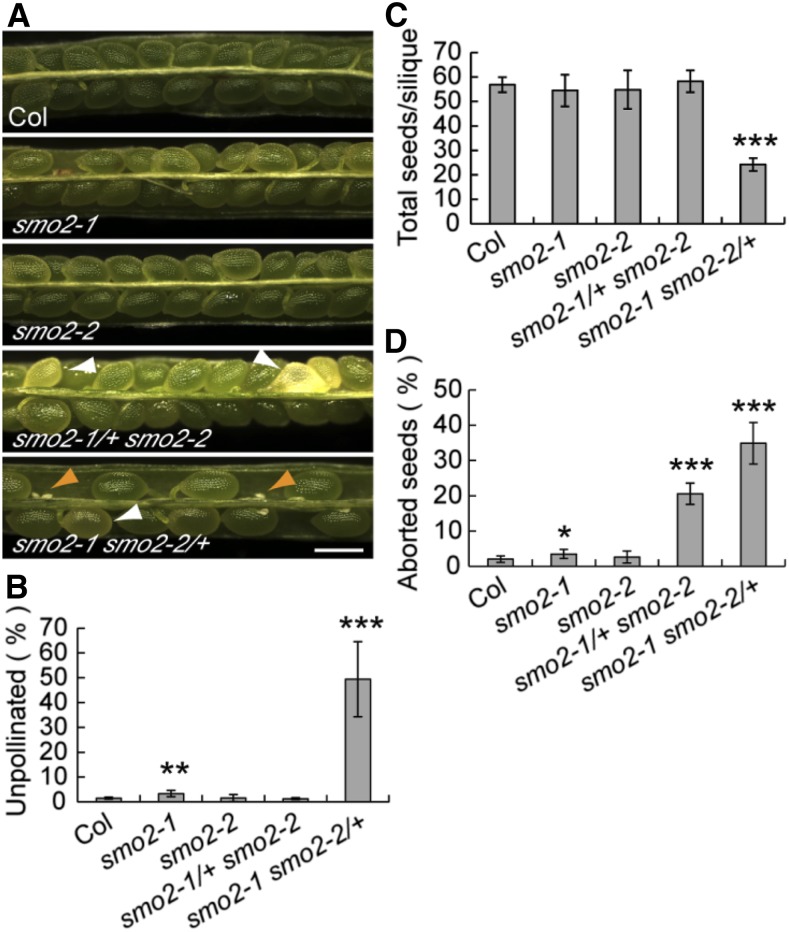
Because no homozygous smo2-1 smo2-2 double mutant plants were recovered, we examined siliques from both the smo2-1 smo2-2/+ and smo2-1/+ smo2-2 mutants. Compared with the normal seed development of wild-type plants, we observed white wrinkled seeds in both smo2-1/+ smo2-2 and smo2-1 smo2-2/+ siliques (Fig. 3A). This finding suggested that the smo2-1 smo2-2 double mutant was embryonic lethal. Additionally, siliques from smo2-1 smo2-2/+ plants were found to have approximately 49.4% undeveloped ovules compared with less than 3.3% in wild-type, smo2-1, smo2-2, and smo2-1/+ smo2-2 siliques (means of five experiments, n = 289–617 for each experiment; Fig. 3, A and B). Consistently, approximately 57 seeds on average were identified in siliques of wild-type, smo2-1, smo2-2, and smo2-1/+ smo2-2 plants; however, the smo2-1 smo2-2/+ siliques contained only approximately 24 seeds (Fig. 3C). These results suggest that gametophytic development or transmission is affected in the smo2-1 smo2-2/+ mutant. Self-fertilized smo2-1/+ smo2-2 plants segregated approximately 2:1 for smo2-1/+ smo2-2 and SMO2-1 smo2-2 progeny (424:221; P = 0.9088 by χ2 test). Similarly, self-fertilized smo2-1 smo2-2/+ plants segregated approximately 2:1 for smo2-1 smo2-2/+ and smo2-1 SMO2-2 progeny (491:249; P = 0.9989 by χ2 test). These results suggest that gametophytic transmission of the smo2-1 smo2-2 double mutation was not affected. Together, these data indicate that gametophytic development was affected in the smo2-1 smo2-2/+ plants. We subsequently determined the frequency of aborted seeds in smo2-1/+ smo2-2 and smo2-1 smo2-2/+ siliques. Approximately 20.6% (n = 2,772) of aborted seeds were observed in siliques from smo2-1/+ smo2-2 plants, which was lower than the expected 25% ratio for the smo2-1 smo2-2 double mutant, although not significantly different (P = 0.252 by χ2 test; Fig. 3D). Although the smo2-1/+ smo2-2 mutant had no gametophytic defects, it is possible that the double mutant gametophyte was less competitive at fertilization, resulting in a slightly lower segregation ratio for the smo2-1 smo2-2 embryos. By contrast, the seed abortion rate in smo2-1 smo2-2/+ siliques deviated significantly from the expected 25% ratio (34.9%; n = 1,098, P < 0.001 by χ2 test; Fig. 3D). This segregation ratio distortion was likely due to the gametophytic defect of the smo2-1 smo2-2/+ mutant. To confirm that the observed embryonic developmental defects were caused by SMO2 loss of function, we introduced SMO2 genomic fragment-GFP translational fusions (SMO2-1-EGFP and SMO2-2-EGFP) into the smo2-1 smo2-2/+ and smo2-1/+ smo2-2 mutants, respectively. These transgenes rescued all the defects of the smo2-1 smo2-2/+ and smo2-1/+ smo2-2 mutants, and smo2-1 smo2-2 double mutant plants with normal seed setting were recovered (Supplemental Fig. S6).
Figure 3.
Altered seed set in smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants. A, Siliques 9 to 10 d after pollination from wild-type Columbia-0 (Col), smo2-1, smo2-2, smo2-1/+ smo2-2, and smo2-1 smo2-2/+ plants. White arrowheads indicate aborted seeds, and orange arrowheads indicate undeveloped ovules. Bar = 500 μm. B to D, Quantification of unpollinated ovules and total and aborted seeds in siliques from the wild type and smo2 single and double mutants. The data were derived from five experiments and are presented as means ± sd. For each experiment, six to 10 siliques (289–617 seeds plus unpollinated ovules) from five plants were examined. Significant differences were analyzed using Student’s t test (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
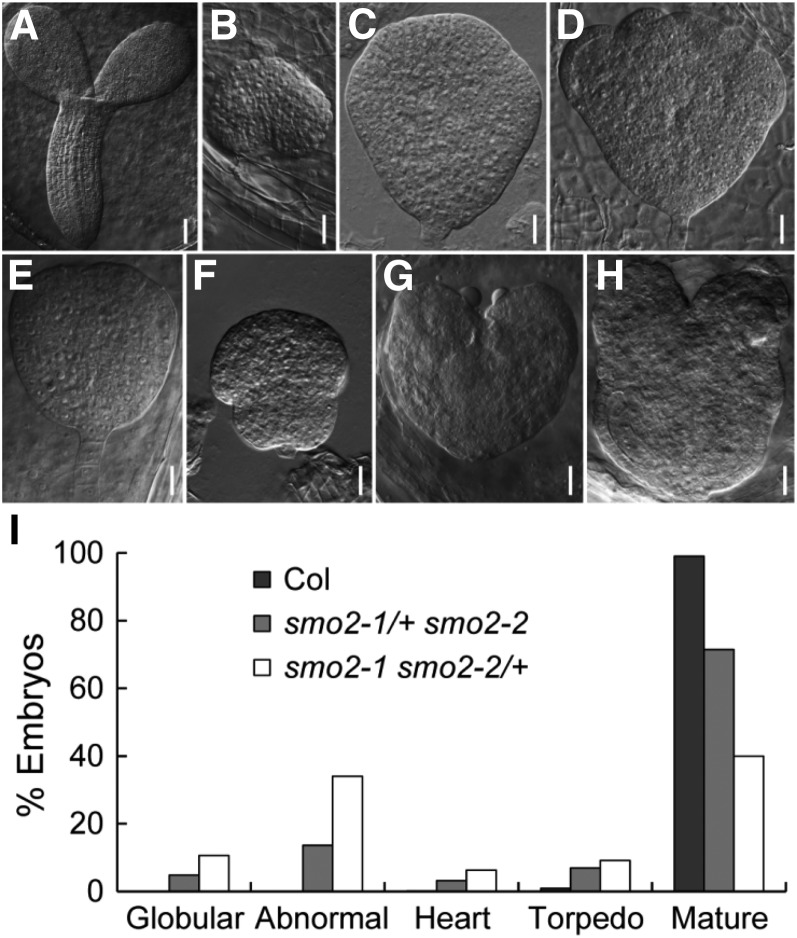
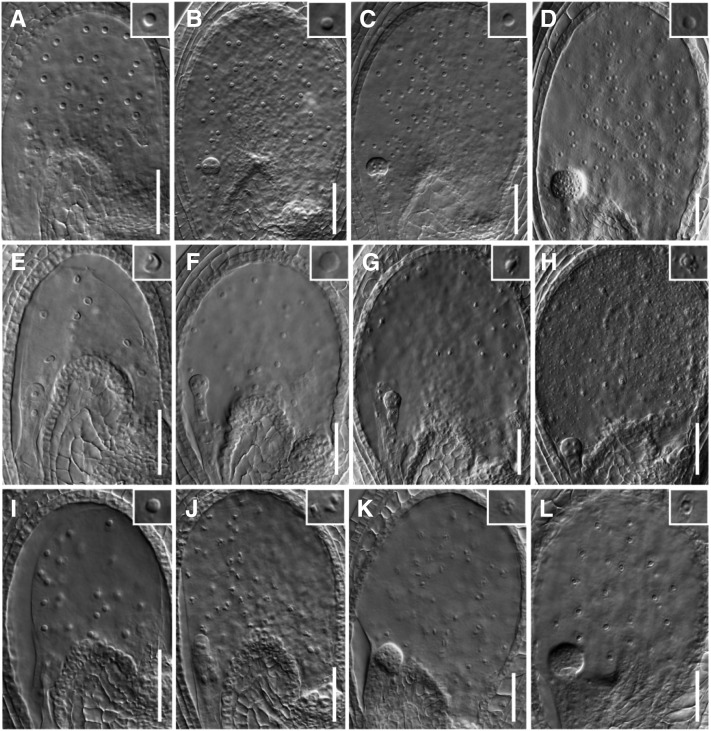
To identify the stage at which smo2-1 smo2-2 embryonic development was arrested, we first analyzed cleared seeds from mature wild-type, smo2-1 smo2-2/+, and smo2-1/+ smo2-2 siliques. Compared with the mature wild-type embryos (Fig. 4A), the smo2-1 smo2-2 embryos were arrested at the globular to heart-like stages (Fig. 4, B–H). These arrested embryos showed patterning defects similar to those of the fk, hyd1, and smt1/cph mutants (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002; Willemsen et al., 2003) and did not develop the characteristic heart shape (Supplemental Fig. S7A). Instead, the arrested embryos exhibited an abnormal round shape without cotyledon protrusion (Fig. 4, C and F), had one or more cotyledon protrusions (Fig. 4D; Supplemental Fig. S7B), or had two very small cotyledon protrusions (Fig. 4, G and H). The statistical analysis indicated that in the wild-type siliques, only 0.1% and 0.9% of seeds bore embryos delayed at the heart and torpedo stages, respectively (n = 1,631). In the smo2-1/+ smo2-2 siliques, 4.9% of seeds contained embryos arrested at the globular stage, 13.6% of seeds contained abnormal embryos, and 3.1% and 6.9% of seeds contained embryos delayed at the heart and torpedo stages, respectively (n = 1,561). In the smo2-1 smo2-2/+ siliques, 10.6% of seeds contained embryos arrested at the globular stage, 34% of seeds contained abnormal embryos, and 6.3% and 9.1% of seeds contained embryos delayed at the heart and torpedo stages, respectively (n = 635; Fig. 4I). The frequency of defective embryos in the smo2-1/+ smo2-2 plants was close to 25%, the expected rate for the homozygous smo2-1 smo2-2 double mutant. However, for smo2-1 smo2-2/+, the rate of defective embryos was much higher than expected. This segregation ratio distortion may have been due to the gametophytic defect of the smo2-1 smo2-2/+ plants.
Figure 4.
Phenotypes of arrested smo2-1 smo2-2 double mutant embryos. A to H, Differential interference contrast images of cleared seeds obtained from mature siliques of wild-type plants (A), self-pollinated smo2-1 smo2-2/+ plants (B–D), and self-pollinated smo2-1/+ smo2-2 plants (E–H). The smo2-1 smo2-2 double mutant embryos were arrested at the late globular/transition developmental stages (B, C, E, and F) and at the heart-like stage (D, G, and H). Bars = 50 μm (A) and 20 μm (B–H). I, Embryonic stage quantification of wild-type Columbia-0 (Col), smo2-1/+ smo2-2, and smo2-1 smo2-2/+ seeds from siliques 12 d after pollination. Note that embryos displaying an abnormal shape were counted as abnormal. In total, 1,631, 1,561, and 635 seeds were scored for the wild type, smo2-1/+ smo2-2, and smo2-1 smo2-2/+, respectively.
We then analyzed cleared seeds from siliques of the smo2-1 smo2-2/+ and smo2-1/+ smo2-2 mutants at different developmental stages. Consistent with the above observations, the segregated embryos of the smo2-1 smo2-2 double mutant grew more slowly than those of wild-type or single mutant siblings, but most of them still showed normal morphology until the late globular stage (Supplemental Fig. S8, A–D). After the late globular stage, the smo2-1 smo2-2 double mutant embryos arrested their development by failing to transition from the triangular to the heart stage or from the heart to the torpedo stage (Supplemental Fig. S8, E–J). This finding is in agreement with the expression patterns of SMO2 genes during embryonic development (Fig. 1, I–K; Supplemental Fig. S3). Together, these data indicate that loss of function of both SMO2 genes delays embryonic development and impairs the embryonic patterning and elongation processes.
The smo2-1 smo2-2 Double Mutant Is Defective in Endosperm Development
We also noticed that endosperm development was abnormal in seeds segregated from smo2-1 smo2-2/+ and smo2-1/+ smo2-2 siliques. In wild-type siliques, most seeds had properly proliferating endosperm nuclei, which were round and surrounded by a dense cytoplasm (Fig. 5, A–D; Table I). However, in the smo2-1 smo2-2/+ siliques, approximately 47% of seeds had defects in endosperm development. There were five types of seeds in the smo2-1 smo2-2/+ siliques at the early developmental stage. Type I seeds were as normal as wild-type seeds (approximately 53%; Supplemental Fig. S9, A and E). In type II seeds, although the embryo was normal, the endosperm proliferation was delayed, and fewer larger endosperm nuclei were observed (approximately 13%; Fig. 5, E and F; Table I; Supplemental Fig. S9, B and F). In type III seeds, both embryonic and endosperm development was delayed (approximately 28%; Supplemental Fig. S9, C and G). In type IV seeds, the endosperm nuclei were not round but showed a triangular shape, and the surrounding cytoplasm was not as dense as in the wild type but appeared collapsed and dotted instead (Fig. 5, G and H), suggesting that they were degenerating (approximately 11%). Type V seeds were embryoless (approximately 5%; Fig. 5I; Supplemental Fig. S9, D and H). In the smo2-1/+ smo2-2 seeds, the endosperm defects were similar to those of the smo2-1 smo2-2/+ mutant, except that there were substantially fewer embryoless seeds (approximately 1%) and a more prominent endosperm nuclear degeneration defect (approximately 32%; Fig. 5, J–L). These data indicate that the Arabidopsis SMO2 family is essential for endosperm nuclear proliferation and viability.
Figure 5.
Endosperm development is defective in smo2-1 smo2-2 embryo sacs. A to D, Wild-type seeds with normal proliferating endosperm nuclei in a one-cell embryo (A), four-cell embryo (B), eight-cell embryo (C), and globular embryo (D). E and F, smo2-1 smo2-2/+ seeds contain fewer and larger endosperm nuclei at the one-cell (E) and four-cell (F) embryo stages. G and H, smo2-1 smo2-2/+ seeds containing an eight-cell embryo and degenerating endosperm nuclei. Note that the endosperm nuclei are collapsed and dotted, rather than round. I, smo2-1 smo2-2/+ seeds containing proliferating endosperm nuclei but no embryo. J to L, smo2-1/+ smo2-2 mutant seeds containing a two-cell embryo and degenerating endosperm nuclei (J) and a globular-stage embryo and degenerating endosperm nuclei (K and L). Three experiments were performed, and seven to 10 siliques were observed for each genotype in each experiment. Bars = 50 μm.
Table I. Endosperm nuclear proliferation in early developmental stage ovules.
Siliques at 1 to 3 DPA were collected from wild-type, smo2-1 smo2-2/+, and smo2-1/+ smo2-2 plants and dissected. Seeds were cleared using chloral hydrate solution, and endosperm nuclei were scored using a differential interference contrast microscope. One to nine seeds from three to six siliques were examined. ND, Not determined.
| Genotype | Embryonic Developmental Stage |
||||
|---|---|---|---|---|---|
| 1 Cell | 2 Cell | 4 Cell | 16 Cell | 32 Cell | |
| Wild type | 26.1 ± 1.6 | 45 ± 2.1 | 73.8 ± 15.9 | 95.7 ± 8.7 | 129.8 ± 10 |
| smo2-1 smo2-2a | 14.5 ± 0.7 | 23.8 ± 6.2 | 37.3 ± 18.4 | 34.8 ± 19 | 49 |
| smo2-1 smo2-2b | ND | ND | 34 ± 3.7 | 38.1 ± 9.7 | 46.4 ± 11 |
smo2-1 smo2-2 double mutant seeds segregated from smo2-1 smo2-2/+ siliques.
smo2-1 smo2-2 double mutant seeds segregated from smo2-1/+ smo2-2 siliques.
The Auxin Response and PIN1 Auxin Efflux Facilitator Expression Are Impaired in smo2-1 smo2-2 Embryos
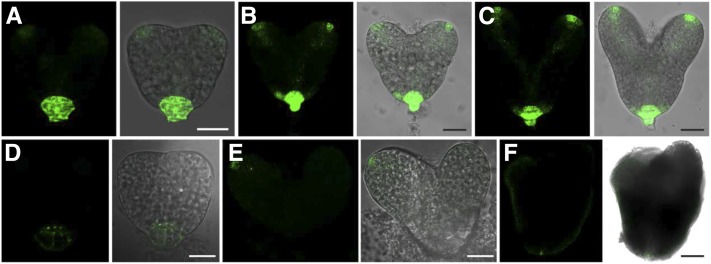
The patterning defects of the smo2-1 smo2-2 double mutant embryos suggest inappropriate auxin distribution. To address this possibility, we crossed DR5rev:GFP (an auxin-responsive reporter whose expression correlates with high free auxin levels; Friml et al., 2003) into the heterozygous smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants and analyzed the GFP signals in the segregating smo2-1 smo2-2 double mutant embryos. In triangular-stage wild-type embryos, the strongest signal of DR5rev:GFP was observed in the uppermost suspensor cell (hypophysis), and a strong signal also was detected in the two emerging cotyledon primordia (Fig. 6A). In heart-stage wild-type embryos, DR5rev:GFP expression was readily detected in the hypophysis, cotyledon primordial tips, and prevascular cells (Fig. 6, B and C). Compared with the wild type, smo2-1 smo2-2 embryos displayed weak and abnormal DR5rev:GFP expression (Fig. 6, D–F), suggesting reduced free auxin levels and abnormal auxin distribution. At the triangular stage, very weak DR5rev:GFP activity was observed in the hypophysis, and the auxin maxima indicated by DR5rev:GFP corresponding to the two emerging cotyledon primordia was absent (Fig. 6D). At the heart stage, the DR5rev:GFP signal in the hypophysis and the cotyledon primordia was either absent or very weak (Fig. 6, E and F). Sometimes, only one cotyledon primordium showed a DR5rev:GFP signal (Fig. 6E), which may have caused asymmetric cotyledon growth (Supplemental Fig. S7, B and D). At the late heart-like stage, polar auxin distribution at the cotyledons disappeared; instead, the DR5rev:GFP signal was evenly distributed throughout the entire protoderm (Fig. 6F).
Figure 6.
Expression of DR5rev:GFP during embryogenesis. A to C, DR5rev:GFP distribution in wild-type embryos at the triangular stage (A), early heart stage (B), and late heart stage (C). D to F, DR5rev:GFP distribution in embryos dissected from smo2-1 smo2-2/+ siliques at the triangular stage (D), early heart stage (E), and late heart-like stage (F). Bars = 25 μm (A and C–E) and 10 μm (B and F).
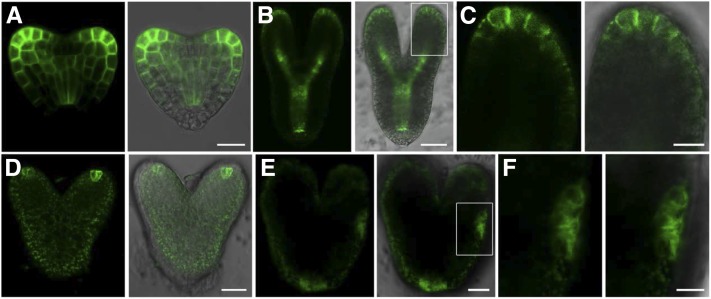
The correct establishment and maintenance of auxin maxima in Arabidopsis embryos rely on the polar localization of the PIN1 auxin efflux facilitator. Changes in the sterol component of the plasma membrane can lead to the incorrect localization of PIN proteins (Willemsen et al., 2003; Men et al., 2008). VIGS silencing of the SMO2 genes altered the quantitative and qualitative sterol profiles of the infected plants (Darnet and Rahier, 2004). Therefore, we speculated that loss of function of SMO2-1 and SMO2-2 might affect the correct localization of PIN1. To investigate this possibility, we examined PIN1-GFP (Benková et al., 2003) expression in the double mutant embryos. In wild-type embryos, a strong PIN1-GFP signal was observed at the tip of the developing cotyledons and the vascular initials (Fig. 7, A–C). However, in the segregated smo2-1 smo2-2 embryos, only a very weak PIN1-GFP signal was detected at the tip of the cotyledons (Fig. 7D) and abnormal distribution of PIN1 was observed (Fig. 7E). With a polar plasma membrane localization in the wild-type embryos (Fig. 7C), PIN1-GFP was internalized to the cytosol in the smo2-1 smo2-2 embryos (Fig. 7F), suggesting that the smo2-1 smo2-2 embryos were defective in the polar localization of PIN1. Taken together, these data indicate that reduced or absent auxin maxima and defective PIN1 expression/localization coincide with the developmental defects of the smo2-1 smo2-2 embryos.
Figure 7.
Expression of PIN1-GFP during embryogenesis. A and B, PIN1-GFP distribution in wild-type embryos at the heart stage (A) and the torpedo stage (B). C, Higher magnification image of the boxed region in B. D and E, PIN1-GFP distribution in embryos dissected from smo2-1/+ smo2-2 siliques at the heart stage (D) and the late heart-like stage (E). F, Higher magnification image of the boxed region in E. Bars = 10 μm (A, C, and F) and 25 μm (B, D, and E).
Exogenous Application of Auxin Partially Rescued smo2-1 smo2-2 Embryonic Lethality and the Root Growth Defects of the smo2-1 smo2-2/+ Mutant
The data described above demonstrated that reduced auxin levels and abnormal auxin distribution and transport were correlated with the smo2-1 smo2-2 embryonic patterning and elongation defects, and auxin is known to be essential for embryonic pattern formation and cell expansion (Friml et al., 2003). Therefore, we wondered whether auxin could rescue the smo2-1 smo2-2 embryonic patterning defects. We performed auxin application on pistils of smo2-1/+ smo2-2 and smo2-1 smo2-2/+ plants and examined the embryos 10 d later. Compared with the mock treatment, 1-naphthaleneacetic acid (NAA) application significantly reduced the embryo abortion rate of both the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ plants (Fig. 8), indicating that auxin application partially rescued the smo2-1 smo2-2 embryonic lethality.
Figure 8.
NAA partially rescues smo2-1 smo2-2 embryonic lethality. Average results of three experiments are shown. For each experiment, pistils of 10 opening flowers from five plants of each genotype were either treated with 300 μm NAA or underwent mock treatment, and embryos were scored 10 d after application. Note that embryos displaying an abnormal shape were counted as abnormal. Bars represent means ± sd. Bars in each column with different letters differ significantly (Student’s t test, P < 0.05). Col, Wild-type Columbia-0.
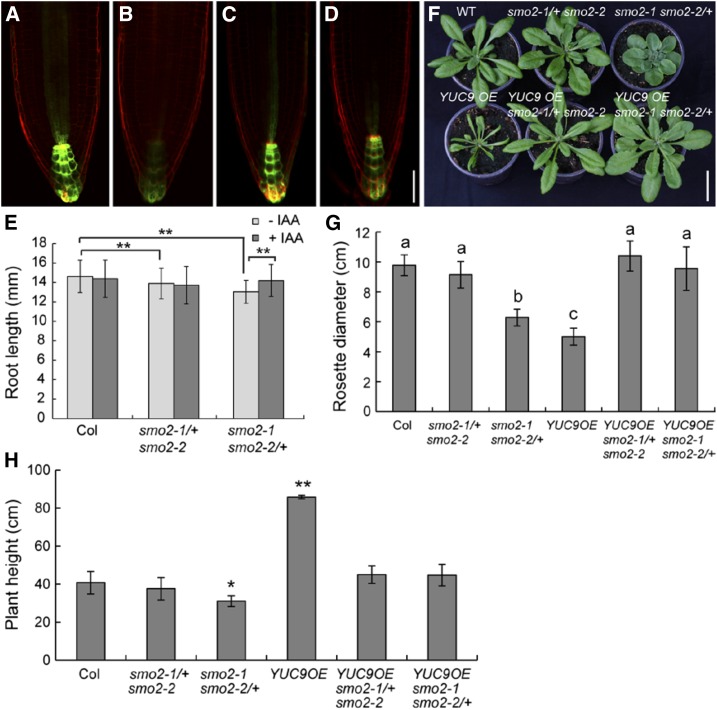
We also examined DR5rev:GFP expression in the smo2-1 smo2-2/+ roots. Compared with the wild type, smo2-1 smo2-2/+ roots showed a weak DR5rev:GFP signal (Fig. 9, A–D). Therefore, we also examined the effect of auxin on root growth in the smo2-1 smo2-2/+ mutant. When grown on MS medium with 50 nm indole-3-acetic acid (IAA), wild-type root growth was inhibited. However, this concentration of IAA promoted the growth of the smo2-1 smo2-2/+ mutant root (Fig. 9E), suggesting that, similar to the smo2-1 smo2-2 embryonic lethality, the root growth defects of the smo2-1 smo2-2/+ mutant also were due to an auxin defect.
Figure 9.
Exogenously applied auxin and endogenous auxin overproduction rescue smo2-1 smo2-2/+ mutant phenotypes. A and B, DR5rev:GFP expression in wild-type (A) and smo2-1 smo2-2/+ (B) seedling root tips grown on MS plates without IAA. C and D, DR5rev:GFP expression in wild-type (C) and smo2-1 smo2-2/+ (D) seedling root tips grown on MS plates with the addition of 50 nm IAA. Bar = 50 μm. E, Root lengths of 6-d-old wild-type Columbia-0 (Col), smo2-1/+ smo2-2, and smo2-1 smo2-2/+ seedlings grown on either MS plates (−IAA) or MS plus 50 nm IAA plates (+IAA); n = 32 to 60. Asterisks indicate significant differences (Student’s t test, P < 0.01). Note that the application of 50 nm IAA promoted smo2-1 smo2-2/+ root growth but inhibited wild-type and smo2-1/+ smo2-2 root growth. F, Rosette phenotypes of 4-week-old plants. WT, Wild type. Bar = 3 cm. G, Quantification of rosette diameter of 4-week-old plants. Bars in each column with different letters differ significantly (Student’s t test, P < 0.05). H, Height of mature plants. *, P < 0.05; and **, P < 0.01 (Student’s t test). The data presented in G and H represent means ± sd of three experiments; n = 10 for each experiment.
Endogenous Auxin Overproduction Partially Rescued smo2-1 smo2-2 Embryonic Lethality and Fully Rescued smo2-1 smo2-2/+ Mutant Phenotypes
Previous studies in Arabidopsis have indicated that YUC9 OE contributes to the overproduction of free IAA (Hentrich et al., 2013). To examine the effects of endogenously overproduced auxin on smo2 double mutants, we crossed YUC9 OE transgenic plants with smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants. To our surprise, the dwarf phenotype of the smo2-1 smo2-2/+ heterozygous double mutant was fully rescued by YUC9 OE (Fig. 9, F–H). Because root growth is sensitive to auxin levels, the YUC9 OE seedlings had very short roots with long root hairs (Supplemental Fig. S10). However, the roots of both the smo2-1/+ smo2-2 YUC9 OE and smo2-1 smo2-2/+ YUC9 OE seedlings were markedly longer than those of YUC9 OE itself (Supplemental Fig. S10). These results suggest that loss of function of SMO2s in Arabidopsis reduces auxin levels in the root, which is consistent with the weak DR5rev:GFP signal detected in these mutants (Figs. 6 and 9, A–D).
To explore whether endogenous auxin overproduction could rescue smo2-1 smo2-2 embryonic lethality, we examined siliques from both smo2-1 smo2-2/+ YUC9 OE and smo2-1/+ smo2-2 YUC9 OE plants. Compared with an average of approximately 27 seeds in siliques of the smo2-1 smo2-2/+ mutant, there were approximately 32 seeds in siliques of the smo2-1 smo2-2/+ mutant when the YUC9 gene was overexpressed (Supplemental Fig. S11A). Although the embryonic-lethal phenotype of the smo2-1 smo2-2 double mutant was not fully rescued, YUC9 OE significantly reduced the embryo abortion rate of both the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ plants (Supplemental Fig. S11, B and C). These data indicate that endogenous auxin overproduction also partially rescued the smo2-1 smo2-2 embryonic lethality.
smo2-1/+ smo2-2 and smo2-1 smo2-2/+ Mutants Accumulate the 4α-Methylsterols 24-Ethylidene Lophenol and 24-Ethyl Lophenol
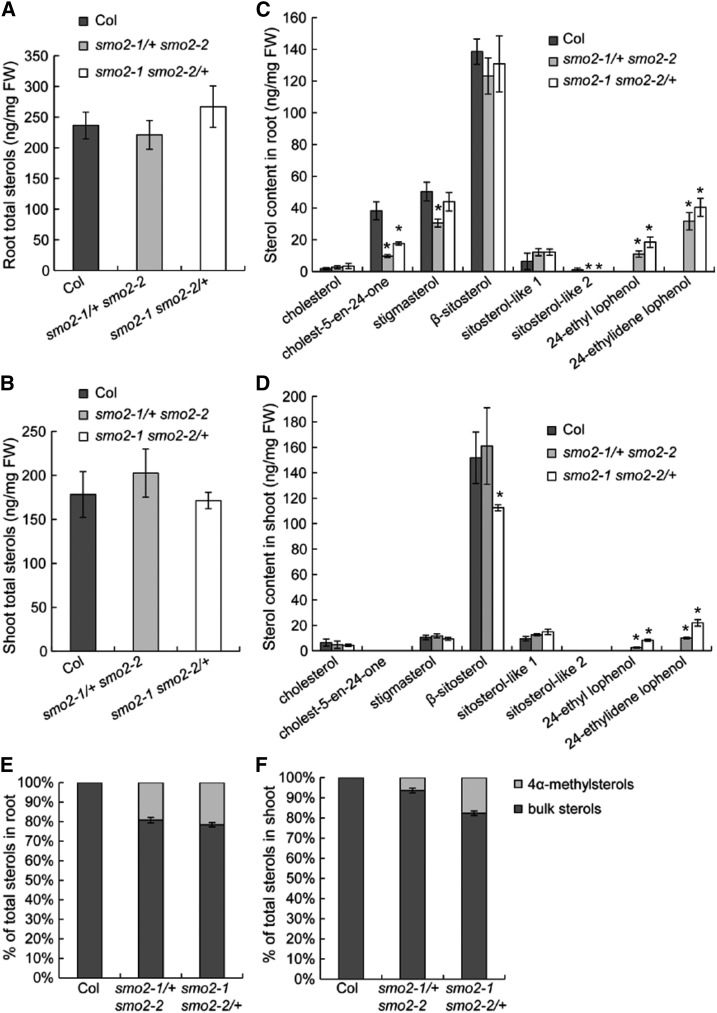
To determine whether sterol profiles were affected in the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants, we analyzed the sterol composition in roots and shoots of 8-d-old wild-type, smo2-1/+ smo2-2, and smo2-1 smo2-2/+ seedlings. Our results indicated that the total quantity of the pool constituted by the different sterols was not altered significantly in either the roots or shoots of the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants compared with the wild type (Fig. 10, A and B). However, the sterol compositions in both the roots and shoots of the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants were profoundly altered. We found that the amounts of the bulk sterols, including β-sitosterol and stigmasterol, varied according to mutants and organs. In smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutant roots, there was no decrease in sitosterol, although there was a decrease in stigmasterol in the smo2-1/+ smo2-2 mutant (Fig. 10C). In shoots, there was a decrease in sitosterol, but not stigmasterol, in the smo2-1 smo2-2/+ mutant (Fig. 10D). We also detected cholest-5-en-24-one in roots, and there was a decrease in this sterol in both the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants (Fig. 10C). These results indicate that the quantity of bulk sterols is decreased in the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants but that, depending on the organs (roots and shoots), the shuffling toward one or the other bulk sterol might be different. More strikingly, we measured an accumulation of the 4α-methylsterols 24-ethylidene lophenol and 24-ethyl lophenol in both roots and shoots of the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants compared with the wild type (Fig. 10, C and D; Supplemental Fig. S12). Given that the total quantity of sterols is not modified in the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants, we could conclude that the accumulation of 4α-methylsterols in these mutants represent around 20% of the total quantity of sterols found compared with the wild type (Fig. 10, E and F). These sterols are substrates for the SMO2 enzyme (Supplemental Fig. S1) and were not detected in wild-type samples under our experimental conditions. Furthermore, 24-ethyl lophenol was accumulated to a lesser extent than 24-ethylidene lophenol in both the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants (Fig. 10, C and D; Supplemental Fig. S12). This result likely reflects the substrate preference of SMO2 enzymes. These results also indicate that the accumulation of both 24-ethylidene lophenol and 24-ethyl lophenol varied according to organs. The roots of smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants contained higher quantities than the shoots (Fig. 10, C and D; Supplemental Fig. S12). These variations in substrate accumulation were likely due to the different expression patterns of the SMO2-1 and SMO2-2 genes in roots and shoots (Fig. 1).
Figure 10.
Sterol contents of wild-type Columbia-0 (Col), smo2-1/+ smo2-2, and smo2-1 smo2-2/+ seedlings. Samples were prepared from roots and shoots of 8-d-old seedlings grown on one-half-strength MS and 1% Suc plates and were analyzed by gas chromatography-mass spectrometry (GC-MS). A and B, Total sterol contents in roots (A) and shoots (B). C and D, Composition analysis of sterols found in roots (C) and shoots (D). E and F, Percentage of bulk sterols and 4α-methylsterols in the total pool of root (E) and shoot (F) sterols. The data represent means of four independent experiments, and error bars indicate sd. Statistically significant changes are indicated by asterisks (P < 0.05 by Student’s t test). FW, Fresh weight. Sitosterol-like 1 and 2 are likely α- and γ-sitosterol.
Together, our data suggest that the cause of the smo2-1 smo2-2/+ mutant dwarf phenotype may have been the accumulation of 24-ethylidene lophenol and 24-ethyl lophenol (which may be deleterious to Arabidopsis) or the decrease of bulk sterols. In an attempt to identify a possible role of 24-ethylidene lophenol and 24-ethyl lophenol, we tested the effects of 24-ethylidene lophenol (BioBioPha) on wild-type and YUC9 OE seedlings. We found that 24-ethylidene lophenol neither inhibited root elongation in the wild-type seedlings nor suppressed the short-root phenotype of the YUC9 OE seedlings (Supplemental Fig. S13). However, it is difficult to draw conclusions based on these experiments, since we do not know whether Arabidopsis seedlings have integrated the 24-ethylidene lophenol in their membranes. Because we were unable to obtain 24-ethyl lophenol, we could not test its effects on Arabidopsis growth. In any case, modification of the sterol composition observed in smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants is probably responsible for the observed phenotype, because the proper membrane sterol composition has already been shown to be essential for correct PIN auxin efflux facilitator positioning and auxin-dependent developmental processes (Souter et al., 2002; Willemsen et al., 2003; Men et al., 2008; Pullen et al., 2010).
DISCUSSION
SMO2-1 and SMO2-2 Play Unequally Redundant Roles in Plant Development
SMO2-1 and SMO2-2 share 91% amino acid sequence identity, and both can rescue the yeast erg25 mutant (Darnet et al., 2001; Darnet and Rahier, 2004), suggesting that they exert similar functions in yeast. Consistently, we observed no obvious phenotype in single homozygous smo2-1 or smo2-2 mutants, but the smo2-1 smo2-2 double mutant was embryonic lethal, indicating that SMO2-1 and SMO2-2 share essential but redundant functions in Arabidopsis. Our study also revealed unequally redundant roles for SMO2-1 and SMO2-2 in plant development. The smo2-1/+ smo2-2 mutant showed moderate phenotypes in vegetative tissues and displayed no fertility defect, whereas the smo2-1 smo2-2/+ mutant was dwarf and had gametophytic defects. These observations indicate that, in the absence of SMO2-2, a single copy of SMO2-1 is sufficient for vegetative growth and gametogenesis, whereas a single copy of SMO2-2 is insufficient in the absence of SMO2-1. This unequal redundancy may be due to the differences in the expression patterns and transcriptional levels between SMO2-1 and SMO2-2. SMO2-1 is broadly expressed at high levels, whereas the expression of SMO2-2 is lower and restricted (Fig. 1).
SMO2s Are Essential for Embryogenesis
Although previous studies clearly revealed that SMO2s function biochemically as 4α-methyl-∆7-sterol-4α-methyl oxidases (Darnet et al., 2001; Darnet and Rahier, 2004), the physiological functions of SMO2s remain elusive. Silencing of SMO2s in Nicotiana benthamiana using the VIGS approach profoundly altered the sterol profiles of the infected plants but caused no phenotype (Darnet and Rahier, 2004). This result may have occurred because the expression level of the SMO2 genes had not been reduced sufficiently. Small interfering RNA-mediated gene-silencing methods, such as VIGS, may not be efficient in knocking down sterol biosynthesis pathway genes, because a previous study found that intact sterol biosynthesis is required for the activity of plant small interfering RNAs (Brodersen et al., 2012). A recent study of CYCLOARTENOL SYNTHASE (CAS) also found that the expression of the CAS gene was only moderately reduced (approximately 35%) by the use of artificial microRNA (Gas-Pascual et al., 2015). In this respect, SMO2 gene knockout mutants should provide important clues to elucidate the developmental roles of SMO2s. In this study, we isolated two smo2-1 T-DNA insertion mutant alleles and one smo2-2 T-DNA insertion allele. We found that single smo2 mutants displayed no obvious phenotypic defects (Fig. 2; Supplemental Fig. S5), but the smo2-1 smo2-2 double mutant was embryonic lethal (Figs. 3 and 4) and the smo2-1 smo2-2/+ heterozygous double mutant was dwarf (Fig. 2). The development of smo2-1 smo2-2 double mutant embryos was arrested during the transition from the globular to the heart stage or from the heart to the torpedo stage (Fig. 4; Supplemental Fig. S8), indicating that SMO2s are essential for embryonic development and patterning. Consistent with their roles in embryogenesis, both SMO2 genes are expressed in embryos, with SMO2-2 expressed at lower levels than SMO2-1 (Fig. 1, I–K; Supplemental Fig. S3).
Previous studies in Arabidopsis indicated that mutations of genes in early sterol biosynthetic steps (i.e. upstream of HYDRA1, which encodes a sterol C-8,7-isomerase) produced embryonic defects, whereas downstream mutations did not (Clouse, 2002; Souter et al., 2002). Downstream mutants, such as dwf7/ste1, dwf5, and dwf1/dim, displayed phenotypes similar to BR-deficient mutants and could be rescued by BR application (Klahre et al., 1998; Choe et al., 1999a, 1999b, 2000). By contrast, upstream mutants, such as smt1/cph, fk, and hyd1, could not be rescued by either end-pathway regular sterols or BR (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002). These data suggest that a steroid molecule, rather than BR, is required for embryogenesis, and this yet-unidentified sterol could be an SBI derived from one of the biosynthetic steps between HYDRA1 and DWF7. Based on our studies, we propose that, if a steroid molecule is required for embryogenesis, it may be an SBI derived from the SMO2-catalyzed reaction (Fig. 11). The following lines of evidence support our speculation. First, our data showed that loss of function of both SMO2s, which catalyze the reaction step immediately after HYDRA1 (Supplemental Fig. S1), also caused embryonic defects. Furthermore, the phenotypes of smo2-1 smo2-2 double mutant embryos (Fig. 4; Supplemental Figs. S7 and S8) were very similar to those of the fk, hyd1, and smt1/cph mutants (Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002). Second, a previous study showed that loss of function of both Arabidopsis CSDs, which are directly downstream of SMO2 (Supplemental Fig. S1), caused no phenotype. However, overexpression of either CSD caused growth defects (Kim et al., 2012). Because CSDs catalyze the decarboxylation of SMO2 products (4α-carboxysterols), overexpression of CSDs may cause a reduction in these molecules, leading to growth defects. Third, CMMC, which is the product of SMO1, has been demonstrated to inhibit PAT when applied to wild-type Arabidopsis plants (Mialoundama et al., 2013). Notably, SMO2 and SMO1 both belong to the sterol C-4 demethylase multienzyme complex, which catalyzes the removal of C-4 methyl groups (Rahier, 2011), and smo2-1 smo2-2 double mutant embryos had auxin-related patterning defects. Therefore, it is plausible that one of the SMO2 products may similarly affect PAT and cause PAT-related embryonic patterning defects. Future studies will focus on identifying the specific SMO2-related SBI and evaluating its activities. However, we cannot exclude the possibility that the accumulation of 24-ethylidene lophenol and/or 24-ethyl lophenol was deleterious to Arabidopsis embryos, although we demonstrated that 24-ethylidene lophenol was not deleterious to Arabidopsis seedlings. It is also possible that the embryogenesis defects of these upstream sterol mutants are due to a basic requirement for wild-type membrane sterol composition for proper PIN auxin efflux facilitator positioning and auxin-dependent gene expression. Several reports have emphasized the importance of correct membrane sterol composition for PIN polar localization and auxin signaling (Jang et al., 2000; Schrick et al., 2000, 2002; Souter et al., 2002; Willemsen et al., 2003; Men et al., 2008). However, the results from the cpi1-1 mutant do not support this view. Although sterol composition was profoundly altered in the cpi1-1 mutant, cpi1-1 embryos displayed no obvious defect (Men et al., 2008).
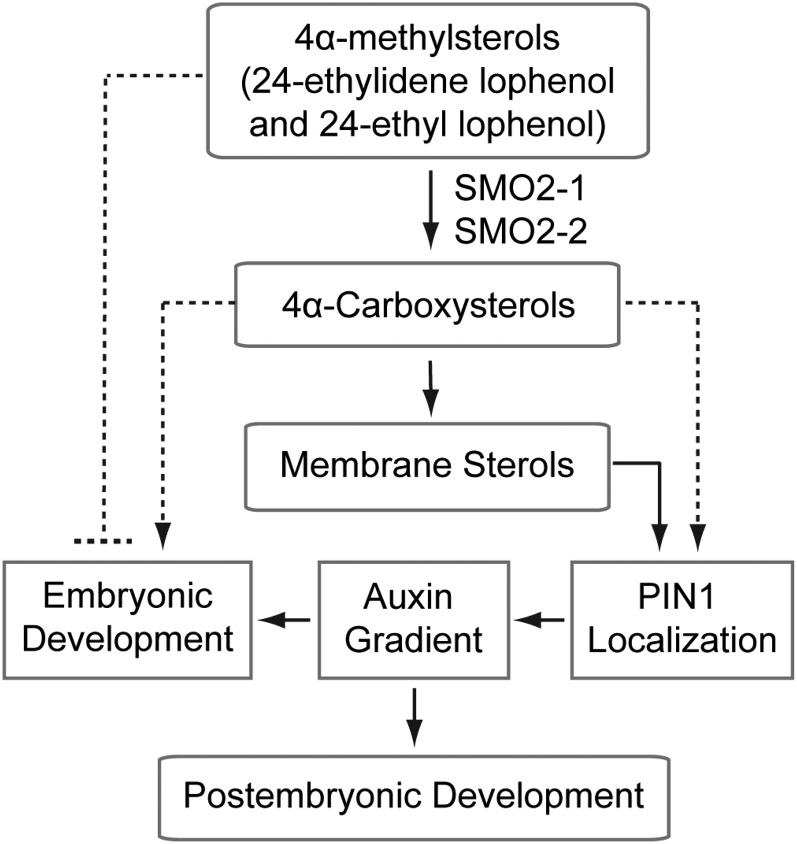
Figure 11.
Model of the functions of SMO2 proteins in Arabidopsis embryonic and postembryonic development. The SMO2-1 and SMO2-2 enzymes catalyze the conversion of 4α-methylsterols (mainly 24-ethylidene lophenol and 24-ethyl lophenol) to 4α-caboxysterols, which are further converted into membrane sterols. Normal membrane sterol composition is required for correct PIN1 auxin efflux facilitator positioning and PIN1-dependent auxin gradient formation, which are essential for embryonic and postembryonic development. It is also possible that 4α-caboxysterols are needed for PIN1 localization and embryonic development and that 24-ethylidene lophenol and 24-ethyl lophenol are deleterious to Arabidopsis embryos.
SMO2s Are Required for Endosperm Development
Plant sterols play important roles in reproductive developmental processes such as embryogenesis and male fertility (Clouse, 2002; Schaller, 2003; Suzuki et al., 2004, 2009). However, it has not been shown that sterols are required for endosperm development. In this study, we found that endosperm nuclear proliferation was defective in smo2-1 smo2-2 double mutant seeds. Compared with the wild type, there were significantly fewer endosperm nuclei in the smo2-1 smo2-2 double mutant early-developing seeds (Fig. 5; Table I), indicating that SMO2s are required for endosperm nuclear division. Consistent with these observations, SMO2-1 was expressed in the endosperm at high levels (Fig. 1). Although previous studies did not observe endosperm developmental defects in upstream sterol mutants, they found cell division defects in embryos and roots (Schrick et al., 2000; Souter et al., 2002; Carland et al., 2010). Detailed analyses of endosperm development in those upstream mutants will clarify whether the endosperm nuclear proliferation defects are specific to the smo2-1 smo2-2 double mutant.
SMO2s Function Partially through Effects on Auxin Accumulation and PIN1 Expression
Previous studies have shown profound interactions between sterols and the auxin pathway. On the one hand, the expression of sterol biosynthetic pathway genes such as FK was up-regulated by auxin (He et al., 2003), concentrations of the major plant sterols were significantly reduced in the axr1-12 auxin signaling mutant (Pan et al., 2009), and sterol mutants such as fk and cvp1 displayed altered responses to auxin (Souter et al., 2002; Carland et al., 2010). On the other hand, sterols were required for auxin signaling and polar transportation. The polar localization of auxin efflux carriers such as PIN1, PIN2, and PIN3 was impeded in sterol biosynthetic mutants, including smt1, cpi1-1, hyd1, and fk (Willemsen et al., 2003; Men et al., 2008; Pullen et al., 2010). Further analysis showed that sterol-dependent endocytosis was required for the postcytokinetic establishment of PIN2 polarity (Men et al., 2008). Additionally, sterols were found to be essential for auxin-inhibited PIN2 protein endocytosis (Pan et al., 2009; Carland et al., 2010). A previous study found that sterols also are required for the trans-Golgi network-to-plasma membrane trafficking of the ATP-binding cassette B19 auxin transporter (Yang et al., 2013). However, it remains unknown whether a specific sterol molecule contributes to these functions. A recent study found that CMMC, an SBI derived from the first C-4 demethylation step, was accumulated in erg28 mutants; CMMC can inhibit PAT when applied to wild-type Arabidopsis plants (Mialoundama et al., 2013). However, CMMC was not detectable in wild-type plants, and its role in plants is unclear. Our results showed that loss of function of both SMO2 proteins, which catalyze the second C-4 demethylation step, caused embryonic defects characteristic of auxin-deficient mutants (Fig. 4; Supplemental Figs. S7 and S8); auxin responses and PIN1 expression were indeed abnormal in smo2-1 smo2-2 embryos (Figs. 6 and 7).
In agreement with these observations, the embryonic-lethal phenotype of smo2-1 smo2-2 double mutants was partially rescued by either exogenously applied auxin or endogenously overproduced auxin (YUC9 OE; Fig. 8; Supplemental Fig. S11). Moreover, the developmental defects of the smo2-1 smo2-2/+ heterozygous double mutant were completely rescued by YUC9 OE (Fig. 9, F–H). On the other hand, the short-root phenotype of YUC9 OE seedlings was suppressed by the smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutations (Supplemental Fig. S10). A sterol analysis showed that the sterol composition, rather than the sterol quantity, was altered in the smo2 heterozygous double mutants, with the major characteristic being an accumulation by 20%, compared with the wild type, of the 4α-methylsterols 24-ethylidene lophenol and 24-ethyl lophenol, which are substrates of the SMO2 enzyme. We further demonstrated that 24-ethylidene lophenol, the major substrate of SMO2 (Supplemental Fig. S12), had no effects on the root growth of either wild-type or YUC9 OE seedlings (Supplemental Fig. S13). However, this last result has to be considered with caution, due to the absence of evidence regarding whether 24-ethylidene lophenol was effectively integrated in membranes of Arabidopsis cells. From our results, it could equally be that the diminution of bulk sterols (decreases of β-sitosterol in shoots of the smo2-1 smo2-2/+ mutant and stigmasterol in roots of the smo2-1/+ smo2-2 mutant) caused the observed phenotypes. Although we cannot distinguish whether the accumulation of 4α-methylsterol precursors or the decrease of bulk sterols causes the phenotype, it appears clear that sterol composition rather than sterol quantity is responsible for the phenotype, since we did not detect any modification of global sterol quantity in our mutants. Hence, our study specifically indicates that the balance between bulk sterols and 4α-methylsterol precursors, for the same quantity of sterols, is critical for plant development.
In summary, we demonstrated that SMO2 family genes are essential for Arabidopsis embryogenesis and postembryonic development. Our data reveal that SMO2s are important for correct sterol composition, but not quantity, and function partially through effects on auxin accumulation, auxin response, and PIN1 expression to regulate embryonic and postembryonic development.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown either on MS medium or in soil in a greenhouse with a 16-h-light/8-h-dark cycle at 22°C. Prior to in vitro culture, seeds were surface sterilized for 5 min in 70% (v/v) ethanol and for 10 min in 1% (v/v) Clorox bleach before being washed three times with sterile water. The T-DNA insertion lines SALK_105017 (smo2-1.1) and SALK_030719 (smo2-2) were obtained from the Nottingham Arabidopsis Stock Centre, and FLAG_522B02 (smo2-1.2) was obtained from the Versailles Arabidopsis Stock Center. The DR5rev:GFP (Friml et al., 2003) and PIN1-GFP (Benková et al., 2003) lines were crossed with the smo2 single and double mutants.
Chemicals
NAA, IAA, and 24-epibrassinolide were obtained from Sigma-Aldrich. Commercial 24-ethylidene lophenol, known as citrostadienol, was purchased from BioBioPha.
Plasmid Construction and Plant Transformation
To create the ProSMO2-1:GUS and ProSMO2-2:GUS constructs, 742 bp of DNA upstream of the Arabidopsis SMO2-1 gene start codon and 992 bp of DNA upstream of the SMO2-2 gene start codon were PCR amplified and inserted into pGreenII0229-GUS (Men et al., 2008) upstream of the GUS gene. Then, 1,117 bp of DNA downstream of the SMO2-1 stop codon and 642 bp of DNA downstream of the SMO2-2 stop codon were PCR amplified and cloned into the above constructed vectors downstream of the GUS gene.
To generate the SMO2-1-EGFP and SMO2-2-EGFP constructs, the EGFP coding sequence was inserted into the pGreenII0229 vector (from the John Innes Centre; http://www.pgreen.ac.uk). Then, genomic sequences including the 5′ promoter, exons, and introns were amplified. The sequence lengths were 2,863 bp for SMO2-1 (nucleotides −742 to 2,121 from ATG) and 3,021 bp for SMO2-2 (nucleotides −992 to 2,039 from ATG). The PCR products were cloned into the pGreenII0229-EGFP construct. Finally, the terminator sequences, 1,117 bp of sequence downstream of the SMO2-1 stop codon and 642 bp of sequence downstream of the SMO2-2 stop codon, also were cloned into the above constructed pSMO2-1-EGFP and pSMO2-2-EGFP, respectively. The primer sequences are listed in Supplemental Table S1.
All of the constructed vectors were introduced into Agrobacterium tumefaciens strain C58C1 (pMP90/pJIC Sa-Rep) and transformed into Columbia-0 plants by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected by spraying with 0.001% (v/v) Basta and PCR amplification. Analyses were performed on segregating T2 or homozygous T3 lines from several independent transformants.
Histology and Microscopy
Seeds were cleared for visualization using a clearing solution of 8:3:1 chloral hydrate:distilled water:glycerol as described by Men et al. (2008). Then, the seeds were placed on a glass slide in a drop of clearing solution and were covered with a coverslip. Observations were performed with an Olympus BX63 microscope under differential interference contrast.
GUS Histochemical Staining
GUS activity was analyzed by staining various tissues of ProSMO2-1:GUS transgenic plants for 4 h and staining tissues of ProSMO2-2:GUS plants for 36 h at 37°C in staining solution (1.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-GlcA, 50 mm sodium phosphate buffer, pH 7, 0.1% (v/v) Triton X-100, 0.5 mm potassium ferricyanide, and 0.5 mm potassium ferrocyanide). Ten independent transgenic lines were analyzed, and they all showed similar patterns. Samples were examined using a Leica M165FC dissection microscope and an Olympus BX63 microscope.
Immunocytochemistry and Confocal Laser Scanning Microscopy
Whole-mount immunofluorescence was performed on roots of 5-d-old ProSMO2-1:SMO2-1-EGFP and ProSMO2-2:SMO2-2-EGFP transgenic plants as described previously (Men et al., 2008). Two different transgenic lines for each construct were used, and the immunolocalization experiments were performed in two independent sessions, each involving 10 to 15 roots. A mouse monoclonal anti-BiP (an ER-intrinsic chaperone used as an ER marker) antibody (Enzo Life Sciences) was used at a 1:1,000 dilution. A donkey anti-mouse tetramethylrhodamine-coupled secondary antibody (Jackson ImmunoResearch) was used at a 1:200 dilution.
Fluorescence Detection of Embryos
Wild-type and mutant embryos expressing PIN1-GFP or DR5rev:GFP were dissected from seeds at different developmental stages and mounted in an 8% (v/v) glycerol solution. Signals were detected by confocal laser scanning microscopy (Leica TCS SP5).
RT-PCR
For RT-PCR of SMO2-1 and SMO2-2 loss-of-function mutants, total RNA was extracted from 6-d-old seedlings using Trizol reagent. Total RNA (1 μg) was digested with RNase-free DNase (TaKaRa) and was reverse transcribed using EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech). A 1-μL aliquot of the synthesized complementary DNA was used as a template for PCR. The ACTIN2 gene was used as an internal control for equal loading. The primer sequences are listed in Supplemental Table S1.
Auxin Application
To examine the effects of auxin on smo2-1 smo2-2 embryonic development, opening flowers were dipped in a 300 μm NAA solution for a few seconds, and embryos were scored 10 d after treatment. To examine the effect of auxin on root growth, wild-type and mutant seeds were germinated on plates containing MS medium supplemented with 50 nm IAA and were cultured for 5 d; then, the root length was scored. IAA and NAA were first dissolved in 70% (v/v) ethanol. A mock treatment was performed using either distilled water or MS medium supplemented with an equal volume of 70% (v/v) ethanol.
Sterol Measurements
For sterol characterization, Arabidopsis roots and cotyledons were scaled and ground in a Potter-Elvehjem glass device in 2 mL of chloroform:methanol (2:1; implemented with the standard 25 µg of α-cholestanol in 2 mL) for 2 h at room temperature. The lipid extract was then washed with 1 mL of 0.9% (w/v) NaCl, shaken vigorously, and centrifuged at 700g for 5 min at room temperature. The organic (lower) phase was harvested and evaporated. Then, saponification was performed on the lipid extract by incubating it with 1 mL of 99% (v/v) ethanol and 100 µL of 11 n KOH for 1 h at 80°C. After incubation, 1 mL of 99% (v/v) hexane and 2 mL of water were added. After vigorous shaking and centrifugation at 700g for 5 min at room temperature, the upper phase was harvested, placed in a new tube, and buffered with 1 mL of 100 mm Tris, 0.09% (w/v) NaCl, pH 8, with HCl. After evaporation, the sterols were incubated with 200 µL of N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA) + 1% (v/v) trimethyl chlorosilane (TMCS) at 110°C for 20 min. After evaporation, the sterols were resuspended in 100 µL of 99% (v/v) hexane and analyzed using GC-MS. The fragmentation spectrum of 24-ethylidene lophenol was characterized by injecting commercially available 24-ethylidene lophenol (BioBioPha) into the GC-MS device. For 24-ethylidene lophenol, the main mass-to-charge ratio peaks (after BSTFA + TMCS treatment) were at 357 and 400. Concerning 24-ethyl lophenol, its mass was identified (after BSTFA + TMCS treatment) at 500, which correspond to its real mass (427) plus a trimethylsilyl (73). The fragmentation spectrum of 24-ethyl lophenol gave two main mass-to-charge ratio peaks at 269 and 500, which correspond to what was published before (Darnet and Rahier, 2004).
Sequence data from this article can be found in the GenBank/EMBL or Arabidopsis Genome Initiative data libraries under the following accession numbers: SMO2-1 (AT1G07420) and SMO2-2 (AT2G29390). T-DNA insertions in the SMO2-1 gene were smo2-1.1 (SALK_105017) and smo2-1.2 (FLAG_522B02) and in the SMO2-2 gene was smo2-2 (SALK_030719).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sterol biosynthetic pathway in higher plants.
Supplemental Figure S2. Sequence alignment and protein topology of SMO2 proteins.
Supplemental Figure S3. Relative transcript levels of the SMO2-1 and SMO2-2 genes in developing Arabidopsis seeds and embryos.
Supplemental Figure S4. ER localization of the SMO2-1 and SMO2-2 proteins.
Supplemental Figure S5. Phenotypes of smo2 mutants.
Supplemental Figure S6. smo2-1/+ smo2-2 and smo2-1 smo2-2/+ complementation experiments.
Supplemental Figure S7. Embryos of smo2-1 smo2-2/+ plants have different cotyledon sizes.
Supplemental Figure S8. Embryonic development of smo2-1 smo2-2/+ mutants.
Supplemental Figure S9. Embryonic and endosperm defects of smo2-1 smo2-2/+ seeds.
Supplemental Figure S10. smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants suppressed the short-root phenotype of YUC9 OE seedlings.
Supplemental Figure S11. YUC9 OE partially rescues smo2-1 smo2-2 embryonic lethality.
Supplemental Figure S12. Accumulation of SMO2 substrates in smo2-1/+ smo2-2 and smo2-1 smo2-2/+ mutants.
Supplemental Figure S13. The SMO2 substrate 24-ethylidene lophenol has no effect on root growth of either wild-type or YUC9 OE seedlings.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Jirí Friml for DR5rev:GFP and PIN1-GFP seeds, Stephan Pollmann for the YUC9 OE line, the Nottingham Arabidopsis Stock Centre and the Versailles Arabidopsis Stock Center for the T-DNA insertions, the John Innes Centre for the pGREENII0229 vector, Jiehua Wang for helpful discussions, and Ruming Liu for technical assistance in the use of the confocal laser scanning microscope.
Glossary
- BR
brassinosteroid
- PAT
polar auxin transport
- SBI
sterol biosynthetic intermediate
- CMMC
4-carboxy-4-methyl-24-methylenecycloartanol
- VIGS
virus-induced gene silencing
- RT
reverse transcription
- ER
endoplasmic reticulum
- T-DNA
transfer DNA
- NAA
naphthylacetic acid
- MS
Murashige and Skoog
- IAA
indole-3-acetic acid
- BSTFA
N,O-bis(trimethylsilyl) trifluoroacetamide
- TMCS
trimethyl chlorosilane
- GC-MS
gas chromatography-mass spectrometry
Footnotes
This work was supported by the National Science Foundation of China (grant nos. 31570247, 91417308, 91017009, and 31460453), the Natural Science Foundation of Tianjin (grant no. 12JCZDJC23200), the 111 Project (grant no. B08011), and the Ph.D. Candidate Research Innovation Fund of Nankai University.
References
- Bard M, Bruner DA, Pierson CA, Lees ND, Biermann B, Frye L, Koegel C, Barbuch R (1996) Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc Natl Acad Sci USA 93: 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Benveniste P. (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Biol 55: 429–457 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Frescatada-Rosa M, Men S, Chow CM, Ebine K, Gustavsson A, Johansson L, Ueda T, Moore I, Jürgens G, et al. (2010) Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J 29: 546–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Schaller H, Khafif M, Schott G, Bendahmane A, Voinnet O (2012) Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc Natl Acad Sci USA 109: 1778–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland F, Fujioka S, Nelson T (2010) The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol 153: 741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, et al. (1999a) The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol 119: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, et al. (1999b) The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11: 207–221 [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA (2000) Lesions in the sterol ∆7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J 21: 431–443 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD. (2002) Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 14: 1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnet S, Bard M, Rahier A (2001) Functional identification of sterol-4α-methyl oxidase cDNAs from Arabidopsis thaliana by complementation of a yeast erg25 mutant lacking sterol-4α-methyl oxidation. FEBS Lett 508: 39–43 [DOI] [PubMed] [Google Scholar]
- Darnet S, Rahier A (2004) Plant sterol biosynthesis: identification of two distinct families of sterol 4α-methyl oxidases. Biochem J 378: 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gas-Pascual E, Simonovik B, Schaller H, Bach TJ (2015) Inhibition of cycloartenol synthase (CAS) function in tobacco BY-2 cells. Lipids 50: 761–772 [DOI] [PubMed] [Google Scholar]
- He JX, Fujioka S, Li TC, Kang SG, Seto H, Takatsuto S, Yoshida S, Jang JC (2003) Sterols regulate development and gene expression in Arabidopsis. Plant Physiol 131: 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Kratz LE, Michel JJ, Vallejo AN, Ferris L, Kelley RI, Hoover JJ, Jukic D, Gibson KM, Wolfe LA, et al. (2011) Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J Clin Invest 121: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich M, Böttcher C, Düchting P, Cheng Y, Zhao Y, Berkowitz O, Masle J, Medina J, Pollmann S (2013) The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J 74: 626–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J (2000) A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev 14: 1485–1497 [PMC free article] [PubMed] [Google Scholar]
- Kim B, Kim G, Fujioka S, Takatsuto S, Choe S (2012) Overexpression of 3β-hydroxysteroid dehydrogenases/C-4 decarboxylases causes growth defects possibly due to abnormal auxin transport in Arabidopsis. Mol Cells 34: 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Lee H, Oh CJ, Lee HY, Eum HL, Kim HS, Hong YP, Lee Y, Choe S, An CS, et al. (2010) Postembryonic seedling lethality in the sterol-deficient Arabidopsis cyp51A2 mutant is partially mediated by the composite action of ethylene and reactive oxygen species. Plant Physiol 152: 192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Schaller H, Goh CH, Kwon M, Choe S, An CS, Durst F, Feldmann KA, Feyereisen R (2005) Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol 138: 2033–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH (1998) The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10: 1677–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SW, Glover BJ, Davies JM (2005) Lipid microdomains: plant membranes get organized. Trends Plant Sci 10: 263–265 [DOI] [PubMed] [Google Scholar]
- Men S, Boutté Y, Ikeda Y, Li X, Palme K, Stierhof YD, Hartmann MA, Moritz T, Grebe M (2008) Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol 10: 237–244 [DOI] [PubMed] [Google Scholar]
- Mialoundama AS, Jadid N, Brunel J, Di Pascoli T, Heintz D, Erhardt M, Mutterer J, Bergdoll M, Ayoub D, Van Dorsselaer A, et al. (2013) Arabidopsis ERG28 tethers the sterol C4-demethylation complex to prevent accumulation of a biosynthetic intermediate that interferes with polar auxin transport. Plant Cell 25: 4879–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo C, Valachovic M, Randall SK, Nickels JT, Bard M (2002) Protein-protein interactions among C-4 demethylation enzymes involved in yeast sterol biosynthesis. Proc Natl Acad Sci USA 99: 9739–9744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, Bonneu M, Simon-Plas F, Lessire R, Bessoule JJ (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Mongrand S, Stanislas T, Bayer EM, Lherminier J, Simon-Plas F (2010) Membrane rafts in plant cells. Trends Plant Sci 15: 656–663 [DOI] [PubMed] [Google Scholar]
- O’Brien M, Chantha SC, Rahier A, Matton DP (2005) Lipid signaling in plant cloning and expression analysis of the obtusifoliol 14α-demethylase from Solanum chacoense Bitt., a pollination- and fertilization-induced gene with both obtusifoliol and lanosterol demethylase activity. Plant Physiol 139: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Fujioka S, Peng J, Chen J, Li G, Chen R (2009) The E3 ubiquitin ligase SCFTIR1/AFB and membrane sterols play key roles in auxin regulation of endocytosis, recycling, and plasma membrane accumulation of the auxin efflux transporter PIN2 in Arabidopsis thaliana. Plant Cell 21: 568–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D, Castanedo I, Borsani O, Nieto B, Rosado A, Taconnat L, Ferrer A, Dolan L, Valpuesta V, Botella MA (2009) Identification of the Arabidopsis dry2/sqe1-5 mutant reveals a central role for sterols in drought tolerance and regulation of reactive oxygen species. Plant J 59: 63–76 [DOI] [PubMed] [Google Scholar]
- Pullen M, Clark N, Zarinkamar F, Topping J, Lindsey K (2010) Analysis of vascular development in the hydra sterol biosynthetic mutants of Arabidopsis. PLoS ONE 5: e12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahier A. (2011) Dissecting the sterol C-4 demethylation process in higher plants: from structures and genes to catalytic mechanism. Steroids 76: 340–352 [DOI] [PubMed] [Google Scholar]
- Schaller H. (2003) The role of sterols in plant growth and development. Prog Lipid Res 42: 163–175 [DOI] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jürgens G (2000) FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev 14: 1471–1484 [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Martin G, Bellini C, Kuhnt C, Schmidt J, Jürgens G (2002) Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J 31: 61–73 [DOI] [PubMed] [Google Scholar]
- Souter M, Topping J, Pullen M, Friml J, Palme K, Hackett R, Grierson D, Lindsey K (2002) hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14: 1017–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kamide Y, Nagata N, Seki H, Ohyama K, Kato H, Masuda K, Sato S, Kato T, Tabata S, et al. (2004) Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. Plant J 37: 750–761 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Nakagawa S, Kamide Y, Kobayashi K, Ohyama K, Hashinokuchi H, Kiuchi R, Saito K, Muranaka T, Nagata N (2009) Complete blockage of the mevalonate pathway results in male gametophyte lethality. J Exp Bot 60: 2055–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Senthil-Kumar M, Ryu CM, Kang L, Mysore KS (2012) Phytosterols play a key role in plant innate immunity against bacterial pathogens by regulating nutrient efflux into the apoplast. Plant Physiol 158: 1789–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D, Venglat P, Tibiche C, Yang H, Risseeuw E, Cao Y, Babic V, Cloutier M, Keller W, Wang E, et al. (2011) Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol 156: 346–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Richter GL, Wang X, Młodzińska E, Carraro N, Ma G, Jenness M, Chao DY, Peer WA, Murphy AS (2013) Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J 74: 37–47 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.