Plants of the usually perennial autotetraploid Arabidopsis arenosa that colonized railways became vernalization insensitive, early and perpetually flowering, and constitutively heat and cold stress tolerant.
Abstract
Weediness in ephemeral plants is commonly characterized by rapid cycling, prolific all-in flowering, and loss of perenniality. Many species made transitions to weediness of this sort, which can be advantageous in high-disturbance or human-associated habitats. The molecular basis of this shift, however, remains mostly mysterious. Here, we use transcriptome sequencing, genome resequencing scans for selection, and stress tolerance assays to study a weedy population of the otherwise nonweedy Arabidopsis arenosa, an obligately outbreeding relative of Arabidopsis thaliana. Although weedy A. arenosa is widespread, a single genetic lineage colonized railways throughout central and northern Europe. We show that railway plants, in contrast to plants from sheltered outcrops in hill/mountain regions, are rapid cycling, have lost the vernalization requirement, show prolific flowering, and do not return to vegetative growth. Comparing transcriptomes of railway and mountain plants across time courses with and without vernalization, we found that railway plants have sharply abrogated vernalization responsiveness and high constitutive expression of heat- and cold-responsive genes. Railway plants also have strong constitutive heat shock and freezing tolerance compared with mountain plants, where tolerance must be induced. We found 20 genes with good evidence of selection in the railway population. One of these, LATE ELONGATED HYPOCOTYL, is known in A. thaliana to regulate many stress-response genes that we found to be differentially regulated among the distinct habitats. Our data suggest that, beyond life history regulation, other traits like basal stress tolerance also are associated with the evolution of weediness in A. arenosa.
Life history traits differ between and within plant species and commonly reflect the requirements of the habitats in which they are found (Baker, 1974; Weinig et al., 2003; Grime, 2006). Depending on abiotic and biotic conditions, a variety of strategies can be favored, and accordingly, weeds are phenotypically diverse. In environments that are unpredictable, with frequent occurrences of stresses like drought, temperature fluctuations, or human-associated perturbations, rapid cycling and early flowering are common (Hall and Willis, 2006; Sherrard and Maherali, 2006; Franks et al., 2007; Wu et al., 2010). Life history adaptations can help mediate tradeoffs between resource accumulation and stress avoidance and are important for wild species as well as for crops (Jung and Müller, 2009). Comparing results among species, as well as the correlates of these traits with other fitness-related traits, promises new insights into the mechanisms of adaptation to unpredictable habitats.
A common phenotype of plants in unpredictable habitats is early and prolific flowering relative to related populations in more stable habitats (Baker, 1965; Grotkopp et al., 2002; Blair and Wolfe, 2004; Burns, 2004; Hall and Willis, 2006; Sherrard and Maherali, 2006; Franks et al., 2007). The complex genetic architecture of flowering has been well studied in the annual Arabidopsis thaliana (Andrés and Coupland, 2012), where independent changes in two genes in particular, FLOWERING LOCUS C (FLC) and FRIGIDA (FRI), have been repeatedly found to underlie natural variations in flowering time and vernalization responsiveness (Michaels and Amasino, 1999; Johanson et al., 2000; Le Corre et al., 2002; Gazzani et al., 2003; Lempe et al., 2005; Shindo et al., 2005; Werner et al., 2005; Brachi et al., 2010; Méndez-Vigo et al., 2011, 2016; Salomé et al., 2011; Song et al., 2013). The same genes also are important in the closely related Arabidopsis lyrata (Kuittinen et al., 2008) and in other species in the Brassicaceae (Slotte et al., 2009; Wang et al., 2009; Guo et al., 2012). Active FRI alleles enhance the expression of FLC, which in turn represses floral activators including FLOWERING LOCUS T and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1; Michaels and Amasino, 1999; Searle et al., 2006). Arabidopsis thaliana accessions with functional alleles of both FRI and FLC are late flowering in the laboratory, but prolonged exposure to cold (vernalization) epigenetically represses FLC expression, allowing flowering upon the return to warm temperatures (Song et al., 2013). Many independent disruptions of FRI or FLC have been identified in A. thaliana (Johanson et al., 2000; Le Corre et al., 2002; Gazzani et al., 2003; Shindo et al., 2005; Werner et al., 2005; Méndez-Vigo et al., 2011) that result in reduced or abrogated FLC expression, leading to earlier flowering and reduced need for vernalization. However, nonfunctional FRI alleles are associated with negative pleiotropic effects on branching and fitness, likely limiting the adaptive potential of the FRI locus (Scarcelli et al., 2007). Natural variation in FLC and FRI also affects other important life history traits, including seed germination (Chiang et al., 2009), water use efficiency, which is a major dehydration avoidance mechanism (McKay et al., 2003), and even flower tolerance to heat shocks (Bac-Molenaar et al., 2015). FLC also plays a role in more long-lived plants. In the perennial Arabis alpina, an ortholog of FLC, PERPETUAL FLOWERING1 (PEP1), contributes to late flowering and the vernalization requirement as it does in A. thaliana but it also promotes a return to vegetative development after each flowering episode, which is an important feature of perennial life cycles (Wang et al., 2009). Variation in PEP1 activity is associated with distinct life histories in different A. alpina accessions (Albani et al., 2012).
Arabidopsis arenosa is a close relative of A. thaliana and A. lyrata (O’Kane, 1997; Clauss and Koch, 2006). In contrast to A. thaliana A. arenosa is a perennial obligate outcrosser with high genetic diversity and both diploid and tetraploid variants (Hollister et al., 2012; Schmickl et al., 2012). The autotetraploid A. arenosa arose from a single diploid population closely related to populations found today in the Carpathian Mountains of Slovakia around 19,000 generations ago; by 15,000 generations ago, autotetraploid lineages had begun radiating across the landscape into the distinct types found in diverse habitats across Europe today (Arnold et al., 2015). Distinct genetic lineages correlate with geography and habitat. Rocky outcrops are generally populated by a perennial mountain form, while ruderal settings, especially railways, are colonized by an annual, flatland form (Scholz, 1962). Although the mountain form comprises at least four distinct genetic lineages associated with geography, we found previously that railway populations from geographically distant locations are extremely closely related, suggesting that this habitat was colonized just once by a single genetic lineage that subsequently spread along this habitat (Arnold et al., 2015).
Here, we study representative populations of the perennial mountain form and the flatland form of A. arenosa. Specifically, we use phenotypic, genomic, and transcriptomic experiments to assess flowering time, vernalization responsiveness, and stress resilience. We found that populations from ruderal sites are rapid cycling, do not require vernalization, and do not resume vegetative growth after a single flowering episode, while mountain populations remain vernalization responsive. We compared transcriptomes of early-flowering (railway) and late-flowering (mountain) plants across time series that were either vernalized or not. We found that rapid-cycling plants from railway populations have very low FLC expression and a sharply abrogated vernalization response, while plants from a mountain population show transient repression of FLC by vernalization similar to what was described in A. alpina (Wang et al., 2009; Albani et al., 2012). We also found constitutive differences in the expression of cold and heat stress-response genes. Consistent with the expression data, we found that railway plants had higher basal heat and cold stress tolerance than mountain plants. A genome-resequencing scan for divergence identified 20 loci with evidence of positive selection in the weedy railway lineage. Among these is the circadian clock regulator LATE ELONGATED HYPOCOTYL (LHY), which regulates many of the cold- and heat-responsive genes we found to be differentially expressed in these two A. arenosa types. Our data suggest that, in addition to flowering behavior, traits like flowering induction and heat and cold stress tolerance that are environmentally inducible in mountain plants became constitutive in the weedy railway plants.
RESULTS
Flowering Time and Vernalization Response in A. arenosa
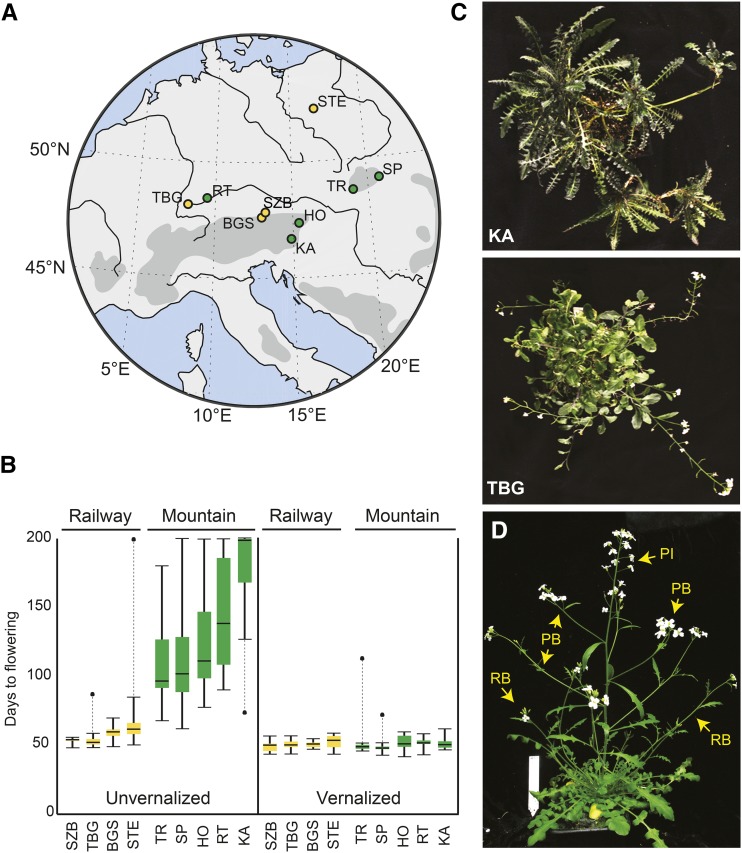
We grew plants from seeds sampled from five mountain and four railway populations of A. arenosa in controlled conditions from seeds collected from wild plants (Fig. 1A). We measured flowering time (as days from germination to first open flower) for plants grown with or without an 8-week vernalization period that consisted of a cold treatment (4°C) under short-day conditions (8 h of light instead of 16 h; see “Materials and Methods”). All mountain populations flowered significantly later than all railway populations when unvernalized (Fig. 1B), but all populations flowered similarly when vernalized. This shows that all sampled railway populations are rapid cycling and have lost vernalization responses, while all mountain populations retain them and flower late without cold treatments. We previously showed that the railway plants are all extremely closely related, suggesting a single colonization event that is consistent with the genetic similarity of the different railway populations (Arnold et al., 2015).
Figure 1.
Phenotypes of railway and mountain A. arenosa plants. A, Map of central Europe with locations of A. arenosa populations sampled from railway (yellow) and mountain (green) sites. TBG = Triberg, Germany; RT = Upper Danube Valley, Germany; BGS = Berchtesgaden, Germany; SZB = Salzburg, Austria; HO = Hochlantsch, Austria; KA = Kasparstein, Austria; TR = Trencin, Slovakia; SP = Spisska, Slovakia. B, Box plots showing flowering phenotypes of plants grown from seeds collected from railway and mountain populations. Flowering time was quantified as the time from germination to the first open flower for vernalized (left) and nonvernalized (right) plants from both accessions. Plants that did not flower by the end of the experiment (200 d) were assigned 200 d as their flowering date. C, Images of two representative vernalized individuals taken at 38 weeks. TBG plants flower continuously, while KA plants revert to vegetative growth after an episode of flowering. The development of secondary rosettes along branched stems of KA plants can then be observed. D, Representative greenhouse-grown A. arenosa indicating scored phenotypes of primary inflorescence branches (PB) and rosette branches (RB). PI indicates the primary inflorescence.
We selected a single railway population (TBG) and a single mountain population (KA) as representative of the two types to analyze in more depth the molecular basis of their phenotypic differences. TBG is from a railway at Triberg railway station in southwest Germany. KA is from a limestone outcrop on Kasparstein Mountain, near Loschental, Austria (Fig. 1A). These populations are members of genetically distinct mountain and railway lineages with no evidence of recent gene flow between them (Arnold et al., 2015). Among unvernalized plants, those from KA flowered much later than those from the TBG (Wilcoxon P < 10e−6): time to open flower averaged 56 d for unvernalized TBG plants, while 67% of KA plants had not yet flowered by 200 d, at which point we ended the experiment (Fig. 1B). There was no significant difference in flowering times of cold-treated (vernalized) plants from the two populations (Fig. 1B; Wilcoxon P > 0.08). We confirmed the similarity of flowering behavior of the two vernalized populations using a Mann-Whitney U test (P > 0.6). Vernalization had no significant effect on the mean flowering time of TBG, although there was a reduction in the sd from 12 d (nonvernalized) to 4 d (vernalized), implying that TBG plants, although lacking a true vernalization response, still show a residual response to prolonged cold treatment. The difference in flowering behavior persists in subsequent generations in the laboratory, showing that this is not merely a maternal environmental effect resulting from differences in conditions in wild populations (data not shown).
After flowering, vernalized KA plants reverted to vegetative growth while TBG flowered continuously until senescence (Fig. 1C), paralleling the distinction between perpetual and episodic flowering described previously in A. alpina (Wang et al., 2009). Furthermore, from our initial phenotyping of 13 KA plants and 20 TBG plants, it was also clear that TBG plants grew more rosette inflorescence branches (RB in Fig. 1D; average rosette inflorescences longer then 5 cm at 20 d after flowering = 14.35) compared with KA, which usually had none or only one (average rosette branches longer then 5 cm at 20 d after flowering = 0.54; Student’s t test P = 8 × 10−7). The number of inflorescence branches (PB in Fig. 1D) also differs similarly dramatically (0.23 branches in KA versus 18.15 branches in TBG; P = 1.7 × 10−9). Thus, TBG plants show common weedy phenotypic characteristics, including rapid cycling and bushy and abundant inflorescence growth (Baker, 1965; Grotkopp et al., 2002; Blair and Wolfe, 2004; Burns, 2004; Hall and Willis, 2006).
Abrogated Vernalization Responsiveness and Loss of FLC Expression in a Railway Accession
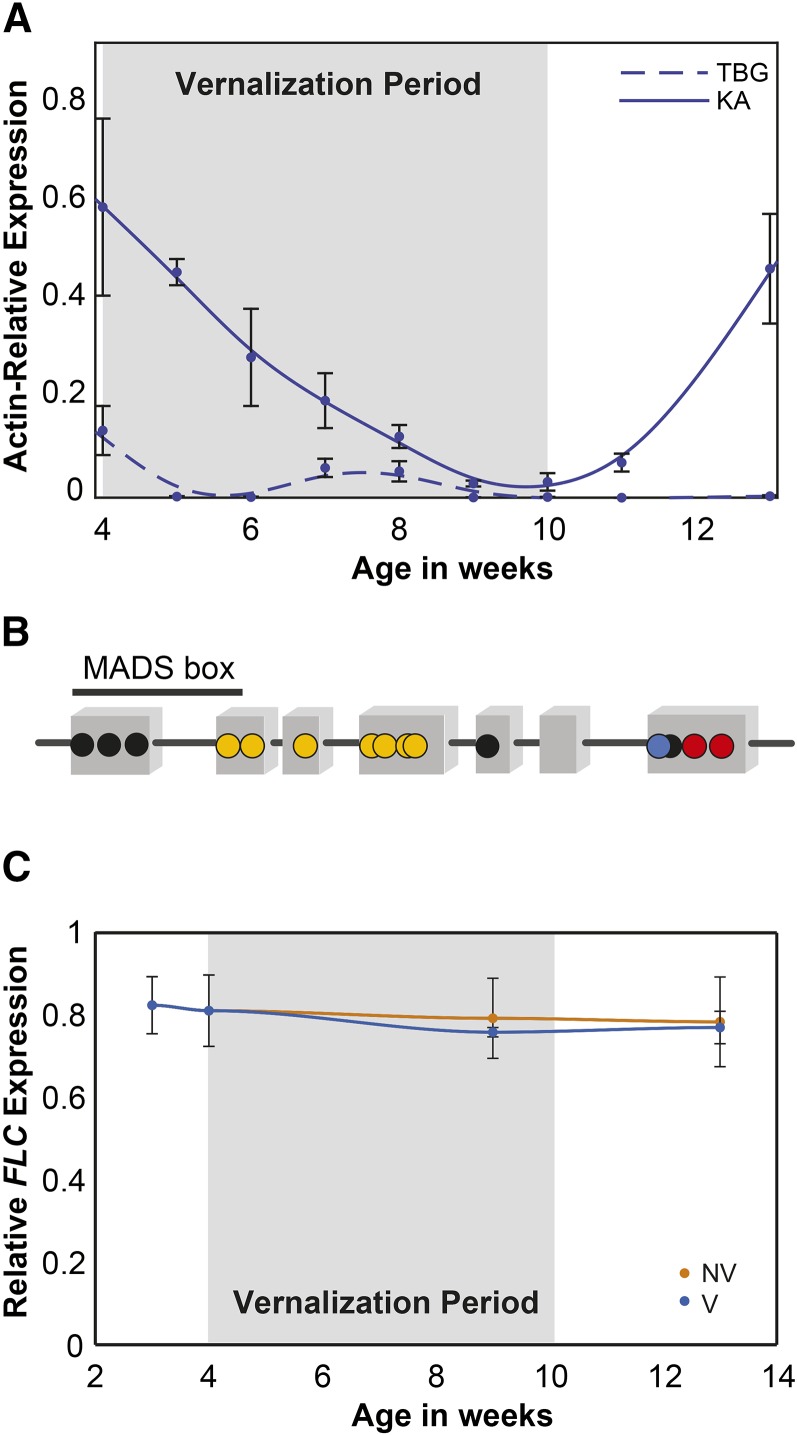
To compare the vernalization responses of KA and TBG, we analyzed the transcriptomes of plants from these two populations by transcriptome sequencing (RNA-seq) at four time points (3, 4, 9, and 13 weeks) with or without a 6-week vernalization period at weeks 4 to 10 (see “Materials and Methods”). We quantified expression by read depth after aligning to the A. lyrata reference genome (Hu et al., 2011), which includes 32,670 annotated genes (see “Materials and Methods”). We first examined the expression of 151 A. lyrata homologs of 174 genes associated with flowering regulation in A. thaliana (Fornara et al., 2010). Among these, FLC was the most differentially expressed between the two vernalized time series of KA and TBG, with virtually undetectable expression in TBG. FLC showed initially strong expression in KA followed by a clear suppression by vernalization but a subsequent return to prevernalization levels after plants were returned to warmer conditions. We confirmed this expression profile by quantitative reverse transcription-PCR with a finer sampling resolution (Fig. 2A).
Figure 2.
Differential FLC expression and responsiveness to vernalization. A, Quantitative reverse transcription-PCR of FLC expression relative to ACTIN (ACT) in vernalized KA and TBG plants. While undetectable in TBG, FLC is suppressed by vernalization in KA but comes back to unvernalized levels after plants are returned to warm conditions. B, Single-nucleotide differences between the coding sequences of the two AaFLC1 and AaFLC2 paralogs in our sample. Gray boxes are exons. Yellow indicates differences between AaFLC1 and AaFLC2 present in our samples as well as the published BAC sequence (Nah and Chen, 2010). Black indicates differences between AaFLC1 and AaFLC2 in the BAC not found in our accessions. Red indicates differences between AaFLC1 and AaFLC2 in our samples but not present in the BAC. Blue indicates differences between AaFLC1 and AaFLC2 where both paralogs differ from the BAC. C, Expression levels of AaFLC1 as a proportion of total AaFLC locus expression across the vernalized (V) and nonvernalized (NV) time series, showing that the relative expression of the two FLC copies does not change by treatment or through the time series.
In the A. arenosa genome, FLC is present in two full-length copies and one truncated duplicate copy (Nah and Chen, 2010), but only one copy was annotated in the version of the A. lyrata genome we used for aligning RNA-seq reads (scaffold 6: 4,040,170–4,045,798). Since finalizing this work, a newer annotation has been published (Rawat et al., 2015) that recognizes two FLC genes in A. lyrata. Because of the very close similarity between the two genes, we could not use our coverage estimates from read alignments to A. lyrata or to the A. arenosa bacterial artificial chromosome (BAC) to differentiate the expression of the two copies. Thus, to estimate relative expression levels, we genotyped our read alignments for polymorphisms that distinguish the two full-length FLC copies. We found a total of nine single-nucleotide polymorphisms that differ between the paralogs in our transcriptome samples (Fig. 2B). Seven of these single-nucleotide polymorphisms had been identified previously in a BAC sequence of the A. arenosa FLC locus from the Care-1 strain (Nah and Chen, 2010). Only three were among the 16 polymorphisms differentiating the two A. lyrata paralogs. At all nine positions, the nucleotides characteristic of AaFLC2 were at low frequency in our transcriptome alignments relative to those characteristic of AaFLC1, indicating low expression of AaFLC2 relative to AaFLC1.
To more finely quantify the differential expression of AaFLC1 and AaFLC2, we mapped our RNA-seq data on the BAC sequence and followed a previously described method for the detection of allelic expression differences (Perez et al., 2015), estimating the expression of the two transcripts with MMSEQ (Turro et al., 2011). This approach confirmed the differential expression of AaFLC1 and AaFLC2 in the KA samples (where sufficient levels of expression were detected). In the two time series of this population, AaFLC1 contributed to an average of 77% of the total AaFLC expression. The relative ratios of the duplicates remain consistent in our time series, indicating that, although AaFLC1 dominates in terms of total expression, both paralogs respond similarly to vernalization in KA (Fig. 2C).
As expected, positive regulators of flowering showed opposite trends to the floral repressor FLC, including SOC1, SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE15 (SPL15), and SPL4, which were all expressed throughout the time course in TBG but only after vernalization in KA. The expression profile of SOC1, a flowering promoter, was especially strongly anticorrelated with FLC expression (Supplemental Fig. S1; Pearson correlation, −0.87). VERNALIZATION INSENSITIVE3 (VIN3), which encodes a component of the POLYCOMB REPRESSIVE COMPLEX2 responsible for establishing the repression of FLC during vernalization, is up-regulated by vernalization in A. thaliana (Sung and Amasino, 2004). VIN3 is similarly up-regulated in both TBG and KA (Supplemental Fig. S1), but the magnitude of the response is lower in TBG than in KA, consistent with the hypothesis that TBG maintains some vernalization responsiveness, albeit in a strongly abrogated form, and that the cause of this abrogation may lie upstream of VIN3.
Vernalization-Response Differences
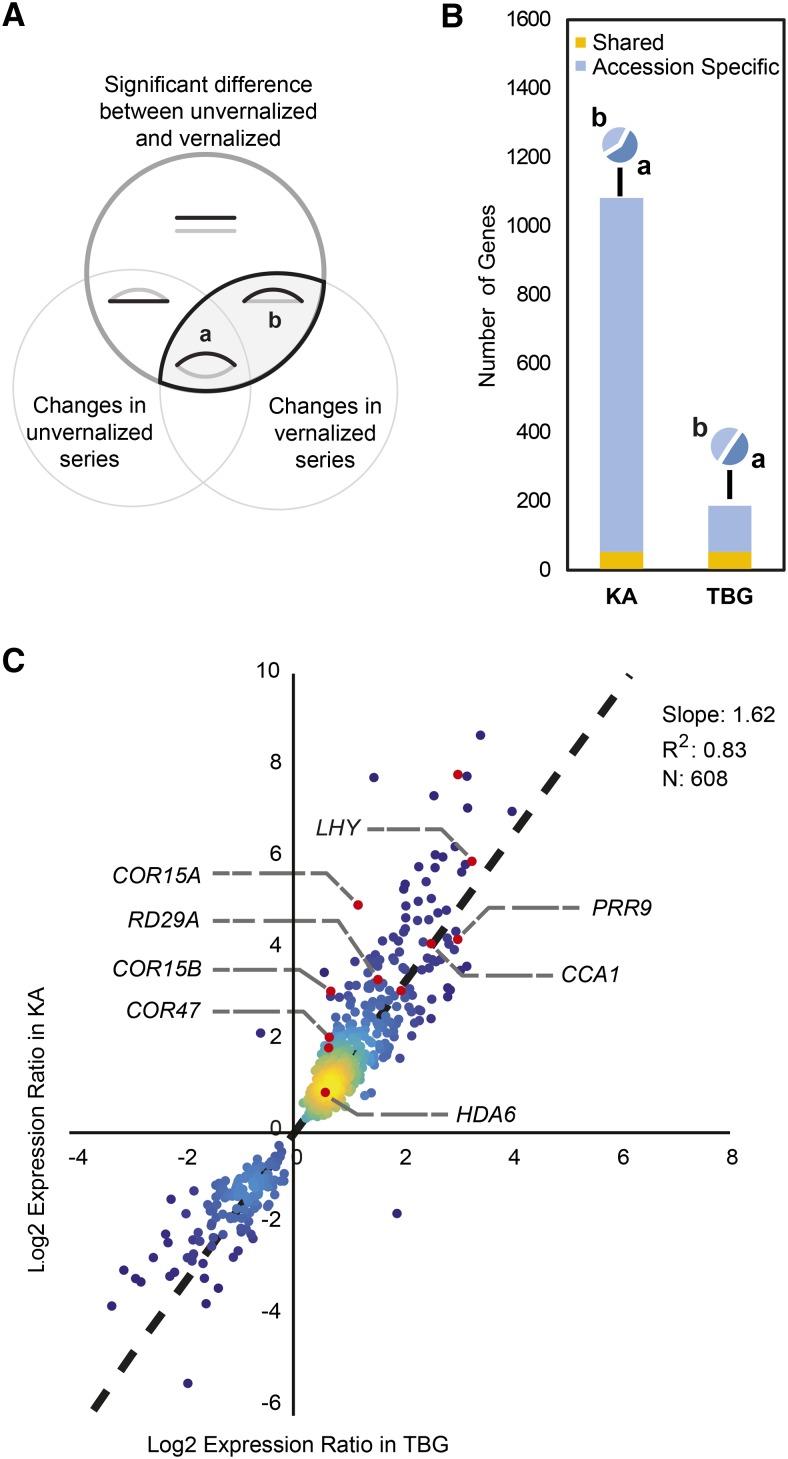
We next set out to characterize the vernalization-responsive subset of genes within the entire transcriptomes of KA and TBG with two goals: (1) to understand the vernalization response in A. arenosa, and (2) to compare the responsiveness of the two accessions qualitatively and quantitatively. Within each genotype, we identified vernalization-response genes as the intersection of genes that differ between growth conditions (vernalized versus nonvernalized) and those that change expression through the time course within each of the conditions. We used a nonparametric ranking test (see “Materials and Methods”) to identify significantly differentially expressed vernalization-responsive genes. We considered genes to be vernalization responsive if they showed a differential expression during the vernalized time series and had a significant growth condition interaction. This category includes genes that are (Fig. 3A, a) or are not (Fig. 3A, b) differentially expressed at different time points in the nonvernalized time series.
Figure 3.
Vernalization response differences between KA and TBG mainly due to a reduced magnitude of response in TBG. A, Venn diagram representing the seven categories of differential responses for each accession. The top circle (dark gray) includes genes with significant differential expression when comparing the vernalized and unvernalized time series. The bottom left circle contains genes with significant differential expression among time points in the unvernalized time series. The bottom right circle contains genes with significantly different expression among time points in the vernalized time series. Cartoon curves show schematic vernalized (black) and unvernalized (gray) expression profiles of genes found in each category. Vernalization-responsive genes are found for each accession at the intersection between the top and bottom right circles. Within this pattern, a represents genes that change across the time series in both vernalized and unvernalized plants, but in distinct ways, while b includes genes that show changes in the vernalized time series but not in the unvernalized time series. B, Decomposition of each vernalization response showing which genes change in both accessions (yellow), which are accession specific (blue), and how they are partitioned between a- and b-category patterns (camembert diagrams). Almost 6 times more genes are identified as vernalization responsive in KA compared with TBG. C, Comparison of vernalization responsiveness in KA versus TBG. The responsiveness of a gene is calculated for each accession as the log2 ratio of vernalized over nonvernalized expression levels. Only ratios significantly different from 1 (log2 ratios different from 0) in both accessions are displayed. Several genes discussed here are highlighted. Colors signify plot density. The dotted line indicates the linear regression fit line based on n = 608 data points. The slope and r2 values for the fit are given on the chart.
Using our criteria, we identified 1,088 genes as vernalization responsive in KA, almost 6 times more than the 187 found in TBG (Supplemental Table S1). Only a small percentage of transcripts (representing 53 genes) are vernalization responsive in both accessions (Fig. 3B), but when we compared expression ratios between vernalized and nonvernalized time points in KA and TBG, 60% of these genes showed a significant correlation (r2 = 0.83) between the two accessions. The slope of the log regression was significantly higher than 1 (1.62), suggesting that TBG still has some vernalization responsiveness, but the magnitude of the response is strongly dampened relative to that in KA (Fig. 3C). This reduced responsiveness likely explains the lower number of genes passing our thresholds of detection for vernalization responsiveness in TBG.
Gene Ontology (GO) analysis of KA vernalization-responsive genes showed a very strong representation of genes implicated in light sensitivity and abiotic stress responses. The light-related GO terms included response to UV light (GO:0009411; P = 0.002), response to light stimulus (GO:0009416; P = 0.007), and long-day photoperiod flowering (GO:0048574; P = 2 × 10−5); these were equally divided between the two categories (a and b) of the KA vernalization response (Fig. 3B). On the other hand, the cold stress terms, including cold response (GO:0009409; P = 1 × 10−8), water deprivation response (GO:0009414; P = 0.03), salt stress response (GO:0009651; P = 5 × 10−8), and hyperosmotic stress response (GO:0006972; P = 6 × 10−5), were mainly present in the b category, meaning that their expression shifts over the vernalized time course but not over the nonvernalized time course (Fig. 3B), suggesting that they might be coregulated with or by the vernalization response. In particular, the cold acclimation term was only enriched in the b category (GO:0009631; P = 0.003) and included well-known cold-regulated genes like COLD-REGULATED47 (COR47), EARLY RESPONSIVE TO DEHYDRATION, LOW TEMPERATURE-INDUCED, and KINASE1 (Maruyama et al., 2004). The b category also included other known components of the cold response of A. thaliana such as COR15A, COR15B, COR78, and PSEUDO-RESPONSE REGULATOR9. These genes have been shown to confer different degrees of freezing tolerance (Artus et al., 1996; Jaglo-Ottosen et al., 1998; Steponkus et al., 1998) as well as acclimation, the capacity to increase freezing tolerance after exposure to nonfreezing cold temperatures (Thomashow, 1999).
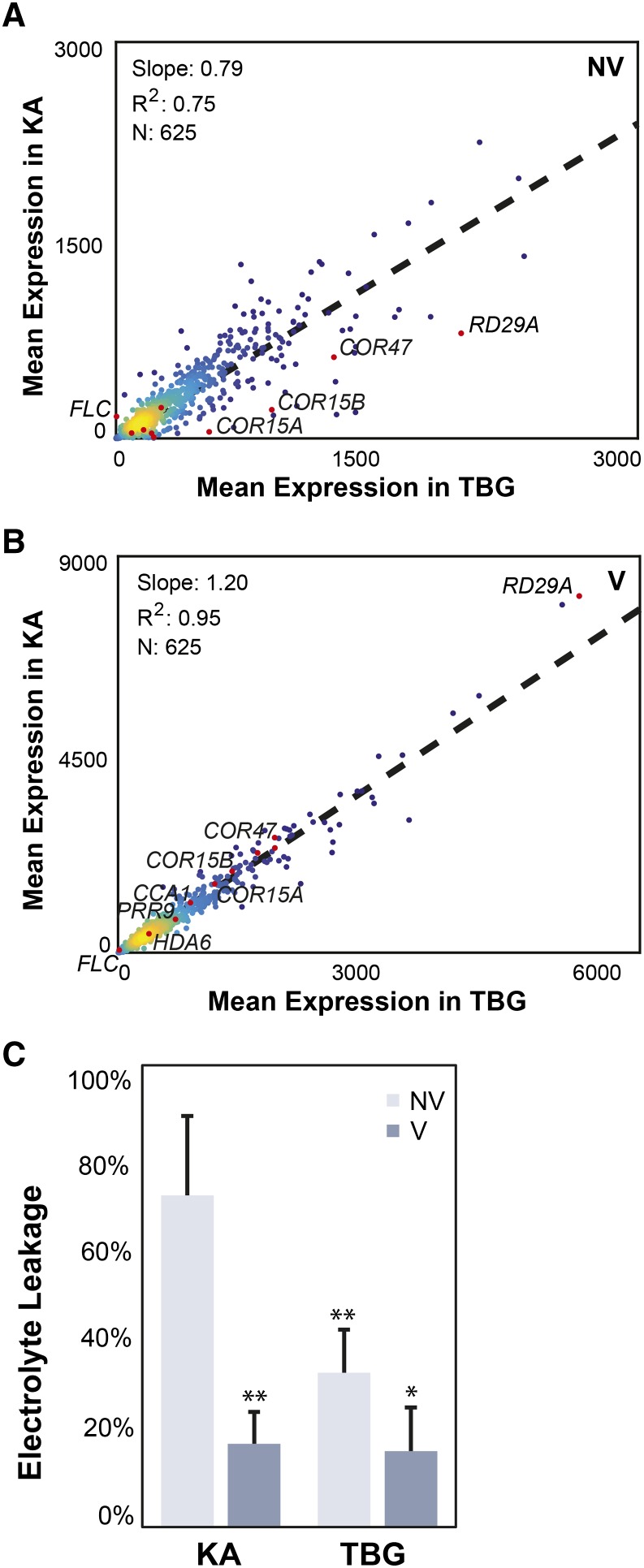
In our two A. arenosa populations, the magnitude of the response of most of the vernalization genes is amplified in KA relative to TBG, but the nonvernalized basal levels of expression of stress-responsive genes are higher in TBG (slope of 0.79 in nonvernalized expression levels, r2 = 0.75; Fig. 4A). The core cold-responsive genes highlighted previously are outliers from this correlation, with an even stronger tendency to be highly expressed in TBG. This relationship is inverted by cold treatment, due to the stronger magnitude of the KA response to vernalization treatment (slope of 1.2 in vernalized expression levels, r2 = 0.95; Fig. 4B).
Figure 4.
Expression of vernalization-responsive genes in KA versus TBG. A, Expression levels of vernalization-responsive genes in unvernalized KA plotted against their levels in unvernalized TBG. The slope of the linear regression (0.79 with r2 = 0.75) indicates that vernalization-responsive genes have a higher expression in TBG in unvernalized plants. Known cold-responsive genes are highlighted in red, and in particular, COR15A, COR15B, COR47, and RD29A show even stronger bias toward higher constitutive expression in TBG. B, Expression levels of vernalization-responsive genes in vernalized KA versus vernalized TBG. Due to the stronger vernalization response in KA, the relationship is inversed compared with A (slope of 1.2 with r2 = 0.95). C, Electrolyte leakage measured after freezing at −6°C of leaves from 7-week-old KA and TBG plants vernalized for 1 week (V; dark blue bars) or not vernalized (NV; light blue bars). Two asterisks indicate significant differences of vernalized KA and nonvernalized TBG compared with the high leakage of unvernalized KA plants (Student’s t test P < 1%). One asterisk denotes a significant difference of vernalized TBG from the controlled leakage of unvernalized TBG (Student’s t test P < 5%).
The expression results led us to hypothesize that there might be a difference in the constitutive and acquired freezing tolerance of KA and TBG. To test this, we quantified the freezing tolerance of these two accessions by measuring electrolyte leakage from detached leaves of vernalized and nonvernalized plants exposed to freezing (Sukumuran and Weiser, 1972). Leaves from nonvernalized KA plants showed a very high electrolyte leakage at −6°C (72%), while TBG leaves were significantly lower, at 33% (one-tailed Student’s t test P < 0.005). Both accessions showed significant reductions of leakage after 1 week of vernalization, to 18% and 16% for KA and TBG, respectively (Fig. 4C). This suggests that, while tolerance is constitutively higher in TBG than in KA, after prior exposure to 4°C, KA can achieve levels of freezing tolerance comparable to TBG. In addition, the tolerance of TBG also increases further with cold exposure, again consistent with the hypothesis that it retains some ability to respond to cold.
Global TBG-KA Expression Differences
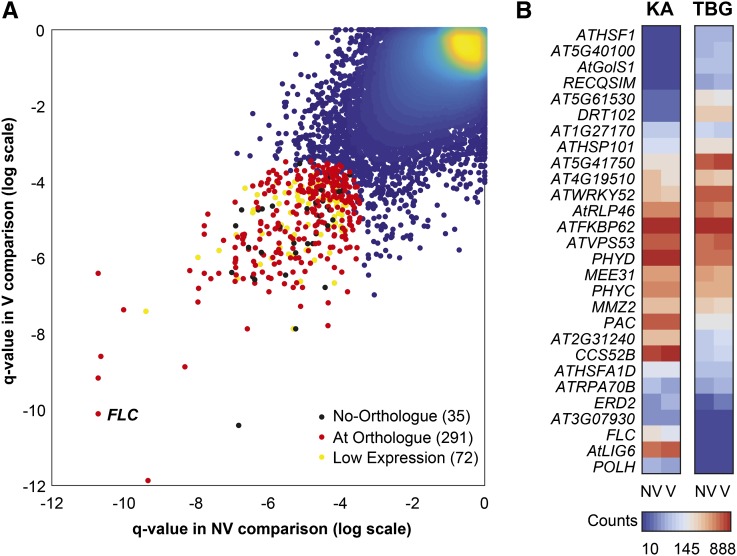
We next compared the overall transcriptomes of the KA and TBG time series for both the nonvernalized and vernalized samples (Fig. 5). For each comparison, we obtained a q value (false discovery rate corrected; see “Materials and Methods”) for every gene annotated in the A. lyrata reference genome (Hu et al., 2011). Out of the 326 genes that constitute the 1% most strongly differentially expressed genes between TBG and KA, 35 have no orthologs in A. thaliana (Fig. 5A; Supplemental Table S2), even though orthologless genes more frequently display null mean and variance of expression levels than genes with orthologs (Supplemental Fig. S2). Over the whole genome, the expression difference of FLC ranked third in comparisons of TBG and KA in both vernalized and unvernalized time series, making it the most differentially expressed gene between these accessions overall. The only genes more highly ranked for differential expression between TBG and KA in the nonvernalized series are ALPHA TUBULIN1, a pollen specific α-tubulin gene (Kim and An, 1992), and BTB AND TAZ DOMAIN PROTEIN4. In the vernalized series, the top two differentially expressed genes are At4g10320 (which encodes a tRNA synthetase) and g932613 (which has no A. thaliana homolog).
Figure 5.
Overall transcriptome differential expression between KA and TBG. A, Genome-wide distribution of q values comparing KA and TBG nonvernalized (NV; x axis) and vernalized (V; y axis) time series. FLC is highlighted, as it is ranked third in both comparisons, which makes it the most differentiated expression pattern overall. The top 1% (326 genes) overall (sum of both q values) are colored in red if a homolog is known in A. thaliana and in black if not. Yellow dots represent genes with low expression across all time points, genotypes, and conditions and, therefore, excluded from further consideration in the top 1% subset. B, Expression heat map of genes that are both within the 1% most differentiated pattern and associated with the GO category response to stimulus (GO:0050896). For each gene, the mean expression of both vernalized and nonvernalized time series is given for KA and TBG. Expression is given in normalized gene counts.
We next asked if there are any GO terms for biological processes enriched in the 1% top differentially expressed genes between KA and TBG in both time series. The significantly overrepresented categories include several stimulus-response gene categories, including heat response (GO:0009408; P = 0.023), protein unfolding (GO:0043335; P = 0.015), Golgi vesicle transport (GO:0048193; P = 0.02), detection of visible light (GO:0009584; P = 0.001), and DNA repair (GO:0006281; P = 0.044). Overall, these categories point to a generalized elevation of stress-associated genes in TBG. The GO category detection of visible light (GO:0009584) has the most significant P value in our analysis. Among differentially expressed genes in this category are PHYTOCHROME C (PHYC) and PHYD, two of the four phytochromes annotated in the A. lyrata genome. Both are involved in light signaling (Hu et al., 2013) and are expressed at lower levels in TBG. Mutation of PHYC leads to early flowering in A. thaliana (Monte et al., 2003), while polymorphisms in PHYC are thought to be involved in climatic adaptation (Balasubramanian et al., 2006; Samis et al., 2008; Méndez-Vigo et al., 2011). PALE CRESS, which in A. thaliana is important in chloroplast development and regulating levels of carotenoids and chlorophyll (Reiter et al., 1994), is more highly expressed in KA relative to TBG. Carotenoids are known to be important in the chloroplast capacity to respond to high-light stress (Havaux, 1998). In parallel, several genes implicated in DNA repair and recombination also show significant differential expression between KA and TBG in both time series, including DNA LIGASE6, MMS ZWEI HOMOLOG2, AT3G07930, REPLICATION PROTEIN A1B (RPA1B), RECQ HELICASE SIM (RECQSIM), and Y-FAMILY DNA POLYMERASE H (POLH). All but RECQSIM have lower expression in TBG than in KA. POLH and RPA1B, in particular, are known to be involved in the DNA repair response to UV light damage (Ishibashi et al., 2005; Anderson et al., 2008).
A number of heat-responsive proteins are differentially expressed in TBG and KA, including HEAT SHOCK FACTOR1 (HSF1) and HEAT SHOCK PROTEIN101 (HSP101), which are both expressed at higher levels in TBG, and HSFA1D, which is expressed at lower levels in TBG than in KA (Fig. 5B). HSF1 and HSFA1D are members of a four-gene family of class A heat shock factors that are regulators of the heat shock response and other abiotic stress responses (Liu et al., 2011; Yoshida et al., 2011). Overexpression of HSF1 in A. thaliana induces tolerance to heat shocks (Qian et al., 2014), but its absence does not completely impede acquired thermotolerance (Liu and Charng, 2012). HSP101 also plays a role in acquired thermotolerance (Gurley, 2000; Hong and Vierling, 2000, 2001; Queitsch et al., 2000). Additional modulators of thermotolerance, FK506-BINDING PROTEIN62, GALACTINOL SYNTHASE1 (GOLS1), and VPS53 (Lobstein et al., 2004; Lee et al., 2006; Wang et al., 2011), are also among our top differentially expressed genes.
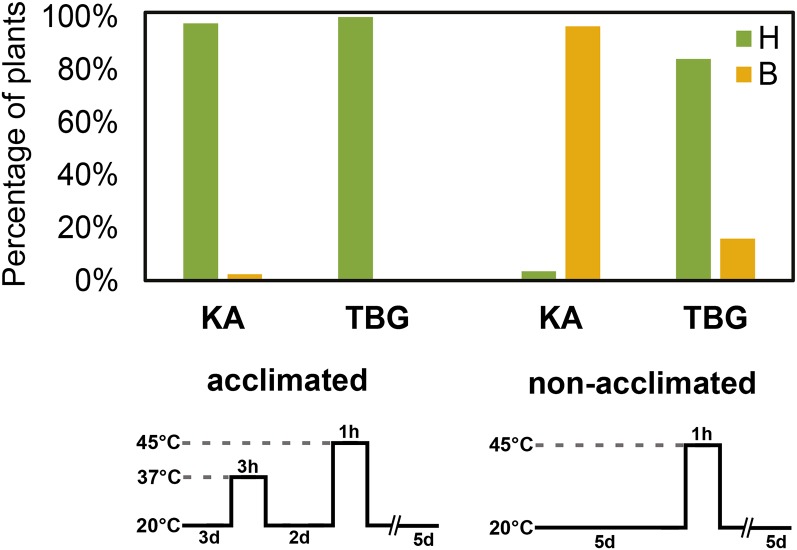
The generally high constitutive expression of heat-responsive genes in TBG led us to hypothesize that the heat tolerance of TBG might be elevated relative to KA. To test this, we exposed 5-d-old KA and TBG seedlings to a 45°C heat shock for 1 h, with and without an acclimation treatment at 37°C for 3 h (see “Materials and Methods”), which allows us to assay basal and acquired tolerance, respectively. After a 5-d recovery period at 20°C, we screened seedlings for partially or totally bleached cotyledons. After heat shock without prior acclimation, fewer than 20% of TBG seedlings showed signs of bleaching after 5 d, while more than 95% of KA seedlings were partially or entirely bleached (Fig. 6). When acclimated at 37°C 2 d before heat shock, both lines performed similarly with little or no bleaching. This fits with constitutive differences in the expression of heat response genes between KA and TBG associated with higher basal tolerance to heat shock by the railway line, while KA retains a capacity for acquired tolerance.
Figure 6.
Constitutive heat shock tolerance in railway plants. The percentage of seedlings exhibiting partial or total bleaching of their cotyledons and leaves after heat shock treatment is shown. H indicates healthy after treatment (green), and B indicates bleached after treatment (orange). Acclimated seedlings were subjected to a 3-h 37°C pretreatment 2 d prior to a 1-h heat shock at 45°C, while nonacclimated seedlings were incubated directly for 1 h at 45°C. Bleaching was often nonlethal even for KA plants.
Evidence of Selection in the Railway Population
We next looked for evidence of genetic differentiation that might be responsible for the abrogation of the vernalization response and/or constitutive induction of the stress responses in TBG relative to KA. To do so, we first complemented our previously generated genome sequence data for A. arenosa (Hollister et al., 2012) by sequencing a total of eight KA and seven TBG individuals, which samples 32 and 28 copies of each chromosome, respectively. To identify candidate genomic regions showing habitat-associated genetic differentiation, we used FST (a measure of differentiation; Weir, 1990), which is generally low among tetraploid A. arenosa populations (0.047–0.063; Hollister et al., 2012), as well as Fay and Wu’s H, a statistic sensitive to an excess of high-frequency variants compared with neutral expectations (Fay and Wu, 2000).
We identified 20 genes that showed evidence of strong differentiation between TBG and KA (Table I). With respect to the expression differences in cold and heat responses, one candidate stood out: LHY, which plays a central role in the regulation of the plant circadian clock (Alabadí et al., 2001) but is also known to broadly affect downstream cold-response genes (Vogel et al., 2005; Bieniawska et al., 2008). LHY is an outlier for Fay and Wu’s H in TBG but not in KA; nucleotide diversity measured by the number of pairwise differences is lower in TBG than in KA (Supplemental Fig. S3A), and 22 derived polymorphisms have significantly higher frequencies in TBG than in KA (average 82% versus 37%) in a region concentrated around exons 6 and 7 (Supplemental Fig. S3B). These patterns are consistent with the hypothesis that TBG experienced positive selection on LHY. Twenty of these polymorphisms fall within the coding sequence of LHY, and 14 are nonsynonymous. These 14 polymorphisms are all distributed along exons 6 and 7, with a dense cluster positioned at the beginning of exon 7 (Supplemental Fig. S3C). Of these 14 polymorphisms, two induce a strong change of hydrophobicity of the amino acids they encode (acidic Glu versus hydrophobic Gly) and one induces a charge change (Glu versus basic Lys; Supplemental Fig. S3C).
Table I. List of 1% genomic outliers for FST and Fay and Wu’s H.
| TAIR Identifier | Common Name |
|---|---|
| AT3G03510 | |
| AT3G56670 | |
| AT1G01060 | LHY |
| AT1G08135 | AtCHX6B |
| AT1G48090 | |
| AT1G72300 | PSY1R |
| AT1G78770 | APC6 |
| AT3G14980 | IDM1 |
| AT2G17140 | |
| AT2G31260 | APG9 |
| AT2G41700 | ABCA1 |
| AT3G56900 | |
| AT5G10560 | |
| AT5G21160 | AtLARP1a |
| AT4G32410 | ANY1 |
| AT4G25970 | PSD3 |
| AT2G24680 | |
| AT3G20660 | AtOCT4 |
| AT2G15620 | AtHNIR |
| AT3G57590 |
To determine whether the amino acid changes in TBG all lie on the same haplotype, we first cloned and sequenced a fosmid of the LHY locus from a TBG individual. The fosmid clone carried the derived alleles at all 22 positions, showing that the derived polymorphisms are all found together on a single haplotype that seems to have been the target of selection in TBG. We then cloned and sequenced additional PCR products from another TBG plant and found the same to be true for the exon 6/7 region in this individual as well. Since the derived polymorphisms were all present also in KA, albeit at low frequency, we asked if they were also all found on a single haplotype in KA identical to the haplotype found in KA. We cloned and sequenced PCR products from the exon 6/7 region and confirmed, first, that the derived polymorphisms were rare in KA but, also, that no KA haplotype within our sampling had all of the derived polymorphisms together. The rare derived polymorphisms in KA are in blocks of up to 18 of the 22 derived polymorphisms (Supplemental Fig. S4). In each case, a single recombination event between two haplotypes would suffice to toggle between the haplotype found in TBG and the rare haplotypes present in KA.
DISCUSSION
Here, we investigated the molecular basis of phenotypic differences between two A. arenosa populations, one of which has weedy traits associated with the colonization of railways in flatland Europe. The ancestral form usually inhabits shaded sites on hills or mountains, often in forests. These two distinct types correspond to a previously recognized distinction between a flatland and a mountain type within A. arenosa (Scholz, 1962). We previously showed that all railway plants we have sampled are members of a single genetically distinct lineage within tetraploid A. arenosa that diverged from mountain lineages fewer than 15,000 generations ago (Arnold et al., 2015). This suggests that the weedy phenotype of railway plants evolved once within A. arenosa, followed by the spread of this lineage throughout flatland Europe. We show here that the weedy phenotype of A. arenosa includes rapid cycling, abundant inflorescence growth, loss of the vernalization requirement, loss of other traits associated with perenniality, and constitutive stress tolerance. These are hallmarks of an all-in phenotype commonly observed in weedy colonizers (Baker, 1965; Grotkopp et al., 2002; Blair and Wolfe, 2004; Burns, 2004; Hall and Willis, 2006). The phenotypes observed in weedy A. arenosa likely reflect that railway sites are sunnier and more exposed to other abiotic stresses, including more rapid temperature fluctuations than would generally be experienced in the usually forested outcrop sites where most other A. arenosa populations are found. Although these types of adaptations are not uncommon for plants found in unpredictable and often human-associated habitats, their molecular basis has remained largely unknown.
Using transcriptome and phenotypic analyses, we found evidence that a major feature of the colonization of ruderal railway sites in A. arenosa is that responses to heat and cold that are inducible in mountain accessions became constitutive in railway accessions. These features include reproductive initiation, heat shock resistance, and freezing tolerance. Railway plants also exhibit perpetual flowering and are heavily branched, while mountain plants flower more modestly and return to vegetative growth after flowering, paralleling a distinction described previously in A. alpina of episodic versus perpetual flowering (Wang et al., 2009). This fits with the description of mountain accessions as perennial and ruderal railway accessions as rapid-cycling annuals, suggesting that the loss of perennial life history traits also accompanied the transition to weediness in A. arenosa.
One of the most differentially expressed genes when comparing railway and mountain plants was FLC, which is almost undetectable at any time point in the rapid-cycling railway plants. This gene has been identified frequently as a cause for independent transitions to rapid cycling and loss of vernalization sensitivity in A. thaliana (Méndez-Vigo et al., 2011; Guo et al., 2012). In A. alpina, an ortholog of FLC, PEP1, has been linked to a switch between late-flowering perennial life habits and rapid and perpetual flowering (Wang et al., 2009; Albani et al., 2012), but whether this FLC homolog is causal in A. arenosa remains to be tested. The low expression of FLC in the railway form, or its down-regulation in the mountain form upon vernalization, is strongly correlated with an up-regulation of floral promoters, including SOC1. This is also consistent with the known repression exerted by the FLC-SVP complex on SOC1 in A. thaliana (Michaels and Amasino, 2001; Li et al., 2008). Similarly, SPL transcription factors, which redundantly promote both vegetative phase change and flowering in A. thaliana (Schwarz et al., 2008; Wang et al., 2008), are constitutively expressed in railway plants and are initially absent but up-regulated during the vernalization period in mountain plants. Thus, in general, floral promoters are constitutively expressed in the railway accession and induced by vernalization in the mountain form, where their expression is anticorrelated with FLC expression. This suggests that the vernalization and flowering responses in A. arenosa, as well as the implication of FLC in life history changes, are consistent with findings in A. thaliana and related species.
In plants from the KA mountain site, FLC is initially expressed at very high levels and rapidly repressed during exposure to prolonged cold, just as it is in A. thaliana (Sung and Amasino, 2004; Coustham et al., 2012). However, as the plants begin to flower after a return to warm conditions, FLC expression returns to prevernalization levels within about 3 weeks. This suggests a mechanism similar to what is found in A. alpina, where meristems switch from vegetative to reproductive fate during vernalization, and any meristem arising during or after vernalization remains vegetative, leading to episodic flowering cycles characteristic of perennials (Wang et al., 2009). This cyclical FLC expression could explain the formation of secondary rosettes as branches from basal rosettes in A. arenosa (Fig. 1). We also observed this phenotype in diploid A. arenosa, suggesting that it is ancestral (data not shown). Paralleling phenotypes reported in A. alpina (Albani et al., 2012), these secondary rosettes require an additional vernalization treatment to flower. These secondary rosettes can form roots and allow the plants to make use of vegetative reproduction. In A. alpina, there are two PEP1 transcripts, and these show different expression patterns thought to be associated with the perennial life cycle of this species (Albani et al., 2012). In A. arenosa, there are also two full-length FLC genes (Nah and Chen, 2010), but we detected no significant difference in the response of the two paralogs to vernalization: AaFLC1 was expressed at higher levels than AaFLC2, but this difference remains consistent over our time course.
From our transcriptome analyses, we found that the railway plants have a generally strongly dampened vernalization response. Almost 6 times more genes were vernalization responsive in the mountain accession KA than in the railway accession TBG, but for 60% of them, the stronger magnitude of response in KA is nonetheless log-linearly correlated with a weaker response in railway plants, suggesting that the vernalization response is strongly abrogated but not completely absent in the railway plants.
We also found that the reduced vernalization responsiveness in railway plants was coupled with a constitutively high expression of a number of cold- and heat-response genes. A cross talk between flowering and the cold response has been described in A. thaliana and is associated with the flowering regulators SOC1, FLC, and FVE (Kim et al., 2004; Seo et al., 2009; Richter et al., 2013). Several COR genes are induced during vernalization in mountain A. arenosa plants, including COR15A, COR15B, COR47, COR314, and COR78, and reach higher levels after vernalization in mountain than in railway plants. COR genes are an essential component of the cold-acclimation response (Thomashow, 1999). The same genes, however, show higher expression before vernalization in railway plants than in mountain plants. The ability to cold acclimate increases the freezing tolerance of a plant after exposure to low temperature, and freezing tolerance has been shown to correlate with winter temperatures in natural accessions of A. thaliana (Hannah et al., 2006). For A. arenosa, we found that (fitting with COR gene expression trends) the mountain accession showed a much greater capacity to cold acclimate than the railway accession, but among unacclimated plants, those from railways had a higher basal tolerance to freezing than mountain plants. This is consistent with results from a CBF1-overexpressing A. thaliana mutant, which has high expression of the COR genes as well as enhanced freezing tolerance (Jaglo-Ottosen et al., 1998), as well as findings that naturally increased nonacclimated freezing tolerance can result from the constitutive activation of the CBF pathway (Hannah et al., 2006).
The COR genes are also implicated in the response to dehydration (Liu et al., 1998; Shinozaki and Yamaguchi-Shinozaki, 2000). AtGOLS1, which is constitutively overexpressed in railway plants, is involved in the accumulation of galactinol and raffinose (Panikulangara et al., 2004), two compounds known for their protective properties against oxidative stress (Nishizawa et al., 2008), and is involved in drought, cold, and high-salinity stresses (Taji et al., 2002). Therefore, constitutive overexpression of the COR genes, as well as AtGOLS1, could possibly also reflect selection for higher drought tolerance in the railway environment, where soil is drier and more exposed to water loss in the heat of summer. Perhaps constitutive freezing and drought tolerance could both have arisen as pleiotropic effects of alterations of COR gene expression during adaptation to railway habitats.
In addition to cold tolerance genes, we saw a similar shift toward constitutive expression in railway plants of heat response-associated genes. This includes HSF1, known in A. thaliana to be involved in thermotolerance (Qian et al., 2014), and HSP101, a key component of acquired thermotolerance (Gurley, 2000; Hong and Vierling, 2000, 2001; Queitsch et al., 2000). HSPs are molecular chaperones rapidly activated by the binding of HSFs in response to environmental stresses such as heat stress or other proteotoxic stresses such as drought and freezing (Schöffl et al., 1998). Twenty-one HSF genes have been annotated in A. thaliana (Nover et al., 2001), but the HSFA1s (a–d) constitute the main transcriptional activators in response to heat shock (Yoshida et al., 2011). Overexpression of HSF1 leads to stronger induction of HSPs by heat shock and increased stress tolerance but not to the acquisition of basal thermotolerance, as observed in HSFA1b-overexpressing plants (Prändl et al., 1998; Qian et al., 2014). Indeed, HSF1 requires the stress-induced formation of homotrimers to be activated and accumulate in the nucleus (Yoshida et al., 2011; Liu et al., 2013). Here, we observed in the railway accession constitutively high expression of HSF1 and HSP101 correlated with an increased basal thermotolerance. Together, these patterns of overexpression of heat shock genes suggest a constitutive activation of the heat shock response pathway in the railway accession, which was corroborated by the high basal heat shock tolerance of these plants: seedlings of railway plants show stronger heat shock tolerance than those of mountain plants, but we detected no difference in the acquired thermotolerance of the two types. The acquisition of constitutive thermotolerance, thus bypassing the requirement for a priming event, may be a significant advantage for populations colonizing habitats like railways, where they are more likely to be exposed to abrupt temperature fluctuations (Zerebecki and Sorte, 2011).
Given that the mountain and railway A. arenosa plants show clearly distinct environmental response phenotypes that are likely adaptive to their respective habitats, we used a genome-scanning approach to determine if any genomic regions show evidence of selection. By our criteria, 20 genes in the genome showed evidence of having been under selection in the railway population. The encoded proteins are important for a variety of processes, and a few have possible implications for the differences between railway and mountain types: CATION/H+ EXCHANGER6B (AtCHX6B) and ORGANIC CATION TRANSPORTER4 (AtOCT4) are both members of gene families involved in cation transmembrane transport (Remy et al., 2012; Ye et al., 2013), and NITRITE REDUCTASE1 (AtHNIR) is involved in the nitrate response (Konishi and Yanagisawa, 2010), which together hint at possible adaptation to substrate differences. This may be relevant, as KA is from a high-pH limestone site (which is likely the ancestral habitat for A. arenosa) and TBG is from railway ballast in the Black Forest, an acidic silicaceous region. LA-RELATED PROTEIN1A (AtLARP1A) is required for the heat-dependent degradation of mRNA involved in the thermotolerance of A. thaliana with consequences for its acclimation capacity (Merret et al., 2013). AUTOPHAGY9 (APG9) mutants have accelerated seed set and senescence (Hanaoka et al., 2002), which may be related to the senescence difference between the rapid-cycling TBG and the perennial mountain type KA.
For the overall control of stress responsiveness, one candidate target of selection stood out: the circadian clock regulator LHY. In A. thaliana LHY is known to affect flowering, albeit indirectly (Schaffer et al., 1998; Mizoguchi et al., 2002), but also cold responsiveness (Vogel et al., 2005; Bieniawska et al., 2008) and freezing tolerance (Bieniawska et al., 2008; Espinoza et al., 2008; Nakamichi et al., 2009; Dong et al., 2011). Double mutants for LHY and another circadian regulator, CCA1, show a greatly diminished induction of cold responses and the COR genes, including COR15A, COR47, and COR78 (Dong et al., 2011), that were differentially expressed among our A. arenosa strains. Thus, it is enticing to hypothesize that the derived variant of LHY that seems to have been under selection pressure in railway plants may somehow lead to a change in the regulation of downstream stress tolerance as well as possibly flowering responses. Whether LHY variation is actually responsible for the phenotypic differences in stress tolerance or flowering we observe between these mountain and railway populations remains to be tested.
Overall, we found that an A. arenosa lineage that adapted to railway sites in flatland Europe evolved features commonly observed in weedy plants found in human-associated habitats, including rapid cycling, perpetual flowering, extensive inflorescence growth, and constitutive heat and cold tolerance (Baker, 1965; Grotkopp et al., 2002; Blair and Wolfe, 2004; Burns, 2004; Hall and Willis, 2006). These phenotypes are all inducible in the mountain accession. Thus, there is a general trend that normally inducible phenotypes have become constitutive in the weedy A. arenosa form, which was likely important in the colonization of a volatile and risky human-associated habitat. The fact that these A. arenosa populations are autotetraploids appears to be in line with a strong potential for habitat colonization and the transition to weediness by polyploids (Soltis and Soltis, 2000; Pandit et al., 2006; Prentis et al., 2008). However, it is clear that, in this case, neither weediness nor other traits associated with the ruderal railway habitats were immediate consequences of genome duplication. Among five tetraploid A. arenosa lineages we have sampled, only one successfully colonized ruderal habitats (Arnold et al., 2015), and we show here that it has distinct phenotypic traits consistent with specific adaptations to that habitat not shared by other tetraploids. Thus, while colonization of the railway habitat was perhaps facilitated by the increased genetic diversity available in tetraploids (Otto et al., 2007; Hollister et al., 2012; Arnold et al., 2015), the autotetraploids as a group are not globally tolerant of ruderal habitats, and adaptation was required postpolyploidy for this colonization.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
We collected seeds from natural populations in June and July, 2009 to 2011. All populations are autotetraploid; all originate from regions where only autotetraploids occur (Jørgensen et al., 2011), and ploidy of a subset of individuals from each population was confirmed using flow cytometry (Hollister et al., 2012). For flowering time phenotyping, we grew 48 single progeny from each of several individuals per population. Plants were grown as described previously (Hollister et al., 2012) in Conviron MTPC-144 chambers with 8 h of dark at 12°C, 4 h of light (Cool-White fluorescent bulbs) at 18°C, 8 h of light at 20°C, and 4 h of light at 18°C. A subset (24) of 4-week-old plants were transferred to a chamber with a constant temperature of 4°C and short days (8 h of light) for 8 weeks and then returned to warm and long-day conditions.
For the transcriptomic time series, we grew arrays of 48 siblings from seeds harvested from single individuals growing in nature. Plants were grown under similar growth conditions in three flats in which we mixed TBG and KA plants in order to avoid flat effects. Half of the plants from each flat were vernalized for 6 weeks before being returned to their flat of origin. For our RNA extractions, we used tissue from young rosette leaves from both vernalized and unvernalized plants using one biological replicate from each flat. Leaves were harvested every 7 d after 9 to 10 h from Zeitgeber time 0 in long-day conditions and after 5 to 6 h in short days. Zeitgeber time 0 is defined as the time of lights on.
Phenotyping of Flowering Time
Using germination date recorded as the first date when root tip emergence was evident on agar plates, we measured flowering time as the time to first open flower. We calculated flowering time for vernalized plants based on the total number of growing days excluding the cold treatment. For plants that had not flowered by experiment end (200 d), we assigned these cutoff values. Phenotypic values for time to first open flower were not normally distributed even after transformation (Shapiro-Wilk W test P < 0.0001), so we used nonparametric Wilcoxon tests to assess the significance of differences among populations.
RNA Isolation, Sequencing, and Expression Analysis
We extracted RNA using the RNeasy Plant Mini Kit (Qiagen). We synthesized single-stranded complementary DNA (cDNA) from 500 ng of total RNA using VN-anchored poly-T(23) primers with MuLV Reverse Transcriptase (Enzymatics) according to the manufacturer’s recommendations. Quantitative PCR was carried out on a Stratagene Mx3005P machine with an annealing temperature of 55°C using Taq DNA polymerase (New England Biolabs). Reactions were carried out in triplicate, and we normalized FLC expression against the expression of ACT using the 2–ΔΔCT method taking into account each primer’s efficiency as described in the Bio-Rad Real-Time PCR Applications Guide. The sd of each biological replicate was calculated using a first-order propagation of error formula on the variance of the technical replicates. We used cDNA-specific primers 5′-CAGCTTCTCCTCCGGCGATAACCTGG-3′ and 5′-GGCTCTGGTTACGGAGAGGGCA-3′ for FLC (87% efficiency) and 5′-CGTACAACCGGTATTGTGCTGGAT-3′ and 5′-ACAATTTCCCGCTCTGCTGTTGTG-3′ for ACT (91% efficiency).
We prepared RNA-seq libraries using the TruSeq RNA Sample Prep Kit version 2 (Illumina) on RNA samples from both vernalized and unvernalized plants at four time points (3, 4, 9, and 13 weeks). Libraries were sequenced on an Illumina HiSeq 2000 with 50-bp single-end reads. We sequenced between 9.8 and 18.8 million reads (average, 13.6 million). We aligned reads to the Arabidopsis lyrata genome (Hu et al., 2011) using TopHat2 (Kim et al., 2013) and realigned unmapped reads using Stampy (Lunter and Goodson, 2011). We acquired read counts for each of the 32,670 genes using HTSeq-count (Anders et al., 2015) with A. lyrata gene models (Hu et al., 2011). We normalized for sequencing depth using DEseq2 in R (Anders and Huber, 2010). Further analyses were performed in MATLAB (MathWorks) except GO enrichment analyses, realized with the Bioconductor package GOstats (Falcon and Gentleman, 2007) in RStudio. RNA-seq read data have been deposited in the National Center for Biotechnology Information Short-Read Archive database under accession number SRP070489 within the National Center for Biotechnology Information BioProject PRJNA312410.
We obtained estimates of differential expression through the time series using a combination of two tests, a Kruskal-Wallis test and a two-way ANOVA coded on MATLAB. We used the first as a nonparametric ranking test within each condition and accession, and this allowed us to detect significant effects of time on the expression level of each gene within a time series without assuming a normal distribution of gene counts. We then used a two-way ANOVA to account for paired data, screen for time series effects, and generate comparisons of gene expression profiles between time series. We combined the results of these tests into seven categories of gene expression profiles for each pairwise comparison between time series, of which two reflect the response to vernalization (Fig. 3A, a and b).
Global expression differences were estimated using a Kruskal-Wallis test between TBG and KA within each condition (vernalized and nonvernalized). The P values obtained were then corrected for false discovery rate using the linear step-up procedure originally introduced by Benjamini and Hochberg (1995). For each gene, the two q values thus obtained for each condition were then summed in order to establish the 1% most differentiated genes with the lowest sum (QQ50 in Supplemental Table S2). Genes with a normalized gene count below 50 across all time points, genotypes, and conditions were excluded in order to filter from this 1% subset genes with high relative but low absolute differences.
Heat Stress
We adapted a protocol by Meiri and Breiman (2009) to Arabidopsis arenosa. Seedlings were germinated on plates containing Murashige and Skoog medium for 5 to 14 d. After 1 week at 4°C in the dark for stratification, seeds were grown on plates at 20°C under long-day conditions (16 h of light and 8 h of dark). Three-day-old seedlings were incubated for 3 h at 37°C in an incubator and then returned to recovery conditions of 22°C under long days for 2 d (see figure legends). For thermotolerance bioassays, the plates were then incubated for 60 min at 45°C under light. After the 45°C treatment, the plates were incubated at 20°C for recovery under long-day conditions for another 5 d.
Freezing Tolerance
Cold tolerance was assessed after 1 week of acclimation at 4°C. Fully expanded leaves of 7-week-old plants were harvested and placed in glass test tubes containing 0.4 mL of deionized water. The tubes were placed on ice, and extracellular freezing of the leaf tissues was initiated by the addition of deionized ice chips to each tube. After transfer to the controlled freezing bath set at 0°C and a 1-h equilibration period, the samples were cooled at 2°C h−1 to −6°C. The tubes were withdrawn after 30 min at −6°C, placed on ice, and thawed overnight at 4°C. After thawing, 12 mL of deionized water was added to each tube, and tubes were shaken gently (200 rpm) at room temperature for 3 h. The conductivity of the extraction solution was measured with an Orion conductimeter (model 105), and the leaves were frozen at −80°C for 1.5 h. The same extraction solution was readded to each tube after 30 min of reequilibration at room temperature and shaken for 2.5 h, and the conductivity of the solution was measured once again to normalize by the total amount of electrolytes.
Fosmid Libraries
We extracted DNA from 3-week-old TBG plants using a large-scale cetyl-trimethyl-ammonium bromide protocol (Porebski et al., 1997) including treatment with pectinase (Rogstad et al., 2001). We constructed a fosmid library using the Copy Control Fosmid Library Production Kit (Epicentre) and screened it as described previously (Bomblies et al., 2007) using digoxigenin-labeled (Roche) PCR probes to LHY (primers 5′-ACGCGGTTCAAGATGCTCCCA-3′ and 5′-GCTGCAGCATGAGCAGCAGGA-3′). We bar coded positive clones as described (Peterson et al., 2012) and sequenced 100-bp paired-end reads on an Illumina HiSeq 2000. We assembled reads de novo using Velvet (Zerbino and Birney, 2008).
Differentiation Analysis
To test for genetic differentiation, we used our previously published genomic short-read sequences for A. arenosa (Hollister et al., 2012; Yant et al., 2013) that we complemented with similarly processed genomes to reach a total of eight KA and seven TBG individuals. We aligned reads to the A. lyrata genome (Hu et al., 2011) using BWA (Li and Durbin, 2009) and realigned unmapped reads using Stampy (Lunter and Goodson, 2011). We calculated FST (Weir, 1990) and Fay and Wu’s H (Fay and Wu, 2000) after genotyping the alignments with GATK (McKenna et al., 2010). LHY haplotypes were sequenced from one TBG fosmid (see “Fosmid Libraries”) and PCR clones obtained from cDNA of three KA and one TBG individuals. The PCR products amplified with primers 5′-TTTCCACGCGGGTATTGTGA-3′ (forward) and 5′-TGTGTTCCCAACTTGGCTCT-3′ (reverse) were then ligated in pBluescript and sequenced from both ends with M13 forward and reverse primers. Where more than four clones were sequenced per individual, only four different haplotypes were reported.
Sequence data from this article can be found in the NCBI SRA data libraries under accession number SRP070489 within the NCBI BioProject PRJNA312410.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RNA-seq expression of SOC1 and VIN3.
Supplemental Figure S2. Marks of selection on a derived haplotype of LHY in the railway accession.
Supplemental Figure S3. High frequency of null expression levels and/or variance among genes without orthologs.
Supplemental Figure S4. LHY haplotypes in KA and TBG.
Supplemental Table S1. Vernalization response genes (a) + (b).
Supplemental Table S2. The 1% most differentially expressed genes.
Supplementary Material
Acknowledgments
We thank Mary Gehring and Elena Kramer for comments and discussion on this article, Missy Holbrook and colleagues for support with stress tolerance assays, Nimrod Rubinstein for help with paralog-specific expression analyses, and Kevin Wright for helpful advice across all dimensions of this work.
Glossary
- RNA-seq
RNA sequencing
- BAC
bacterial artificial chromosome
- GO
Gene Ontology
- cDNA
complementary DNA
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–1146465 to K.B.) and the Jean Gaillard Memorial Fellowship Fund (to P.B.).
Articles can be viewed without a subscription.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Albani MC, Castaings L, Wötzel S, Mateos JL, Wunder J, Wang R, Reymond M, Coupland G (2012) PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genet 8: e1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson HJ, Vonarx EJ, Pastushok L, Nakagawa M, Katafuchi A, Gruz P, Di Rubbo A, Grice DM, Osmond MJ, Sakamoto AN, et al. (2008) Arabidopsis thaliana Y-family DNA polymerase eta catalyses translesion synthesis and interacts functionally with PCNA2. Plant J 55: 895–908 [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Arnold B, Kim ST, Bomblies K (2015) Single geographic origin of a widespread autotetraploid Arabidopsis arenosa lineage followed by interploidy admixture. Mol Biol Evol 32: 1382–1395 [DOI] [PubMed] [Google Scholar]
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF (1996) Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA 93: 13404–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bac-Molenaar JA, Fradin EF, Becker FFM, Rienstra JA, van der Schoot J, Vreugdenhil D, Keurentjes JJB (2015) Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis thaliana. Plant Cell 27: 1857–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker HG. (1965) Characteristics and modes of origin of weeds. In Baker HG, Stebbins GL, eds, The Genetics of Colonizing Species. Academic Press, New York, pp 147–168 [Google Scholar]
- Baker HG. (1974) The evolution of weeds. Annu Rev Ecol Syst 5: 1–24 [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA (2008) Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol 147: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair AC, Wolfe LM (2004) The evolution of an invasive plant: an experimental study with Silene latifolia. Ecology 85: 3035–3042 [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F (2010) Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet 6: e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JH. (2004) A comparison of invasive and non-invasive dayflowers (Commelinaceae) across experimental nutrient and water gradients. Divers Distrib 10: 387–397 [Google Scholar]
- Chiang GCK, Barua D, Kramer EM, Amasino RM, Donohue K (2009) Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc Natl Acad Sci USA 106: 11661–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss MJ, Koch MA (2006) Poorly known relatives of Arabidopsis thaliana. Trends Plant Sci 11: 449–459 [DOI] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C (2012) Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337: 584–587 [DOI] [PubMed] [Google Scholar]
- Dong MA, Farré EM, Thomashow MF (2011) Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C, Bieniawska Z, Hincha DK, Hannah MA (2008) Interactions between the circadian clock and cold-response in Arabidopsis. Plant Signal Behav 3: 593–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R (2007) Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258 [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141: 550–550.e2 [DOI] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE (2007) Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci USA 104: 1278–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP. (2006) Plant Strategies, Vegetation Processes, and Ecosystem Properties. John Wiley & Sons, New York [Google Scholar]
- Grotkopp E, Rejmánek M, Rost TL (2002) Toward a causal explanation of plant invasiveness: seedling growth and life-history strategies of 29 pine (Pinus) species. Am Nat 159: 396–419 [DOI] [PubMed] [Google Scholar]
- Guo YL, Todesco M, Hagmann J, Das S, Weigel D (2012) Independent FLC mutations as causes of flowering-time variation in Arabidopsis thaliana and Capsella rubella. Genetics 192: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley WB. (2000) HSP101: a key component for the acquisition of thermotolerance in plants. Plant Cell 12: 457–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Willis JH (2006) Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution 60: 2466–2477 [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK (2006) Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol 142: 98–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M. (1998) Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci 3: 147–151 [Google Scholar]
- Hollister JD, Arnold BJ, Svedin E, Xue KS, Dilkes BP, Bomblies K (2012) Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa. PLoS Genet 8: e1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27: 25–35 [DOI] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC (2013) Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc Natl Acad Sci USA 110: 1542–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Koga A, Yamamoto T, Uchiyama Y, Mori Y, Hashimoto J, Kimura S, Sakaguchi K (2005) Two types of replication protein A in seed plants. FEBS J 272: 3270–3281 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Jørgensen MH, Ehrich D, Schmickl R, Koch MA, Brysting AK (2011) Interspecific and interploidal gene flow in Central European Arabidopsis (Brassicaceae). BMC Evol Biol 11: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Müller AE (2009) Flowering time control and applications in plant breeding. Trends Plant Sci 14: 563–573 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, Kim MD, Kim HJ, Lee MH, Moon J, Lee I, et al. (2004) A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet 36: 167–171 [DOI] [PubMed] [Google Scholar]
- Kim Y, An G (1992) Pollen-specific expression of the Arabidopsis thaliana alpha 1-tubulin promoter assayed by beta-glucuronidase, chloramphenicol acetyltransferase and diphtheria toxin reporter genes. Transgenic Res 1: 188–194 [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S (2010) Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J 63: 269–282 [DOI] [PubMed] [Google Scholar]
- Kuittinen H, Niittyvuopio A, Rinne P, Savolainen O (2008) Natural variation in Arabidopsis lyrata vernalization requirement conferred by a FRIGIDA indel polymorphism. Mol Biol Evol 25: 319–329 [DOI] [PubMed] [Google Scholar]
- Le Corre V, Roux F, Reboud X (2002) DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol 19: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Lee CF, Pu HY, Wang LC, Sayler RJ, Yeh CH, Wu SJ (2006) Mutation in a homolog of yeast Vps53p accounts for the heat and osmotic hypersensitive phenotypes in Arabidopsis hit1-1 mutant. Planta 224: 330–338 [DOI] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D (2005) Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet 1: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Charng YY (2012) Acquired thermotolerance independent of heat shock factor A1 (HsfA1), the master regulator of the heat stress response. Plant Signal Behav 7: 547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang C, Chen J, Guo L, Li X, Li W, Yu Z, Deng J, Zhang P, Zhang K, et al. (2013) Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiol Biochem 64: 92–98 [DOI] [PubMed] [Google Scholar]
- Lobstein E, Guyon A, Férault M, Twell D, Pelletier G, Bonhomme S (2004) The putative Arabidopsis homolog of yeast vps52p is required for pollen tube elongation, localizes to Golgi, and might be involved in vesicle trafficking. Plant Physiol 135: 1480–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M (2011) Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21: 936–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- McKay JK, Richards JH, Mitchell-Olds T (2003) Genetics of drought adaptation in Arabidopsis thaliana. I. Pleiotropy contributes to genetic correlations among ecological traits. Mol Ecol 12: 1137–1151 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri D, Breiman A (2009) Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J 59: 387–399 [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B, Picó FX, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C (2011) Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol 157: 1942–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Vigo B, Savic M, Ausín I, Ramiro M, Martín B, Picó FX, Alonso-Blanco C (2016) Environmental and genetic interactions reveal FLOWERING LOCUS C as a modulator of the natural variation for the plasticity of flowering in Arabidopsis. Plant Cell Environ 39: 282–294 [DOI] [PubMed] [Google Scholar]
- Merret R, Descombin J, Juan YT, Favory JJ, Carpentier MC, Chaparro C, Charng YY, Deragon JM, Bousquet-Antonelli C (2013) XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep 5: 1279–1293 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, Young J, Austin-Phillips S, Quail PH (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15: 1962–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nah G, Chen ZJ (2010) Tandem duplication of the FLC locus and the origin of a new gene in Arabidopsis related species and their functional implications in allopolyploids. New Phytol 186: 228–238 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane SL. (1997) A synopsis of Arabidopsis (Brassicaceae). Novon 7: 323–327 [Google Scholar]
- Otto CRV, Snodgrass JW, Forester DC, Mitchell JC, Miller RW (2007) Climatic variation and the distribution of an amphibian polyploid complex. J Anim Ecol 76: 1053–1061 [DOI] [PubMed] [Google Scholar]
- Pandit MK, Tan HTW, Bisht MS (2006) Polyploidy in invasive plant species of Singapore. Bot J Linn Soc 151: 395–403 [Google Scholar]
- Panikulangara TJ, Eggers-Schumacher G, Wunderlich M, Stransky H, Schöffl F (2004) Galactinol synthase1: a novel heat shock factor target gene responsible for heat-induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiol 136: 3148–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JD, Rubinstein ND, Fernandez DE, Santoro SW, Needleman LA, Ho-Shing O, Choi JJ, Zirlinger M, Chen SK, Liu JS, et al. (2015) Quantitative and functional interrogation of parent-of-origin allelic expression biases in the brain. eLife 4: e07860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7: e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15: 8–15 [Google Scholar]
- Prändl R, Hinderhofer K, Eggers-Schumacher G, Schöffl F (1998) HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol Gen Genet 258: 269–278 [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ (2008) Adaptive evolution in invasive species. Trends Plant Sci 13: 288–294 [DOI] [PubMed] [Google Scholar]
- Qian J, Chen J, Liu YF, Yang LL, Li WP, Zhang LM (2014) Overexpression of Arabidopsis HsfA1a enhances diverse stress tolerance by promoting stress-induced Hsp expression. Genet Mol Res 13: 1233–1243 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat V, Abdelsamad A, Pietzenuk B, Seymour DK, Koenig D, Weigel D, Pecinka A, Schneeberger K (2015) Improving the annotation of Arabidopsis lyrata using RNA-Seq data. PLoS ONE 10: e0137391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter RS, Coomber SA, Bourett TM, Bartley GE, Scolnik PA (1994) Control of leaf and chloroplast development by the Arabidopsis gene pale cress. Plant Cell 6: 1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy E, Cabrito TR, Batista RA, Teixeira MC, Sá-Correia I, Duque P (2012) The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol 195: 356–371 [DOI] [PubMed] [Google Scholar]
- Richter R, Bastakis E, Schwechheimer C (2013) Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol 162: 1992–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogstad SH, Keane B, Keiffer CH, Hebard F, Sisco P (2001) DNA extraction from plants: the use of pectinase. Plant Mol Biol Rep 19: 353–359 [Google Scholar]
- Salomé PA, Bomblies K, Laitinen RAE, Yant L, Mott R, Weigel D (2011) Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samis KE, Heath KD, Stinchcombe JR (2008) Discordant longitudinal clines in flowering time and phytochrome C in Arabidopsis thaliana. Evolution 62: 2971–2983 [DOI] [PubMed] [Google Scholar]
- Scarcelli N, Cheverud JM, Schaal BA, Kover PX (2007) Antagonistic pleiotropic effects reduce the potential adaptive value of the FRIGIDA locus. Proc Natl Acad Sci USA 104: 16986–16991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schmickl R, Paule J, Klein J, Marhold K, Koch MA (2012) The evolutionary history of the Arabidopsis arenosa complex: diverse tetraploids mask the Western Carpathian center of species and genetic diversity. PLoS ONE 7: e42691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl F, Prändl R, Reindl A (1998) Regulation of the heat-shock response. Plant Physiol 117: 1135–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz H. (1962) Nomenklatorische und systematische Studien an Cardaminopsis arenosa (L.) Hayek. Wildenowia 3: 137–149 [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E, Lee H, Jeon J, Park H, Kim J, Noh YS, Lee I (2009) Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrard ME, Maherali H (2006) The adaptive significance of drought escape in Avena barbata, an annual grass. Evolution 60: 2478–2489 [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol 138: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Slotte T, Huang HR, Holm K, Ceplitis A, Onge KS, Chen J, Lagercrantz U, Lascoux M (2009) Splicing variation at a FLOWERING LOCUS C homeolog is associated with flowering time variation in the tetraploid Capsella bursa-pastoris. Genetics 183: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE (2000) The role of genetic and genomic attributes in the success of polyploids. Proc Natl Acad Sci USA 97: 7051–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Irwin J, Dean C (2013) Remembering the prolonged cold of winter. Curr Biol 23: R807–R811 [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF (1998) Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumuran N, Weiser C (1972) An excised leaflet test for evaluating potato frost tolerance. HortScience 7: 467–468 [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Thomashow MF. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Turro E, Su SY, Gonçalves Â, Coin LJM, Richardson S, Lewin A (2011) Haplotype and isoform specific expression estimation using multi-mapping RNA-seq reads. Genome Biol 12: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Tsai MC, Chang KY, Fan YS, Yeh CH, Wu SJ (2011) Involvement of the Arabidopsis HIT1/AtVPS53 tethering protein homologue in the acclimation of the plasma membrane to heat stress. J Exp Bot 62: 3609–3620 [DOI] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC (2009) PEP1 regulates perennial flowering in Arabis alpina. Nature 459: 423–427 [DOI] [PubMed] [Google Scholar]
- Weinig C, Dorn LA, Kane NC, German ZM, Halldorsdottir SS, Ungerer MC, Toyonaga Y, Mackay TFC, Purugganan MD, Schmitt J (2003) Heterogeneous selection at specific loci in natural environments in Arabidopsis thaliana. Genetics 165: 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS. (1990) Genetic Data Analysis: Methods for Discrete Population Genetic Data. Sinauer Associates, Sunderland, MA: [DOI] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D (2005) FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CA, Lowry DB, Nutter LI, Willis JH (2010) Natural variation for drought-response traits in the Mimulus guttatus species complex. Oecologia 162: 23–33 [DOI] [PubMed] [Google Scholar]
- Yant L, Hollister JD, Wright KM, Arnold BJ, Higgins JD, Franklin FCH, Bomblies K (2013) Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr Biol 23: 2151–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye CY, Yang X, Xia X, Yin W (2013) Comparative analysis of cation/proton antiporter superfamily in plants. Gene 521: 245–251 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim JM, Seki M, Todaka D, et al. (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 286: 321–332 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerebecki RA, Sorte CJB (2011) Temperature tolerance and stress proteins as mechanisms of invasive species success. PLoS ONE 6: e14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.