Rescuing the feedback inhibition of fatty acid synthesis in castor hydroxylase-expressing Arabidopsis seeds increases ricinoleic acid accumulation.
Abstract
Previous attempts at engineering Arabidopsis (Arabidopsis thaliana) to produce seed oils containing hydroxy fatty acids (HFA) have resulted in low yields of HFA compared with the native castor (Ricinus communis) plant and caused undesirable effects, including reduced total oil content. Recent studies have led to an understanding of problems involved in the accumulation of HFA in oils of transgenic plants, which include metabolic bottlenecks and a decrease in the rate of fatty acid synthesis. Focusing on engineering the triacylglycerol assembly mechanisms led to modest increases in the HFA content of seed oil, but much room for improvement still remains. We hypothesized that engineering fatty acid synthesis in the plastids to increase flux would facilitate enhanced total incorporation of fatty acids, including HFA, into seed oil. The transcription factor WRINKLED1 (WRI1) positively regulates the expression of genes involved in fatty acid synthesis and controls seed oil levels. We overexpressed Arabidopsis WRI1 in seeds of a transgenic line expressing the castor fatty acid hydroxylase. The proportion of HFA in the oil, the total HFA per seed, and the total oil content of seeds increased to an average of 20.9%, 1.26 µg, and 32.2%, respectively, across five independent lines, compared with 17.6%, 0.83 µg, and 27.9%, respectively, for isogenic segregants. WRI1 and WRI1-regulated genes involved in fatty acid synthesis were up-regulated, providing for a corresponding increase in the rate of fatty acid synthesis.
Modified fatty acids (FA) such as hydroxy fatty acids (HFA), cyclopropane FA, epoxy FA, and many more have physical and chemical properties that make them useful in industry or human health (Badami and Patil, 1980). However, the plants that make these unusual modified FA are generally unsuitable for large-scale, industrialized agriculture (Voelker and Kinney, 2001; Dyer et al., 2008). It was initially proposed that by identifying and expressing the enzymes responsible for synthesizing the desired FA in high-yield crop plants, many of the difficulties involved in obtaining large quantities of modified FA could be overcome; however, most transgenic plants that were engineered to produce the desired modified FA accumulated them at low levels compared with the native plant (Broun and Somerville, 1997; Cahoon et al., 2006). The lack of selectivity for modified FA substrates by the host plant enzymes, the presence of metabolic bottlenecks, and feedback inhibition of de novo FA synthesis have all been identified as factors limiting oilseed engineering (Knutzon et al., 1999; Cahoon et al., 2007; Burgal et al., 2008; Li et al., 2010; Bates and Browse, 2011; Kim et al., 2011; van Erp et al., 2011; Bates et al., 2014). HFA are an important class of modified FA and are very useful in industrial applications as lubricants and in the making of plastics, nylons, synthetic resins, rubber, dyes, food additives, cosmetics, etc. They are produced at high levels in seeds of castor (Ricinus communis). Similar to most plants that make modified FA, castor is unsuitable for large-scale, industrialized agriculture. The seeds of castor contain an extremely toxic protein, ricin, to which there is no known antidote. Also, additional toxic compounds present in castor seeds can cause allergic reactions in humans. Most of the cultivation of castor around the world occurs in places with tropical climates, especially India, China, and Brazil, and is done using manual labor. Our focus is on engineering of HFA in Arabidopsis (Arabidopsis thaliana) to explore factors affecting the accumulation of HFA in nonnative plant species. Identifying strategies to improve the accumulation of HFA in seeds of heterologous plant systems will be helpful in creating solutions for large-scale, reliable production of HFA and, likely, other unusual FA as well.
The FA hydroxylase enzyme from castor (RcFAH12) hydroxylates the Δ12 position of oleic acid esterified to the sn-2 position of phosphatidylcholine to produce ricinoleic acid (Δ12-hydroxy-9-cis-octadecanoic acid). FAH12 is a homolog of the oleoyl-phosphatidylcholine desaturase FAD2 (van de Loo et al., 1995). Castor RcFAH12 complementary DNA (cDNA) has been expressed under the control of several seed-specific promoters in Arabidopsis. This led to HFA accumulation of only 17% of total seed FA (Broun and Somerville, 1997; Smith et al., 2000, 2003; Lu et al., 2006) compared with nearly 90% in triacylglycerol (TAG) of castor seeds (Badami and Patil, 1980). Some of the strategies used to engineer Arabidopsis seeds to increase HFA accumulation in transgenic plants include the coexpression of cDNAs from castor encoding acyltransferases such as ACYL-COENZYME A:DIACYLGLYCEROL ACYLTRANSFERASE2 (RcDGAT2; Burgal et al., 2008) or PHOSPHOLIPID:DIACYLGLYCEROL ACYLTRANSFERASE1A (RcPDAT1A; Kim et al., 2011; van Erp et al., 2011) to increase the specificity for HFA incorporation into TAG. Individually, RcDGAT2 and RcPDAT1A each increased HFA levels from 17% to approximately 26%. Stacking RcDGAT2 and RcPDAT1A further increased HFA levels to 28% of total seed FA, thus supporting the hypothesis that castor enzymes coevolved to accumulate HFA. In a further improvement to introducing the castor acyltransferases in Arabidopsis, the endogenous AtDGAT1 gene was knocked down to reduce competition with RcDGAT2 for common FA. This strategy further increased the HFA level to 31% (van Erp et al., 2015). However, introducing the castor electron transport system for efficient electron transport to RcFAH12 did not yield any additional increase (Wayne and Browse, 2013).
Further research into the metabolic changes in Arabidopsis seeds expressing RcFAH12 that limit the accumulation of HFA in the oil has revealed that inefficient utilization of HFA induces feedback inhibition of acetyl-CoA carboxylase, leading to reduced FA synthesis and decreased seed oil content of the transgenic seed (Bates et al., 2014). We hypothesized that engineering FA synthesis to increase flux might overcome the feedback inhibition and facilitate the flow of FA into TAG, thus restoring seed oil content and increasing HFA content on a weight basis. In wild-type Arabidopsis and in rapeseed (Brassica napus), it has proved possible to increase FA synthesis and seed oil content by the overexpression of transcription factors such as WRINKLED1 (WRI1; Cernac and Benning, 2004; Shen et al., 2010; Kelly et al., 2013; van Erp et al., 2014; Wu et al., 2014). WRI1, an APETALA2/ethylene-responsive element-binding protein-type transcription factor (Cernac and Benning, 2004; Masaki et al., 2005), is a master regulator that controls the expression of genes involved in FA synthesis and embryogenesis in many plant species (Ruuska et al., 2002; Baud et al., 2009; Maeo et al., 2009; To et al., 2012; Ma et al., 2013). WRI1 binds the conserved ASML1/WRI1 (AW) box, [CnTnG](n)7[CG] (where n is any nucleotide), present in the proximal upstream region of WRI1 target genes and activates transcription. Mutations in the conserved [CnTnG] and [CG] core motifs abolish activation by WRI1 (Maeo et al., 2009). A loss-of-function mutation, wri1, in Arabidopsis causes an 80% reduction in the level of FA in seeds, which leads to a wrinkled appearance (Focks and Benning, 1998; Cernac and Benning, 2004). Both in seeds and in vegetative tissues, overexpression of WRI1 leads to an increase in the level of FA accumulating in TAG (Cernac and Benning, 2004; Shen et al., 2010; Sanjaya et al., 2011; Kelly et al., 2013; Vanhercke et al., 2013, 2014; van Erp et al., 2014; Wu et al., 2014; Grimberg et al., 2015; Reynolds et al., 2015; Zale et al., 2016). WRI1 overexpression elevates the transcript level of its target genes involved in FA synthesis (Maeo et al., 2009; To et al., 2012), and this is assumed to lead to an increase in protein level and enzyme activity. WRI1 transcript levels are tightly regulated by many transcription factors, including LEAFY COTYLEDON1 (LEC1; Mu et al., 2008) and LEC2 (Baud et al., 2007a), and also by feedback regulation by the WRI1 protein (Cernac and Benning, 2004). WRI1 protein levels also are regulated by interaction with CULLIN3-based E3 ligases, which target WRI1 for degradation via the 26S proteasome (Chen et al., 2013). WRI1 is constantly degraded, but the rate of turnover varies, possibly in response to unknown signals (Chen et al., 2013).
To test our hypothesis that feedback inhibition of FA synthesis in HFA-producing seeds could be overwhelmed, we chose WRI1 to specifically target and enhance the expression of genes involved in FA synthesis in seeds. We overexpressed Arabidopsis WRI1 using a seed-specific promoter in an RcFAH12-expressing line. We observed an increase in WRI1 transcript and the transcripts of WRI1 target genes involved in FA synthesis. The rate of FA synthesis, total oil, HFA content, and TAG-containing multiple HFA all increased compared with the RcFAH12-expressing line.
RESULTS
WRI1 Overexpression in fatty acid elongase1 RcFAH12 Seeds Increases HFA Accumulation
RcFAH12 expressed in an Arabidopsis fatty acid elongase1 (fae1) background (Kunst et al., 1992) led to the accumulation of approximately 17% HFA in seed oil (Lu et al., 2006). The fae1 mutation blocks the elongation of 18:1 to 20:1 (Kunst et al., 1992), with the result that no 20:1-OH is produced in the hydroxylase-expressing line (Broun and Somerville, 1997; Lu et al., 2006), simplifying HFA analysis by gas chromatography (GC).
The rate of FA synthesis is reduced 30% to 50% in developing seeds of the fae1 RcFAH12 line CL37 due to feedback inhibition, resulting in reduced seed oil content compared with fae1 (Bates et al., 2014). To test our hypothesis that increasing flux through FA synthesis in seeds would rescue feedback inhibition and increase HFA and total FA contents of seed oil, we overexpressed Arabidopsis WRI1 in the fae1 RcFAH12 background. We cloned a WRI1 cDNA under the control of the strong seed-specific phaseolin promoter from Phaseolus vulgaris (Sengupta-Gopalan et al., 1985) in a plant transformation vector with the fluorescent DsRed selection marker as described previously (Lu et al., 2006).
After transformation of fae1 RcFAH12 with the WRI1 expression construct, fluorescent red seeds were selected and propagated. Segregating T2 seeds from 69 independent T1 transformants were analyzed by GC to assess the proportion of HFA in seed lipids and the total HFA per seed. The difference in HFA between red (transformed) and brown (nontransformed) segregating T2 seeds from each independent T1 line allowed a quantitative measurement of changes in HFA attributable to the overexpression of WRI1. Of the 69 lines tested, red seeds from 62 lines exhibited increases in both the proportion and total amount of HFA. Red seeds from seven lines had substantially reduced HFA compared with segregating brown controls, possibly due to the cosuppression of WRI1 in these transgenics. From an initial set of 37 lines, six lines with 20% or more HFA in their red seeds also segregated in a 3:1 ratio of red to brown seeds, indicating a single site of insertion for the WRI1/DsRed transgene construct. These are lines 9, 13, 22, 27, 28, and 30 as shown in Figure 1. Of these, lines 9, 13, and 22 exhibited more than 30% increase in the proportion of HFA and more than 70% increase in total HFA per seed over their respective brown segregating seeds (Fig. 1). Analysis of a second set of 32 lines identified 19 lines segregating in a 3:1 red:brown ratio. These 19 lines are the remaining lines shown in Figure 1 and include five lines (38, 49, 55, 61, and 69) with significantly more than 20% HFA. Among the 25 lines shown in Figure 1, lines 9, 13, 22, 42, and 61 had large increases in the proportion of HFA in red seeds compared with brown seed controls, accompanied by substantial increases in total HFA per seed. These five lines were chosen for propagation and further analysis in the T3 generation.
Figure 1.
FA analysis of fae1 RcFAH12 WRI1 transgenics. Primary transformants were grown to maturity, then T2 seed from individual T1 plants was separated into transgenic seeds expressing the DsRed marker (red) and nontransgenic siblings (brown) and analyzed by GC. A, Percentage HFA, sorted by highest level. B, HFA per seed, sorted by highest level. One to three replicates were analyzed per line. Error bars represent sd.
WRI1 Overexpression Increases the Level of HFA and Total Oil in Seeds
Between 50 and 60 transgenic T2 plants from each of the lines 9, 13, 22, 42, and 61 were grown along with their corresponding nontransgenic fae1 RcFAH12(−) segregants and fae1 RcFAH12 in the same growth conditions for accurate comparison of total seed oil, which varies considerably depending on growth conditions (Li et al., 2006). Two red, two brown, and one fae1 RcFAH12 were planted in random positions per pot. Pots were randomly distributed within the growth chamber and rotated on a regular basis to uniformly expose each plant to the growth conditions. Between eight and 11 homozygous fae1 FAH12 WRI1 segregants were identified (based on 100% red T3 seeds) for each of the four lines. Table I summarizes the data for FA content and composition for these homozygous transgenics compared with segregating fae1 RcFAH12(−) siblings and plants of the parental fae1 RcFAH12 line grown as controls. For all of the parameters measured, each line showed substantial and statistically significant increases relative to both controls. The proportion of HFA in the oil was increased by 8% (line 9) to 31% (line 13), HFA per seed by 22% (line 61) to 80% (line 13), total FA per seed by 22% (line 9) to 49% (line 13), and FA as a proportion of seed weight (percentage oil) by 10% (line 61) to 22% (line 13). The large increases in the proportion of HFA and in total HFA per seed (Table I) indicate that the Phas:WRI1 transgene boosts HFA accumulation in fae1 RcFAH12 plants. The increases in total FA per seed and percentage oil (Table I) demonstrate that the transgene increases FA accumulation in fae1 RcFAH12, possibly by partially alleviating the reduced rate of FA synthesis caused by the expression of RcFAH12 (Bates et al., 2014).
Table I. FA analysis of fae1 RcFAH12 WRI1 T3 lines.
fae1 RcFAH12 WRI1 lines 13, 42, 22, 61, and 9 were obtained from independent transformation events. Data for independent lines represent averages of eight to 11 individual plants. Averages at the end of each category represent means of the five independent lines. Error is represented by se; one-way ANOVA, different letters across rows indicate values that are significantly different.
| Category | fae1 RcFAH12 | fae1 RcFAH12(−) | fae1 RcFAH12 WRI1 |
|---|---|---|---|
| Percentage HFA | |||
| Line 13 | 17.1 ± 0.46 b | 16.8 ± 0.58 b | 22.1 ± 0.46 a |
| Line 42 | 16.6 ± 0.37 b | 16.2 ± 0.40 b | 20.3 ± 0.35 a |
| Line 22 | 18.7 ± 0.49 b | 18.3 ± 0.47 b | 22.5 ± 0.58 a |
| Line 61 | 18.5 ± 0.19 b | 18.6 ± 0.16 b | 20.1 ± 0.44 a |
| Line 9 | 18.2 ± 0.17 b | 18.2 ± 0.34 b | 19.7 ± 0.30 a |
| Average | 17.8 ± 0.41 b | 17.6 ± 0.46 b | 20.9 ± 0.56 a |
| Total HFA per seed (µg) | |||
| Line 13 | 0.74 ± 0.07 b | 0.71 ± 0.08 b | 1.38 ± 0.05 a |
| Line 42 | 0.67 ± 0.04 b | 0.67 ± 0.05 b | 1.14 ± 0.07 a |
| Line 22 | 0.82 ± 0.07 b | 0.92 ± 0.06 b | 1.42 ± 0.10 a |
| Line 61 | 0.96 ± 0.02 b | 1.01 ± 0.03 b | 1.20 ± 0.04 a |
| Line 9 | 0.87 ± 0.04 b | 0.85 ± 0.05 b | 1.14 ± 0.05 a |
| Average | 0.81 ± 0.05 b | 0.83 ± 0.06 b | 1.26 ± 0.06 a |
| Total FA per seed (µg) | |||
| Line 13 | 4.29 ± 0.32 b | 4.10 ± 0.39 b | 6.25 ± 0.21 a |
| Line 42 | 4.04 ± 0.19 b | 4.02 ± 0.22 b | 5.50 ± 0.24 a |
| Line 22 | 4.38 ± 0.19 b | 4.95 ± 0.17 b | 6.19 ± 0.31 a |
| Line 61 | 5.11 ± 0.10 b | 5.44 ± 0.14 b | 5.99 ± 0.17 a |
| Line 9 | 4.75 ± 0.16 b | 4.75 ± 0.18 b | 5.79 ± 0.22 a |
| Average | 4.52 ± 0.19 b | 4.65 ± 0.27 b | 5.96 ± 0.13 a |
| Percentage oil (as a fraction of dry seed weight) | |||
| Line 13 | 26.6 ± 1.82 b | 25.9 ± 1.58 b | 32.1 ± 0.69 a |
| Line 42 | 26.9 ± 1.01 b | 25.6 ± 0.97 b | 30.6 ± 0.68 a |
| Line 22 | 27.2 ± 1.01 b | 28.5 ± 0.84 b | 32.8 ± 1.12 a |
| Line 61 | 29.8 ± 0.37 b | 30.2 ± 0.54 b | 33.0 ± 0.66 a |
| Line 9 | 28.2 ± 0.31 b | 29.2 ± 0.71 b | 32.3 ± 0.66 a |
| Average | 27.7 ± 0.57 b | 27.9 ± 0.90 b | 32.2 ± 0.43 a |
Developing Seeds of fae1 RcFAH12 WRI1 Have Higher Transcript Levels of WRI1 Compared with fae1
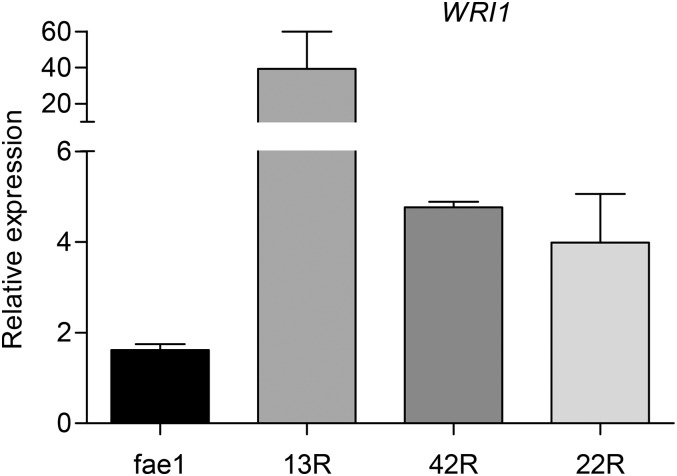
In our analysis of T3 seed samples (Table I), lines 13, 42, and 22 showed the largest increases in percentage HFA and oil per seed weight. To find out the extent of WRI1 expression in these lines, we harvested developing seeds at 11 to 12 d after flowering (DAF) from homozygous transgenics of each line, together with seeds from fae1 plants grown alongside as a control. RNA samples prepared from four biological replicates of each line were used to assess WRI1 transcript levels by quantitative reverse transcription (qRT)-PCR, using the transcript of protein phosphatase 2A subunit (PP2A) as a reference (Czechowski et al., 2005). The transcript level of WRI1 in line 13 was 24-fold higher than in the fae1 control, while increases of 2.5- and 3-fold were found for lines 42 and 22, respectively (Fig. 2). All the increases were statistically significant (P < 0.05). These results are consistent with the increases in HFA and FA in fae1 RcFAH12 WRI1 seeds being a consequence of increased expression of the WRI1 transcription factor.
Figure 2.
Transcriptional analysis of WRI1 gene expression. WRI1 transcript level was measured by qRT-PCR in 11- to 12-DAF developing seeds of fae1 and fae1 RcFAH12 WRI1 T3 lines 13, 42, and 22. n = 4, and error bars represent se. Two-tailed Student’s t test, P ≤ 0.05 for all lines compared with fae1.
Seed Oil Content of Line 13 Is Intermediate between fae1 RcFAH12 and fae1
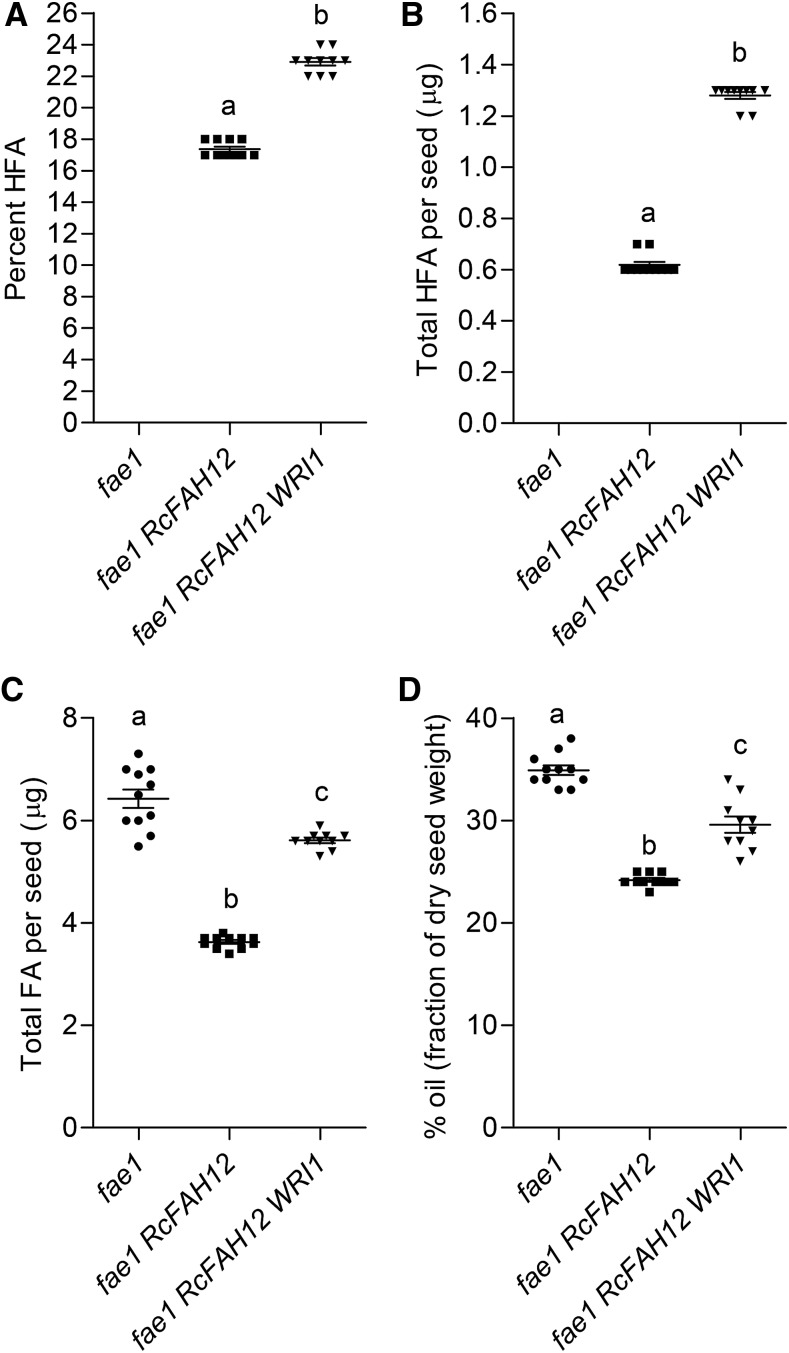
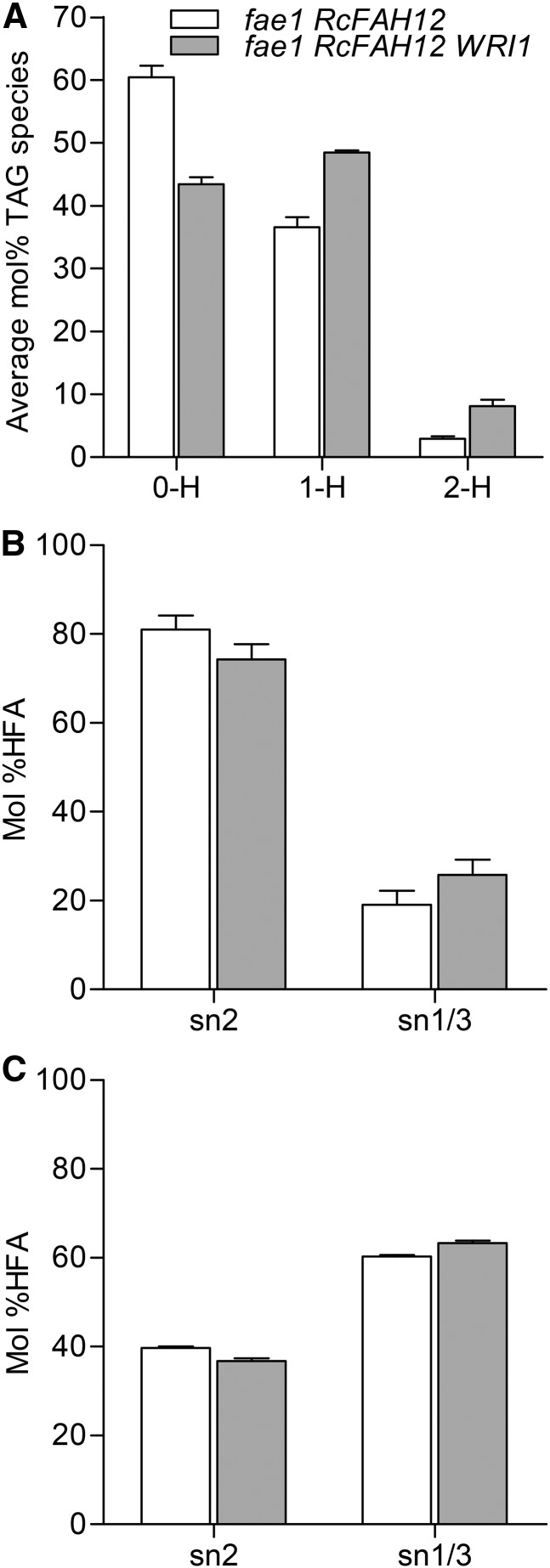
Because seeds of fae1 RcFAH12 WRI1 line 13 had the highest expression of WRI1 (Fig. 2) and the largest increases in both HFA and total FA in our analysis of homozygous T2 plants (Table I), we carried out additional experiments on this line. Previous studies have found that expression of the castor hydroxylase in Arabidopsis seeds causes substantial reductions in the rate of FA synthesis and the oil content of mature seeds (Bates and Browse, 2011; van Erp et al., 2011; Bates et al., 2014), while our results from five fae1 RcFAH12 WRI1 lines indicate that they all have higher oil contents than fae1 RcFAH12 (Table I). To assess the extent to which WRI1 overexpression had reversed the declines in these parameters, we grew homozygous T3 plants of line 13 along with fae1 RcFAH12 and fae1 plants. Data on the proportion of HFA in seed oil (Fig. 3A) and total HFA per seed (Fig. 3B) were similar to those in our previous experiment (Table I) in showing large increases associated with the overexpression of WRI in line 13. In this experiment, fae1 seeds contained 6.43 ± 0.18 µg seed−1, corresponding to 34.9% ± 0.5% oil (Fig. 3, C and D). Consistent with previously published results (Bates and Browse, 2011; Bates et al., 2014), total FA were decreased to only 3.62 ± 0.04 µg seed−1, or 24.2% ± 0.2% of seed weight, in the fae1 RcFAH12 (CL37) line. In line 13 overexpressing WRI1, the decrease in total FA per seed was substantially (70%) reversed to 5.61 ± 0.05 µg seed−1 (Fig. 3C). Recovery of oil content, 29.6% ± 0.8% (Fig. 3D), was somewhat less, but this also indicates that the effect of the bottleneck in lipid metabolism caused by expression of the castor hydroxylase (Bates et al., 2014) is partially alleviated by overexpression of the WRI1 transcription factor. As indicated by the results in Figure 3A and the complete analysis of seed FA composition (Supplemental Fig. S1), HFA were increased proportionally more than total FA by overexpression of WRI1 in the CL37 (fae1 RcFAH12) line. This effect is also apparent from the analysis of TAG molecular species shown in Figure 4A. Compared with TAG in fae1 RcFAH12 seeds, the TAG from seeds in fae1 RcFAH12 WRI1 line 13 contain higher proportions of molecular species with either one or two HFA and a substantial reduction in molecular species with no HFA. As determined by Student’s t test, these differences were statistically significant (P < 0.008). We used lipase assays (van Erp et al., 2011) to determine the distribution of HFA between the sn-2 position and the sn-1/3 positions of 1-HFA-TAG (Fig. 4B) and 2-HFA-TAG (Fig. 4C). In 1-HFA-TAG, the percentage of HFA at the sn-2 position decreased from 81% to 74% in fae1 RcFAH12 WRI1 compared with fae1 RcFAH12, and HFA at the sn-1/3 positions correspondingly increased from 19% to 26% in fae1 RcFAH12 WRI1 compared with fae1 RcFAH12 (Fig. 4B). However, these differences were not statistically significant. In 2-HFA-TAG, HFA at the sn-2 position decreased from 40% to 37% in fae1 RcFAH12 WRI1 relative to fae1 RcFAH12 controls, with an increase from 60% to 63% at the sn-1/3 positions, and were statistically significant, as determined by Student’s t test (P < 0.05). Taken together, these results suggest little to no difference between the distribution of HFA at the sn-2 or sn-1/3 positions between fae1 RcFAH12 and fae1 RcFAH12 WRI1.
Figure 3.
Extended analysis of FA from T4 seeds of fae1 RcFAH12 WRI1. Seed samples were analyzed from fae1, fae1 RcFAH12, and T4 seeds of fae1 RcFAH12 WRI1 line 13. A, Percentage HFA. B, HFA per seed. C, Total FA per seed. D, Seed oil content. Horizontal bars represent average values of 10 to 11 individuals. Each symbol represents an individual plant, and error bars represent se. One-way ANOVA, different letters above each average indicates values that are significantly different.
Figure 4.
Characterization of TAG species. A, HFA-containing molecular species of TAG. TAG molecular species from fae1 RcFAH12 and fae1 RcFAH12 WRI1 line 13 were separated by thin-layer chromatography (TLC) and quantified by GC. Columns indicate TAG molecular species containing zero, one, or two HFA (0-H, 1-H, and 2-H). n = 3, and error bars represent se. B, Regiochemical analysis of 1-HFA-TAG. n ≥ 3, and error bars represent se. C, Regiochemical analysis of 2-HFA-TAG. n ≥ 3, and error bars represent se.
WRI1 Overexpression Elevates Transcripts of Genes Involved in FA Synthesis
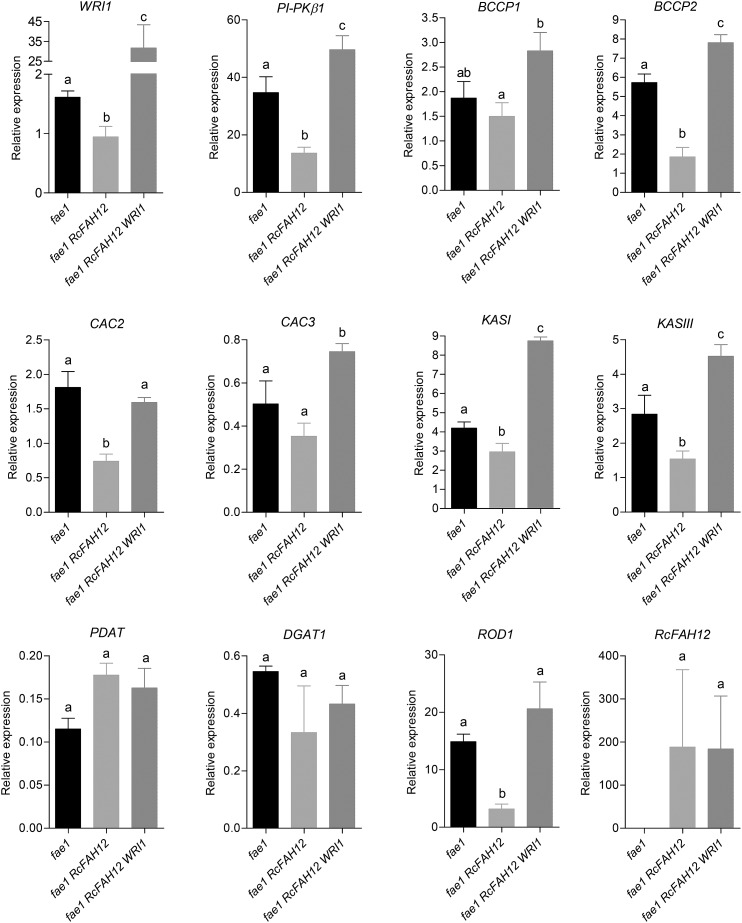
WRI1 was shown previously to regulate glycolysis and FA synthesis genes, and overexpression of WRI1 increases the transcript levels of many of these genes (Ruuska et al., 2002; Maeo et al., 2009; Sanjaya et al., 2011; To et al., 2012). To test if the same was true in our hydroxylase-expressing line 13 overexpressing WRI, we tested the expression of known WRI1 targets as well as nontargets involved in TAG assembly. PYRUVATE KINASE SUBUNIT (PI-PKβ1; AT5G52920), BIOTIN CARBOXYL CARRIER PROTEIN1 (BCCP1; AT5G16390), BCCP2 (AT5G15530), ACCASE BIOTIN CARBOXYLASE SUBUNIT (CAC2; AT5G35360), CAC3 (AT2G38040), 3-KETOACYL ACP SYNTHASE I (KASI; AT5G46290), and KASIII (AT1G62640) are key genes in glycolysis and FA synthesis and are known to be up-regulated by the overexpression of WRI1 (Maeo et al., 2009; To et al., 2012). We measured transcript levels of these genes and WRI1 in developing seeds of the same three lines grown for the analysis of mature seeds shown in Figure 3. Interestingly, the expression of RcFAH12 alone is associated with a significant 43% decrease in WRI1 transcript relative to the fae1 parental line (Fig. 5). The lower WRI1 expression is reflected in substantial decreases in transcripts for most of the FA synthesis genes measured, ranging from 32% (for KASI) to over 3-fold (for BCCP2). There is also a 60% decrease in the level of transcript for PI-PKβ1 (Fig. 5). In developing seeds of fae1 RcFAH12 WRI1 line 13, WRI1 expression was 22-fold higher than in fae1 seeds and 34-fold higher than in the fae1 RcFAH12 parental line. Consistent with the characterized activity of the WRI1 transcription factor, transcripts of all the FA synthesis genes measured and of PI-PKβ1 are increased 2- to 4-fold in line 13 relative to fae1 FAH12, and all the genes except CAC2 have transcript levels significantly (P < 0.05) higher than those in the fae1 control (Fig. 5).
Figure 5.
Analysis of gene expression in developing seeds. Transcript levels of genes involved in FA synthesis and TAG accumulation were analyzed by qRT-PCR in 11- to 12-DAF developing seeds of fae1, fae1 RcFAH12, and fae1 RcFAH12 WRI1 T3 line 13. n = 4 biological replicates, and error bars represent se. One-tailed Student’s t test, columns with different letters are significantly different.
Genes participating in TAG assembly are not up-regulated by WRI1 overexpression based on previous research (Maeo et al., 2009; To et al., 2012), and similarly, we did not observe any changes in transcript levels of DGAT1 (AT2G19450) and PDAT (AT3G44830) among the three lines (Fig. 5). Significantly, however, we measured a 4.6-fold reduction between fae1 and fae1 RcFAH12 in transcript of the ROD1 gene (AT2G19450) that encodes phosphatidylcholine:diacylglycerol cholinephosphotransferase (PDCT), an important enzyme in seed TAG synthesis (Lu et al., 2009). Overexpression of WRI1 in the fae1 RcFAH12 background resulted in a 6.4-fold increase in ROD1 expression (Fig. 5). These results suggest the regulation of ROD1 by WRI1. Upon scanning the region upstream of the ROD1 transcription start site, we detected an AW box, CtTgGaaatctcCG, in the 5′ untranslated region (UTR) at positions −83 to −69 bp relative to the ATG start codon (Supplemental Fig. S2), similar to the location of AW boxes in 5′ UTRs and promoters of FA synthesis genes (Supplemental Fig. S3). Genes containing the AW box upstream of their transcription start site have been shown to be direct targets of WRI1 (Maeo et al., 2009), so the changes in ROD1 expression shown in Figure 5 may well be the result of changes in WRI1 expression among the three lines we tested. Activation of ROD1 by WRI1 appears to be a conserved mechanism across plants, as we were able to identify AW boxes in the proximal upstream region of the ROD1 transcription start site from at least 10 different plant species (Supplemental Fig. S4).
WRI1 Overexpression Increases FA Synthesis in fae1 RcFAH12
Previously, we demonstrated that the production of HFA in fae1 RcFAH12 results in a bottleneck in seed lipid metabolism and reduces the rate of de novo FA synthesis, measured as tritiated water incorporation into FA, compared with the fae1 parental line (Bates et al., 2014). We hypothesized that the increased total oil observed in seeds of fae1 RcFAH12 WRI1 line 13 (Table I; Fig. 3D) may be due to an increased rate of FA production from the up-regulation of FA synthesis genes in fae1 RcFAH12 WRI1 developing seeds. To test if the overexpression of WRI1 increased the rate of de novo FA synthesis, 11- to 12-DAF seeds from fae1, fae1 RcFAH12, and fae1 RcFAH12 WRI1 line 13 plants were incubated in medium containing tritiated water for 30 min, and the amount of radiolabel incorporated into newly synthesized FA was determined. Consistent with previous results, we observed a significant, 21% ± 3% decrease in radiolabeled FA in fae1 RcFAH12 compared with fae1 (Fig. 6). In seeds of fae1 RcFAH12 WRI1 line 13, radiolabel in FA increased significantly to values comparable to the fae1 seeds. Therefore, overexpression of WRI1 restored the reduced rate of FA synthesis within the fae1 RcFAH12 background.
Figure 6.
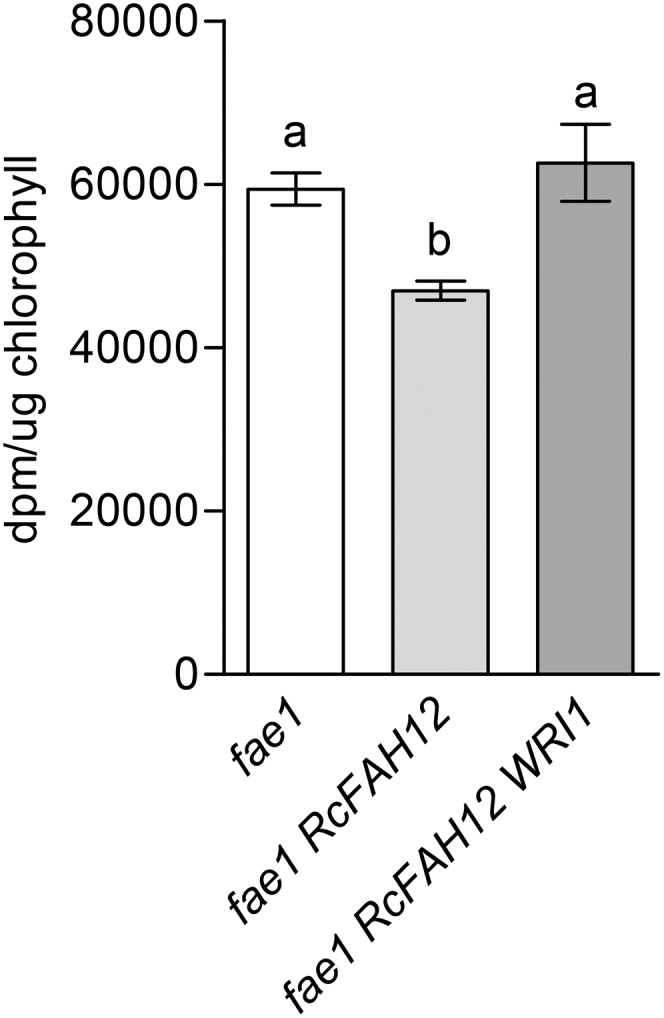
Comparison of FA synthesis rate in developing seeds. Seeds of fae1, fae1 RcFAH12, and fae1 RcFAH12 WRI1 line 13 were harvested 11 to 12 DAF and incubated in tritiated water for 30 min. n = 5, and error bars represent se. One-way ANOVA, columns with different letters are significantly different.
Seed Germination and Phenotypic Characteristics of WRI1 Overexpressors
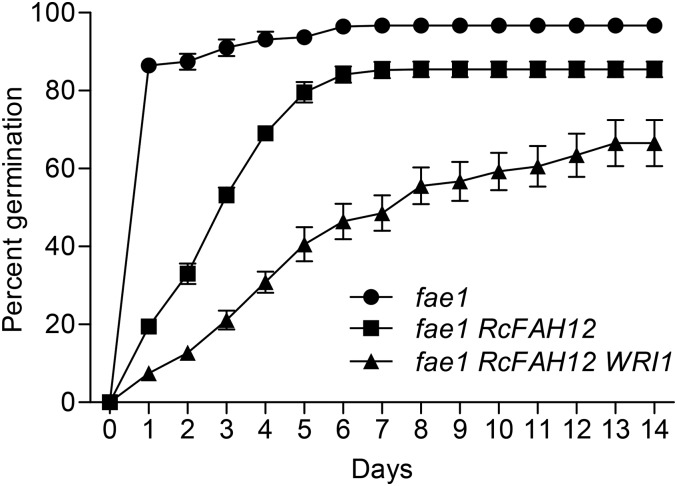
WRI1 expression in seeds is tightly regulated, and perturbing the expression level and/or pattern of this gene can negatively affect seed germination (Cernac et al., 2006; Shen et al., 2010). We expressed WRI1 in fae1 RcFAH12 under the control of a seed-specific promoter in an attempt to avoid germination phenotypes. Seeds of fae1 RcFAH12 WRI1 line 13 were tested for germination and seedling establishment rate relative to fae1 controls and the fae1 RcFAH12 parental line. Emergence of the radicle was counted as germination, and the appearance of roots and green cotyledons was scored as establishment of the germinated seedlings. Germination in fae1 RcFAH12 background lines was delayed and reduced compared with fae1 (Fig. 7). The germination rate of fae1 RcFAH12 WRI1 was reduced to 67% ± 5.9% compared with 86% ± 2% for fae1 RcFAH12 and 98% ± 2.2% for fae1. Seedling establishment in all genotypes (as a percentage of seeds that germinated), including fae1 RcFAH12 WRI1, was above 95%, indicating that the seed-specific expression of RcFAH12 and WRI1 reduced germination but had no substantial effect on the establishment of the newly germinated seedlings. We noted no visible differences in vegetative growth among the three lines, and at maturity, plants were comparable in height (Fig. 8A). However, the average seed yield from fae1 RcFAH12 plants was less than half of that obtained from plants of both fae1 and fae1 RcFAH12 WRI1 line 13 (Fig. 8B), and seeds were also an average 20% smaller by weight (Fig. 8C). Thus, overexpression of WRI1 in the fae1 RcFAH12 background substantially overcame the deficiencies in average seed weight and seed yield caused by the expression of RcFAH12, but it did result in further reduction and delay of seed germination.
Figure 7.
Analysis of germination rate. Seeds from plants of fae1, fae1 RcFAH12, and fae1 RcFAH12 WRI1 line 13 were plated onto 1× Linsmaier and Skoog plates containing 2% Suc. Each data point represents an average of seven replicates of 59 to 75 seeds per genotype. n = 7, and error bars represent se.
Figure 8.
Plant growth and seed production. Plants of fae1, fae1 RcFAH12, and fae1 RcFAH12 WRI1 line 13 were grown side by side, and seeds were collected at maturity. A, Plant height. B, Seed yield per plant. C, Average seed weight. n = 5 to 11, and error bars represent se. One-way ANOVA, columns with different letters are significantly different.
DISCUSSION
Castor oil is an important raw material with many industrial applications, including plastics, nylons, and lubricants. Many new industrial applications for castor oil continue to be discovered, thus making this a high-value commodity. Transgenic expression of the castor hydroxylase, RcFAH12, in nonnative plant species only provides for 17% HFA in the seeds compared with 90% for castor. The level in transgenic plants is too low to make this an efficient strategy for producing HFA in transgenic crops. Moreover, the synthesis of HFA in seeds of Arabidopsis leads to a feedback inhibition of FA synthesis (Bates et al., 2014) and substantial reductions in the levels of HFA and total oil (Bates and Browse, 2011; van Erp et al., 2011). Our data for the fae1 RcFAH12 line CL37 versus the fae1 control are in agreement with these earlier reports (Fig. 3).
Studies of fae1 RcFAH12 indicate that feedback inhibition of FA synthesis in these plants occurs at the ACCase step, as determined from feeding of radiolabeled substrates to developing seeds (Bates et al., 2014). ACCase converts acetyl-CoA to malonyl-CoA, the first committed intermediate in FA biosynthesis. ACCase is also a key regulatory step in FA synthesis and is thought to be subject to several different forms of regulation (Ohlrogge and Jaworski, 1997; Sasaki and Nagano, 2004). ACCase has been identified as the major enzyme targeted during the feedback inhibition of FA synthesis both in embryo-derived cell cultures of B. napus and in developing seeds of fae1 RcFAH12 (Andre et al., 2012; Bates et al., 2014). Both of those studies concluded that the inhibition of ACCase occurred by a posttranslational mechanism. In the study of Bates et al. (2014), transcriptional profiling of developing fae1 and fae1 FAH12 seeds failed to detect any statistically significant difference in the expression of genes encoding components of the multimeric chloroplast ACCase or other enzymes of FA synthesis. By contrast, we did observe reductions in the transcript levels of genes that encode several components of the plastid ACCase as well as key genes involved in glycolysis and FA synthesis in fae1 RcFAH12 developing seeds (Fig. 5). In the glycolysis pathway, PI-PKβ1 encodes a subunit of the plastidial pyruvate kinase, which is a key regulated enzyme in the supply of acetyl-CoA for FA synthesis (Andre et al., 2007; Baud et al., 2007b). BCCP1, BCCP2, CAC2, and CAC3 encode the subunits of the heteromeric ACCase localized in the plastids (Sasaki and Nagano, 2004). KASI and KASIII encode the condensing enzymes, responsible for the synthesis of FA up to 16 carbons (Ohlrogge and Browse, 1995). Transcripts for all of these genes except BCCP1 were reduced significantly in fae1 RcFAH12 seeds relative to fae1 controls. Importantly, the expression of RcFAH12 in the CL37 line (and the attendant synthesis of HFA) results in a 43% reduction in transcript for WRI1, a transcription factor that is known to regulate the expression of many genes involved in glycolysis and FA synthesis, including those investigated in this study. This suggests a signaling cascade in response to HFA production that results in the down-regulation of WRI1 and WRI1-controlled genes. Down-regulation of the transcript level suggests a decrease in protein abundance and, thus, enzyme activity. Consistent with this notion, the reduction in transcripts of genes involved in FA synthesis in fae1 RcFAH12 compared with fae1 was associated with a corresponding decrease in the rate of FA synthesis (Fig. 6), similar to that observed previously (Bates et al., 2014). These data suggest that, in addition to biochemical mechanisms, the feedback inhibition of ACCase also involves transcriptional regulation by WRI1. In future studies, it will be important to understand the complex mechanisms of ACCase regulation during oilseed biosynthesis.
Previous attempts to improve HFA accumulation in TAG of fae1 RcFAH12 seeds focused on engineering TAG assembly genes, including the expression of RcDGAT2 and RcPDAT1A. These effects resulted in substantial increases in the rate of FA synthesis and improvement in HFA levels in TAG from 17% to approximately 30% (Burgal et al., 2008; van Erp et al., 2011, 2015; Bates et al., 2014). Our goal here was to test a strategy that focuses on increasing the rate of FA synthesis as a means to improve the accumulation of HFA in fae1 RcFAH12 seeds. We hypothesized that, by targeting glycolysis, ACCase, and condensing steps of FA synthesis to increase the flux of carbon into FA, we could alleviate, at least partially, the feedback inhibition and increase HFA accumulation in TAG.
We chose to engineer seed lipid metabolism by the overexpression of WRI1 in fae1 RcFAH12 because several earlier studies have shown that WRI1 overexpression causes increased expression of our target genes (Baud et al., 2009; Maeo et al., 2009; To et al., 2012) and increased seed oil in Arabidopsis and other plant species (Cernac and Benning, 2004; Shen et al., 2010; Kelly et al., 2013; van Erp et al., 2014; Wu et al., 2014). We transformed a WRI1 cDNA, under the control of the strong seed-specific phaseolin promoter from P. vulgaris (Sengupta-Gopalan et al., 1985), into the fae1 RcFAH12 line CL37. We observed significant increases in the transcript level of WRI1 in 11- to 12-DAF developing seeds of WRI1 transgenics compared with fae1 and fae1 RcFAH12 (Figs. 2 and 5). In the line chosen for detailed study, fae1 RcFAH12 WRI line 13, transcripts of PI-PKβ1, BCCP1, BCCP2, CAC2, CAC3, KASI, and KASIII were all increased significantly at 11 to 12 DAF compared with the fae1 RcFAH12 parental line (Fig. 5). In this line, we observed a 33% increase in the rate of FA synthesis (Fig. 6) and significant increases in the levels of both HFA and total oil (Table I; Fig. 3). Notably, the most increased FA were HFA, as observed by an analysis of FA composition (Table I; Supplemental Fig. S1). WRI1 overexpression up-regulates many genes involved in FA synthesis, among which we have tested the key ones. Interpretation of the changes in FA profile thus becomes complex, and it is not so straightforward to attribute effects to specific genes (Supplemental Fig. S1). We consistently observed increases in both the proportion of HFA in the oil and total HFA per seed in WRI1 transgenic seeds compared with fae1 RcFAH12 segregants in T2 seeds from 25 independent transgenics (Fig. 1). More detailed analyses of T3 seeds of five transgenic lines (Table I) showed that, relative to controls, WRI1-overexpressing plants had a 30% increase in total FA per seed, a 54% increase in HFA per seed, and thus an 18% increase in the proportion of HFA in oil. Taken together, these results demonstrate real and consistent increases in both total FA and HFA caused by overexpressing WRI1.
Since WRI1 overexpression has not been found previously to increase the transcript levels of TAG assembly genes, we did not expect to see an increase in the proportion of HFA in TAG but only an increase in total HFA, so the increases in percentage HFA (Table I; Figs. 1, 3, and 4) were unanticipated. Consistent with previous work (Maeo et al., 2009), we did not observe any significant change in transcript levels of DGAT1 and PDAT between the lines we tested (Fig. 5). However, we did observe a significant, 5-fold reduction in ROD1 transcript in fae1 RcFAH12 compared with fae1, and ROD1 expression was restored in plants overexpressing WRI1 (Fig. 5). ROD1 transcript levels have been observed previously to be lower in developing seeds of a wri1 wri3 wri4 triple mutant line compared with the wild type (To et al., 2012), suggesting at least a partial regulation by WRI1, but it is not known if ROD1 transcript levels also are reduced in developing seeds of wri1 mutants. In Nicotiana benthamiana leaves, transient overexpression of WRI1 significantly up-regulated the transcript level of ROD1 (Grimberg et al., 2015). Consistent with a positive regulation of ROD1 by WRI1, we discovered an AW box in the 5′ UTR of ROD1 (Supplemental Fig. S2). The location and sequence of this AW box are comparable to those of AW boxes found in other genes that are known targets of WRI1 (Supplemental Fig. S3), and the AW box has been demonstrated to be a binding site for WRI1 (Maeo et al., 2009). These results and our identification of AW boxes in ROD1 homologs from different plant species (Supplemental Fig. S4) indicate that ROD1 is a direct target of WRI1. The PDCT enzyme encoded by ROD1 is important for HFA accumulation, as demonstrated previously (Hu et al., 2012). Previously, we demonstrated that HFA released from phosphatidylcholine by acyl editing are inefficiently incorporated into TAG, but HFA-containing phosphatidylcholine-derived diacylglycerol can be efficiently utilized for TAG synthesis (Bates and Browse, 2011). Thus, increased activity of ROD1 due to increased expression may be at least partly responsible for the increased proportion of HFA in fae1 RcFAH12 WRI1 seeds. We hypothesized that an increase in the proportion of HFA at the sn-2 position of 2-HFA-TAG in fae1 RcFAH12 WRI1 compared with fae1 RcFAH12 may be possible. However, our regiochemical analyses revealed instead small decreases in HFA at the sn-2 position of fae1 RcFAH12 WRI1 compared with fae1 RcFAH12 (Fig. 4C). Because we did not observe any change in the transcript level of DGAT1 and PDAT genes in fae1 RcFAH12 (Fig. 5), we hypothesize that the activity of these enzymes remained similar to wild-type levels. Overexpression of WRI1 restored FA synthesis to wild-type levels, but since the FA synthesis rate did not exceed wild-type levels (Fig. 6), these endoplasmic reticulum enzymes functioning at their original capacity are able to handle the increase in metabolite flux, thus leading to increases in FA including HFA (Table I; Figs. 1 and 3).
The expression of RcFAH12 leads to additional undesirable plant phenotypes due to downstream effects resulting from the accumulation of HFA and the feedback inhibition of FA synthesis. For example, germination rates of fae1 RcFAH12 seeds show considerable variation, the exact mechanism of which is not clear. Lu et al. (2006) observed a normal germination rate, whereas Bayon et al. (2015) reported a 24% germination rate of fae1 RcFAH12 seeds. In our experiments, we observed germination rates from 30% to 98% between different batches of seeds. fae1 RcFAH12 seeds exhibited delayed germination compared with fae1 (Fig. 7) and had reduced seed weight (Fig. 8C) and low total seed yield per plant compared with fae1 (Fig. 8B). WRI1 is tightly regulated and has a specific pattern and level of expression (Cernac et al., 2006). Changes to this pattern and level by means of either constitutive overexpression or mutations leading to lower levels have been shown to lead to germination defects and other undesirable growth phenotypes (Focks and Benning, 1998; Cernac et al., 2006; Shen et al., 2010; Sanjaya et al., 2011; Chen et al., 2013). Overexpressing WRI1 in Arabidopsis seeds under the control of the Sucrose Synthase2 (SUS2) promoter (Baud et al., 2004; Angeles-Núñez and Tiessen, 2012), which matches the developmental pattern of expression of WRI1 in developing seeds, was recently shown to increase total oil and not cause a significant reduction in seed germination (van Erp et al., 2014). We hypothesized that, by seed specifically overexpressing WRI1, we could circumvent additional undesirable phenotypes in fae1 RcFAH12. However, we observed both a delay in germination of fae1 RcFAH12 WRI1 and a reduction in germination compared with fae1 RcFAH12 (Fig. 7). However, plant height was unchanged between fae1, fae1 RcFAH12, and fae1 RcFAH12 WRI1 (Fig. 8B), suggesting that plant size is not negatively affected by WRI1 overexpression in fae1 RcFAH12 seeds. In addition, seed weight and total seed yield per plant increased to fae1 levels in fae1 RcFAH12 WRI1 compared with fae1 RcFAH12, improving some of the undesirable phenotypes of fae1 RcFAH12 (Fig. 8, B and C).
Expression of the castor hydroxylase in Arabidopsis seeds leads to the accumulation of relatively low levels of HFA compared with the native castor plant and causes numerous deleterious effects, including feedback inhibition of FA synthesis and reductions in seed oil, weight, yield, and germination. By overexpressing WRI1 seed specifically in a fae1 RcFAH12 background, we have shown that it is possible to substantially restore FA synthesis, increase total oil, improve the efficiency of accumulation of HFA into TAG, and alleviate the undesirable low seed weight and seed yield phenotypes caused by RcFAH12 expression. In future studies, combining this strategy with other successful approaches for increasing HFA may lead to additional benefits for the accumulation of HFA in seeds.
MATERIALS AND METHODS
Cloning Procedures
AT3G54320.3 (splice variant 3), which has been shown to be the predominantly expressed form of WRI1 in Arabidopsis (Arabidopsis thaliana) developing seeds (Ma et al., 2013), was amplified from Arabidopsis cDNA using gene-specific primers and KOD high-fidelity polymerase (Novagen) and cloned into the pENTR-D-TOPO vector (Invitrogen). WRI1 was then transferred into the pGate-Phas-DsRed (pGPD) Gateway-compatible binary vector (Lu et al., 2006) under the control of the seed-specific phaseolin promoter (Slightom et al., 1983) and with the DsRed selection marker (Stuitje et al., 2003). The cloned sequence was verified by sequencing (Eurofins MWG Operon) part of the promoter and the WRI1 coding region. pGPD-WRI1 was then transformed into Agrobacterium tumefaciens GV3101, which was used for plant transformation.
Plant Growth Conditions
Arabidopsis ecotype Columbia was the background in all experimental lines. fae1 mutant AC56 (Kunst et al., 1992) expressing RcFAH12 (line CL37; Lu et al., 2006) was used as a parental line. Seed sterilization was as described (Adhikari et al., 2011). Briefly, seeds were surface sterilized by first mixing in a solution of 95% (v/v) ethanol and 0.5% (v/v) Triton X-100 for 10 min on a tube mixer, followed by 10 min in 95% (v/v) ethanol on a tube mixer. Seeds were then air dried in a laminar flow hood on filter paper rafts soaked with 95% (v/v) ethanol. Seeds were sprinkled uniformly on plates containing 1× Linsmaier and Skoog (Caisson Laboratories) medium supplemented with 2% Suc and 0.5% Phytagel (Sigma) and stratified at 4°C for 3 d. Plates were then incubated at 22°C under approximately 100 µmol m−2 s−1 continuous white light for 10 to 14 d and transplanted to soil. All plants on soil were grown under approximately 120 µmol m−2 s−1 continuous white light from broad-spectrum fluorescent tube lamps at 22°C and 60% to 70% relative humidity until maturity. Plants were transformed by the floral dip method (Clough and Bent, 1998). Transgenic seeds expressing the DsRed marker were identified by exciting with a green light and using a red emission filter attached to a microscope (Stuitje et al., 2003).
Determination of Seed FA and Oil Content, and GC Analysis
Seed FA and oil contents were determined according to the method of Li et al. (2006). Seed weights were determined for 100 seeds picked randomly per sample. A total of 200 µL of toluene containing approximately 20 µg of 17:0 TAG and 0.005% (v/v) butylated hydroxy toluene was added to each sample. FA methyl esters were derivatized from the FA of 20 to 100 whole seeds in 1 mL of 5% (v/v) sulfuric acid in methanol for 1.5 h at 90°C. FA methyl esters were quantified by GC with flame ionization detection on a wax column (EC wax; 30 m × 0.53 mm i.d. × 1.20 µm; Alltech). GC parameters were as follows: 210°C for 1 min followed by a ramp to 250°C at 10°C min−1, with a final 9-min temperature hold.
Radiolabeling Studies
Plant lines were grown randomized under the same growth conditions as described above. For each plant line, 11- to 12-DAF seeds were dissected from approximately 50 siliques and split into five individual samples for incubation with 1 mCi of tritiated water (American Radiolabeled Chemicals; www.arc-inc.com) as described by Bates et al. (2014).
Characterization of TAG Species, and Regiochemical Analysis of 1-HFA-TAG and 2-HFA-TAG
For the characterization of HFA-TAG molecular species, mature T4 seeds of fae1 RcFAH12 WRI1 line 13 were quenched in 1 mL of 85°C isopropanol containing 0.01% (v/v) butylated hydroxy toluene and extracted as described by Bates and Browse (2011). HFA-containing neutral lipids were separated by a previously optimized TLC system (Bates and Browse, 2011) on 20- × 20-cm EMD silica gel TLC plates involving two developments in different mixtures of chloroform:methanol:acetic acid (v/v/v). The first development was 12 cm from the bottom of the plate in 97:3:0.5. This was dried in a vacuum for 15 min, then the second development was to 19 cm in 99:0.5:0.5. All lipids were identified by comigration with lipid standards after staining with 0.005% (w/v) Primulin in 80% acetone and visualization under UV light. Lipids were quantified by GC-flame ionization detection as above. Regiochemical analyses were performed exactly as described by van Erp et al. (2011).
Analysis of Gene Expression
Young siliques were harvested approximately 12 DAF from plants, flash frozen in liquid nitrogen, and stored at −80°C. Developing seeds were released from siliques using the method described (Bates et al., 2013). Siliques were popped open or crushed using a liquid nitrogen-cooled glass rod on Petri plates stored over dry ice. Developing seeds were then collected in 1.5-mL Eppendorf tubes held in liquid nitrogen by filtering through liquid nitrogen-cooled sieves to remove debris from siliques. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). Samples were treated with RNase-free DNase (Qiagen) using on-column DNase digestion. RNA was quantified using a Nanophotometer (Implen). cDNA was synthesized using the SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies). Transcript levels were analyzed by quantitative PCR using Platinum SYBR Green qPCR SuperMix-UDG (Life Technologies) and the Stratagene Mx3005P quantitative PCR system (Agilent Technologies). Primers used for qRT-PCR analysis are listed in Supplemental Table S1. Primer pairs were selected based on 100% efficiency and single peaks. Relative mRNA levels were measured by the comparative threshold cycle method and normalized to PP2A expression (Czechowski et al., 2005).
Promoter Analysis
Promoter and 5′ UTR sequences were obtained from The Arabidopsis Information Resource and the National Center for Biotechnology Information. AW box sequences used for alignments were adapted from Maeo et al. (2009). FIMO software (Grant et al., 2011) was used for scanning DNA sequences to detect the AW box motif. Multiple sequence alignment and shading of conserved residues were done using ClustalW2 and BoxShade.
Germination Rate and Seedling Establishment Rate
Seeds for the germination assay were from plants that were grown randomized in the same growth conditions. Seeds were harvested at maturity and dried in silica beads (Fisher Scientific) for at least 2 d, sterilized using the 95% (v/v) ethanol and 0.5% (w/v) Triton X-100 mixture as described above, and spotted uniformly on plates in sectors for comparison. Germination was tested on 1× Linsmaier and Skoog plates supplemented with 2% (w/v) Suc. Between 411 and 528 seeds per genotype were spotted uniformly across seven independent plates. Plates were incubated under approximately 100 µmol m−2 s−1 continuous white light as described above for 14 d. Seeds that produced a radicle were counted as germinated. Of these, seedlings that produced roots and green cotyledons were scored as being able to establish. Germination was scored every day from day 0 to day 14.
Plant Phenotypes
Plants were grown in the same conditions as described above at the same time. Height was recorded at maturity from the level of the rosette. For analysis of total seed yield, seeds from each plant above were harvested and dried by mixing with silica beads (Fisher Scientific) for at least 2 d before weighing. To measure seed weight, 100 random seeds were picked from each sample and weighed.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. FA composition in line 13 T4 seeds.
Supplemental Figure S2. Location of the AW box in the ROD1/PDCT proximal upstream region.
Supplemental Figure S3. AW boxes in WRI1 targets involved in lipid metabolism.
Supplemental Figure S4. AW boxes in various plant species.
Supplemental Table S1. Primers used in qRT-PCR analysis.
Supplementary Material
Acknowledgments
From the Browse laboratory, we would like to thank Dr. Shuangyi Bai, Dr. Jeremy Jewell, Dr. Amanda Wager, Gracen Smith, and Breana Tate for help with dissecting developing seeds from siliques; Dr. Jim Wallis for helpful discussions; and Dr. Harrie van Erp for donating PDAT q-PCR primers and advice on regiochemistry experiments.
Glossary
- FA
fatty acids
- HFA
hydroxy fatty acids
- cDNA
complementary DNA
- TAG
triacylglycerol
- GC
gas chromatography
- DAF
days after flowering
- qRT
quantitative reverse transcription
- UTR
untranslated region
- TLC
thin-layer chromatography
Footnotes
This work was supported by the U.S. National Science Foundation Plant Genome Research Program (grant no. IOS–1339385) and the Agricultural Research Center at Washington State University.
Articles can be viewed without a subscription.
References
- Adhikari ND, Froehlich JE, Strand DD, Buck SM, Kramer DM, Larkin RM (2011) GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23: 1449–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19: 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C, Haslam RP, Shanklin J (2012) Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. Proc Natl Acad Sci USA 109: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeles-Núñez JG, Tiessen A (2012) Regulation of AtSUS2 and AtSUS3 by glucose and the transcription factor LEC2 in different tissues and at different stages of Arabidopsis seed development. Plant Mol Biol 78: 377–392 [DOI] [PubMed] [Google Scholar]
- Badami RC, Patil KB (1980) Structure and occurrence of unusual fatty acids in minor seed oils. Prog Lipid Res 19: 119–153 [DOI] [PubMed] [Google Scholar]
- Bates PD, Browse J (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 [DOI] [PubMed] [Google Scholar]
- Bates PD, Jewell JB, Browse J (2013) Rapid separation of developing Arabidopsis seeds from siliques for RNA or metabolite analysis. Plant Methods 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Johnson SR, Cao X, Li J, Nam JW, Jaworski JG, Ohlrogge JB, Browse J (2014) Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly. Proc Natl Acad Sci USA 111: 1204–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B (2007a) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Baud S, Vaultier MN, Rochat C (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55: 397–409 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C (2007b) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52: 405–419 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuillème S, To A, Rochat C, Lepiniec L (2009) Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J 60: 933–947 [DOI] [PubMed] [Google Scholar]
- Bayon S, Chen G, Weselake R.J, Browse J (2015) A small phospholipase A2-α from castor catalyzes the removal of hydroxy fatty acids from phosphatidylcholine in transgenic Arabidopsis seeds. Plant Physiol 167: 1259–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Somerville C (1997) Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol 113: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, Browse J (2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 6: 819–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Dietrich CR, Meyer K, Damude HG, Dyer JM, Kinney AJ (2006) Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds. Phytochemistry 67: 1166–1176 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Shockey JM, Dietrich CR, Gidda SK, Mullen RT, Dyer JM (2007) Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr Opin Plant Biol 10: 236–244 [DOI] [PubMed] [Google Scholar]
- Cernac A, Andre C, Hoffmann-Benning S, Benning C (2006) WRI1 is required for seed germination and seedling establishment. Plant Physiol 141: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chen L, Lee JH, Weber H, Tohge T, Witt S, Roje S, Fernie AR, Hellmann H (2013) Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 25: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JM, Stymne S, Green AG, Carlsson AS (2008) High-value oils from plants. Plant J 54: 640–655 [DOI] [PubMed] [Google Scholar]
- Focks N, Benning C (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg Å, Carlsson AS, Marttila S, Bhalerao R, Hofvander P (2015) Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol 15: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Ren Z, Lu C (2012) The phosphatidylcholine diacylglycerol cholinephosphotransferase is required for efficient hydroxy fatty acid accumulation in transgenic Arabidopsis. Plant Physiol 158: 1944–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, van Erp H, Quettier AL, Shaw E, Menard G, Kurup S, Eastmond PJ (2013) The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol 162: 1282–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Lee KR, Go YS, Jung JH, Suh MC, Kim JB (2011) Endoplasmic reticulum-located PDAT1-2 from castor bean enhances hydroxy fatty acid accumulation in transgenic plants. Plant Cell Physiol 52: 983–993 [DOI] [PubMed] [Google Scholar]
- Knutzon DS, Hayes TR, Wyrick A, Xiong H, Maelor Davies H, Voelker TA (1999) Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn-2 position of triacylglycerols in lauric rapeseed oil and can increase total laurate levels. Plant Physiol 120: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Taylor DC, Underhill EW (1992) Fatty-acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol Biochem 30: 425–434 [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Li R, Yu K, Hatanaka T, Hildebrand DF (2010) Vernonia DGATs increase accumulation of epoxy fatty acids in oil. Plant Biotechnol J 8: 184–195 [DOI] [PubMed] [Google Scholar]
- Lu C, Xin Z, Ren Z, Miquel M, Browse J (2009) An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci USA 106: 18837–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fulda M, Wallis JG, Browse J (2006) A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J 45: 847–856 [DOI] [PubMed] [Google Scholar]
- Ma W, Kong Q, Arondel V, Kilaru A, Bates PD, Thrower NA, Benning C, Ohlrogge JB (2013) Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE 8: e68887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Masaki T, Mitsui N, Tsukagoshi H, Nishii T, Morikami A, Nakamura K (2005) ACTIVATOR of Spomin:LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol 46: 547–556 [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, et al. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48: 109–136 [DOI] [PubMed] [Google Scholar]
- Reynolds KB, Taylor MC, Zhou XR, Vanhercke T, Wood CC, Blanchard CL, Singh SP, Petrie JR (2015) Metabolic engineering of medium-chain fatty acid biosynthesis in Nicotiana benthamiana plant leaf lipids. Front Plant Sci 6: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjaya, Durrett TP, Weise SE, Benning C (2011) Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol J 9: 874–883 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Nagano Y (2004) Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem 68: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Sengupta-Gopalan C, Reichert NA, Barker RF, Hall TC, Kemp JD (1985) Developmentally regulated expression of the bean beta-phaseolin gene in tobacco seed. Proc Natl Acad Sci USA 82: 3320–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC (2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom JL, Sun SM, Hall TC (1983) Complete nucleotide sequence of a French bean storage protein gene: phaseolin. Proc Natl Acad Sci USA 80: 1897–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Moon H, Kunst L (2000) Production of hydroxy fatty acids in the seeds of Arabidopsis thaliana. Biochem Soc Trans 28: 947–950 [PubMed] [Google Scholar]
- Smith MA, Moon H, Chowrira G, Kunst L (2003) Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis thaliana. Planta 217: 507–516 [DOI] [PubMed] [Google Scholar]
- Stuitje AR, Verbree EC, van der Linden KH, Mietkiewska EM, Nap JP, Kneppers TJ (2003) Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol J 1: 301–309 [DOI] [PubMed] [Google Scholar]
- To A, Joubès J, Barthole G, Lécureuil A, Scagnelli A, Jasinski S, Lepiniec L, Baud S (2012) WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24: 5007–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Loo FJ, Broun P, Turner S, Somerville C (1995) An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci USA 92: 6743–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp H, Bates PD, Burgal J, Shockey J, Browse J (2011) Castor phospholipid:diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic Arabidopsis. Plant Physiol 155: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp H, Kelly AA, Menard G, Eastmond PJ (2014) Multigene engineering of triacylglycerol metabolism boosts seed oil content in Arabidopsis. Plant Physiol 165: 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp H, Shockey J, Zhang M, Adhikari ND, Browse J (2015) Reducing isozyme competition increases target fatty acid accumulation in seed triacylglycerols of transgenic Arabidopsis. Plant Physiol 168: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke T, El Tahchy A, Liu Q, Zhou XR, Shrestha P, Divi UK, Ral JP, Mansour MP, Nichols PD, James CN, et al. (2014) Metabolic engineering of biomass for high energy density: oilseed-like triacylglycerol yields from plant leaves. Plant Biotechnol J 12: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke T, El Tahchy A, Shrestha P, Zhou XR, Singh SP, Petrie JR (2013) Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett 587: 364–369 [DOI] [PubMed] [Google Scholar]
- Voelker T, Kinney AJ (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol 52: 335–361 [DOI] [PubMed] [Google Scholar]
- Wayne LL, Browse J (2013) Homologous electron transport components fail to increase fatty acid hydroxylation in transgenic Arabidopsis thaliana. F1000 Res 2: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XL, Liu ZH, Hu ZH, Huang RZ (2014) BnWRI1 coordinates fatty acid biosynthesis and photosynthesis pathways during oil accumulation in rapeseed. J Integr Plant Biol 56: 582–593 [DOI] [PubMed] [Google Scholar]
- Zale J, Jung JH, Kim JY, Pathak B, Karan R, Liu H, Chen X, Wu H, Candreva J, Zhai Z, et al. (2016) Metabolic engineering of sugarcane to accumulate energy-dense triacylglycerols in vegetative biomass. Plant Biotechnol J 14: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.