Seed production in the monocarpic plant Arabidopsis thaliana promotes shoot senescence and suppresses shoot growth, affecting similar processes as apical dominance does.
Abstract
Correlative control (influence of one organ over another organ) of seeds over maternal growth is one of the most obvious phenotypic expressions of the trade-off between growth and reproduction. However, the underlying molecular mechanisms are largely unknown. Here, we characterize the physiological and molecular effects of correlative inhibition by seeds on Arabidopsis (Arabidopsis thaliana) inflorescences, i.e. global proliferative arrest (GPA) during which all maternal growth ceases upon the production of a given number of seeds. We observed transcriptional responses to growth- and branching-inhibitory hormones, and low mitotic activity in meristems upon GPA, but found that meristems retain their identity and proliferative potential. In shoot tissues, we detected the induction of stress- and senescence-related gene expression upon fruit production and GPA, and a drop in chlorophyll levels, suggestive of altered source-sink relationships between vegetative shoot and reproductive tissues. Levels of shoot reactive oxygen species, however, strongly decreased upon GPA, a phenomenon that is associated with bud dormancy in some perennials. Indeed, gene expression changes in arrested apical inflorescences after fruit removal resembled changes observed in axillary buds following release from apical dominance. This suggests that GPA represents a form of bud dormancy, and that dominance is gradually transferred from growing inflorescences to maturing seeds, allowing offspring control over maternal resources, simultaneously restricting offspring number. This would provide a mechanistic explanation for the constraint between offspring quality and quantity.
The production of new flowers is a central mechanism to determine the total reproductive output of a flowering plant (Lloyd, 1980). In plant species that exhibit indeterminate growth, the initiation of new flowers can temporally overlap with the development of offspring, the seeds. In principle, the maternal plant then has to decide on how to balance current and future investment into offspring so that the seeds in production can be provided with adequate nutrients. Furthermore, developing offspring and the mother plants should have conflicting interests regarding the production of new reproductive structures. Interestingly, control mechanisms exerted by developing seeds over maternal growth and life span have been observed in plants (Murneek, 1926; Molisch, 1929; Noodén et al., 2003). In legumes such as soybean (Glycine max), for example, both reproductive growth and the onset of leaf senescence are under correlative control (Noodén and Penney, 2001) by developing seeds so that plants grow and live considerably longer when developing fruits are continuously removed (Leopold et al., 1959; Lindoo and Noodén, 1977). Such a pattern is readily found in monocarpic plants (i.e. plants that die after a single reproductive period) and can be viewed as an evolutionary adaptation that ensures efficient nutrient reallocation from the vegetative to the reproductive parts of the plant (Noodén et al., 2003).
In the monocarpic species Arabidopsis (Arabidopsis thaliana), correlative control of developing fruits over maternal growth and longevity has also been reported (Hensel et al., 1994; Noodén and Penney, 2001). Under constant growth conditions, an Arabidopsis plant produces a certain number of flowers before its reproductive meristems arrest. This process is termed global proliferative arrest (GPA) as correlative inhibition through offspring affects all maternal aboveground meristematic activity (Hensel et al., 1994). At the same time, the senescence of Arabidopsis rosette leaves has been reported to be uncoupled from reproduction (Hensel et al., 1993; Noodén and Penney, 2001), but some results from experimental removal of reproductive bolts have suggested a negative feedback from shoots to leaves affecting chlorophyll levels (Ye et al., 2000). The effects that fruits have on plant longevity were proposed to be primarily caused by the prevention of further growth and continuous regeneration of new photosynthetic tissues (Robinson and Hill, 1999; Noodén and Penney, 2001). However, the latter model would lead to the counterintuitive prediction that offspring number can be uncoupled from offspring quality in Arabidopsis since correlative control seems to influence offspring number but not source-sink relationships between vegetative and reproductive tissues. Indeed, an Arabidopsis mutant isolated in a forward genetic screen for increased seed number did seem to produce seeds that were of normal size (L. Hensel, personal communication). Unfortunately, this mutant could not be cloned because of unstable inheritance of the phenotype (L. Hensel, personal communication). Thus, we currently have little understanding of the potential trade-offs that exist between seed production and other maternal traits, or of the molecular mechanisms that mediate GPA in Arabidopsis.
Much controversy exists around the causal agent(s) underlying correlative control exerted by seeds. Mainly, two competing hypotheses have received the most attention in the past, one that invokes a simple source-sink relationship (Molisch, 1929) and one that invokes offspring-derived signals (Wilson, 1997). By far the most extensive research examining the causes of monocarpic senescence has been performed in legumes such as pea (Pisum sativum) and soybean (Leopold et al., 1959; Lindoo and Noodén, 1977; Noodén and Murray, 1982), but even in these species the debate has not been settled. A potential role of the hormones auxin and abscisic acid (ABA) in correlative control has been discussed (Tamas and Engels, 1981). It is also likely that different mechanisms operate in different species, and multiple evolutionary origins of monocarpic senescence seem likely, especially given the various ways in which correlative control and senescence patterns are expressed in different species (Noodén, 1980; Kelly et al., 1988; Noodén et al., 2003).
Even less is known about the exact molecular and physiological effects that correlative control has on maternal organs, such as the shoot apex. It has been proposed that growth arrests upon fruit production should be termed “mitotic senescence” as meristematic cells lose the ability to undergo mitotic cell division (Murneek, 1926; Molisch, 1929; Gan, 2003; Noodén et al., 2003). Peas seem to be a good example of such apex senescence as DNA fragmentation (a sign of cell death) was detected in mitotically arrested apices (Noodén and Penney, 2001; Wang et al., 2007). In previous work, levels of hormones such as GA and auxin were shown to be affected during monocarpic senescence in pea shoots (Leopold et al., 1959; Lindoo and Noodén, 1977; Zhu and Davies, 1997). The role and the causality of multiple correlated events during monocarpic senescence are, however, still under debate (Noodén et al., 2003; Davies and Gan, 2012).
In recent years, research efforts focusing on the causes and consequences of correlative control have declined, possibly because of the complexity of the phenomenon and the difficulties to interpret seemingly contradictory experimental data. Here, we examined GPA (i.e. the most obvious phenotypic expression of correlative control of seeds over maternal growth) and the effects of fruits on maternal shoots in Arabidopsis using recently developed molecular approaches. We focused primarily on the comparison of molecular and physiological differences in sterile, fruit-producing growing, and fruit-producing arrested plants. Our goal was to generate a dataset that allows a new view on the associated phenomena in this species, more specifically, (1) to identify genes and processes that are under correlative control in maternal apices, (2) to resolve the question whether maternal growth inhibition by fruits indeed occurs without changes in source-sink relationships between tissues (i.e. without promotion of maternal senescence), and (3) to evaluate various hypotheses regarding the nature of the signals involved in correlative inhibition by seeds.
RESULTS
Plant Fertility Influences Meristematic Activity and Eventually Leads to GPA
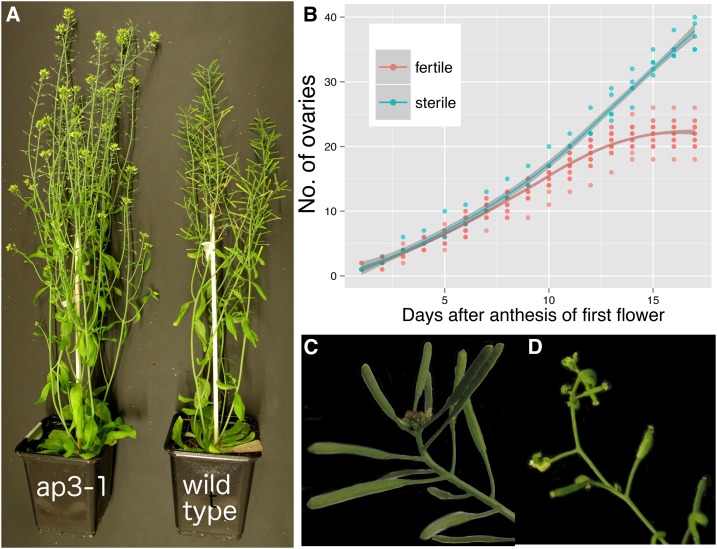
The sequential production of flowers in Arabidopsis eventually stops once a certain number of fruits (under given growth conditions) have been produced (Hensel et al., 1994; Noodén and Penney, 2001). We found that sterile plants produced many more ovaries and axillary branches than fertile ones over their lifetimes (Fig. 1; see also Hensel et al. [1994] and Noodén and Penney [2001]). Under given growth conditions, fruit production leads to a characteristic arrest of growing inflorescences visible through the production of terminal clusters of nonmaturing flower buds (Fig. 1C). The inflorescences of sterile plants, i.e. male sterile1-1 (ms1-1) or apetala3-1 (ap3-1) mutants, did not exhibit such a characteristic growth arrest, but eventually growth also halted in these genotypes through a process previously named “terminal differentiation” (Fig. 1D; Hensel et al., 1993, 1994; Noodén and Penney, 2001).
Figure 1.
Effect of reproduction on maternal growth in Arabidopsis. A, Sterile (Ler ap3-1/ap3-1) and fertile (Ler wild type) plants grown under identical conditions eventually exhibit different branching patterns and ovary numbers (as described by Hensel et al. [1994]). B, The numbers of mature ovaries produced on the main inflorescence of sterile (ms1-1/ms1-1) and fertile (wild-type segregants from the same family) plants produced over time. Flower production rates were taken from the day of anthesis of the first flower of an individual. The proliferative activity of sterile plants had not stopped by the end of the time course shown here. C, An arrested inflorescence of a fertile plant showing a cluster of flower buds that have ceased development. D, A growth-arrested inflorescence of a sterile plant. The last few flowers exhibit homeotic transformations of organs, and the inflorescence apex has terminally differentiated into a flower-like structure.
Since reproductive status and developmental age of the plant are normally correlated (which can lead to confounding effects in our experiments), we first tested if and how quickly growth-arrested inflorescences can be reactivated. Upon removal of most fruits from plants with arrested apices, meristematic activity readily resumed (Fig. 2). Signs of new growth became visible within 3 to 4 d through the appearance of new floral buds from previously arrested inflorescences. These developed into new mature flowers and fruits (not shown). However, previously arrested floral buds never recommenced development and senesced (Fig. 2A; described also in spontaneously reactivated inflorescences in Hensel et al. [1994] and Ye et al. [2000]). Furthermore, we observed that fruit removal readily promoted the outgrowth of previously dormant axillary shoots (data not shown). In summary, our data together with previous reports demonstrate that fruits promote the cessation of maternal growth and limit the number of ovaries produced by an Arabidopsis plant.
Figure 2.
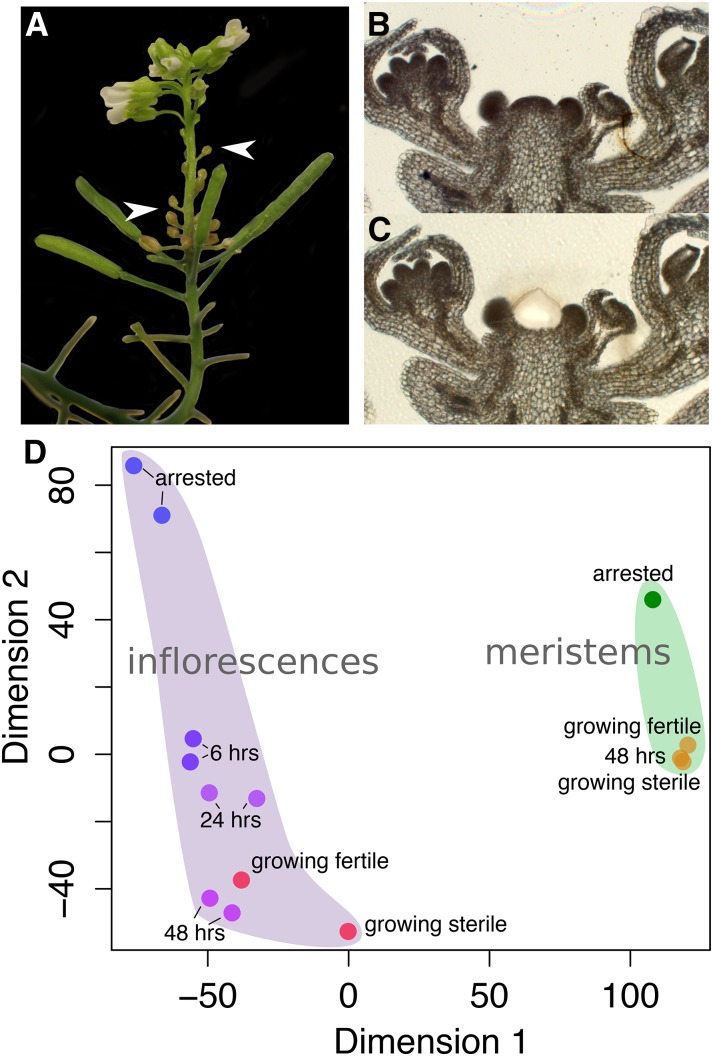
Distinct transcriptomes of meristems and inflorescences before, during, and after GPA. A, An inflorescence photographed 8 d after GPA and subsequent fruit removal. Newly developing fertile flowers with mature petals are visible but develop only after several senescent floral buds (white arrowheads) emerge. B, A 10-μm thin section of an arrested inflorescence (wild-type segregant). C, LAM of the meristematic region from the shoot apex. D, Multidimensional scaling (using Euclidean distance) of Affymetrix GeneChip datasets taken either from inflorescences (left) or meristems isolated by LAM (right). 6 hrs, 24 hrs, and 48 hrs denote samples at 6, 24, and 48 h after fruit removal from arrested plants, respectively.
Transcriptomes of Meristems and Whole Inflorescences Are Distinct, But Both Show Extensive Changes during GPA
Having seen clear differences in meristem behavior depending on plant fertility, we next studied the molecular effects associated with correlative control by the use of transcriptomics. We performed a time-series analysis of inflorescences reactivated by fruit removal but before clear morphological signs of growth were visible. To this aim, we harvested and pooled inflorescences that were arrested, or at 6, 24, and 48 h after fruit removal, to extract RNA and perform microarray experiments using the Affymetrix ATH1 array. Additionally, inflorescences of growing plants that were either sterile (ms1-1/ms1-1) or fertile (wild-type segregants from the same family) were harvested and processed in the same manner. As shown by multidimensional scaling of the gene expression data, arrested inflorescences exhibited a transcriptional profile distinct from growing inflorescences (Fig. 2D). However, upon fruit removal, transcriptional profiles of reactivated inflorescences rapidly became very similar to those of growing inflorescences. We also checked the expression of core cell cycle genes in our dataset and found that, overall, they were expressed at low levels in arrested inflorescences, at slightly elevated levels at 6 h after fruit removal, and at high levels at 24 and 48 h after fruit removal (Supplemental Fig. S1).
Taken together, these results suggest that GPA leads to mitotic quiescence rather than senescence of arrested meristems since they are fully revertible to growing meristems upon the removal of correlative inhibition by fruits. In contrast, older differentiated flower buds in arrested inflorescences never continued development upon fruit removal and appear to be senescent. Therefore, these two inflorescence tissues appear to show distinct responses to correlative inhibition.
To examine the differences between sterile or fertile meristems and whole inflorescences before or after GPA in more detail, we used laser-assisted microdissection (LAM) to isolate meristems from fixed and sectioned inflorescences of (1) growing sterile, (2) growing fertile, (3) arrested, and (4) fully reactivated (i.e. 48 h after fruit removal from arrested plants) inflorescences (Fig. 2, B and C). We extracted total RNA from the microdissected meristems and performed mRNA amplification and either microarray hybridizations or RNA sequencing (RNA-Seq). RNA-Seq was performed with three independent biological replicates to enable more detailed statistical analyses. In both transcriptome datasets, we found the largest differences between growth-arrested and growing tissues regardless of plant age (Fig. 2D; Supplemental Fig. S2). However, the multidimensional scaling of the microarray data suggested that differences between arrested and growing meristems were smaller than between arrested and growing inflorescences (Fig. 2D).
These findings show that gene expression profiles from inflorescences and isolated meristems are distinct and that differences between more homogeneous tissues, i.e. meristems isolated by LAM, are smaller than between complex samples consisting of various organs and tissues.
Meristems But Not Whole Inflorescences Retain Basic Functional Properties during GPA
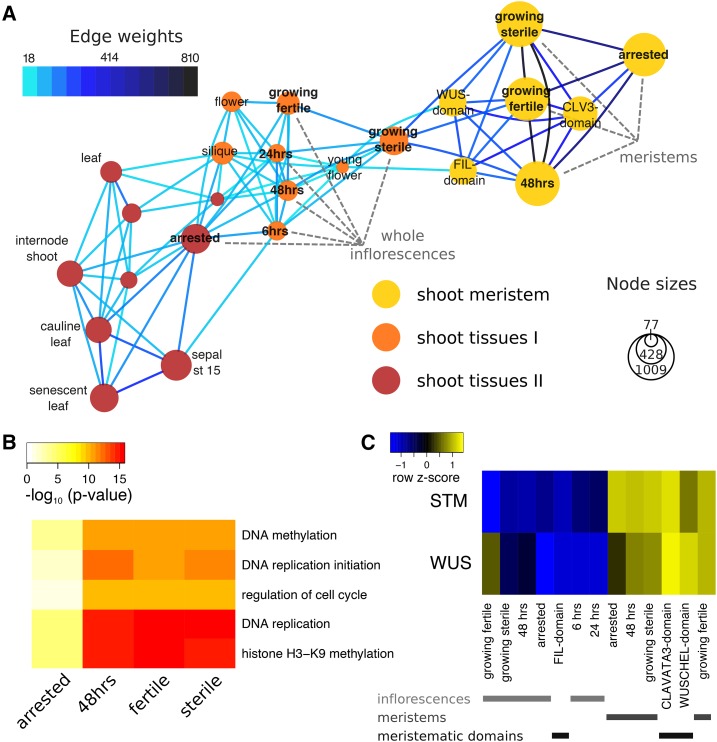
To gain more insight into the developmental context of GPA, we used the microarray data generated in our study to construct gene-sharing networks (Li et al., 2012) by adding publicly available datasets from various developmental stages and plant parts (Fig. 3; datasets described by Ó’Maoiléidigh et al. [2013]). This method first identified genes specifically expressed in one or a subset of tissues or cell types. Genes shared between tissues also enabled the construction of a network of tissues. Connections (edges) between tissues (nodes) that contain more genes than expected were identified (see Fig. 3A and Supplemental Fig. S3, and “Materials and Methods” for details on the computations).
Figure 3.
Gene-sharing network of Arabidopsis tissues. A, Gene-sharing network visualizing shared subsets of genes between tissues. For clarity, only a subset of all tissues in the gene-sharing network is represented; please refer to Supplemental Figure S3 for the full network. Nodes represent tissues, with areas proportionate to the number of genes specific to a given tissue. Edges are drawn only if they contain more genes shared between tissues than expected by chance, and the number of genes represented by an edge is color-coded. Node colors indicate the grouping into network communities. The LAM-isolated “meristem” as well as the “whole inflorescence” samples produced in this study are labeled (gray). B, Example GO terms significantly enriched in meristem nodes. The heatmap represents transformed P values from a test for enrichment of gene-GO term associations within the set of genes in a node. Only five of the terms that vary most in P values between different nodes are shown (see Supplemental Dataset S1 for a full list of significant GO terms). C, Relative expression of the shoot meristem stem cell markers WUS and STM across whole inflorescence samples, as well as LAM-isolated meristems and cell-sorted meristematic domains (Yadav et al., 2009). Colors in the heatmap represent scaled expression estimates, with blue denoting low and yellow denoting high expression, respectively.
Next, a random walk-based algorithm determined seven communities within our gene-sharing network, i.e. seven subgroups of tissues that exhibit more edges between nodes within the subgroup than between subgroups. The communities represented intuitive groups of tissues, such as root, shoot (two separate communities), seed, shoot meristem, and male and female gametophytes (Supplemental Fig. S3). All of the meristem datasets generated in this study formed nodes within a shoot meristem community (Fig. 3A), together with previously published data from cell-sorted domains of the shoot apical meristem, i.e. domains expressing the CLAVATA3 or WUSCHEL (WUS) protein (Yadav et al., 2009). The whole inflorescence nodes from this study were only loosely connected to the shoot meristem nodes. The former were strongly interconnected and formed part of the two separate shoot communities (Fig. 3A). One of the shoot communities contained mainly growing tissues, such as young flowers and floral organs, including young petals, carpels, and ovules, as well as all our inflorescence samples with the exception of arrested inflorescences. Another shoot community contained seedlings and leaves of different ages (including cauline leaves and sepals), older flowers and floral organs, as well as shoot internodes and axillary buds. The arrested whole inflorescence node was contained within this second shoot community, and exhibited strong connections to cauline and senescent leaves, older flowers, and vegetative floral organs (sepals and older petals).
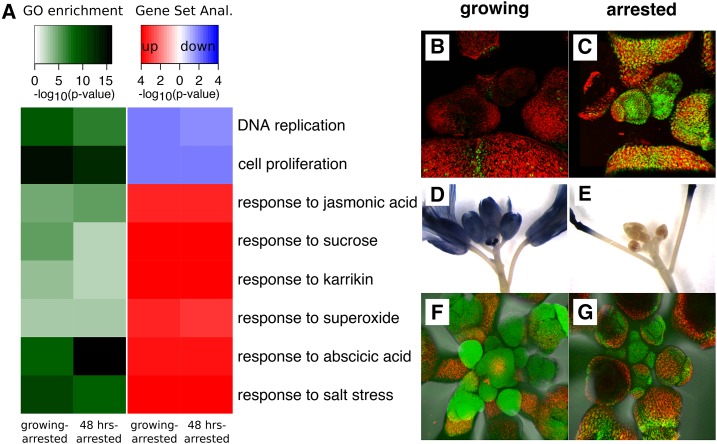
We performed a gene ontology (GO) analysis on genes contained in the nodes of the gene-sharing network, and found 52 terms enriched among genes contained in any of the whole-inflorescence nodes (adjusted enrichment P < 0.01) and 66 terms enriched in any of the meristem nodes from our study. These GO terms were related to developmental processes and growth as well as hormonal responses or specific biosynthetic activities (Supplemental Dataset S1). For example, GO terms enriched in all of our meristem nodes, including arrested meristem nodes, were related to functions typical of meristematic processes such as chromatin remodeling, production of small RNAs, DNA repair processes, etc. GO terms that exhibited largest variation between nodes within subnetworks are shown in Figure 3B and Supplemental Figure S4. They include terms that relate to the mitotic activity enriched especially in growing but less in arrested meristems (Fig. 3B), or terms relating to pollen development and redox processes (enriched especially in growing whole inflorescences), as well as response to salicylic acid, flavonoid synthesis, and the hypersensitive response (enriched especially in arrested whole inflorescences; Supplemental Fig. S4).
Both the assignment of all meristem nodes to a single community as well as the GO terms enriched in all our meristem samples suggest that basic functional properties in meristems were maintained throughout our experiments. To assess whether meristem identity was also maintained, we checked the expression of the shoot meristem marker genes WUS and SHOOTMERISTEMLESS (STM) in both our microarray and RNA-Seq datasets. Both markers were readily detectable in all meristematic samples, though WUS expression levels were slightly reduced in arrested meristems (Fig. 3C; Supplemental Fig. S4).
From the gene-sharing network analysis, we conclude that (1) meristems and whole inflorescences are distinct regarding functional properties and responses to fruit production, (2) arrested meristems seem primarily affected by GPA in their mitotic activities but not in other meristem-specific functional properties, and (3) arrested whole inflorescences share gene expression properties with older and differentiated plant tissues, possibly indicating altered resource allocation patterns in mother plants.
Nonmeristematic Shoot Tissues Exhibit Signs of Senescence upon Fruit Production
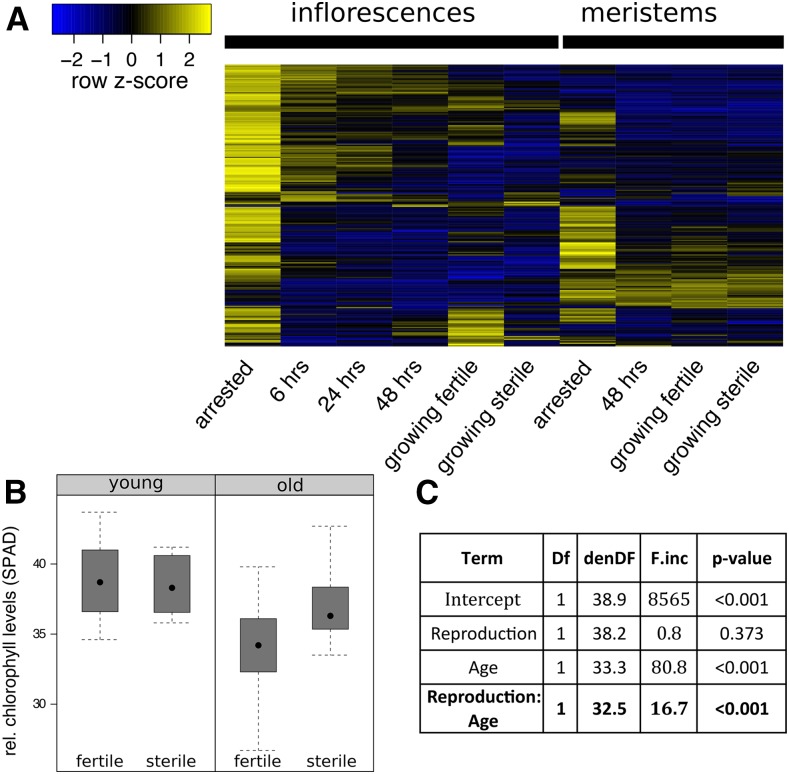
To investigate the effect of fruit production on maternal resource allocation patterns in more detail, we first checked the extent of senescence-associated gene expression in our whole inflorescence and meristem samples. Previous work has established a set of genes that are consistently up-regulated in the course of different senescence programs (Buchanan-Wollaston et al., 2005). As shown in Figure 4, these genes exhibited strongest expression in arrested inflorescences, but also showed some expression in growing inflorescences of fruit-bearing plants and in arrested meristems. Expression of the senescence marker gene SAG12, for example, was not detected in arrested meristems but was strongly expressed in arrested whole inflorescences.
Figure 4.
Signs of senescence upon fruit production in maternal tissues. A, Heatmap showing the relative expression of previously identified senescence-associated genes in whole inflorescence and meristem samples. Expression values were scaled per row (z score transformation); yellow values denote high expression and blue values denote low expression. Strongest expression of genes can generally found in arrested inflorescences. B, Boxplot of relative chlorophyll levels (SPAD measurements) in cauline leaves of young (few fruits produced; left) and old (at the time of GPA of fertile plants; right) fertile and sterile plants. Measurements of 23 fertile (wild-type segregants) and 11 sterile (ms1-1/ms1-1) plants are shown. C, ANOVA of a linear mixed model with SPAD measurements as explained variable, plant fertility (Reproduction) and developmental stage (Age) as fixed explanatory variables, and plant individual as random variable.
Because we had noted some differences in the onset of leaf senescence in cauline leaves of sterile and fertile plants in our previous experiments, we measured chlorophyll absorption levels in basal cauline leaves of the primary inflorescence using a SPAD spectrometer. We compared chlorophyll absorption in sterile (ms1-1/ms1-1) to fertile (wild-type segregants) plants, and repeated the measurement at an early stage of inflorescence growth (“young”) and around the time where most fertile plants had just arrested growth upon fruit production (“old”). Indeed, fruit-bearing wild-type segregants exhibited lower chlorophyll levels of cauline leaves than homozygous sterile ms1-1 plants but only at later stages of development (Fig. 4, B and C, interaction F1,32.5 = 16.7, P < 0.005).
In summary, both the induction of senescence-associated gene expression and the drop in chlorophyll absorption levels in cauline leaves suggest that fruit production leads to the reallocation of resources in maternal shoot tissues.
GPA Is Associated with Responses to Stress and to Growth-Inhibitory Hormones, But a Reduction of the Level of Reactive Oxygen Species in the Shoot
Since the hormone(s) or signal(s) that underlies the correlative inhibition of fruits over maternal growth and resources remains unknown (but should lead to strong responses in shoot tissues upon GPA), we examined differentially expressed genes in our RNA-Seq dataset. We determined differentially expressed genes in the contrasts of fertile growing versus arrested meristems and of arrested meristems versus reactivated meristems upon fruit removal from arrested plants. A GO analysis supported the notion that cellular components necessary for mitotic activity were down-regulated upon the production of maturing fruits (Fig. 5; Supplemental Fig. S5). At the same time, various expression signatures of hormonal responses were up-regulated in arrested meristems. Furthermore, a large proportion of up-regulated functional terms could be interpreted as a response to abiotic and biotic stress in arrested meristems (Fig. 5A; Supplemental Fig. S5), e.g. genes responding to wounding, to salt or cold stress, or to the oxygen radical superoxide, respectively. Genes that respond to ABA were very strongly induced, e.g. the HISTONE H1.3 gene, known to be highly ABA responsive (Fujita et al., 2005; Yao and Finlayson, 2015), was among the top differentially expressed genes in both contrasts. Indeed, using confocal scanning laser microscopy, we observed strong fluorescence differences in growing and arrested shoot apices in a previously described H1.3-GFP expression marker line (Fig. 5, B and C; Rutowicz et al., 2015).
Figure 5.
Growth-inhibitory and stress-responsive gene expression increases but ROS levels decrease during GPA. A, Heatmap showing the results of a GO term and subsequent gene set enrichment analysis on genes found to be differentially expressed between arrested and growing or reactivated meristems. The gene set enrichment analysis determines the directionality of gene expression changes within selected GO terms: red colors indicate that genes within a term are up-regulated in arrested meristems, and blue colors indicate that genes within a term are down-regulated. B and C, Confocal imaging of growing (B) or arrested (C) shoot apices of the H1.3-GFP expression marker line. D and E, NBT staining for superoxide in growing fertile (D) or arrested (E) inflorescences. F and G, Fluorescein staining for ROS in either growing fertile (F) or arrested (G) inflorescences. Only representative figures are shown in B to G.
Based on these observations, we hypothesize that a physiological trade-off between current and future reproduction could arise if increased stress levels upon reproduction and GPA would lead to decreased integrity of cellular structures or macromolecules in meristematic cells. To test this idea, we monitored cellular reactive oxygen species (ROS) and superoxide levels of whole inflorescences using the dyes dichlorofluorescein and nitrotetrazolium blue (NBT), respectively. Since ROS have a wide role in plant cell death and stress response pathways (Gechev et al., 2006), we expected to see increased levels in arrested inflorescence tissues. Contrary to our expectations, we detected little or no staining in arrested inflorescences, but very strong staining with both dyes in growing meristems and developing flowers (Fig. 5, D–G). To our knowledge, such extreme differences in ROS levels have not been described in Arabidopsis inflorescences upon growth transitions. Instead, they are reminiscent of effects previously described in transitions in to and out of bud dormancy in other species, such as kiwifruit, grapevine, and poplar (Considine and Foyer, 2014).
GPA Represents a Form of Shoot Dormancy
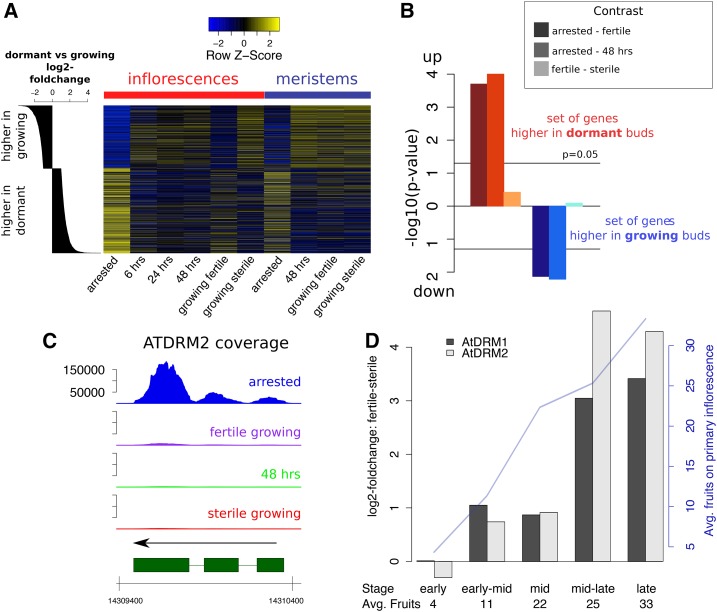
Based on the unexpected results with regard to ROS and superoxide in fertile and arrested meristems, we explored the parallels between bud dormancy and GPA in more detail. In Arabidopsis, dormancy occurs e.g. in axillary buds that are suppressed by apical dominance (Shimizu-Sato and Mori, 2001). Previous work has established patterns of gene expression changes in lateral buds upon release of apical dominance, and identified two expression markers that are highly expressed in dormant buds, the DORMANCY-ASSOCIATED PROTEIN1 and 2 (AtDRM1 and AtDRM2) genes (Tatematsu et al., 2005). Using our microarray dataset, we visualized the expression of genes that were previously determined to respond in axillary buds to the removal of dominant shoots (Fig. 6). We found that genes more highly expressed in suppressed compared with growing axillary buds were also more highly expressed in arrested inflorescences or meristems. Furthermore, these genes generally exhibited high expression in whole inflorescences of fertile growing plants. Conversely, genes previously determined as more highly expressed in growing axillary buds were also more highly expressed in sterile and reactivated inflorescences and all growing meristems. A gene set enrichment analysis of gene expression changes in arrested versus growing meristems added statistical support to the idea that changes in growing versus suppressed axillary buds were reflected in similar expression changes in meristems upon state transitions (Fig. 6B). Similarly, the AtDRM1 and AtDRM2 transcripts were both strongly up-regulated in arrested meristems as compared with growing meristems and up-regulated in fertile as compared with sterile meristems (Fig. 6C; Supplemental Fig. S6A). We therefore compared the expression levels of the two AtDRM genes in inflorescences of sterile and fertile plants at different developmental stages using quantitative reverse transcription PCR (qRT-PCR). We found that expression of both genes changed throughout shoot development (Supplemental Fig. S6, B–D). Furthermore, there were no expression differences between fertile and sterile plants as the fruit development began, but expression of both genes became higher in shoots of fertile plants as fruit production continued (Fig. 6D; Supplemental Fig. S6, B–D).
Figure 6.
Fruit- and GPA-induced changes resemble axillary bud dormancy. A, Heatmap showing relative expression of genes either more highly expressed in dormant axillary buds (bottom rows) or more highly expressed in activated axillary buds (top rows). Genes are sorted according to log2 fold changes reported by Tatematsu and colleagues (histogram on the left; Tatematsu et al., 2005). Blue colors represent low, yellow colors represent high expression in our microarray dataset. B, Results of a gene set enrichment analysis testing for directionality of expression changes in our meristem RNA-Seq dataset. The gene sets chosen are the groups of genes previously reported to be up- or down-regulated in dormant compared with activated axillary buds (as also seen in A). The three different bars for each test represent different contrasts, e.g. contrasting meristems of arrested versus fertile growing plants, etc. Negative decadal logarithms are represented. C, RNA-Seq normalized coverages across the AtDRM2 genic region in the four meristematic states. D, Estimated expression differences of the AtDRM1 and AtDRM2 genes in fertile versus sterile plants at different stages of development (from “early,” first fertilized fruits emerging from the primary inflorescences, to “late,” proliferative arrest; with n = 3 for each time point and reproductive status). Average numbers of fruits on the primary inflorescence of the fertile plants at each stage are given. A full statistical analysis of the data is shown in Supplemental Figure S6B.
Overall, GPA and bud dormancy exhibit striking similarities at changes in gene expression and ROS levels. Furthermore, expression of dormancy-related genes was already higher in fruit-bearing compared with sterile shoot apices before GPA.
DISCUSSION
Correlative inhibition of reproductive structures over maternal growth has been described previously in Arabidopsis (Hensel et al., 1994; Noodén and Penney, 2001), but the underlying developmental principles and molecular changes have remained elusive (Gan, 2003). Furthermore, previous work has established that rosette leaf senescence is uncoupled from reproduction (Hensel et al., 1993; Noodén and Penney, 2001). In such a case, offspring would have control over the production of flower numbers (i.e. restricting maternal seed production) but not over maternal resource allocation patterns, leaving the question open as to how the seed number-seed quality relationship is constrained in this species.
We assessed the molecular responses of meristems and whole inflorescences to either fruit production or GPA. We aimed to identify genes and processes that are under correlative control in maternal apices, but also detect clues that allow for an evaluation of different hypotheses about the nature of the correlative signal. We used a combination of genetic manipulations and fruit removal experiments, and measured genome-wide gene expression changes and changes in physiological variables. We distinguished effects on mature maternal tissues and on maternal meristems since these two types exhibit different phenotypic responses to fruit production and to GPA.
At the gene expression levels, meristems (isolated by LAM from maternal shoot apices) and whole inflorescences were distinct in their responses to fruit production and to GPA: the responses in meristems were less extensive than responses of whole inflorescences. Our data suggest that the meristem retained identity and functional properties throughout all experimental conditions, and responded to GPA largely by a reduction in cellular components necessary for mitotic activity. However, increases in stress levels and up-regulation of some senescence-associated genes (including AtNAP, as described in a previous publication [Sablowski and Meyerowitz, 1998]) were also readily detectable in arrested meristems. At the same time, mitotic activity and transcriptional states rapidly reverted to prearrested levels upon fruit removal. Thus, our results suggest that the term “mitotic senescence” does not appropriately describe GPA.
In contrast to meristematic tissue and young floral buds (the latter of which regenerated after GPA and subsequent fruit removal), older floral buds that had been formed just prior to GPA did not resume development and exhibited signs of senescence. Transcriptomes of all our growing inflorescences grouped with those of other young and growing (sink) tissues in our gene-sharing network, whereas whole inflorescences undergoing GPA grouped with older (source) tissues, the latter including cauline and senescent leaves. Furthermore, extensive stress- and senescence-associated gene expression was detectable in whole inflorescences upon GPA. Chlorophyll levels of cauline leaves were lower in fruit-bearing old plants than in sterile old plants. Together, these observations support the notion that the production of fruits leads to extensive reallocation of resources in maternal shoot tissues. As mentioned above, previous reports have shown that rosette leaf senescence and fertility are uncoupled in Arabidopsis (Hensel et al., 1993, 1994; Noodén and Penney, 2001). At the same time, removal of reproductive bolts has been shown to delay chlorophyll degradation in rosette leafs in previous reports (Ye et al., 2000). Therefore, it is possible that altered resource allocation upon the commitment to reproduction occurs in multiple steps, e.g. resources might (1) be translocated from rosette leaves to shoots upon the floral transition and (2) be remobilized from shoot tissues into developing seeds as the latter mature. Consistent with this idea and our results, a delay in cauline leaf senescence was proposed to occur in abi3-1 mutant plants, in which the late seed-filling process and maternal resource allocation is altered in the shoot (Robinson and Hill, 1999). However, in Arabidopsis, correlative control over leaf senescence might be expressed only relatively weakly because it is superimposed on other fast (but possibly passive) aging mechanisms that render leaves of this species very short-lived (Noodén, 2013). Furthermore, since soil nutrient availability or light quality influence senescence patterns (Gan and Amasino, 1997), correlative control might express differentially under different environmental conditions.
Finally, we observed that seed production enhances shoot senescence and stress-related gene expression, but that GPA is associated with a drop of ROS levels in growing shoots, as measured by NBT and dichlorofluorescein staining. It is thus likely that growing shoot tissues and meristems exhibit anoxic conditions: multiple reports have shown a relationship between ROS levels and root development, but less attention has been dedicated to the link between ROS levels and shoot meristematic functions. However, work in perennial species had suggested that bud break after winter dormancy is associated with strong increases in ROS levels (Considine and Foyer, 2014). We explored the similarities between dormancy and GPA: gene expression changes from growing to arrested meristems and vice versa are consistent with the idea that GPA represents a shoot dormancy. For example, two of the genes with highest gene expression changes during GPA are the AtDRM1 and AtDRM2 genes, considered markers for dormant axillary buds (Tatematsu et al., 2005; Rae et al., 2014). Furthermore, genes responding to ABA, which have recently been proposed to regulate axillary bud growth (Yao and Finlayson, 2015), are strongly up-regulated during GPA. Thus, GPA might rely on the same signaling and execution mechanisms that regulate axillary bud growth inhibition. It would be interesting to assess how GPA is changed in mutants that exhibit distorted apical dominance patterns, but the interpretation of such experiments is difficult because of developmental epistasis (i.e. changes in apical dominance will also alter the reproductive allocation patterns that lead to GPA) and subsequent pleiotropic effects of such mutations. In this context it is interesting to note that the dormancy-associated expression markers AtDRM1 and AtDRM2 are induced already upon fruit production in growing maternal inflorescences, before any phenotypic expression of GPA. Also, growth-kinetic differences between sterile and fertile plants are visible before arrest of fertile mothers (see also Fig. 1B). Together, these observations suggest that the acquisition of a dormancy-related expression program in growing shoots happens somewhat gradually upon fruit production.
We propose a model that describes the gradual transfer of dominance from the growing shoot to the seeds (as the latter mature), which then leads to a reduction of shoot growth and eventually the arrest of all its meristematic activities. Such a model would not necessarily need the postulation of new signaling components that mediate correlative control of offspring over maternal allocation strategies (e.g. a “death hormone”; Wilson, 1997; Noodén et al., 2003). Rather, both the changes in apical growth patterns and the promotion of senescence in cauline leaves would be explained if seeds interfere with or alter preexisting pathways that normally mediate apical dominance. Sachs already noted that dominant shoots exert correlative control over leaf senescence (Sachs, 1966), and application of auxin to defruited peduncles or deseeded fruits in peas has been shown to result in increased nutrient accumulation at the site of application, senescence, and bud growth inhibition (Seth and Wareing, 1967; Tamas and Engels, 1981). Auxin production in seeds could be important for such correlative control, but testing this hypothesis will require carefully designed experiments that go beyond the application of hormones to different tissues (see Noodén et al. [2003] for critical considerations in such empirical studies). Furthermore, interactions with other hormones are likely to be part of correlative control, which will complicate the matter (Seth and Wareing, 1967; Zhu and Davies, 1997).
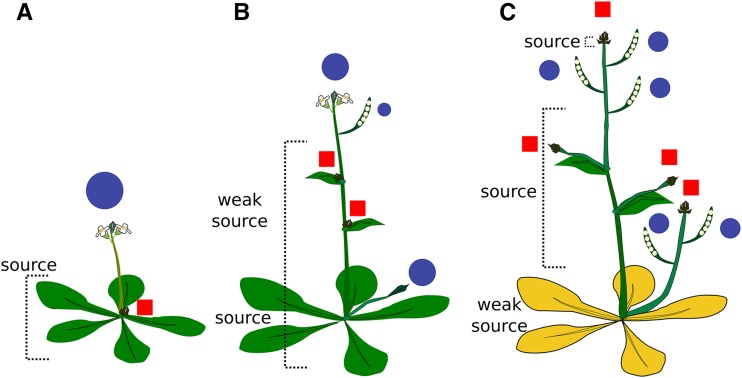
A useful working model for correlative control in Arabidopsis (Fig. 7) could be the following: (1) Growing apices after the floral transition exert apical dominance over dormant axillary buds. (2) Concomitant with this inhibition of nondominant branches, apical dominance controls source-sink relationships and promotes senescence in leaves. (3) Dominance is gradually transferred from the initially dominant shoot to the seeds as the latter mature. This leads to a (first gradual) reduction of growth and eventually to the arrest of all shoot proliferative activities. Seed production also suppresses axillary growth through the same principle, but only at later stages of development as dominance is transferred from the main apex to seeds. Some shoot tissues predominantly act as source tissues for the developing seeds, and sterility delays shoot senescence (e.g. senescence of cauline leaves). Removal of fruits at this point leads to a reactivation of the arrested shoot meristem and the outgrowth of axillary buds. Senescence in rosette leaves is already at an advanced stage and is therefore not influenced by fruit production.
Figure 7.
Model of dominance patterns and source-sink relationship changes upon fruit production. Blue circles denote points of dominance and sink tissues; red rectangles denote suppressed shoot axes. Green leaf color represents living tissue, and yellow leaf color represents senescing tissue. A, Following the transition to flowering, a single dominant shoot suppresses axillary buds and acts as a resource sink. The rosette leaves become source tissues. Removal of bolts at this point delays rosette leaf senescence. B, Following fruit set, developing seeds gradually become dominant and act as sink tissues. Resources have partially been translocated into shoots so that some shoot tissues gradually become source tissues for seeds and growing apices. C, Seeds increasingly become dominant over maternal growth so that a GPA suppresses growth of previously dominant shoot apices and of axillary buds.
Such a working model leads to the following predictions:
-
-
The relationship between seed number and quality is constrained by a relatively direct mechanism in Arabidopsis, i.e. plant longevity is not purely achieved by continuous regeneration of new photosynthetic tissues. Genetic or transgenic methods that interfere with seed sink strength will affect branching and fruit production from the main shoots. Methods that interfere with leaf senescence will interfere with source-sink relationships that provision maturing seeds with nutrients.
-
-
Genetic or environmental changes that affect apical dominance also affect seed quality or size. For instance, an effect of auxin on seed dormancy and on seed size has recently been shown (Liu et al., 2013; Liu et al., 2015), but, to our knowledge, changes in seed size or quality have not explicitly been described in mutants with altered apical dominance.
-
-
If patterns of apical dominance and fertility influence resource partitioning in Arabidopsis mother plants, they should also influence other functions that rely on resources, e.g. resistance to biotic or abiotic stresses. Such relationships should follow a trade-off function, e.g. fruit production should alter maternal defense against pathogens or stress. Effects should first become apparent in rosette leaves upon bolting and in cauline leaves upon fruit production.
-
-
There exists a direct link between apical dominance and seed size. If this is so, it should also lead to constrained evolution of inflorescence architecture and seed size due to the genetic correlation between these traits. It is predicted that large-seeded relatives of Arabidopsis (if of similar size) have fewer branches and vice versa.
The work presented here establishes a molecular and physiological framework to test such a model and the predictions arising from it.
MATERIALS AND METHODS
Plants and Growth Conditions
For all the molecular analyses, plants of Arabidopsis (Arabidopsis thaliana) accessions Landsberg erecta (Ler), Columbia-0, ap3-1 (NASC Code N3085; see www.arabidopsis.info), and ms1-1 (NASC Code N75) were grown on soil (ED73; Universalerde) covered with a thin quartz sand layer, in a growth chamber at the Department of Evolutionary Biology and Environmental Studies under 16 h light (at 20°C and 5 kLux) and 8 h dark (at 16°C). For the analyses of leaf senescence, the plants were grown in a growth chamber at the Department of Plant and Microbial Biology under 16 h light and 8 h dark (at 22°C and 60% humidity). Different genotypes were grown in individual pots placed randomly on trays, and pot and tray locations were rerandomized regularly (at least every 10 d). For the analysis of flower production of fertile and sterile plants as shown in Figure 1, the plants were grown on nutrient-poor soil, i.e. a mixture consisting of one-quarter ED73 Universalerde and three-quarters quartz sand.
Tissue Collection for Molecular Analyses
Whole inflorescences containing flowers up to approximately floral stage 10 were collected under a dissecting microscope using ultrafine tweezers. Inflorescences from primary and secondary branches were either pooled from several plants (between three and 10, for microarray analyses) or collected individually (for qRT-PCR) and immediately frozen in liquid nitrogen and stored at −80°C until further use.
For LAM, inflorescences from at least 20 individual plants were collected on ice and fixed in 3:1 (v/v) ethanol:acetic acid under vacuum (∼500 mBar) for 2 × 10 min at 4°C and then left in the fixative overnight at 4°C. The tissue was embedded into Paraplast X-tra Embedding Media (Sigma-Aldrich) using an automated protocol and the Leica ASP200 embedding machine as follows: 1 h 70% (v/v) ethanol, 3 × 1 h 90% (v/v) ethanol, 3 × 1 h 100% ethanol, 2 × 1 h 100% xylol, and 1 × 15 min 100% xylol, all at room temperature, and then 2 × 1 h Paraplast X-tra and 1 × 3 h Paraplast X-tra at 56°C. Inflorescences were then poured into paraplast blocks and cooled at 4°C until use. Embedded samples were sectioned on a Leica RM2145 microtome to 10 μm thickness and mounted on membrane slides (Molecular Machines and Industries) using sterile water for mounting. The slides were then dried overnight at 42°C on a heating table and laser dissected immediately. LAM was performed on a Molecular Machines and Industries CellCut Plus after dewaxing the tissue sections for 2 × 10 min in xylol as described by Wuest et al. (2010).
RNA-Seq and Transcriptional Profiling
For laser-microdissected samples, the total RNA was extracted using the PicoPure RNA isolation kit (Life Technologies), including the DNAseI treatment as recommended in the technical manual. Two to four caps containing tissue sections were pooled on-column. For each RNA-Seq replicate, between 16 and 31 embedded inflorescences were sectioned and mounted onto slides, which resulted in 58 to 91 laser-dissected meristem sections that were pooled per replicate. For RNA-Seq, total RNA was amplified using the NuGEN Ovation 3′DGE amplification kit, according to the technical manual. The resulting amplified DNA was sequenced on the Illumina HiSeq at Functional Genomics Center Zurich (FGCZ). The four samples of each block were multiplexed and sequenced on a single lane per block. The sequencing and microarray data have been deposited in the National Center for Biotechnology Information GEO database (series GSE74386 and GSE79287).
For whole tissue samples (i.e. inflorescences) and microarray profiling, total RNA was extracted using the Sigma Plant Mini RNA extraction kit as described in the technical manual and included a DNAseI treatment (Roche). RNA labeling for microarray hybridizations was performed using the GeneAtlas 3′IVT Express Kit (Affymetrix). RNA extracted from laser-dissected tissues and used for microarray hybridization was amplified using the Arcturus RiboAmp Plus kit (Applied Biosystems) according to the manufacturer’s instruction, but with the second in vitro transcription performed using Affymetrix 3′IVT labeling kit reagents. Hybridization of the Affymetrix ATH1 microarrays and subsequent washing and scanning were performed at FGCZ or Biozentrum Basel according to the manufacturer’s instructions.
For qRT-PCR, total RNA was extracted from whole inflorescence samples using TRIZOL reagent (Ambion) according to the manufacturer’s instructions. A DNAse treatment was subsequently performed using the Ambion RNAse-free DNAse I (Life Technologies Europe B.V.), and cDNA was synthesized using the Superscript III first-strand synthesis kit (Invitrogen). The qPCR reactions were performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories AG) on the Applied Biosystems 7500 Fast Real-Time PCR System. In addition to the two assays for the AtDRM1 (forward primer: ATTCTCCCTCCGTCTACGACTG; reverse primer: CTTGAGTCACCGCTGTACAACC) and AtDRM2 genes (forward primer: TTCTCACCCAAACTCTCCCAC; reverse primer: TCACTCTACCCTCTCGGCTC), assays for two reference genes (Czechowski et al., 2005) AT4G34270 (forward primer: ATCTGCGAAAGGGTATCCAGTTGAC; reverse primer: TGGAAGCCTCTGACTGATGGAGC) and AT2G28390 (forward primer: CACTCTTCTATGTTGGGTCACACCAG; reverse primer: TTATCGCCATCGCCTTGTCTGC) were used.
Design of Molecular Analyses
For the laser-dissected meristem samples from sterile growing plants, fruit-bearing growing plants, fruit-bearing arrested plants, and arrested plants that had their fruit removed 48 h prior to sample collection, RNA-Seq experiments were performed in three blocks (i.e. independently grown batches of plants). The positions of plants from different treatment groups were fully randomized within each block. For the first block and also for the microarray experiment of meristems, the sterile plants were conditional male-sterile ap3-1 mutants (in a Ler background), and all other treatment groups were Ler wild-type plants. For blocks 2 and 3 of the RNA-Seq experiment, the genotype used was the ms1-1 male-sterile mutant. Thereby, seeds from single heterozygous plants were used so that the fruit-bearing plants were segregants either heterozygous or wild type for the ms1-1 allele and sterile plants were homozygous for the ms1-1 allele.
For the microarray profiling of whole inflorescences, Ler plants were allowed to arrest and collected at 0, 6, 24, and 48 h after removal of all but three or four fruits closest to the inflorescence. Several inflorescences (including flowers up to approximately Stage 10 according to Smyth et al. [1990]) of at least three plants were pooled.
Analyses of RNA-Seq Dataset
Sequencing reads were mapped against the Arabidopsis genome (TAIR version 10) using the subread software and default parameter (Liao et al., 2013), and sorted and indexed using the samtools software (Li et al., 2009). All subsequent analyses were performed in the statistical software R (R Core Team, 2015) and software packages implemented in the Bioconductor project (Gentleman et al., 2004). Reads were assigned to genes using the featureCounts function implemented in the R package Rsubread (Liao et al., 2014) and gene definitions from TAIR version 10 (www.arabidopsis.org). Coverage values were calculated from aggregated reads over all three replicates and standardized using the relative number of mapped reads for each condition. The GenomeGraphs package (Durinck et al., 2009) was used to plot coverages, and the Rsamtools package (Morgan et al., 2015) was utilized as an interface with indexed BAM files. Differentially expressed genes were identified using the edgeR package (Robinson et al., 2010). Normalization factors were calculated using the weighted trimmed mean of M values (Robinson and Oshlack, 2010). Then, a negative binomial generalized log-linear model (McCarthy et al., 2012) was fitted that included block (1–3) and developmental stage (“arrested,” “fruits removed,” “fertile growing,” and “sterile growing”). Contrasts were used to compare different developmental stages (Supplemental Dataset S2).
GO Term Enrichment and Gene Set Analyses
GO term enrichment analysis was performed on a subset of all terms relating to a “biological process.” Redundant terms were filtered as described previously (Wuest et al., 2012), that is, removing parent terms in cases where they shared more than 80% of the genes with all respective children terms. GO analyses were performed using the goseq package with default parameters (Young et al., 2010). The significant terms were then used in a gene set enrichment analysis using the mroast function implemented in the limma package (Wu et al., 2010) with “msq” set statistic and 10,000 rotations.
Analyses of Microarray Datasets
Microarrays were normalized (invariant set normalization) and preprocessed (PM-only model) using dChip software version 2010 (Li and Wong, 2001). Gene-sharing networks were constructed as described (Li et al., 2012), and edges between tissues were retained if they contained more genes than expected from the sizes of the respective nodes (as determined by a one-sided Fisher’s exact test and P value cutoff < 1e-10). The resulting gene-sharing network was visualized using the Rgraphviz package (Hansen et al., 2015). A GO analysis of genes contained within nodes of the gene-sharing networks was also performed using the goseq package but assuming no bias in power to detect differentially expressed genes in the experiment. A random-walk-based algorithm (Pons and Latapy, 2006) was used to detect communities in our network with the help of the walktrap.community function implemented in the igraph package (Csardi and Nepusz, 2006). Heatmaps were drawn using the heatmaps.2 function implemented in the gplots package (Warnes et al., 2015).
Staining of ROS and Confocal Imaging
For NBT (Sigma-Aldrich Switzerland) staining, inflorescences were covered in a 2-mL Eppendorf tube with freshly made NBT solution (0.5 mg/mL NBT in 0.01 m potassium phosphate, pH 7.4) and then vacuum infiltrated for 2 min. Tubes were left for 2 to 3 h at 20°C in the dark for the color to develop. Then, the NBT solution was removed and the samples washed twice with potassium phosphate buffer. Finally, samples were destained using 100% ethanol for 24 h, and pictures were taken immediately under a dissecting microscope.
For 2′,7′-dichlorofluorescein diacetate (catalog no. D6883; Sigma-Aldrich Switzerland) staining, inflorescences were incubated for 2 h in buffer containing 50 mm KCl, 50 mm CaCl2, 10 mm MES-Tris, pH 6.15. Then, H2DCF-DA was added to a concentration of 50 mm. Inflorescences were further incubated for 40 min at room temperature in the dark, and excess dye was removed by washing the samples three times for 5 min using the mentioned buffer. Fluorescence was then detected under a Leica SP5 confocal microscope with a HCX PL APO lambda blue 20.0 × 0.70 IMM UV objective (excitation: 488 nm, 19%; emission: 510–540 nm; emission autofluorescence: 600–800 nm, laser 30%). For analysis of the H1.3-GFP marker line, the same confocal microscope was used (excitation: 488 nm, 10%; emission GFP [green]: 490–530 nm; emission autofluorescence (red): 600–800 nm, laser [argon] 30%).
Analysis of Senescence
Offspring of ms1-1 heterozygous plants were sown directly on ED73 soil covered with a thin layer of quartz sand and grown under fully randomized conditions on two trays (with rearrangements of pots within trays at least every 10 d). To avoid between-plant variation, we measured leaf chlorophyll levels using the nondestructive chlorophyll meter SPAD-502Plus (Minolta) repeatedly from the same plant. SPAD measurements were taken for the first cauline leaf on the primary axis of each plant after 4 weeks (early measurement) and 8.5 weeks (late measurement) after sowing. The early measurement took place as plants had bolted, produced cauline leaves, and produced several flowers. The second measurement was taken when inflorescences of the fertile plants had arrested but before any cauline leaves showed visible signs of senescence. For the statistical analysis using the software asreml-R, we fitted a mixed model with developmental stage (early versus late measurements) and fertility as crossed fixed factors, and individual plant as random factor.
Sequence and microarray data from this article can be found in the National Center for Biotechnology Information GEO database under accession numbers GSE74386 and GSE79287.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Heatmap representing expression of core cell cycle genes in growing whole inflorescences, and in arrested whole inflorescences at 0, 6, 24, and 48 h after fruit removal.
Supplemental Figure S2. Sample clustering of RNA-Seq dataset, based on the top 1000 most variable genes, log-transformed counts per million.
Supplemental Figure S3. Gene-sharing network of all tissues considered in this study.
Supplemental Figure S4. Meristems retain identity throughout state transitions and respond differently than whole inflorescences to GPA.
Supplemental Figure S5. GO and gene set enrichment analysis on gene differentially expressed in growing and arrested meristems.
Supplemental Figure S6. Expression of the AtDRM1/2 genes upon the production of fruits and proliferative arrest.
Supplemental Dataset S1. Results of the GO enrichment analysis in selected nodes of the gene-sharing network.
Supplemental Dataset S2. RNA-Seq analysis of laser-dissected meristem samples using the edgeR package.
Supplementary Material
Acknowledgments
We thank Anna Bratus, Catharine Aquino (Functional Genomic Center Zurich), and Philippe Demougin (University of Basel) for help with producing the transcriptomic datasets, Andreas Portmann (University of Zurich [UZH]) for help with tissue collection and Song Li (Duke University) for gene-sharing network scripts. Ulrike Bätz (UZH) kindly provided the NBT staining protocol, Kinga Rutowicz (UZH) the H1.3-GFP expression marker line, and Stefan Hörtensteiner (UZH) the SPAD measurement device. We are grateful to Marion Wood (The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand) and Linda Hensel (Mercer University, Macon, GA) for helpful discussions.
Glossary
- GPA
global proliferative arrest
- LAM
laser-assisted microdissection
- GO
gene ontology
- ROS
reactive oxygen species
- ABA
abscisic acid
- NBT
nitrotetrazolium blue
- qRT-PCR
quantitative reverse transcription PCR
Footnotes
This work was supported by the University of Zürich, a Marie Curie Intra European Fellowship within the 7th Framework Programme (PIEF-GA-2011-302684) and an Ambizione Fellowship (PZ00P3_148223) of the Swiss National Science Foundation (to S.E.W.), and a grant from the Swiss National Science Foundation (31003A_141245 to U.G.).
Articles can be viewed without a subscription.
References
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin J-F, Wu S-H, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Considine MJ, Foyer CH (2014) Redox regulation of plant development. Antioxid Redox Signal 21: 1305–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, Nepusz T (2006) The igraph software package for complex network research. InterJournal Complex Systems 1695
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ, Gan S (2012) Towards an integrated view of monocarpic plant senescence. Russ J Plant Physiol 59: 467–478 [Google Scholar]
- Durinck S, Bullard J, Spellman PT, Dudoit S (2009) GenomeGraphs: Integrated genomic data visualization with R. BMC Bioinformatics 10: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S. (2003) Mitotic and postmitotic senescence in plants. Sci SAGE KE 2003: RE7. [DOI] [PubMed] [Google Scholar]
- Gan S, Amasino RM (1997) Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Gentry J, Long L, Gentleman R, Falcon S, Hahne F, Sarkar D (2015) Rgraphviz: Provides plotting capabilities for R graph objects. R package version 2.10.0, http://www.bioconductor.org/packages/release/bioc/html/Rgraphviz.html
- Hensel LL, Grbić V, Baumgarten DA, Bleecker AB (1993) Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in arabidopsis. Plant Cell 5: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB (1994) The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol 106: 863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MO, Davies PJ, Woolhouse HW (1988) The control of whole plant senescence. Crit Rev Plant Sci 7: 139–173 [Google Scholar]
- Leopold AC, Niedergang-Kamien E, Janick J (1959) Experimental modification of plant senescence. Plant Physiol 34: 570–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Pandey S, Gookin TE, Zhao Z, Wilson L, Assmann SM (2012) Gene-sharing networks reveal organizing principles of transcriptomes in Arabidopsis and other multicellular organisms. Plant Cell 24: 1362–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Lindoo SJ, Noodén LD (1977) Studies on the behavior of the senescence signal in anoka soybeans. Plant Physiol 59: 1136–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Tong H, Xiao Y, Che R, Xu F, Hu B, Liang C, Chu J, Li J, Chu C (2015) Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc Natl Acad Sci USA 112: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang H-Q, Luan S, Li J, He Z-H (2013) Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci USA 110: 15485–15490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd DG. (1980) Sexual strategies in plants. I. An hypothesis of serial adjustment of maternal investment during one reproductive session. New Phytol 86: 69–79 [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molisch H. (1929) Die Lebensdauer der Pflanze. Verlag von Gustav Fischer, Jena, Germany [Google Scholar]
- Morgan M, Pagès H, Obenchain V, Hayden N (2015) Rsamtools: binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R package version 1.18.3, http://bioconductor.org/packages/release/bioc/html/Rsamtools.html
- Murneek AE. (1926) Effects of correlation between vegetative and reproductive functions in the tomato (Lycopersicon esculentum Mill.). Plant Physiol 1: 3–56.7, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noodén LD. (1980) Senescence in the whole plant. In Thimann KV, ed, Senescence in Plants, Ed 1. CRC Press, Boca Baton, FL, pp 219–258 [Google Scholar]
- Noodén LD. (2013) Defining senescence and death in photosynthetic tissues. In Biswal B, Krupinska K, Biswal UC, eds, Plastid Development in Leaves during Growth and Senescence. Springer, Berlin, Germany, pp 283–306 [Google Scholar]
- Noodén LD, Guiamét JJ, John I (2003) Whole plant senescence. In Noodén LD, ed, Plant Cell Death Processes, Ed 1. Academic Press, San Diego, CA, pp 227–244 [Google Scholar]
- Noodén LD, Murray BJ (1982) Transmission of the monocarpic senescence signal via the xylem in soybean. Plant Physiol 69: 754–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noodén LD, Penney JP (2001) Correlative controls of senescence and plant death in Arabidopsis thaliana (Brassicaceae). J Exp Bot 52: 2151–2159 [DOI] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Wuest SE, Rae L, Raganelli A, Ryan PT, Kwasniewska K, Das P, Lohan AJ, Loftus B, Graciet E, et al. (2013) Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25: 2482–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons P, Latapy M (2006) Computing communities in large networks using random walks. JGAA 10: 191–218 [Google Scholar]
- R Core Team (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org
- Rae GM, Uversky VN, David K, Wood M (2014) DRM1 and DRM2 expression regulation: potential role of splice variants in response to stress and environmental factors in Arabidopsis. Mol Genet Genomics 289: 317–332 [DOI] [PubMed] [Google Scholar]
- Robinson CK, Hill SA (1999) Altered resource allocation during seed development in Arabidopsis caused by the abi3 mutation. Plant Cell Environ 22: 117–123 [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutowicz K, Puzio M, Halibart-Puzio J, Lirski M, Kroteń MA, Kotliński M, Kniżewski Ł, Lange B, Muszewska A, Śniegowska-Świerk K, et al. (2015) A specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in Arabidopsis. Plant Physiol 169: 2080–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RW, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Sachs T. (1966) Senescence of inhibited shoots of peas and apical dominance. Ann Bot (Lond) 30: 447–456 [Google Scholar]
- Seth AK, Wareing PF (1967) Hormone-directed transport of metabolites and its possible role in plant senescence. J Exp Bot 18: 65–77 [Google Scholar]
- Shimizu-Sato S, Mori H (2001) Control of outgrowth and dormancy in axillary buds. Plant Physiol 127: 1405–1413 [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas IA, Engels CJ (1981) Role of indoleacetic acid and abscisic acid in the correlative control by fruits of axillary bud development and leaf senescence. Plant Physiol 68: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E (2005) Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D-Y, Li Q, Cui K-M, Zhu Y-X (2007) Gibberellin is involved in the regulation of cell death-mediated apical senescence in G2 pea. J Integr Plant Biol 49: 1627–1633 [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman RC, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, et al (2015) gplots: various R programming tools for plotting data. R package version 2.16.0, http://CRAN.R-project.org/package=gplots
- Wilson JB. (1997) An evolutionary perspective on the “death hormone” hypothesis in plants. Physiol Plant 99: 511–516 [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK (2010) ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26: 2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, O’Maoileidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F (2012) Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci USA 109: 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV (2009) Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Finlayson SA (2015) Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol 169: 611–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Rodriguez R, Tran A, Hoang H, de los Santos D, Brown S, Vellanoweth RL (2000) The developmental transition to flowering represses ascorbate peroxidase activity and induces enzymatic lipid peroxidation in leaf tissue in Arabidopsis thaliana. Plant Sci 158: 115–127 [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YX, Davies PJ (1997) The control of apical bud growth and senescence by auxin and gibberellin in genetic lines of peas. Plant Physiol 113: 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.