Abstract
Purpose
Uveal melanoma (UM) is a rare melanoma variant with no effective therapies once metastases develop. Although durable cancer regression can be achieved in metastatic cutaneous melanoma (CM) with immunotherapies that augment naturally existing anti-tumor T cell responses, the role of these treatments for metastatic UM remains unclear. We sought to define the relative immunogenicity of these two melanoma variants and determine whether endogenous anti-tumor immune responses exist against UM.
Experimental Design
We surgically procured liver metastases from UM (n=16) and CM (n=35) patients and compared the attributes of their respective tumor cell populations and their infiltrating T cells (TIL) using clinical radiology, histopathology, immune assays and whole exomic sequencing.
Results
Despite having common melanocytic lineage, UM and CM metastases differed in their melanin content, tumor differentiation antigen expression, and somatic mutational profile. Immunologic analysis of TIL cultures expanded from these divergent forms of melanoma revealed CM TIL were predominantly composed of CD8+ T cells, while UM TIL were CD4+ dominant. Reactivity against autologous tumor was significantly greater in CM TIL compared to UM TIL. However, we identified TIL from a subset of UM patients which had robust anti-tumor reactivity comparable in magnitude to CM TIL. Interestingly, the absence of melanin pigmentation in the parental tumor strongly correlated with the generation of highly reactive UM TIL.
Conclusions
The discovery of this immunogenic group of UM metastases should prompt clinical efforts to determine whether patients who harbor these unique tumors can benefit from immunotherapies that exploit endogenous anti-tumor T cell populations.
Keywords: Uveal melanoma, cutaneous melanoma, T cells, liver metastases
INTRODUCTION
Uveal melanoma (UM) is a rare and aggressive variant of melanoma that has specific origin within the vascular layers of the eye including the choroid, ciliary body, and iris (collectively known as the uvea) (1). Although UM is the most common intraocular tumor in adults, it accounts for only 3% of all melanomas (2). With an annual incidence of 5.1 per million in the U.S, UM is significantly less common than cutaneous melanomas (CM). Interestingly, UM and CM have a shared lineage, with each arising from neural crest derived melanocytes that are resident to their respective tissues of origin (3). Both forms of melanoma, consequently, share prominent expression of prototypic melanocytic differentiation antigens (MDAs) such as MART-1, gp100, and tyrosinase (4–6). Despite these similarities, UM can be distinguished from CM by characteristic cytogenetic changes (7) and an unusual predilection to primarily metastasize to the liver (1). Further, there exists a striking dichotomy between the clinical management of patients with advanced UM and CM. Immunotherapies have become the main treatment modality for metastatic CM based upon substantial evidence that tumor antigens expressed by CM can be vigorously recognized by T cell populations endogenous to the host immune system (8). By clinically augmenting these immune responses with either systemic cytokines (9), antibodies targeting T cell checkpoint molecules (10, 11), or adoptive transfer of autologous tumor infiltrating lymphocytes (TIL) (12), significant and potentially curative cancer regression can now be achieved in advanced CM patients. However, the role of these immune based therapies for the treatment of metastatic UM remains unclear. Patients with UM are frequently excluded from metastatic melanoma immunotherapy clinical trials because UM is generally thought to be an immunotherapy resistant subtype of melanoma. It has been speculated that since the primary tumor arises in the eye, an immune privileged site, the tumor and its metastases harbor local immunosuppressive or cellular immuno-evasive factors that render immunotherapies unsuccessful (13–16). Another theory proposes that since UM tumors have far fewer somatic mutations compared to sun-exposed CM tumors (17), there are consequentially fewer potential mutated neo-epitope targets for effective anti-tumor immunity. The poor immunogenicity of UM has been further suggested based upon the comparatively low response rates seen in UM patients enrolled into small pilot trials of immune modulating agents such as interleukin-2 (18) and anti-CTLA-4 antibody (19–21). Collectively, these observations have fostered the prevalent belief that UM, in distinction to CM, is a non-immunogenic form of melanoma. However, this hypothesis has largely been based upon inference without formal comparative studies performed directly upon UM and CM metastases to accurately assess their relative immunogenicity. In this study, we aimed to address this deficiency by comparing tumor antigen expression, tumor mutational load, and endogenous anti-tumor immunologic reactivity found in fresh surgically resected UM versus CM metastases. By determining whether tumor specific immune responses naturally exist against UM metastases, we sought to provide insight into the management of this rare melanoma variant with immunotherapies that can exploit these endogenous T cell populations.
METHODS
Study population
A retrospective review of a prospectively maintained database identified 49 patients who underwent liver metastatectomy with a diagnosis of metastatic melanoma at the Surgery Branch of the National Cancer Institute between 2004 and 2014. All patients signed an institutional review board approved consent for tumor tissue procurement and participation in subsequent immunotherapy protocols if the patient required further systemic therapy. Inclusion criteria included pathologically confirmed melanoma, 16 years of age or older, negative serology for HIV, Hepatitis B and C, good performance status (Eastern Cooperative Oncology Group ≤2) and life expectancy greater than 3 months. Patients were stratified into two cohorts based upon the anatomic origin of their primary melanoma. The cutaneous melanoma (CM) cohort included 35 patients; 33 of these patients had documented primary tumors arising from the cutaneous epithelium and 2 additional patients had primary tumors of unknown origin. The uveal melanoma (UM) cohort included 14 patients who had ophthalmologic documentation that their primary melanoma tumors arose specifically from the uveal tract. Patients with documented primary tumors arising from mucosal and conjunctival sites were excluded from analysis.
Tumor procurement
Patients typically underwent resection of a single metastatic liver deposit or a closely approximated cluster of tumors using standardized hepatobiliary surgical techniques. Immediately upon resection, the fresh tumor underwent pathologic assessment, dissection, and processing in the Surgery Branch Cell Production facility in conjunction with a clinical surgical pathologist and research staff. Tumor tissue was assigned a unique liver metastasis identification number (ID #) and allocated for gross and histopathologic analysis, mutational analysis, and TIL culture establishment using methods as described below. Although the main study exclusively focused upon liver metastases, a set of extrahepatic metastases from 8 additional UM patients were incorporated into the tumor driver mutational analysis, as described below.
In situ MRI assessment of tumor melanin content
All patients underwent pre-operative MRI liver imaging as part of their radiographic tumor staging. Quantitative T1-weighted signal intensity measurements (without gadolinium enhancement) of the in situ liver metastases and adjacent normal tissue were obtained using clinical radiology imaging software (Carestream Vue Solutions, version 11.3). Mean tumor and normal intensity were calculated by averaging three separate signal intensity measurements. Hyperintense tumors were defined as having a mean tumor/normal (T/N) intensity ratio > 1.5. Hypointense tumors had a mean T/N ratio < 0.7. Mixed intensity tumors had both hyperintense and hypointense components. The T/N signal intensity ratio for each liver metastasis was objectively calculated for each metastasis and scored as either hyperintense (2+), mixed intensity (1+), or hypointense (0), as illustrated in Supplementary Figure 1.
Gross pathologic assessment of tumor melanin pigmentation
After surgical resection, all liver metastases underwent independent gross pathological assessment and photo documentation by a board certified pathologist who was blinded to the comparative analysis. Each metastasis underwent serial sectioning to assess their melanin pigmentation. Tumors were scored based on their level of pigmentation as either hyperpigmented (2+), mixed pigmented (1+), or hypopigmented (0).
Immunohistochemical staining analysis of tumor metastases
Surgically resected tumor specimens were fixed in 10% neutral buffered formalin for up to 24 hours and routinely processed. Paraffin-embedded tissue sections of 5 mm were deparaffinized through xylene and graded series of alcohols. Immunohistochemical staining was performed following heat-induced epitope retrieval with target retrieval solution (low pH; DAKO, Carpinteria, CA). Slides were incubated in Tris with 3% goat serum for 15 minutes and then incubated at room temperature with primary antibody for 1 to 2 hours. Immunohistochemical stainining was carried out using the Dako Autostainer or Ventana BenchMark XT Slide Stainer (for CD3 antibody) using manufacture supplied reagents and standard protocols with the following primary antibodies: MART-1 (no. CMC756, 1:200; Cell Marque, Rocklin, CA); HMB45 (no. 30930, 1:4; Enzo Life Sciences, Farmingdale, NY); Tyrosinase (no. NCL-TYROS, 1:20; Novocastra Division, Leica Microsystems, Buffalo Grove, IL); MHC Class I (HC-10, 1:1000; provided by Dr. Soldano Ferrone); HLA-DR (TAL.1B5, 1:200; DAKO); CD20 (L26, 1:500; DAKO); CD8 (CD8/144B, 1:50; DAKO); CD4 (1F6, 1:80; Novacastra); CD3 (2GV6, prediluted; Ventana). Detection was carried out using an automated slide stainer (Autostainer; DAKO) with either horseradish peroxidase/3,3′-diaminobenzidine polymer-based detection system (Envision+; DAKO) or a red chromogen (Liquid Permanent Red Substrate-Chromogen; DAKO) for darkly pigmented tumors. The immunohistochemical staining was prospectively assessed and quantitated by 2 board certified pathologists who were blinded to the comparative analysis of the study. The percentage of viable tumor cells expressing a given marker was quantified as 0–5%, 6–50%, or >50%. Staining intensity for each marker was graded on a scale of 0 (no staining), 1+, 2+, or 3+ (high intensity staining). Lymphocytic infiltrate was assessed with CD4, CD8, CD3, and CD20 staining and quantified based upon the percentage of tumor occupied by infiltrating lymphocytes as 0 (no lymphocytes detected), 1+ (<5% of tumor field), 2+ (5–50% of tumor field), or 3+ (extensive lymphoid aggregation occupying over 50% of the tumor field).
Generation and assessment of TIL cultures
Geographically discrete 1 to 2 mm3 tumor fragments (n=24) were freshly dissected from each tumor metastasis and placed individually in wells of a 24-well culture plate containing complete media with human AB serum and recombinant IL-2 (3000IU/ml) as previously described(22). After approximately 2 weeks of culturing, each of the wells was assessed for successful TIL expansion based upon cell count and visual inspection. Expanded TIL cultures underwent flow cytometric phenotypic analysis after staining with anti-human CD3, CD8, and CD4 monoclonal antibodies and their respective isotype controls (BD Biosciences). Immunofluorescence, analyzed as the relative log fluorescence of live cells, was measured using a FACSCanto II flow cytometer with FACSDiva software (BD Biosciences) and FlowJo software (Tree Star, Inc.). The specific anti-tumor reactivity of individual TIL cultures was assessed by co-culture with autologous tumor digest which had been freshly cryopreserved at the time of surgical procurement. Briefly, TIL cells (1x105 cells) and autologous tumor digest (1x105 cells) were co-incubated in a 0.2-ml volume in individual wells of a 96-well plate. Supernatants were harvested from duplicate wells after 20–24 hours and IFN-γ secretion was measured in culture supernatants using commercially available IFN-γ ELISA kits (Endogen). All data is presented as a mean of duplicate samples. Cultures with IFN-γ production greater than 100 pg/ml and twice background of unstimulated TIL and autologous tumor digest alone were considered as having specific anti-tumor reactivity.
Whole exomic sequencing and driver mutational analysis
Exome libraries were prepared from paired UM metastasis and normal samples using Agilent SureSelectXT Human All Exon V5+UTR target enrichment kit as per the manufacturer’s protocols (Agilent, Santa Clara, USA). The samples were pooled 3 samples per lane and sequenced on an Illumina HiSeq2000 sequencer with TruSeq V3 chemistry (paired-end, 101bp read length). Basecalling was carried out using Illumina RTA 1.12.4.2 run-time analysis software and demultiplexing was carried out using Casava 1.8.2. Each sample had >99 million pass filtered reads, with >92% bases having a base quality value >Q30 (Q30: The percentage of bases called with an inferred accuracy of 99.9% or above, a measure of basecalling quality). The percentage of unique library fragments was >90% across all samples. The capture efficiency as measured by the percentage of the reads mapping on the target regions, was >60%. The mean coverage on target regions for all samples was between 60X to 90X with >82.9% of the target regions having at least 30X coverage. The quality of the raw reads was assessed using FastQC(23) and NGSQCtoolkit(24). Reads were trimmed and filtered for adapters using Trimmomatic(25). Alignment was carried out to the human Hg19 reference sequence using BWA-0.7.4(26). Alignment files were indexed, sorted and duplicates were removed. Realignment around InDels and base-quality score recalibration was carried out as per the GATK best practices for exome-seq analysis(27). MuTect was used in the high-confidence (HC) mode for calling somatic point mutations(28). The subset of calls that passed the high-confidence filters after the statistical analysis within Mutect were annotated using Annovar(29) to find both the location and the functional significance of the mutations. Mutations were filtered to keep only those that had Mutation_info: exonic or splicing only; Consequence: non-synonymous or stopgain_SNV. To serve as a reference group, WES data was obtained for 278 CM metastases via the “Skin Cutaneous Melanoma” (SKCM) data portal of The Cancer Genome Atlas (TCGA). Somatic mutation counts for the metastatic patient samples for the SKCM cohort were extracted from the Level 2 MAF file downloaded off the GDAC Firehose resource(30). As only somatic point mutations were of interest while comparing the distribution of somatic mutations between CM and UM cohorts, InDel calls (1.2%) were removed from the TCGA-SKCM cohort. As the annotations for the point mutations in the TCGA-SKCM cohort did not have the same terminology as for the UM cohort, the SKCM mutations were re-annotated using Annovar, with the same filters applied based on the combination of ‘Consequence’ and ‘Mutation_info’ columns. The frequency among the CM and UM tumors for specific mutations in BRAF, GNAQ, and GNA11 was determined primarily from WES analysis. As validation in selected samples, library preparation with 10–20ng genomic DNA was performed using Ion AmpliSeq Cancer Hotspot, Panel V2 and Ion AmpliSeq Library Kit 2.0, using the corresponding User Guide (Life Technologies). The amplicon panel includes 207 primer sets covering ~2,800 COSMIC hotspot mutations in 50 genes. Next generation sequencing was performed in an Ion Torrent Personal genome machine (PGM), and analyzed with Torrent Suite Software (Life Technologies). Annotation and interpretation of all variants were performed in Ion Reporter that links to multiple databases such as RefSeq, OMIM, Oncomine, COSMIC, and dbSNP. Reported mutations were confirmed by inspection of alignments using the Integrative Genomics Viewer(31).
Statistical Analysis
Fisher’s exact test was used to determine associations between dichotomous demographic parameters and the Wilcoxon rank sum test was utilized for the comparison of continuous parameters such as patient age. Non-parametric comparisons between the UM and CM cohorts were performed with the Mann-Whitney test. Student’s t test was used to compare the means of parametric variables. Linear regression analysis was used for correlation studies and presented as R2 values. All P values are 2-tailed and have not been adjusted for multiple comparisons. In view of the exploratory analyses performed, p<0.05 would be considered statistically significant while 0.05 < p <0.1 would be considered a trend. Excel and GraphPad Prism (v6.01) were used for analyses.
RESULTS
Patient demographics and procurement of liver metastases
Between 2004 and 2014, a total of 49 patients with metastatic melanoma underwent liver metastasectomy in the context of approved clinical trials in the Surgery Branch, NCI. The current study selectively analyzed liver metastases because of our previous finding that human melanoma metastases demonstrate significant heterogeneity in tumor antigen expression and lymphocytic infiltrate when stratified based upon their anatomic location in the body (6). Further, since UM predominantly metastasizes to the liver, this homogeneous source of metastases would prevent potential site-specific bias in our comparative assessment of tumors. Patients undergoing liver metastasectomy were stratified into two cohorts, CM and UM, based upon the anatomic origin of their primary melanomas. The CM cohort included 35 patients; 33 of whom had documented primary tumors arising from the cutaneous epithelium and 2 additional patients had primary tumors of unknown origin. Patients with melanoma of unknown origin were included in the CM cohort based upon recent molecular genetic studies which revealed these tumors strongly resembled cutaneous melanomas (32). The UM cohort included 14 patients who had ophthalmologic documentation that their primary melanoma arose specifically from the uveal tract. The characteristics for each of the patients who underwent liver metastasectomy are shown in Table 1 and the comparison of the CM and UM cohorts are shown in Table 2. The age (mean and range) and gender distribution of the patients in the two cohorts were similar. At the time of referral to our center, there was a greater trend for the UM patients to have not received prior systemic therapy for their metastatic disease when compared to the CM patients (UM: 71% vs. CM: 37%, P= 0.06). This finding likely reflected the growing availability of approved systemic agents and clinical trial opportunities for patients with metastatic CM during the study period. Of note, 63% of the CM patients had undergone prior systemic immunotherapy, whereas only 21% of UM patients had received such treatments (CM vs. UM, P=0.01). Although both melanoma cohorts demonstrated metastatic spread to extrahepatic sites, the UM patients had metastases more often confined to the liver (UM: 43% vs. CM: 14%, P= 0.05), while the CM patients demonstrated a trend toward more frequent metastases to lymph nodes and soft tissues (CM: 60% vs. UM: 29%, P= 0.06). For surgical tumor procurement, patients typically underwent resection of a single metastatic liver deposit or a closely approximated cluster of tumors. The size (mean/median) of the tumor deposits resected from the patients were not significantly different between the groups (CM: 6.2/6.0cm vs. UM: 7.2/5.5cm, P= 0.50). The cumulative metastatic tissue that was procured at operation was assigned a unique liver metastasis identification number (ID #) for subsequent analysis. Two UM patients (1 and 3) developed metachronous liver metastases during the study period. These patients underwent two independent liver metastasectomy operations and their individual tumor procurements were assigned unique liver metastasis ID #. Thus, in sum, there were 35 CM liver metastases and 16 UM liver metastases that were available for direct comparative analysis.
Table 1.
Patient demographics and procurement of liver metastases
| Cutaneous Melanoma | ||||||
|---|---|---|---|---|---|---|
| Pt ID # | Age | Gender | Primary Site | Sites of Metastasis | Prior Systemic Therapies | Liver metastasis ID # |
| 1 | 35 | M | Neck | Li | Immuno | L-CM1 |
| 2 | 51 | F | Arm | Li, Lu | None | L-CM2 |
| 3 | 46 | F | Scalp | Li, Lu | Immuno, Chemo | L-CM3 |
| 4 | 62 | F | Shoulder | Li, Lu, ST | None | L-CM4 |
| 5 | 44 | F | Hip | Li, Lu, Kidney, LN | Immuno, Chemo | L-CM5 |
| 6 | 66 | M | Back | Li, LN | Immuno | L-CM6 |
| 7 | 61 | M | Back | Li, LN | Immuno | L-CM7 |
| 8 | 44 | M | Sacral | Li, Lu, LN | Immuno | L-CM8 |
| 9 | 64 | M | Back | Li, ST | None | L-CM9 |
| 10 | 37 | F | Back | Li, Lu, Bn, LN | Immuno | L-CM10 |
| 11 | 54 | M | Shoulder | Li | Immuno | L-CM11 |
| 12 | 42 | M | Back | Li, LN | Immuno | L-CM12 |
| 13 | 47 | F | Infra-auricular | Li, Lu | None | L-CM13 |
| 14 | 35 | M | Unknown | Li, Lu, LN | Immuno, Chemo | L-CM14 |
| 15 | 64 | F | Parotid | Li | Immuno | L-CM15 |
| 16 | 62 | M | Ear | Li, Lu | Immuno | L-CM16 |
| 17 | 50 | F | Neck | Li, Spl, Panc | Immuno, Chemo | L-CM17 |
| 18 | 62 | M | Foot | Li, Lu, Spl, LN | None | L-CM18 |
| 19 | 45 | M | Shoulder | Li, LN | None | L-CM19 |
| 20 | 57 | M | Back | Li | Immuno | L-CM20 |
| 21 | 54 | F | Back | Li, ST | None | L-CM21 |
| 22 | 50 | M | Back | Li, Lu | Immuno | L-CM22 |
| 23 | 57 | M | Back | Li, Lu, ST | None | L-CM23 |
| 24 | 69 | F | Scalp | Lu | Immuno | L-CM24 |
| 25 | 48 | M | Unknown | Li, LN, ST, Bn | None | L-CM25 |
| 26 | 55 | M | Leg | Li, Lu, LN | None | L-CM26 |
| 27 | 65 | M | Scalp | Li, Lu, LN | Immuno | L-CM27 |
| 28 | 53 | M | Shoulder | Li, Bn | None | L-CM28 |
| 29 | 34 | M | Jaw | Li, Lu, LN, Bn | Immuno | L-CM29 |
| 30 | 46 | F | Flank | Li | Immuno, Chemo, Targeted | L-CM30 |
| 31 | 52 | M | Thigh | Li, LN, Adrenal | Immuno | L-CM31 |
| 32 | 62 | M | back | Li, LN | None | L-CM32 |
| 33 | 56 | M | Leg | Li, LN | Immuno | L-CM33 |
| 34 | 19 | F | Preauricular | Li, LN, Spl, Bn | Immuno | L-CM34 |
| 35 | 27 | F | Ear | Li, Lu | None | L-CM35 |
| Uveal Melanoma | ||||||
|---|---|---|---|---|---|---|
| Pt ID # | Age | Gender | Primary Site | Sites or Metastasis | Prior Systemic Therapies | Liver metastasis ID # |
| 1 | 56 | F | Uveal tract | Li, Bn | Immuno, TACE | L-UM1a L-UM1b |
| 2 | 56 | F | Uveal tract | Li | TACE | L-UM2 |
| 3 | 47 | F | Uveal tract | Li, Lu, Panc, Bn | None | L-UM3a L-UM3b |
| 4 | 68 | M | Uveal tract | Li | None | L-UM4 |
| 5 | 60 | M | Uveal tract | Li, Lu, ST, LN, Per | Immuno | L-UM5 |
| 6 | 55 | F | Uveal tract | Li | None | L-UM6 |
| 7 | 64 | M | Uveal tract | Li | None | L-UM7 |
| 8 | 56 | M | Uveal tract | Li | None | L-UM8 |
| 9 | 17 | F | Uveal tract | Li, Lu, Heart, LN | Immuno | L-UM9 |
| 10 | 54 | F | Uveal tract | Li, LN | None | L-UM10 |
| 11 | 58 | M | Uveal tract | Li, ST | None | L-UM11 |
| 12 | 53 | F | Uveal tract | Li, Bn, Per, Om | None | L-UM12 |
| 13 | 52 | M | Uveal tract | Li | None | L-UM13 |
| 14 | 62 | M | Uveal tract | Li, Lu | None | L-UM14 |
Abbreviations: Bn, Bone; Li, Liver; LN, Lymph Node; Lu, Lung; Om, Omentum; Panc, Pancreas; Per, Peritoneum; Spl, Spleen; ST, Soft tissue
Table 2.
Comparison of CM and UM patients who underwent liver metastasectomy
| CM | UM | P value | |
|---|---|---|---|
| No. of patients | 35 | 14 | |
| Age (mean) | 51 | 54 | 0.157 |
| (Range) | 19–69 | 17–68 | |
| Gender (no./%) | |||
| F | 13 (37) | 7 (50) | 0.52 |
| M | 22 (63) | 7 (50) | |
| Prior systemic therapy (no./%) | |||
| Immunotherapy | 22 (63) | 3 (21) | 0.01 |
| Chemotherapy | 5 (14) | 0 (0) | 0.30 |
| None | 13 (37) | 10 (71) | 0.06 |
| Extrahepatic disease (no./%) | |||
| Lung | 17 (49) | 4 (29) | 0.21 |
| LN/ST | 21 (60) | 4 (29) | 0.06 |
| None | 5 (14) | 6 (43) | 0.05 |
Radiographic and pathologic comparison of the melanin content between CM and UM liver metastases
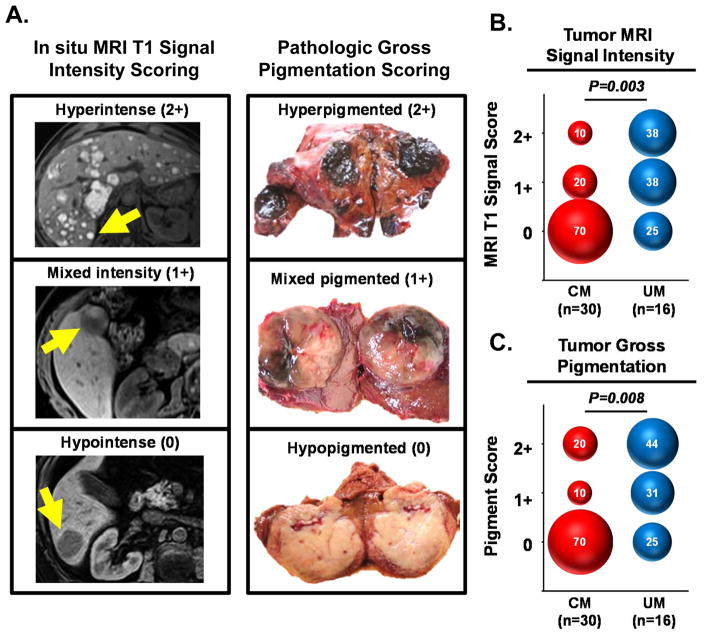
Both CM and UM primary tumors arise from transformed melanocytes. Yet, despite this common origin from melanin-producing cells, the metastases of these tumors can display significant heterogeneity in the quantitative expression of prototypic melanin associated proteins (6). These prior findings prompted us to ask whether CM and UM metastases demonstrated fundamental differences in their melanin pigmentation. To address this question, we utilized pre-operative clinical radiographic imaging and post-operative gross pathologic examination to evaluate the melanin content found in the liver metastases procured from CM and UM patients. A prior study characterizing melanoma metastases with clinical MRI imaging found the in situ tumor signal intensity from T1-weighted sequences strongly correlated with the degree of melanin pigmentation found in those tumors after resection (33). For the current study, we utilized these defined MRI parameters to perform in situ characterization of 30 liver metastases identified in CM patients and 16 liver metastases in UM patients. Quantitative T1-weighted signal intensity measurements (without gadolinium enhancement) of the in situ tumor and adjacent normal tissue were obtained using clinical radiology imaging software (Carestream Vue Solutions, version 11.3). The normalized tumor signal intensity (relative to normal liver) was objectively calculated for each metastasis and scored as either hyperintense (2+), mixed intensity (1+), or hypointense (0), as illustrated in Supplementary Figure 1. Additionally, after surgical resection, metastases underwent independent gross pathologic examination and tumor pigmentation was visually scored as either hyperpigmented (2+), mixed pigmented (1+), or hypopigmented (0). The comparison of pre-operative in situ MRI intensity scoring and postoperative pathologic pigmentation scoring for representative liver metastases is shown in Figure 1A. Analysis of the entire population of liver metastases (n=46) revealed a strong and direct correlation between the MRI and the pathologic scoring of pigmentation (R2= 0.88, P<0.0001) for the individual tumors. Next, we used these two parameters to independently compare the melanin content found in the CM and UM liver metastases. In situ MRI tumor signal intensity was significantly different between the two melanoma cohorts (P = 0.003) (Figure 1B). Whereas, 70% of CM metastases had hypointense (0) MRI signal, only 25% of UM metastases displayed this low signal intensity. Conversely, the UM cohort demonstrated a greater frequency of metastases with hyperintense (2+) MRI signal (UM: 38% vs. CM: 10%). When these same UM and CM metastases underwent pathologic examination after resection, we similarly found a significant difference in their gross pigmentation (P = 0.008) (Figure 1C). CM metastases were more often visually hypopigmented (0) (CM: 70% vs. UM: 25%) and UM metastases were more often hyperpigmentated (2+) (UM: 44% vs. CM: 20%). Thus, we concluded that despite having common lineage from melanin-producing cells, CM and UM metastases displayed significant differences in their overall melanin content.
Figure 1. UM liver metastases have greater melanin pigmentation when compared to CM liver metastases.
(A) Illustrative examples demonstrating the correlation between pre-operative in situ MRI intensity scoring and post-operative gross pathologic scoring of pigmentation in selected melanoma liver metastases. Arrows in the left panel indicate the tumors that underwent metastasectomy; right panel displays photos of the resected tumors. (B) Comparison of CM and UM liver metastases based upon MRI tumor signal intensity score and (C) gross pathologic pigmentation score. Numbers within the bubbles denote the percentage of tumors in the specified cohort with the indicated score. Bubble diameter is proportionate to these percentages. Statistical comparisons between CM and UM cohorts were performed with the Mann-Whitney test.
Comparison of melanocyte differentiation antigens and MHC expression between CM and UM liver metastases
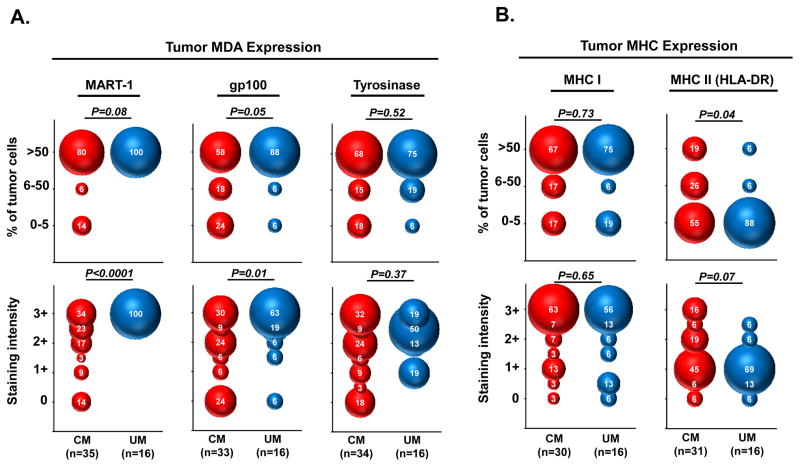
To better understand the differences observed in melanin pigmentation between the CM and UM liver metastases, we next performed immunohistochemistry (IHC) to compare the cellular expression of proteins associated with melanocyte differentiation. The tumor expression (% of viable cells and staining intensity) for MART-1, gp100, and tyrosinase were prospectively assessed by pathologists blinded to the comparative analysis (Figure 2A). Among the CM liver metastases, we found prominent heterogeneity in cellular MART-1 expression between individual metastases (interlesional heterogeneity) and within individual metastases (intralesional heterogeneity). In 20% of CM metastases, MART-1 was either nearly absent (0–5% of tumor cells) or expressed on a fraction of viable tumor cells (6–50% of tumor cells). Further, only 34% of the CM metastases displayed strong cellular staining (3+) for MART-1, while the remaining tumors had weak to intermediate staining (0 to 2+). In contrast, all of the UM liver metastases displayed homogeneous, diffuse, and strong MART-1 staining (>50% of tumor cells with 3+ staining intensity). When the pattern of MART-1 expression was compared between the CM and UM liver metastases, we found UM tumors had significantly stronger MART-1 staining intensity (P<0.0001) and a trend toward a greater percentage of MART-1 stained tumor cells (P=0.08). Expression for gp100 was also greater in the UM metastases compared to the CM metastases based upon percentage of tumor cells stained (P=0.05) and staining intensity (P=0.01). Diffuse gp100 staining (>50% of tumor cells) was seen in 88% of UM tumors versus only 58% of CM tumors. Strong staining intensity (3+) for gp100 was found in 63% of UM tumors versus only 30% of CM tumors. Interestingly, tyrosinase expression (% of tumor cells and staining intensity) was highly variable in both CM and UM metastases and not significantly different between the melanoma cohorts (P=0.52 and 0.37, respectively).
Figure 2. UM liver metastases have greater expression of melanocyte differentiation antigens and lower MHC class II compared to CM liver metastases.
Comparison of CM and UM liver metastases based upon immunohistochemical staining of (A) prototypic melanocyte differentiation antigens and (B) MHC class I and II molecules. Upper panels demonstrate expression based upon percentage of viable tumor cells within individual metastases that stain with the indicated marker; lower panels demonstrate expression based upon tumor cell staining intensity. Numbers within the bubbles denote the percentage of tumors in the specified cohort with the indicated marker expression. Bubble diameter is proportionate to these percentages. Statistical comparisons between CM and UM cohorts were performed with the Mann-Whitney test.
Next, we compared the expression of major histocompatibility complex (MHC) class I and II proteins on tumor cells in the CM and UM liver metastases (Figure 2B). For tumor antigens to be recognized by T cells, antigens must be internally processed into peptides that are presented on the tumor cell surface by these MHC molecules. We found MHC class I expression was equally expressed in both cohorts based upon percentage of tumor cells stained (P=0.73) and staining intensity (P=0.65). Diffuse MHC class I staining (>50% of tumor cells) was observed in the majority of tumors from both cohorts (CM: 67% vs. UM: 75%) with equally strong staining intensity (3+) (CM: 63% vs. UM: 56%). In contrast, we found CM tumors had significantly greater percentage of MHC class II stained tumor cells (P=0.04) and a trend toward stronger MHC class II staining intensity (P=0.07). MHC class II expression was nearly undetectable (0–5% of tumor cells) in 88% of UM tumors versus only 55% of CM tumors. In sum, these IHC studies demonstrated that UM metastases had greater expression of melanocyte lineage antigens and lower expression of MHC class II molecules when compared to CM metastases.
Comparison of tumor infiltrating lymphocytes found in CM and UM liver metastases
High levels of tumor infiltrating lymphocytes (TIL) have been reported to correlate with favorable prognoses in a variety of solid organ malignancies (34–36). Specific immunologic studies of TIL expanded from CM metastases have found that these infiltrating cells can often recognize antigens expressed by the tumor (37). Further, the autologous adoptive transfer of such TIL has shown durable and complete tumor regression in metastatic CM (12). These findings have provided compelling evidence for the natural immunogenicity of CM metastases. However, it is unclear whether UM tumors can similarly elicit adaptive immune responses in vivo. To provide insight, we sought to compare the attributes of TIL found in UM and CM liver metastases. First, the degree of infiltrating T cells (CD3, CD4, and CD8 staining) and B cells (CD20 staining) associated with each of the metastases was prospectively assessed by pathologists blinded to the comparative analysis. From both tumor cohorts, we found significant heterogeneity in the numbers of peripheral and infiltrating T cells which ranged from no lymphocytes detected (0) to extensive lymphoid aggregation occupying over 50% of the tumor field (3+). When the CM and UM metastases were compared, we found no significant differences in the levels of peripheral and infiltrating CD3+, CD4+, or CD8+ T cells between the cohorts (Supplementary Figure 2). Further, B cells (CD20+ cells) were undetectable in the majority of tumors and also not significantly different between the cohorts.
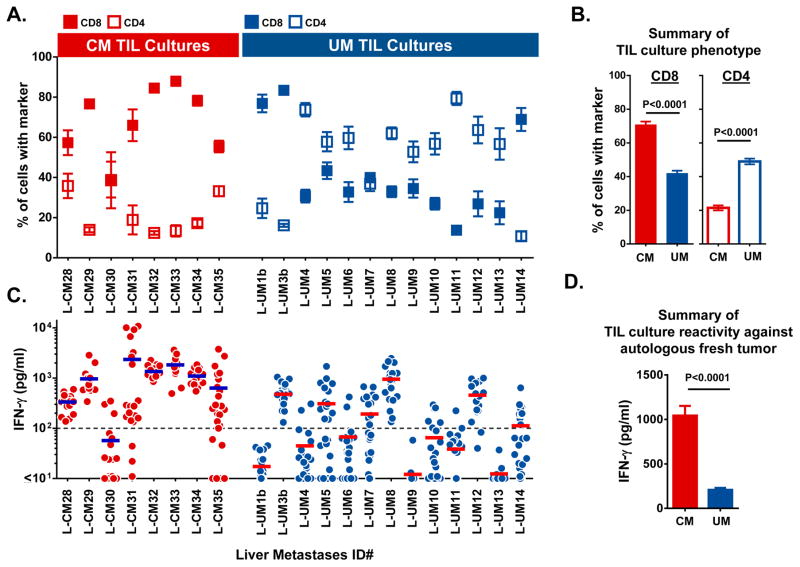
Having observed that the degree of lymphocytic infiltration was similar between the CM and UM liver metastases, we next sought to assess the phenotypic and functional attributes of the TIL after ex vivo expansion. Consecutive metastatic liver tumors were procured from 8 CM and 13 UM patients during a shared time period. To account for intra-tumoral heterogeneity that might influence TIL growth, 24 geographically discrete tumor fragments were freshly dissected from each of the metastases and placed in culture media containing human IL-2 (3000IU/ml). After approximately 2 weeks of culturing, we found that the percentage of tumor fragments that could successfully generate TIL were equivalent between the CM and UM tumors (95% vs. 94%, respectively). Each of these independently expanded TIL cultures were then assessed by flow cytometry to determine their percentage of CD8+ and CD4+ T cells. We observed a significant difference between the CM and UM liver metastases in the ratio of these T cell subsets (Figure 3A). The TIL cultures from 88% of the CM metastases (7 of 8 CM metastases) were composed predominantly of CD8+ T cells. In contrast, only 23% of UM metastases (3 of 13 UM metastases) gave rise to CD8+ enriched TIL. Cumulatively, the mean percentage of CD8+ T cells in the CM derived TIL cultures was significantly greater than in the UM derived TIL (CM: 71% vs. UM: 42%, P<0.0001) (Figure 3B). Conversely, UM derived TIL cultures possessed a greater mean percentage of CD4+ cells when compared with the CM derived TIL cultures (UM: 49% vs. CM: 21%, P<0.0001) (Figure 3B).
Figure 3. Phenotypic and functional comparison of TIL cultures derived from CM and UM liver metastases.
(A) Mean percentage of CD8+ (filled box) and CD4+ (open box) T cells found in TIL cultures derived from individual CM and UM liver metastases. T cell subset frequency as assessed by flow cytometry and CD3 gating. Box denotes the mean of approximately 24 individual TIL cultures with error bars indicating SEM. (B) Cumulative comparison of CM versus UM derived TIL cultures based upon their percentage of CD8+ and CD4+ T cells. Bars demonstrate the mean with error bars indicating SEM. (C) Autologous tumor reactivity of individual TIL cultures derived from CM and UM liver metastases. Each dot represents the IFN-γ production of a single TIL culture in response to overnight co-culture with autologous tumor digest minus the background cytokine levels of unstimulated TIL and tumor digest alone. Bar represents the mean of approximately 24 TIL cultures. Dashed line marks 100pg/ml of IFN-γ on the y-axis. (D) Cumulative comparison of the anti-tumor reactivity of CM and UM derived TIL cultures based upon production of IFN-γ in response to autologous tumor digest. Bars demonstrate the mean with error bars indicating SEM. Statistical comparisons between CM and UM cohorts were performed with the Student’s t test.
Next, we compared the anti-tumor reactivity of the individual CM and UM TIL cultures by overnight co-culture with tumor digests of their respective parental tumors which had been freshly cryopreserved at the time of surgical procurement. Reactive TIL cultures were defined as having tumor induced IFN-γ production ≥100 pg/ml and twice the background of unstimulated TIL and tumor digest alone. The autologous anti-tumor reactivity for each of the TIL cultures from their respective metastases are shown in Figure 3C. The TIL cultures from 88% of the CM metastases (7 of 8 CM metastases) demonstrated mean tumor specific IFN-γ production ≥100 pg/ml. In contrast, 46% of UM metastases (6 of 13 UM metastases) had mean reactivity above this threshold. Cumulatively, CM derived TIL cultures produced higher mean levels of IFN-γ in response to autologous tumor digest when compared to UM derived TIL cultures (CM: 1044 pg/ml vs. UM: 209 pg/ml, P<0.0001) (Figure 3D). Interestingly, however, we identified individual TIL cultures from 46% of UM metastases (6 of 13 UM metastases) (L-UM 3b, 5, 7, 8, 12, and14), with IFN-γ production which was comparable in magnitude to the responses identified from CM TIL (Figure 3C). Thus, although specific autologous anti-tumor T cell responses were more prevalent among the CM liver metastases, there was a subset of UM tumors that could also elicit strong tumor reactive T cell responses.
Metastasis hypopigmentation identifies an immunogenic subset of uveal melanoma
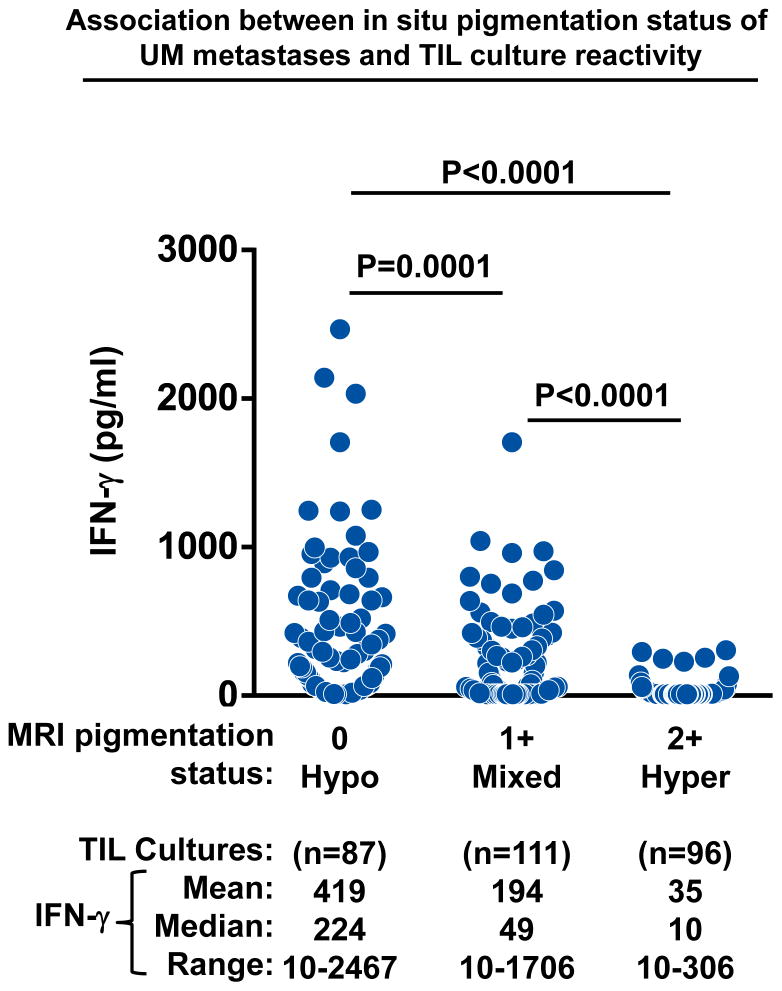
Having found that a subset of UM metastases could naturally elicit auto-reactive TIL responses, we next sought to determine if there was a clinically relevant means to prospectively identify UM patients who harbored these immunogenic tumors. Since the majority of CM metastases possessed TIL with autologous tumor reactivity, we postulated that similar adaptive T cell responses might preferentially be found in UM metastases with attributes akin to CM tumors. Pre-operative MRI and gross pathologic examination had demonstrated that the majority of CM liver metastases lacked melanin pigmentation (Figure 1B). Thus, we investigated whether the in situ melanin content of UM metastases, as determined by pre-operative MRI imaging, might correlate with the subsequent growth of auto-reactive TIL populations. The liver metastases from 13 consecutive UM patients (described in Figure 3C) underwent stratification based upon their pre-operative in situ radiographic attributes. MRI signal intensity scores identified four metastases as hyperpigmented (2+) (L-UM 1b, 4, 9, and10), five metastases as mixed pigmented (1+) (L-UM 3b, 5, 6, 11, and 13), and four metastases as hypopigmented (0) (L-UM 7, 8, 12, 14). Next, the IFN-γ responses from each of the TIL cultures derived from these metastases were assessed based upon the MRI characteristics of their parental tumors (Figure 4). We found that hyperpigmented metastases (2+ MRI signal) uniformly gave rise to TIL cultures (n=96) with low anti-tumor IFN-γ production (mean: 35pg/ml) that did not exceed background control levels. In contrast, the mixed pigmented metastases (1+ MRI signal) generated TIL cultures (n=111) with significantly greater IFN-γ production (mean IFN-γ: 194 pg/ml) (Mixed vs. Hyper P<0.0001); while the hypopigmented metastases (0 MRI signal) were notable for producing TIL cultures (n=87) with the highest anti-tumor reactivity (mean IFN-γ: 419 pg/ml) (Hypo vs. Hyper and Mixed, P<0.0001, respectively). Thus, we concluded that low to absent levels of in situ melanin pigmentation based upon pre-operative clinical MRI could identify a subset of UM metastases capable of eliciting potent immunogenic TIL responses. In contrast, UM metastases with high levels of melanin pigmentation identified a non-immunogenic group of tumors.
Figure 4. Metastasis hypopigmentation identifies an immunogenic subset of uveal melanoma.
Association between the pre-operative melanin content of UM metastases and the autologous anti-tumor reactivity of their derived TIL cultures. UM TIL cultures (n~24) were established from individual liver metastases from 13 UM patients. The cultures were stratified into cohorts based upon the melanin pigmentation status of their parental tumors as assessed by pre-operative in situ MRI signal intensity scoring. Each dot represents the IFN-γ production of a single UM TIL culture in response to overnight co-culture with autologous tumor digest minus the background cytokine levels of unstimulated TIL and tumor digest alone. Statistical comparisons between the stratified pigmentation cohorts were performed with the Student’s t test.
Comparison of tumor mutational profile between CM and UM metastases
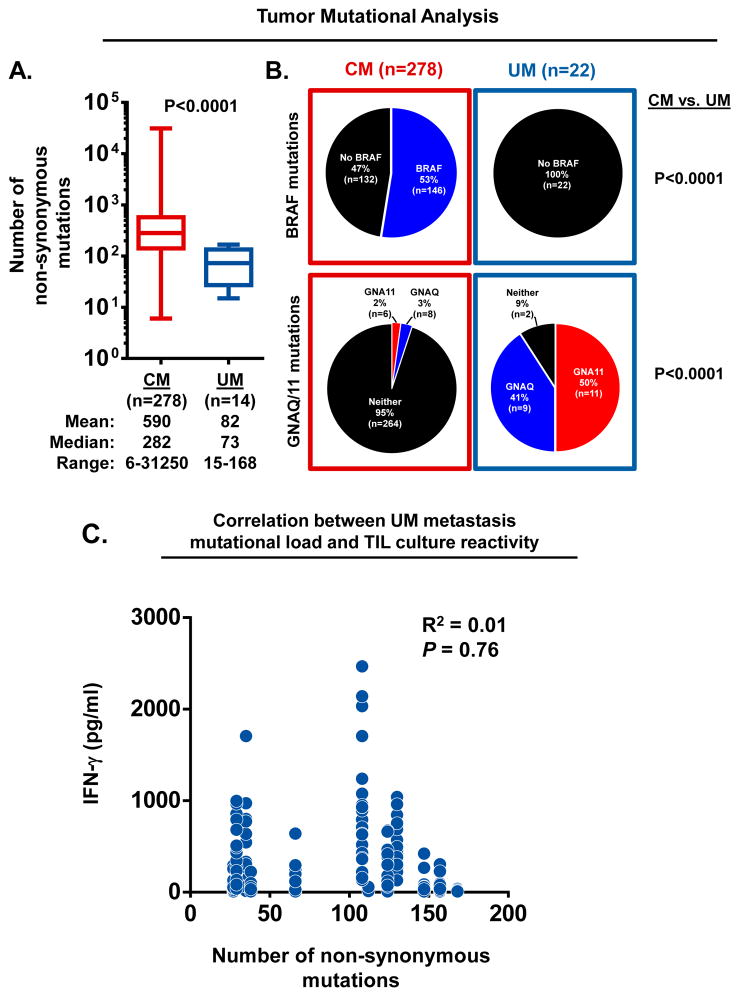
Although normal differentiation antigens are common targets for endogenous T cells in melanoma patients, recent studies have demonstrated that unique somatic mutations expressed by tumors can also elicit autologous T cell responses (37–39). Further, comparative whole exome sequencing (WES) has revealed sun exposed CM tumors to have the highest number of somatic mutations among common malignancies (40). These observations have fostered the theory that the unique responsiveness of metastatic CM to a variety of immunotherapy approaches is a direct consequence of endogenous immune responses against neo-epitopes encoded by these large numbers of mutations. Thus, we next sought to determine if the identified subset of immunogenic UM metastases also harbored a greater mutational load that might explain their enhanced T cell recognition. Previously, it has been reported that sun-shielded melanomas, including UM, have far fewer non-synonymous mutations when directly compared with sun-exposed CM tumors (17). However, these analyses were based upon a limited number of UM samples which included a mixture of primary and metastatic tumors. Thus, we first sought to better determine the frequency and characteristics of the non-synonymous mutations occurring in CM and UM metastases. To provide adequate sample numbers for this analysis we obtained WES data for 278 CM metastases via The Cancer Genome Atlas (TCGA) data portal and compared these against data from 14 UM metastases from our cohort. Of note, since the TCGA database does not denote the anatomic site of the metastases, the CM data represents metastases from a variety of sites. Protein-altering somatic point mutations for each tumor were determined using a common analytical workflow based upon comparison to matched germline DNA. We found CM metastases had a broad range in mutation number (range: 6–31,250) when compared to UM metastases (range: 15–168). Further, as a group, CM metastases had significantly more somatic mutations when compared to UM metastases (median counts; CM: 282 vs. UM: 73, P<0.0001) (Figure 5A).
Figure 5. Tumor mutational profiles of CM and UM metastases.
(A) Comparison of the number of non-synonymous mutations identified in CM and UM metastases. Box extends from 25th to 75th percentile, line through the box indicates median, and bars extend from the smallest to largest values. Statistical comparison between CM and UM cohorts was performed with the Student’s t test. (B) Frequency of BRAF, GNAQ and GNA11 mutations identified in CM and UM metastases. Statistical comparison of oncogene frequency between CM and UM were performed with Fisher’s exact test. (C) Correlation between the number of non-synonymous mutations identified in UM metastases and the autologous anti-tumor reactivity of their derived TIL cultures. Data represents UM TIL cultures (n~24) established from metastases from 12 UM patients. Each dot plots the mutation frequency of the parental tumor (x-axis) versus the IFN-γ production of the derived UM TIL cultures in response to overnight co-culture with autologous tumor digest minus the background cytokine levels of unstimulated TIL and tumor digest alone (y-axis). Linear regression analysis was used to derive R2 values.
Next, we compared the tumor cohorts for the frequency of prototypic melanoma associated oncogenic driver mutations including, BRAF, GNAQ and GNA11 (Figure 5B). We found BRAF mutations in 53% of the CM metastases (n=278). However, BRAF was not mutated in any of the UM tumors (n=22); (BRAF mutation frequency; CM vs. UM metastases, P<0.0001). In contrast, activating mutations in either of the homologous genes, GNAQ or GNA11, were identified in 91% of the UM metastases, but in only 5% of the CM metastases; (GNAQ/GNA11 mutation frequency; CM vs. UM metastases, P<0.0001).
Finally, we investigated in 12 UM patients whether the mutational frequency identified in their metastases correlated with the autologous anti-tumor reactivity of their respectively derived TIL cultures (n~24 cultures/tumor). When the tumor induced IFN-γ production from each of the TIL cultures was assessed against the number of non-synonymous mutations identified in their respective parental tumors, we found no correlation between the parameters (Figure 5C).
DISCUSSION
The last 30 years has provided substantial evidence that the human immune system can naturally generate potent immunologic responses against tumor antigens expressed by metastatic cutaneous melanoma (8). Cancer regression can now be achieved in patients with metastatic CM with mechanistically diverse forms of immunotherapy that augment naturally existing tumor specific T cell responses (9–12). However, the role of these immune based therapies for the treatment of metastatic uveal melanoma patients remains unclear. Patients with UM are frequently excluded from metastatic melanoma immunotherapy clinical trials because UM is generally thought to be a non-immunogenic form of melanoma (13–16). However, there have not been formal comparative studies performed directly upon UM and CM metastases to accurately assess their relative immunogenicity. In this study, we compared the tumor antigen expression, tumor mutational load, and endogenous anti-tumor immunologic reactivity found in fresh surgically resected UM and CM metastases. By defining the tumor specific immune responses that are naturally found in these metastases, we sought to provide insight into the role for immune based therapies for the management of UM patients. We previously reported that melanoma metastases demonstrate significant heterogeneity in tumor antigen expression and lymphocytic infiltrate based upon their anatomic location in the body (6). Thus, to avoid potential site-specific bias in the current study, we focused our comparative analysis selectively upon liver metastases resected from UM and CM patients. Our findings revealed that despite having common melanocytic lineage, UM and CM liver metastases were highly dichotomous in their melanin content, tumor differentiation antigen expression, and somatic mutational profile. The majority of CM liver metastases lacked gross melanin pigmentation, whereas UM liver metastases were more commonly hyperpigmented in appearance. In support of this observation, immunohistochemical profiling revealed that CM metastases had lower cellular expression of proteins associated with melanocyte differentiation, including MART-1 and gp100. Further, we found significant differences in the overall somatic mutational profile between CM and UM liver metastases. Comparative whole exomic sequencing revealed CM metastases had significantly greater mutational burden compared to UM metastases with the melanoma variants also possessing quite different oncogenic driver mutations of the MAPK pathway. Similar to previous reports (41–43), nearly all of the UM metastases had GNAQ and GNA11 mutations, while CM metastases commonly had BRAF mutations. Collectively, these comparative studies demonstrate CM metastases to be far more de-differentiated from their melanocytic origin when compared to UM metastases in terms of their mutational profile, tumor antigen expression, and gross melanin pigmentation.
When endogenous immune responses in these highly divergent forms of melanoma were characterized, we further identified marked differences in the phenotype and anti-tumor reactivity of their respective infiltrating lymphocytes. CM TIL were predominantly composed of CD8+ T cells, while UM TIL were CD4+ dominant. Reactivity against autologous tumor was significantly greater in CM TIL compared to UM TIL. However, we identified TIL from a subset of UM patients which had robust anti-tumor reactivity that was comparable in magnitude to that of CM TIL. The identification of this immunogenic group of UM metastasis has not been previously reported and thus, has fostered our interest in determining the specific antigenic targets that are recognized by these UM derived TIL. Interestingly, the level of in situ melanin pigmentation found in parental tumors strongly correlated with the generation of tumor reactive UM TIL. Hyperpigmented UM metastases generated TIL cultures with poor tumor reactivity, while the metastases that lacked pigmentation produced the most reactive TIL cultures. Although the precise mechanism underlying the relationship between tumor pigmentation and TIL reactivity is not completely understood, the loss of pigment proteins by metastatic tumor cells is thought to be driven by the stochastic genetic instability of tumor cells combined with the non-stochastic selective pressures of the host immune system (immunoediting)(44–48). Animal models have demonstrated the development of vitiligo and the loss of tumor pigment proteins through the action of antigen specific CD8+ T cells (49). Further, it is well known that human CM TIL frequently possess T cells specific for melanocyte pigment proteins, such as MART-1 and gp100 (50). Thus, in the current study, we hypothesize that the loss of tumor pigmentation in this subset of UM metastases could signify the presence of a vigorous immune response targeting pigment antigens. However, beyond the targeting of these normal differentiation antigens, recent analyses have found that unique somatic mutations expressed by tumors can generate neo-epitopes that also elicit robust autologous T cell responses (37–39). Although our study found no correlation between the mutational frequencies identified in UM metastases and the autologous anti-tumor reactivity of their respectively derived TIL cultures, there may still be individual mutations that are recognized. Thus, we have begun studies to assess the tumor reactive UM TIL for recognition of both non-mutated, as well as, mutated antigen targets.
Although not completely validated as a clinical biomarker, the MRI assessment of melanin content in UM metastases was found in this study to accurately identify tumors that can elicit a strong endogenous immune response. We are, thus, interested in determining whether immune based therapies may be more effective in the subset of UM patients who harbor these unique immunogenic tumors. To help address these questions, we are conducting the first inhuman adoptive T cell transfer trial dedicated to patients with metastatic UM (NCT01814046). In this phase II study, patients with metastatic UM undergo surgical metastasectomy to procure tumor tissue for TIL generation. The expanded lymphocytes are then adoptively transferred back into the host in conjunction with a nonmyeloablative lymphodepleting regimen. The primary endpoint of this study is to define the objective response rate of TIL immunotherapy in patients with metastatic UM. The results of this trial should provide valuable insight into the role of immune based therapies for the treatment of metastatic uveal melanoma.
Supplementary Material
Statement of Translational Relevance.
Although remarkable strides have been achieved in the management of metastatic cutaneous melanoma (CM) with T cell based immunotherapies, limited progress has been made with metastatic uveal melanoma (UM), a rare and aggressive variant that is hypothesized to be immunotherapy-resistant. In this study, we sought to formally define the relative immunogenicity of these two melanoma variants and determine whether endogenous anti-tumor immune responses exist against UM. Here, we report the novel identification of TIL from a subset of UM metastases with robust anti-tumor reactivity, comparable in magnitude to that of CM TIL. The discovery of this immunogenic group of UM metastases has important clinical implications for the role of immunotherapies in the treatment of patients who harbor these unique tumors.
Acknowledgments
We thank the Surgery Branch cell production facility and the immunotherapy clinical and support staff for their contributions. We thank Li Jia for assistance in bioinformatics analysis. The Prospective Procurement of Solid Tumor Tissue to Identify Novel Therapeutic study was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services. Whole exome raw data was uploaded to the NIH database for Genotypes and Phenotypes (dbGaP) under accession number phs001003.v1.p1.
Footnotes
Disclosures: There are no commercial or financial disclosures.
Reference List
- 1.Woll E, Bedikian A, Legha SS. Uveal melanoma: natural history and treatment options for metastatic disease. Melanoma Res. 1999;9:575–81. [PubMed] [Google Scholar]
- 2.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118:1881–5. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Woodman SE. Metastatic uveal melanoma: biology and emerging treatments. Cancer J. 2012;18:148–52. doi: 10.1097/PPO.0b013e31824bd256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Vries TJ, Fourkour A, Wobbes T, Verkroost G, Ruiter DJ, van Muijen GN. Heterogeneous expression of immunotherapy candidate proteins gp100, MART-1, and tyrosinase in human melanoma cell lines and in human melanocytic lesions. Cancer Res. 1997;57:3223–9. [PubMed] [Google Scholar]
- 5.de Vries TJ, Trancikova D, Ruiter DJ, van Muijen GN. High expression of immunotherapy candidate proteins gp100, MART-1, tyrosinase and TRP-1 in uveal melanoma. Br J Cancer. 1998;78:1156–61. doi: 10.1038/bjc.1998.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartlett EK, Fetsch PA, Filie AC, Abati A, Steinberg SM, Wunderlich JR, et al. Human melanoma metastases demonstrate nonstochastic site-specific antigen heterogeneity that correlates with T-cell infiltration. Clin Cancer Res. 2014;20:2607–16. doi: 10.1158/1078-0432.CCR-13-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harbour JW. The genetics of uveal melanoma: an emerging framework for targeted therapy. Pigment Cell Melanoma Res. 2012;25:171–81. doi: 10.1111/j.1755-148X.2012.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer. 2014;14:135–46. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 9.Atkins MB, Lotze MT, Dutcher JP, Fisher RI, Weiss G, Margolin K, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol. 1999;17:2105–16. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 10.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–44. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–7. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Chen PW, Li H, Alizadeh H, Niederkorn JY. PD-L1: PD-1 interaction contributes to the functional suppression of T-cell responses to human uveal melanoma cells in vitro. Invest Ophthalmol Vis Sci. 2008;49:2518–25. doi: 10.1167/iovs.07-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna KC, Chen PW. Influence of immune privilege on ocular tumor development. Ocul Immunol Inflamm. 2010;18:80–90. doi: 10.3109/09273941003669950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niederkorn JY. Ocular immune privilege and ocular melanoma: parallel universes or immunological plagiarism? Front Immunol. 2012;3:148. doi: 10.3389/fimmu.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen PW, Mellon JK, Mayhew E, Wang S, He YG, Hogan N, et al. Uveal melanoma expression of indoleamine 2,3-deoxygenase: establishment of an immune privileged environment by tryptophan depletion. Exp Eye Res. 2007;85:617–25. doi: 10.1016/j.exer.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorval T, Fridman WH, Mathiot C, Pouillart P. Interleukin-2 therapy for metastatic uveal melanoma. Eur J Cancer. 1992;28A:2087. doi: 10.1016/0959-8049(92)90266-5. [DOI] [PubMed] [Google Scholar]
- 19.Luke JJ, Callahan MK, Postow MA, Romano E, Ramaiya N, Bluth M, et al. Clinical activity of ipilimumab for metastatic uveal melanoma: a retrospective review of the Dana-Farber Cancer Institute, Massachusetts General Hospital, Memorial Sloan-Kettering Cancer Center, and University Hospital of Lausanne experience. Cancer. 2013;119:3687–95. doi: 10.1002/cncr.28282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maio M, Danielli R, Chiarion-Sileni V, Pigozzo J, Parmiani G, Ridolfi R, et al. Efficacy and safety of ipilimumab in patients with pre-treated, uveal melanoma. Ann Oncol. 2013;24:2911–5. doi: 10.1093/annonc/mdt376. [DOI] [PubMed] [Google Scholar]
- 21.Joshua AM, Monzon JG, Mihalcioiu C, Hogg D, Smylie M, Cheng T. A phase 2 study of tremelimumab in patients with advanced uveal melanoma. Melanoma Res. 2015 doi: 10.1097/CMR.0000000000000175. [DOI] [PubMed] [Google Scholar]
- 22.Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother. 2003;26:332–42. doi: 10.1097/00002371-200307000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bioinformatics.babraham.ac.uk. Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data Internet] 2015 http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. cited March 20, 2015.
- 24.Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Institute G. GATK | Best Practice Workflows [Internet] Broadinstituteorg. 2015 https://www.broadinstitute.org/gatk/guide/best-practices. cited March 30, 2015.
- 28.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gdac.broadinstitute.org. [Internet] 2015 http://gdac.broadinstitute.org/runs/stddata__2014_07_15/data/SKCM/20140715/gdac.broadinstitute.org_SKCM.Mutation_Packager_Calls.Level_3.2014071500.0.0.tar.gz. cited March 20, 2015.
- 31.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egberts F, Bergner I, Kruger S, Haag J, Behrens HM, Hauschild A, et al. Metastatic melanoma of unknown primary resembles the genotype of cutaneous melanomas. Ann Oncol. 2014;25:246–50. doi: 10.1093/annonc/mdt411. [DOI] [PubMed] [Google Scholar]
- 33.Premkumar A, Marincola F, Taubenberger J, Chow C, Venzon D, Schwartzentruber D. Metastatic melanoma: correlation of MRI characteristics and histopathology. J Magn Reson Imaging. 1996;6:190–4. doi: 10.1002/jmri.1880060134. [DOI] [PubMed] [Google Scholar]
- 34.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, Zhang T, Ye J, Li H, Huang J, Li X, et al. Tumor-infiltrating lymphocytes predict response to chemotherapy in patients with advance non-small cell lung cancer. Cancer Immunol Immunother. 2012;61:1849–56. doi: 10.1007/s00262-012-1231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32:2959–66. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–52. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 39.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–5. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De GV, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30:2522–9. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 44.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 46.Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest. 2007;117:1137–46. doi: 10.1172/JCI31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–6. doi: 10.1038/nature11538. [DOI] [PubMed] [Google Scholar]
- 48.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–11. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 49.Overwijk WW, Theoret MR, Finkelstein SE, Surman DR, de Jong LA, Vyth-Dreese FA, et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med. 2003;198:569–80. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawakami Y, Dang N, Wang X, Tupesis J, Robbins PF, Wang RF, et al. Recognition of shared melanoma antigens in association with major HLA-A alleles by tumor infiltrating T lymphocytes from 123 patients with melanoma. J Immunother. 2000;23:17–27. doi: 10.1097/00002371-200001000-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.