Abstract
Semi-synthetic derivatives of the tricyclic diterpene antibiotic pleuromutilin from the basidiomycete Clitopilus passeckerianus are important in combatting bacterial infections in human and veterinary medicine. These compounds belong to the only new class of antibiotics for human applications, with novel mode of action and lack of cross-resistance, representing a class with great potential. Basidiomycete fungi, being dikaryotic, are not generally amenable to strain improvement. We report identification of the seven-gene pleuromutilin gene cluster and verify that using various targeted approaches aimed at increasing antibiotic production in C. passeckerianus, no improvement in yield was achieved. The seven-gene pleuromutilin cluster was reconstructed within Aspergillus oryzae giving production of pleuromutilin in an ascomycete, with a significant increase (2106%) in production. This is the first gene cluster from a basidiomycete to be successfully expressed in an ascomycete, and paves the way for the exploitation of a metabolically rich but traditionally overlooked group of fungi.
Antimicrobial resistance is now universally considered to be one of the greatest threats to human health. Continually emerging resistance has been confounded by a lack of investment into antibiotic research and development over recent decades. The World Health Organization’s (WHO) 2014 report on global surveillance of antimicrobial resistance has recently warned that without urgent, coordinated action, the world is heading towards a post-antibiotic era, in which common infections and minor injuries, which have been treatable for decades, may again be fatal.
A highly promising class of antibiotics for human therapeutics are the semi-synthetic pleuromutilin antibiotics. Pleuromutilin is a tricyclic diterpene produced by the basidiomycete Clitopilus passeckerianus and related basidiomycete fungi1, which was initially isolated and characterized by Kavanagh and co-workers in 19512. The pleuromutilin derivatives tiamulin and valnemulin have been used in veterinary medicine for many years3, but an increasing need for new antibiotics has led to a surge in the development of pleuromutilin derivatives specifically for human use.
In 2007, retapamulin became the first semi-synthetic pleuromutilin antibiotic to be approved by the FDA and EMA and is recommended for the treatment of impetigo and infected small lacerations, abrasions and sutured wounds4. This represents the first antibiotic of a new class to be approved for use against topical infections in more than 30 years.
Nabriva Therapeutics AG, a biotechnology company focused on developing pleuromutilin antibiotics, has been developing novel pleuromutilin derivatives with potent and broad-spectrum activities and the desired pharmacokinetic characteristics for oral and intravenous (IV) administration. One derivative in particular, BC-3781 (lefamulin), is particularly promising and has recently been granted Qualified Infectious Disease Product (QIDP) as well as Fast Track status designation, for the treatment of community-acquired bacterial pneumonia (CABP) and acute bacterial skin and skin structure infections (ABSSSI). Lefamulin is soon to enter phase III clinical trials for the treatment of CABP, and also holds great potential as a treatment for hospital-acquired and ventilator-associated bacterial pneumonia (HABP/VABP), as well as ABSSSI, with potential in several other indications (such as sexually transmitted infections, including multidrug-resistant MDR gonorrhea and osteomyelitis).
There has also been increasing interest in pleuromutilin antibiotics as potential agents to treat multi-drug resistant (MDR-) and extensively drug resistant (XDR-) tuberculosis strains, which are increasingly difficult to treat5,6. Although the biochemical properties and mode of action of pleuromutilins are well understood7,8,9,10, and a partial chemical elucidation of the potential pathway has previously been proposed11, the complete biosynthesis of this increasingly important natural product has never been elucidated. As with most natural products, and particularly fungal natural products, the yields in the native host are very low, and the process of fermentation and isolation of the desired compounds can be logistically challenging. This is particularly true for basidiomycete fungi such as Clitopilus passeckerianus, where traditional strain improvement techniques such as random mutagenesis are rarely successful due to their dikaryotic nature. For these reasons, understanding the biosynthetic pathway of a natural product is often key to developing a strain which can be exploited successfully.
In this work we set out to identify the pleuromutilin gene cluster and identify a strain improvement approach. Initial attempts to improve pleuromutilin yields in C. passeckerianus were followed by the reconstruction of the biosynthetic pathway in the heterologous host Aspergillus oryzae. This represents the first cross-phylum heterologous expression of a basidiomycete gene cluster in an ascomycete host, so will not only increase the potential of the pleuromutilin antibiotics but may also help to inform the future discovery and exploitation of secondary metabolites from the rich but underexploited basidiomycete fungi.
Results
Identification of the candidate pleuromutilin gene cluster
In fungi, it is usual for the gene clusters encoding enzymes for diterpene biosynthesis to include a cluster-specific geranylgeranyl pyrophosphate synthetase (GGS). On this assumption, we used PCR with degenerate primers ggs27 and ggs29 to isolate GGS-related products from the genome of C. passeckerianus. Amplification products were cloned and sequenced. After taking into account allelic differences due to the dikaryotic nature of the fungus, four different GGS-related sequences were obtained and these were screened to determine which followed the predicted expression patterns for pleuromutilin production by RT-PCR and Northern analysis. RT-PCR excluded ggs-1, whilst Northern analysis showed that only ggs-2 was highly expressed under pleuromutilin-producing conditions (Supplementary Figure 1). Southern blot analysis determined that ggs-2 is only present once in the genome (Supplementary Figure 2A).
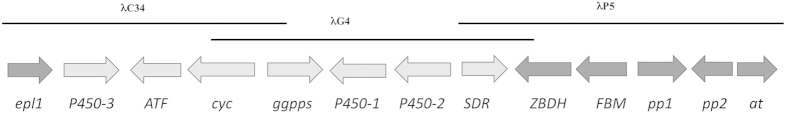
Probing a C. passeckerianus genomic library with this partial ggs-2 PCR product identified lambda clone λG4, and sequencing the full 8.4 kb insert revealed a full-length ggs gene of 1291 bp (Pl-ggs), containing 4 putative introns and encoding a 350 amino-acid protein. Upstream of Pl-ggs was a partial diterpene cyclase (cyc) gene, named Pl-cyc; downstream was a full length cytochrome P450 gene (Pl-p450-1) and a second partial cytochrome P450 gene (Pl-p450-2), showing that Pl-ggs was located within a likely diterpene gene cluster. Rescreening the library to isolate contiguous sequences identified λC34, in which an overlapping region shared 100% homology with λG4, and λP5, whose overlap with λG4 contained several SNPs and indels. This indicated that λG4 and λC34 originate from the same allelic version of the pleuromutilin gene cluster, while λP5 is derived from the other allele. In total a genomic region of 34.6 kb was sequenced and 13 likely genes were identified (Fig. 1 and Table 1).
Figure 1. The pleuromutilin gene cluster of C. passeckerianus.
Genes involved in the biosynthesis of pleuromutilin are light grey. Those in dark grey are considered to lie outside the gene cluster. The horizontal lines above show the regions of the cluster contained within the three lambda phage clones λC34, λG4 and λP5.
Table 1. Genes sequenced during this project.
| Gene | Length (bp) | Number of introns | Predicted protein length (aa) | Putative function | Closest known homologue and accession number | |
|---|---|---|---|---|---|---|
| FDS Locus | Cp-fds | 1487 | 5 | 375 | Farnesyl diphosphate synthase (FDS) | FPPS from Lactarius chrysorrheus (BAD15361) |
| Cp-cdb | 3436 | 21 | 764 | Cellobiose dehydrogenase | cbd from Coniophora puteana (BAD32781) | |
| Cp-ATPase | 4549 | 14 | 1225 | Cation transporter | Cation transporting ATPase from Aspergillus fumigatus Af293 (XP_754245) | |
| Cp-ccp | 435 | 0 | 144 | Cerato-platanin protein | cerato-platanin 6 from Crinipellis campanella (AGL40521) | |
| Pleuromutilin Locus | Pl-p450-3 | 2108 | 10 | 522 | Cytochrome P450 | Pleurotus sapidus putative Cytochrome P450 monooxygenase (CAJ00405) |
| Pl-atf | 1304 | 3 | 377 | Acetyltransferase | Aspergillus fumigatus Acetyltransferase TRI7-like toxin biosynthesis protein (XP_746970) | |
| Pl-cyc | 3040 | 3 | 959 | Terpene Cyclase | Gibberella fujikuroi Ent-kaurene synthase (Ent-copalyl diphosphate synthase) (Q9UVY5) | |
| Pl-ggs | 1291 | 4 | 350 | Geranylgeranyl pyrophosphate synthetase (GGS) | Mucor racemosus Geranylgeranyl pyrophosphate synthetase (Q9P885) | |
| Pl-p450-1 | 2286 | 13 | 523 | P450 monooxygenase | Pleurotus sapidus cytochrome P450-3 monooxygenase (CAL69751) | |
| Pl-p450-2 | 2124 | 11 | 525 | P450 monooxygenase | Pleurotus sapidus cytochrome P450-3 monooxygenase (CAL69751) | |
| Pl-sdr | 945 | 3 | 254 | Short-chain dehydrogenase/reductase (SDR) | Desulfitobacterium hafniense DCB-2 short-chain dehydrogenase/reductase SDR (ZP_01373200) | |
| Cp-zbdh | 1479 | 10 | 321 | Oxidoreductase | Neosartorya fischeri NRRL 181 oxidoreductase, zinc-binding dehydrogenase family (XP_001264189) | |
| Cp-fbm | 2434 | 11 | 616 | Monooxygenase | Aspergillus fumigatus Af293 flavin-binding monooxygenase-like protein (XP_751903) | |
| Cp-pp1 | 2144 | 21 | 338 | Unknown | Predicted protein from Laccaria bicolor S238N-H82 (XP_001884957) | |
| Cp-pp2 | – | – | – | Unknown | predicted protein from Coprinopsis cinerea (EAU81003). | |
| Cp-at (partial) | – | – | – | Putative aminotransferase | Moniliophthora roreri aminotransferase family protein (XP_007868896) |
Whilst Pl-ggs and Pl-cyc could be inferred as the core genes of the diterpene synthesis, from bioinformatic analysis alone, it was not possible to confidently predict the boundaries of the gene cluster. We therefore analysed gene expression by northern blotting to identify which genes were coordinately expressed and correlated with conditions known to induce pleuromutilin biosynthesis.
Genes for GGS, cyclase, acetyl transferase (ATF), short-chain dehydrogenase/reductase (SDR) and three P450 s were shown to be upregulated during pleuromutilin production, whereas flavin binding monooxygenase (fbm), zinc binding dehydrogenase (zbdh), and aminotransferase genes were not, suggesting that they lie outside the pleuromutilin gene cluster (Supplementary Figure 3).
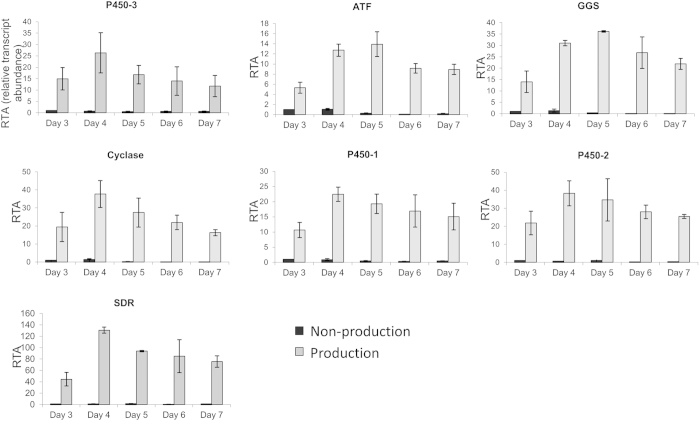
A more thorough expression analysis, combining quantitative RT-PCR with an analysis of pleuromutilin production, demonstrated that the seven putative pleuromutilin biosynthetic genes are not only highly differentially expressed under production and non-production conditions, but also tightly co-regulated (Fig. 2). See Supplementary Figure 4 for the corresponding time-course of pleuromutilin yields.
Figure 2. Real-time PCR data for the seven pleuromutilin biosynthesis genes from C. passeckerianus when grown in production (CGC) and non-production (MM) media.
β-tubulin was used as a reference gene and non-production media on day 3 was used as the calibrator sample (all other expression is shown relative to this data point).
The gene for farnesyl diphosphate synthase (fds) was also isolated and sequenced to assess any role in the pathway. This is another key enzyme in isoprenoid biosynthesis which catalyzes the formation of farnesyl diphosphate, a precursor for several classes of essential metabolites. Degenerate primers (fdsf2A and fdsr4) were used to amplify an fds gene fragment, which was used to pull out a lambda clone that was sequenced in its entirety (Supplementary Figure 5). Southern blot analysis demonstrated that this fds is only present once in the genome, at a different locus to the pleuromutilin cluster, suggesting that it is responsible for the biosynthesis of farnesyl diphosphate for both primary and secondary metabolism. Genes encoding β-tubulin and α- actin were also isolated from a genomic library as reported in Kilaru et al.12, allowing their promoter elements to be exploited for use in expression cassettes.
A gene-silencing approach was used to directly link the 7-gene cluster with pleuromutilin production, where the two core genes Pl-ggs and Pl-cyc were silenced. Plasmids based on pYES-hph-cbxgene (Supplementary Figure 6) were generated containing the full length coding regions of Pl-ggs and Pl-cyc in the antisense direction, under the control of the Agaricus bisporus gpdII promoter. Silencing using antisense constructs has previously been demonstrated to be an efficient approach to genetic manipulation in C. passeckerianus12. Initial screening of transformants via plate-based bioassay revealed reduced clearing zones for both genes (Supplementary Figures 7 to 9), providing direct evidence for their involvement in pleuromutilin production. Further analysis of selected transformants by HPLC demonstrated reduction in pleuromutilin titres. For example the yield of pleuromutilin was reduced by 87% to 103 μg/g in transformant p004-GGSantigene-16, compared to the wild-type C. passeckerianus yield of 765 μg/g.
Attempted increase in pleuromutilin production in C. passeckerianus
Overexpression of the putative pleuromutilin biosynthetic genes was attempted in C. passeckerianus. Initially, the native promoters were used to drive expression. Eight expression vectors were constructed (Supplementary Figure 6B), containing the genes Pl-p450-3, Pl-atf, Pl-cyc, Pl-ggs, Pl-p450-1, Pl-p450-2, Pl-sdr, and Cp-fbm, and transformed into C. passeckerianus (Table 2). At least fifty transformants for each gene were screened via plate-based bioassay with Bacillus subtilis but no set of transformants showed any significant increase in clearing zone compared to untransformed C. passeckerianus (Supplementary Figures 10 to 17). Unexpectedly, however, every set of transformants contained some strains which exhibited a significant reduction of clearing zone, with some transformants expressing Pl-ggs, Pl-cyc or Pl-p450-1 showing a complete disappearance of antibacterial activity. In particular, overexpression of Pl-cyc from its native promoter generated a range of transformants with greatly reduced or complete absence of clearing zones (Supplementary Figure 11). An HPLC analysis of pYES-hph-nativeCycgene transformants 2, 12, 23 and 43 showed a reduction in pleuromutilin titres of 38, 75, 98 and 89% respectively (436, 177, 11 and 78 μg/g) when compared to wild type C. passeckerianus (703 μg/g). This suggests that a form of sense suppression is occurred in these transformants.
Table 2. Transformants obtained with vectors containing pleuromutilin pathway genes under the control of their native promoters.
| Gene Expressed | Plasmid | Total transformants | Transformants assayed | Clearing zone diameter range (% of wild-type) | Average clearing zone diameter (% of wild-type) |
|---|---|---|---|---|---|
| p450-3 | pYES-hph-nativep450-3gene | 221 | 50 | 80–113% | 98.4% |
| atf | pYES-hph-nativeATFgene | 188 | 50 | 76–122% | 106.0% |
| cyc | pYES-hph-nativeCycgene | 189 | 56 | 0–128% | 96.9% |
| ggs | pYES-hph-nativeGGSgene | 179 | 45 | 0–121% | 102.8% |
| p450-1 | pYES-hph-nativep450-1gene | 167 | 56 | 0–142% | 111.1% |
| p450-2 | pYES-hph-nativep450-2gene | 156 | 53 | 60–122% | 104.2% |
| sdr | pYES-hph-nativeSDRgene | 98 | 50 | 84–112% | 98% |
| fbm | pYES-hph-nativeFBMgene | 138 | 52 | 80–113% | 98.4% |
A number of each set were analysed by bioassay to estimate pleuromutilin production. The range and average clearing zone diameters are shown as a percentage of wild-type C. passeckerianus.
As overexpression using native promoters did not enhance pleuromutilin production, the constitutive strong promoter gpdII from A. bisporus was instead used to drive expression of six genes in C. passeckerianus. Twelve constructs were made to include the individual genes Cp-fds, Pl-ggs, Pl-cyc, Pl-p450-1, Pl-p450-2, and Pl-p450-3, either with or without an additional intron (64 bp intron-exon region of A. bisporus gpdII) at the 5′ end of the gene (Supplementary Figure 6C). The presence of a 5′ intron has previously been shown to be essential for successful green fluorescent protein (gfp) and phleomycin resistance (ble) gene expression in C. passeckerianus12. At least twenty transformants were generated for each group using the 12 expression vectors (Table 3). Transformants overexpressing the Cp-fds or Pl-ggs were screened using HPLC for improved pleuromutilin titres. The majority of transformants demonstrated no significant increase in antibiotic titres, but one strain expressing the Pl-ggs without the additional intron showed a 50% increase in pleuromutilin yields when compared to wild-type C. passeckerianus (Table 2 and Supplementary Figure 18). Northern blot analysis showed increased levels of Pl-ggs transcripts when compared to the wild-type (Supplementary Figure 19), suggesting that the improved titre is due to increased GGS expression. The strains overexpressing Pl-cyc, Pl-p450-1, Pl-p450-2, or Pl-p450-3 were screened via plate-based bioassay against B. subtilis, but no significant increases in clearing zones were observed (Table 3 and Supplementary Figures 20 to 26). A reduction in clearing zone diameter was again observed for some transformants from every group, and a complete loss of observable inhibition was seen for one transformant expressing the Cyclase gene without the additional intron (Supplementary Figure 20).
Table 3. Total number of transformants obtained with vectors containing pleuromutilin pathway genes under the control of A. bisporus gpdII promoter.
| Gene Expressed | Plasmid | Total transformants | Pleuromutilin titre range (μg/g) | Clearing zone range (and average) as a percentage of wild-type |
|---|---|---|---|---|
| FDS | p004-CP1-FDSgene | 55 | 595–810 | – |
| FDS | p004-CP1-iFDSgene | 20 | 600–920 | – |
| GGS | p004-CP1-GGSgene | 32 | 495–1,010 | – |
| GGS | p004-CP1-iGGSgene | 22 | 605–795 | – |
| Cyclase | pYES-hph-Cycgene | 31 | – | 0–109.4% (89.0%) |
| Cyclase | pYES-hph-iCycgene | 23 | – | 100.8–122.0% (106.7%) |
| P450-1 | pYES-hph-p450-1gene | 20 | – | 70.4–103.0% (92.4%) |
| P450-1 | pYES-hph-ip450-1gene | 23 | – | 77.0–103.7% (92.5%) |
| P450-2 | pYES-hph- p450-2gene | 22 | – | 68.1–107.3% (89.3%) |
| P450-2 | pYES-hph-ip450-2gene | 30 | – | 83.7–105.2% (94.2%) |
| P450-3 | pYES-hph-ip450-3gene | 32 | – | 49.7–113.8% (98.3%) |
The FDS and GGS lines were analysed by HPLC. All others were analysed by bioassay. The titres of pleuromutilin recorded through HPLC (μg of pleuromutilin per g of dry mycelial mass) are reported for the transformant lines overexpressing the genes fds and ggs. The wild-type levels were 690–815 μg/g.
As a final approach to increase pleuromutilin titres in the native host, the entire putative gene cluster for pleuromutilin – comprising the nine genes Pl-p450-3, Pl-atf, Pl-cyc, Pl-ggs, Pl-p450-1, Pl-p450-2, Pl-sdr, Cp-zbdh and Cp-fbm – was cloned into a yeast shuttle vector (pYES2-hph-pleurocluster – Supplementary Figure 6D), and transformed into C. passeckerianus. One hundred and nineteen transformants were screened for increased antibiotic production by plate-based bioassay. Sixteen transformants showed a 20 to 40% increase in clearing zone diameter, but seven transformants showed complete disappearance of clearing zone (Supplementary Figures 27 and 28).
Heterologous production of pleuromutilin in A. oryzae
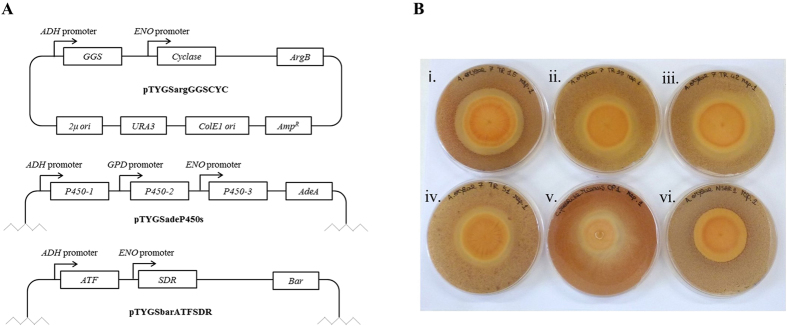
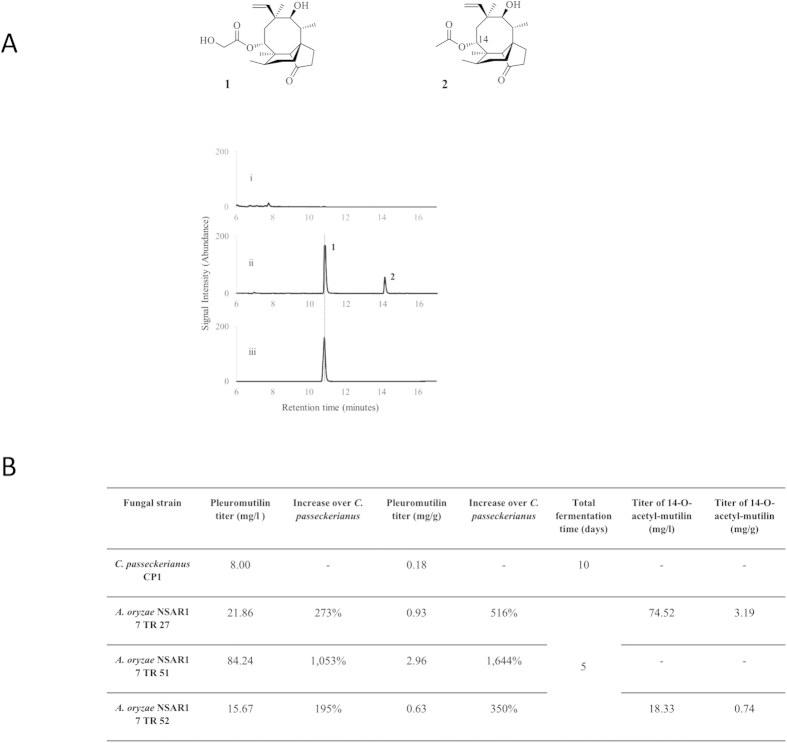
We heterologously expressed the seven genes of the putative gene cluster (Pl-p450-3, Pl-atf, Pl-cyc, Pl-ggs, Pl-p450-1, Pl-p450-2, Pl-sdr) in A. oryzae NSAR1, using our established Aspergillus oryzae multi-gene expression system13. Each of the genes were amplified from cDNA and placed under the control of A. oryzae promoters (Fig. 3A). Twelve independent transformants were screened for their ability to inhibit B. subtilis growth via plate-based bioassay. Eleven strains showed production and diffusion of an antibacterial compound in the medium, witnessed by appearance of clearing zone around the mycelia (Fig. 3B; Supplementary Figure 29). Only one strain (NSAR1 7 TR49) did not inhibit bacterial growth. RT-PCR demonstrated that three out of the seven transgenes were not being expressed in this strain (Pl-p450-1, Pl-p450-2 and Pl-p450-3). Expression of all seven transgenes was confirmed for all of the other transformants (Supplementary Figure 30). To confirm pleuromutilin production, fermentation cultures were extracted with ethyl acetate, and the resulting crude extracts were analysed by HPLC-MS. As expected no production of new metabolites was detected from strain NSAR1 7 TR49, whereas two new compounds (1 and 2) were detected from all other strains (Fig. 4A), with 1 showing the same retention time and mass/charge ratio (m/z) as authentic pleuromutilin. 1 and 2 were purified from a 1 L culture via preparative-HPLC (yielding 12.4 mg and 7.5 mg respectively), and characterized by NMR spectroscopy and ESIHRMS. 1 was determined to be pleuromutilin, by ESIHRMS and by a comparison of the 1H-NMR and 13C-NMR data with that of authentic pleuromutilin and data in the literature14 (Supplementary Figures 31 to 34; Supplementary Table 2). ESIHRMS and a full-range of NMR analyses – including 2D-COSY, HMBC, and HSQC – allowed the identification of 2 as 14-O-acetyl-mutilin (Supplementary Figures 35 to 39; Supplementary Table 3), which is another known product of fermentation of C. passeckerianus15. It has not previously been proven to a precursor of pleuromutilin.
Figure 3.
(A) Plasmid maps of vectors pTYGSargGGSCYC, pTYGSadeP450s, pTYGSbarATFSDR. The features for replication and selection in E. coli and S. cerevisiae are common to the three vectors, therefore shown only in pTYGSargGGSCYC. (B) Plate-based bioassay showing clearing zones produced by (i) A. oryzae NSAR1 7 TR15, (ii) A. oryzae NSAR1 7 TR35, (iii) A. oryzae NSAR1 7 TR42, (iv) A. oryzae NSAR1 7 TR51, (v) C. passeckerianus CP1, and (vi) untransformed A. oryzae NSAR1.
Figure 4.
(A) De novo production of pleuromutilin (1) and 14-O-acetyl-mutilin (2) in A. oryzae. HPLC traces of untransformed A. oryzae NSAR1 (i), A. oryzae with GGS, Cyclase, P450-1, P450-2, P450-3, ATF, and SDR from C. passeckerianus (ii), and authentic pleuromutilin (iii). All traces were monitored through ELSD (Evaporative Light Scattering Detector). (B) Pleuromutilin and 14-O-acetyl-mutilin titres from A. oryzae transformant strains and C. passeckerianus. The titres are expressed either as mg of compound produced per litre of fungal culture (mg/l) or as mg of compound produced per gram of dry mass of mycelium (mg/g). C. passeckerianus required a total of 10 days between inoculation and harvesting, due to the need for a 5-day seed culture in PVS, followed by another 5-day production culture in CGC. A. oryzae transformants were only grown for 5 days in production culture CMP, which was inoculated directly from plates.
Pleuromutilin yields were quantified for the A. oryzae transformants showing the highest production of 1 from the analytical HPLC-MS, and a 195 to 1,053% increase over the native producer was observed, with a maximum production of 84.24 mg/l for NSAR1 7 TR51 (Fig. 4B). Taking into account that C. passeckerianus requires a preliminary seed culture prior to fermentation, the time between initial culture inoculation and harvesting of the antibiotic is also halved in A. oryzae compared to C. passeckerianus (Fig. 4B). This brings the increase in pleuromutilin production in A. oryzae to 2,106% over C. passeckerianus. We also estimated the titre of 14-O-acetyl-mutilin 2, produced by the three A. oryzae transformants. Strain NSAR1 7 TR27 produced the highest titre of 74.52 mg/l, strain NSAR1 7 TR52 produced 18.33 mg/l, whereas no detectable amounts of 2 were observed in NSAR1 7 TR51, the highest producer of pleuromutilin 1 (Fig. 4B).
We carried out Southern blot analysis to detect copy number of the transgenes in these three same transformant strains and detected the presence of multiple copies of the genes Pl-atf and Pl-sdr in the highest producing strain NSAR1 7 TR51 (Supplementary Table 4). Multiple copies of the genes for the three cytochrome P450 s were found in strain NSAR1 7 TR52 (titre of 1: 15.67 mg/l). This does not appear to provide any further increase in antibiotic titre compared to strain NSAR1 7 TR27 (titre of 1: 21.86 mg/l) which has a single copy for each transgene.
Discussion
The emergence of antibiotic resistance is posing a major concern for human health, and the development of robust alternatives to the currently-exploited antimicrobial agents has therefore become of primary importance. As the newest class of antibiotics for use in human therapeutics, and a class with great potential to treat various resistant and highly virulent pathogens such as Methicillin-resistant Staphylococcus aureus (MRSA) and Multi-Drug Resistant (MDR)-tuberculosis, the pleuromutilin antibiotics are becoming increasingly important. Nevertheless, the genetic basis of pleuromutilin production has not before been elucidated. For this reason we set out to identify the pleuromutilin gene cluster in C. passeckerianus, which is currently used to commercially produce pleuromutilin – and investigate potential approaches to increase pleuromutilin yields.
We hypothesized that, pleuromutilin being a diterpene compound, a pleuromutilin gene cluster may contain a pathway specific geranylgeranyl diphosphate synthase gene (ggs), as is the case for many characterized fungal diterpene gene clusters16,17,18. By screening a λ-phage genomic DNA library of C. passeckerianus we isolated four ggs genes. Of these four genes, only ggs-2 was novel, the other three having been previously cloned and sequenced19. ggs-2 was also the only ggs shown to be highly expressed under pleuromutilin production conditions, and was shown to be present only once in the genome, making it highly likely that this was the ggs responsible for providing geranyl-geranyl diphosphate (GGPP) for the biosynthesis of pleuromutilin and referred to thereafter as Pl-ggs.
Genome walking and sequencing of other λ-phage clones allowed the identification of genes adjacent to Pl-ggs with potential roles in secondary metabolite biosynthesis. These included a gene encoding a cyclase (Pl-cyc), which commonly catalyzes the cyclization of GGPP to give the first cyclic intermediate found in the biosynthesis of diterpenes, as well as genes encoding an acetyl transferase (Pl-atf), a short-chain dehydrogenase/reductase (Pl-sdr), three cytochrome P450s (Pl-p450-1, Pl-p450-2 and Pl- p450-3), a zinc-binding dehydrogenase (Cp-zbdh), a flavin-binding monooxygenase-like protein (Cp-fbm), and four other putative proteins (Cp-pp1, Cp-pp2, Cp-at and Cp-epl1). Differential expression of the putative pleuromutilin gene cluster (Pl-ggs, Pl-cyc, Pl-atf, Pl-sdr, Pl-p450-1, Pl-p450-2 and Pl-p450-3), under pleuromutilin production and non-production conditions was assessed through Northern blot analysis. Quantitative PCR analysis confirmed that these seven genes had high expression under production conditions of pleuromutilin, as well as being tightly co-regulated. This allowed the borders of the putative gene cluster to be defined.
By employing the molecular tools that we previously developed for genetic manipulation of C. passeckerianus12, we attempted to increase antibiotic production titres through overexpression of genes from the putative pleuromutilin gene cluster. Lines of the fungus with additional copies of each of the genes Pl-ggs, Pl-cyc, Pl-atf, Pl-sdr, Pl-p450-1, Pl-p450-2, Pl-p450-3 and Cp-fbm under control of their native promoter sequence, were generated. Rather than leading to an increase in pleuromutilin titres, in multiple transformed strains, there was a reduction or loss of pleuromutilin production. This points to C. passeckerianus having a ‘sense-suppression’ mechanism, whereby the introduction of an exogenous copy of a gene will silence both the exogenous and endogenous copies. Such a phenomenon has been seen in other basidiomycete fungi such as Cryptococcus neoformans20 and Schizophyllum commune21.
Overexpression of six selected genes (Cp-fds, Pl-ggs, Pl-cyc, Pl-p450-1, Pl-p450-2 and Pl-p450-3) was also attempted by placing them under the control of the gpdII promoter of Agaricus bisporus, which is known to drive strong expression in C. passeckerianus12. Although not present in the pleuromutilin gene cluster, the farnesyl diphosphate synthase gene (Cp-fds) was included within this set as increasing substrate availability to a pathway can increase production. Only one strain overexpressing Pl-ggs showed approximately 50% increase in pleuromutilin titre, whereas all other strains showed either no change in pleuromutilin production, or a reduction in titres. The whole genomic region containing the putative cluster was also cloned and introduced in additional copies to the genome of C. passeckerianus, but this also failed to provide a consistent and reproducible significant increase in antibiotic production.
Since the manipulation of the pleuromutilin gene cluster in the native host did not achieve a robust increase in antibiotic titre, total biosynthesis of pleuromutilin was recreated in a secondary host, with the aim of providing an alternative platform for improved production of the antibiotic. The ascomycete fungus Aspergillus oryzae was used for this purpose, as it has been used before as recipient organism for total biosynthesis of other fungal SMs, such as the diterpene aphidicolin22 and the polyketide tenellin23. The cDNA sequences of the seven genes identified as constituting the gene cluster in C. passeckerianus – Pl-ggs, Pl-cyc, Pl-atf, Pl-sdr, Pl-p450-1, Pl-p450-2 and Pl-p450-3 – were cloned and placed under control of constitutive promoters for expression in A. oryzae, using a modified version of the expression vectors designed by Pahirulzaman, Williams and Lazarus12. De novo production of pleuromutilin in A. oryzae transformants was assessed via plate-based bioassay, as well as purification and consequent NMR characterization of novel compounds. The metabolite 14-O-acetyl-mutilin, described in literature as a side-product from fermentation of pleuromutilin-producing fungi14, could also be isolated from the same strains. Quantification of pleuromutilin demonstrated that the A. oryzae transformants were producing higher pleuromutilin titres than C. passeckerianus, with one strain giving a remarkable 10-fold increase, which increases to 20-fold if the total fermentation time required is taken into account. Moreover, further improvement of the pleuromutilin-producing A. oryzae strain can now be achieved through classical means of random mutagenesis in a well understood host, as well as by promoting conversion of the putative precursor 14-O-acetyl-mutilin into pleuromutilin. Notably, among the pleuromutilin-producing A. oryzae strains tested, the strain that showed highest production of pleuromutilin also exhibited no detectable amounts of 14-O-acetyl-mutilin, whereas one strain with lower pleuromutilin production showed higher amounts of 14-O-acetyl-mutilin. A higher rate of conversion from 14-O-acetyl-mutilin to pleuromutilin could be achieved through improved expression of the gene responsible for catalyzing the putative last step of the pathway. The use of a well-known host such as A. oryzae, which produces few other secondary metabolites will also increase the efficiency of downstream purification.
The work described here has identified and defined the gene cluster for the important diterpene antibiotic pleuromutilin, from the basidiomycete fungus C. passeckerianus. The lack of consistent enhancement in antibiotic titre obtained through overexpression of the gene cluster in the native host suggests that, despite the availability of molecular tools, C. passeckerianus is not amenable to genetic manipulation aimed at improving metabolite production. Ultimately, seven clustered genes – Pl-ggs, Pl-cyc, Pl-atf, Pl-sdr, Pl-p450-1, Pl-p450-2 and Pl-p450-3 – were identified and confirmed to be sufficient to ensure biosynthesis of pleuromutilin, as shown by de novo production of the antibiotic through heterologous expression in A. oryzae. This also established a potential alternative method for production of the antibiotic in high titres, which will be needed to support large-scale production of pleuromutilin as novel semi-systemic pleuromutilin derivatives are launched.
Beyond the importance of this work to the field of pleuromutilin research, this also represents the first successful heterologous expression of an entire basidiomycete secondary metabolite gene cluster in a well-known ascomycete host. This demonstrates the potential value of this approach for producing natural products from the under exploited and often intractable basidiomycete fungi, in titres which are suitable for commercial exploitation.
Materials and Methods
Bacterial and yeast strains
The Escherichia coli strains routinely used for this study, JM109 and KW251 (both Promega, Southampton, UK), were grown on Luria-Bertani (LB) agar plates, supplemented when necessary with either ampicillin (100 μg/ml) or tetracycline (15 μg/ml). Bacillus subtilis was grown on Tryptic Soy Agar (TSA). E. coli One Shot® ccdB SurvivalTM 2 T1R competent cells (Life Technologies) were used for the propagation of any plasmids containing a gateway cassette.
Saccharomyces cerevisiae BY4742 (genotype MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0)24, was used for homologous recombination-based construction of plasmids, and was maintained on YPDA plates (10 g L−1 yeast extract, 20 g L−1 bactopeptone, 20 g L−1 D-glucose, 15 g L−1 agar) at 28 °C.
Fungal strains and growth conditions
Aspergillus oryzae strain NSAR1 (genotype niaD−, sC−, ΔargB, adeA−)25, was used as heterologous host. This strain was maintained at 28 °C on MEA plates with appropriate supplements (15 g L−1 malt extract, 1.5 g L−1 arginine, 1.5 g L−1 methionine, 0.1 g L−1 adenine, 2 g L−1 ammonium sulphate, 15 g L−1 agar).
Clitopilus passeckerianus strain ATCC 34646 was grown routinely on potato dextrose agar (PDA; Sigma, UK) at 25 oC for five days. For isolation of genomic DNA four 1 cm2 plugs of mycelial mat were used to inoculate 100 ml of CSO1A media (4 ml/L rape seed oil, 50 g/L glucose, 12.5 g/L yeast extract, 1.0 g/L KH2PO4, 0.5 g/L MgSO4⋅ 7H2O, 0.7 g/L Ca(N03)2.4H2O, 0.1 g/L NaCl, 0.05 g/L FeSO4⋅ 7H2O; pH 6.2) and grown at 25 oC for two days with agitation at 230 rpm. Following growth, the mycelium was collected by filtration through Miracloth (Calbiochem Corporation, La Jolla, Calif.), washed with TSE buffer (150 mM NaCl, 100 mM EDTA, 50 mM Tris-HCl pH 8.0), and blotted dry. Samples were ground in liquid nitrogen and used for extraction of genomic DNA.
For the preparation of RNA, C. passeckerianus was grown in 50 ml of PVS seed medium (8 g/L rape seed oil, 35 g/L spray dried corn liquor, 15 g/L glucose, 5 g/L calcium carbonate, pH 5.9) for three to five days at 25 oC with agitation at 230 rpm. Two ml of the resulting seed media was used to inoculate production or non-production media. Production media (CGC) contained 50 g/L glucose, 5 g/L spray dried corn steep liquor and 2 g/L calcium carbonate, pH 6.5. Non-production media (MM) contained 50 mg/L adenine sulphate, 2 g/L L-asparagine, 25 ml/L of stock A [40 g/L KH2PO4, 90 g/L Na2HPO4, 11.6 g/L Na2SO4, 20 g/L Di-Ammonium tartrate], 1 ml/L of stock B [40 mg/L Thiamine], 10 ml/L of stock solution C [25 g/L MgSO4] and 10 g/L glucose, added after autoclaving, pH 6.5.
Cultures were harvested as described above for DNA extraction and stored at −80 oC for the preparation of RNAs. Stock cultures of C. passeckerianus were maintained on potato dextrose agar at 4 oC or as agar plugs in 20% glycerol at −80 oC.
Nucleic acid isolation
Genomic DNA was isolated from C. passeckerianus mycelia by the method of Porebski et al.26. Plasmid and lambda DNA were isolated using Qiagen (Qiagen Ltd, UK) kits. Total RNA was isolated using the RNA plant mini kit (Qiagen).
Southern and Northern Blots
For Southern blots, genomic DNA was digested to completion overnight with a suitable range of restriction enzymes (New England Biolabs), and separated by agarose gel electrophoresis. For Northern blots total RNA (10 μg) was separated on a 1.2% formaldehyde agarose gel. Genomic DNA digests and total RNA were transferred to positively charged nylon membrane (Amersham Life Science) using a vacuum blotter (Bio-rad, UK). Transferred RNA was fixed to membranes by UVP CL-1000 UV cross-linker (Genetic Research Instrumentation Ltd, Essex, England). Filters were probed with [α-32P] (3,000 Ci/mmol; Amersham, Buckinghamshire, United Kingdom). The labelling of DNAs was performed using Ready-To-Go™ DNA Labelling Beads (Amersham Pharmacia Biotech). Unincorporated [α-32P]dCTP was removed using ProbeQuant G-50 microspin columns (Amersham Pharmacia Biotech). Hybridization with 32P-labelled probes were carried out at 65 oC overnight in Church Buffer (1% BSA, 1 mM EDTA, 0.5 M NaPO4 pH 7.2, 7% SDS). The membranes were rinsed in 2 x SSC (20 x SSC per L: 175.3 g NaCl, 88.2 g sodium citrate) at room temperature, followed by a 20 min wash at 65 oC in 2 x SSC and then a wash in 0.5 x SSC at 65 oC for 20 min. Damp filters were placed on Kodak X-Omat AR autoradioagraphy film (Sigma) and allowed to expose for the desired time with intensification at −80 oC to detect hybridization signals.
Degenerate PCR
PCR products amplified with GoTaq DNA polymerase (Promega) were purified with Wizard SV Gel (Promega) and PCR Clean-Up System (Promega). These products were routinely cloned into pGEM-T easy (Promega) and transformed into E. coli JM109. PCRs were carried out in a final volume of 20 μl containing 1 × Green Go-Taq PCR buffer (Promega), 0.4 mM of each primer, 100 μM of dNTPs, 2.5 mM MgCl2 and 1U of Go-Taq DNA polymerase (Promega) and 5 ng of DNA. Reaction conditions for PCR amplification consisted of an initial denaturation step at 94 oC for 2 min followed by 30 cycles of: denaturation at 94 oC for 30 sec, annealing at 50–65 oC for 30 sec and extension at 72 oC for 1 min per kb, with a final extension of 10 min at 72 oC. Reactions were carried out in a MJ Research PTC 200 Thermal Cycler (MJ Research, Watertown, MA). Degenerate primers fdsf2A and fdsr4 were designed by using conserved sequences identified from an alignment of Cryptococcus neoformans var. neoformans JEC21(AAW43830), Lactarius chrysorrheus (BAD15361) and Ustilago maydis 521 (XP_757593) and used to amplify a region of Cp-fds gene. To generate a ggs PCR product, degenerate primers pair ggs27 and ggs29, originally designed by Zhang et al.27 were used.
Real-Time PCR
Maxima SYBR Green/ROX qPCR Master Mix (Fermentas) was used for all reactions (Maxima SYBR Green/ROX qPCR Master Mix: 12.5 μl, Forward primer: 0.3 μM, Reverse primer: 0.3 μM, Template cDNA: 25–50 ng, Water to 25 μl). Reaction conditions were 10 minutes at 95 °C followed by 40 x [95 °C for 15 secs, 60 °C for 30 secs, 72 °C for 30 secs]. This was followed by one cycle of 95 °C for 1 min, 60 °C for 30 secs and 95 °C for 30 secs to calculate the disassociation curves for each reaction. This was used to confirm that the desired amplified product is being detected rather than primer dimers, contamination or misannealed primer product. All reactions were carried out using the Mx3005PTM qPCR instrument (Stratagene).
To analyse the results MX-PRO software, which is part of the Mx3005PTM qPCR system, was used. The threshold fluorescence was set using the amplification-based algorithm according to the software manufacturer’s instructions. The Ct values for each reaction were normalised using β-tubulin as the reference gene. Samples were then compared to a calibrator sample (either wild-type or non-production) to gain relative quantification of transcript levels.
Construction and screening of a C. passeckerianus genomic library
Fifty micrograms of DNA were partially digested with Sau3AI to generate the maximum yield of fragments in the size range 9–23 kb. The DNA was then size-fractionated and cloned in Bam HI-linearized vector Lambda GEM-11 (Promega) following the vector’s instruction manual. In-vitro packaging was performed using the Packagene Lambda DNA System (Promega). Propagation and amplification of the genomic library were performed by infection of E. coli KW251. Aliquots of the amplified library were stored in 7% DMSO at −80 oC. The library was screened by plaque hybridization with C. passeckerianus ggs and fds probes using standard methods28. To confirm number and position of introns in gene sequences, 3′ RACE (Rapid Amplification of cDNA Ends) was performed using the Ambion FirstChoice RLM RACE kit.
DNA sequencing and Bioinformatics
Sequencing reactions, in both directions, were carried out by GlaxoSmithKline (Harlow, United Kingdom) and Agowa (Berlin, Germany). Sequence data was assembled into contigs using Sequencher, version 4.7 (Gene Codes). Database similarity searches were performed using the National Centre for Biotechnological Information (NCBI) online program BLAST (http://www.ncbi.nlm.nih.gov/BLAST)29. The PROSITE database was used to identify motifs and signature sequences in the deduced protein sequences with homology to reported proteins (http://www.expasy.ch/tools/scanprosite/)30. Sequence data were aligned using the ClustalW program (http://www.ebi.ac.uk/clustalw/)31. DNA and protein sequences were analysed using the Sequence Manipulation website (http://www.ualberta.ca/~stothard/javascript/)32.
Construction of C. passeckerianus plasmids
Vectors with the selectable marker genes hph (hygromycin resistance) and cbx (carboxin resistance) were constructed through yeast-based homologous recombination33. The cbx cassettes were amplified from pCbx004 and pCbxi00412 using chimeric primers yeast_AgaricusgpdII_promf and yeast_AspergillustrpC_termr (Supplementary Table 1), then individually recombined into a 5.8 kb XbaI-HindIII fragment of yeast shuttle vector pYES2 (Invitrogen, UK), along with the 1.9 kb fragment of hph cassette, which was amplified from pPHT1 by using chimeric primers yeast_coprinustub_promf and yeast_coprinustub_termr (Supplementary Table 1). This resulted in the plasmids pYES-hph-cbxgene and pYES-hph-icbx respectively (Supplementary Figure 6A). The plasmids are identical except the 64 bp intron-exon region of A. bisporus gpdII gene in pYES-hph-icbx. These plasmids were used as bench plasmids to replace either selectable marker with genes of interest and transform C. passeckerianus.
To produce Pl-cyc antisense silencing cassette, the coding regions of Pl-cyc was amplified in their entirety with suitable primer tails to allow recombination into a XhoI and BamHI fragment of pYES-hph-cbxgene, placing the gene in the antisense orientation under the control of the A. bisporus gpdII promoter.
To produce Pl-cyc antisense silencing cassette, the coding regions of Pl-ggs was amplified in their entirety with suitable primer tails to allow ligation into a NcoI and BamHI fragment of pCbx00412, placing the gene in the antisense orientation under the control of the A. bisporus gpdII promoter.
In order to clone the pleuromutilin pathway genes under the control of their native promoter sequences, individual coding regions of genes with their promoter sequences were amplified from the lambda clones of a genomic library of C. passeckerianus, using corresponding chimeric primers with 30 bp-overlap with the pYES2 vector sequence (Supplementary Table 1). These were individually recombined into a XhoI and BamHI fragment of pYES-hph-cbxgene, resulting in overexpression vectors containing pleuromutilin pathway genes under the control of their native promoters and A. nidulans trpC terminator sequences (Supplementary Figure 6B).
To obtain plasmids with the pleuromutilin pathway genes under the control of the A. bisporus gpdII promoter and A. nidulans trpC terminator, the coding regions of genes were amplified from the corresponding lambda clones of C. passeckerianus genomic library, using respective chimeric primers with 30 bp-overlaps with A. bisporus gpdII promoter and A. nidulans trpC terminator sequence (Supplementary Table 1). For genes Pl-cyc, Pl-p450-1, Pl-p450-2 and Pl-p450-3 the coding region were individually recombined into a XhoI and BamHI fragment of plasmid pYES-hph-cbxgene, resulting in overexpression vectors containing pleuromutilin pathway genes under the control of A. bisporus gpdII promoter and A. nidulans trpC terminator sequences (Supplementary Figure 6C). In order to verify the influence of an intron on gene expression levels, the coding regions were also amplified using a chimeric forward primer with homology to the 64 bp intron-exon region of A. bisporus gpdII. This allowed recombination into pYES-hph-icbxgene, placing the coding regions downstream of a 5′ intron. For genes Cp-fds and Pl-ggs, the coding regions were individually ligated into NcoI and BamHI fragments of pcbx004 and pcbxi00412.
To clone the entire pleuromutilin gene cluster of 25 kb (consisting of coding regions of nine putative genes: Pl-p450-3, Pl-atf, Pl-cyc, Pl-ggs, Pl-p450-1, Pl-p450-2, Pl-sdr, Cp-zbdh and Cp-fbm) under the control of their native regulatory sequences, the whole 25 kb cluster sequence was amplified as five 5 kb fragments. Each fragment was amplified from the corresponding lambda clones, allowing at least 100 bp-overlaps between adjacent fragments. Fragment 1 was amplified from λ42, fragment 2 from λ34, fragment 3 from λG4, fragments 4 and 5 were amplified from λ5. All 5 fragments were recombined into a XhoI and BamHI fragment of plasmid pYES-hph-cbx by yeast-based homologous recombination resulting in pYES-hph-pleurocluster (Supplementary Figure 6D).
PEG-mediated transformation of C. passeckerianus
Recombinant plasmids were transformed into C. passeckerianus protoplasts as described by Kilaru et al.12.
Bio-assay to determine the pleuromutilin production levels
To determine the pleuromutilin production levels, C. passeckerianus transformants and wild type strain were analysed by bio-assay as described in Hartley et al.1. This technique was also employed for initial screening of A. oryzae transformants for antibacterial activity. Wild-type A. oryzae and C. passeckerianus were used as negative and positive controls respectively.
Heterologous production of pleuromutilin
Expression vectors for the heterologous expression of pleuromutilin were constructed through homologous recombination in yeast following the published procedure33. The seven genes were amplified from cDNA using Phusion® High-Fidelity DNA Polymerase with extended primers (Supplementary Table 1). Three expression vectors, pTYGSargGGSCyc, pTYGSadeP450s and pTYGSbarATFSDR (Fig. 3A), were constructed based on those developed by Pahirulzaman, Williams and Lazarus34 and were used to transform A. oryzae NSAR1. Protoplast-mediated transformation of A. oryzae was carried out following the published protocol35.
Screening A. oryzae transformants for expression and production
Aspergillus oryzae transformants were analysed for expression of the transgenes and production of new metabolites. Each transformant strain was grown in 100 mL of CMP medium (35 g L−1 Czapek-dox liquid, 20 g L−1 maltose, 10 g L−1 peptone) at 28 °C for five days prior to proceeding with RNA extraction and RT-PCR for expression analysis. A ten-day culture of each transformant grown under the same conditions was subject to extraction in ethyl acetate, concentrated in vacuo and dissolved in methanol. The crude extract was analysed by HPLC for detection of new metabolites and preparative HPLC was used to purify novel metabolites.
Additional Information
How to cite this article: Bailey, A. M. et al. Identification and manipulation of the pleuromutilin gene cluster from Clitopilus passeckerianus for increased rapid antibiotic production. Sci. Rep. 6, 25202; doi: 10.1038/srep25202 (2016).
Supplementary Material
Acknowledgments
Much of this work was a follow on from BBSRC grant award to develop a Basidio Molecular Toolkit D19266. K.de M.-S. was supported through a BBSRC-G.S.K. CASE Award BB/E528379/1. P.H. and F.A. were both supported through University of Bristol Awards. S.K., C.M.C. and A.J.H. were all supported though funding from G.S.K. to the University of Bristol. Chemical analysis was performed on equipment provided by EPSRC EP/F066104/1 We also wish to thank Dr. Craig Butts for help with acquiring 500 MHz NMR data and Dr. Paul Gates from Bristol University Mass Spectrometry Service for help with obtaining HRMS data.
Footnotes
Author Contributions G.D.F., A.M.B., D.S. and K.O’.D. devised the initial project. G.D.F. and A.M.B. lead the Bristol based team; D.W.S. and K.O’.D. at G.S.K., C.L.W. and R.J.C. contributed to the organic chemistry. C.M.L. contributed to design of Aspergillus strategy. A.M.B., F.A., K.de M.-S., S.K., C.M.C., A.J.H., P.H. and A.G. carried out the experiments and analysis, it should be noted that A.M.B., F.A., S.K., C.M.C. and K.de M.-S. all contributed equally, and author position does not signify any particular role or seniority. A.M.B. supported all computational and chemical analysis. K.de M.-S. and F.A. compiled the first drafts of the manuscript, with final version written by G.D.F. with contributions from other members of the team.
References
- Hartley A. J. et al. Investigating pleuromutilin-producing Clitopilus species and related basidiomycetes. FEMS Microbiol Lett 297, 24–30 (2009). [DOI] [PubMed] [Google Scholar]
- Kavanagh F., Hervey A. & Robbins W. J. Antibiotic substances from Basidiomycetes. VIII. Pleurotus multilus (Fr.) Sacc. and Pleurotus passeckerianus pilat. P Natl Acad Sci USA 37, 570–574 (1951). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen S. M., Karlsson M., Johansson L. B. & Vester B. The pleuromutilin drugs tiamulin and valnemulin bind to the RNA at the peptidyl transferase centre on the ribosome. Mol Microbiol 41, 1091–1099 (2001). [DOI] [PubMed] [Google Scholar]
- Butler M. S. Natural products to drugs: natural product-derived compounds in clinical trials. Nat Prod Rep 25, 475–516 (2008). [DOI] [PubMed] [Google Scholar]
- Ling C. et al. Design, Synthesis, and Structure–Activity Relationship Studies of Novel Thioether Pleuromutilin Derivatives as Potent Antibacterial Agents. J Med Chem 57, 4772–4795 (2014). [DOI] [PubMed] [Google Scholar]
- Dong Y.-J. et al. Synthesis of novel pleuromutilin derivatives. Part I: Preliminary studies of antituberculosis activity. Bioorg Med Chem Lett 25, 1799–1803 (2015). [DOI] [PubMed] [Google Scholar]
- Hodgin L. A. & Högenauer G. The Mode of Action of Pleuromutilin Derivatives. Eur J Biochem 47, 527–533 (1974). [DOI] [PubMed] [Google Scholar]
- Schlünzen F., Pyetan E., Fucini P., Yonath A. & Harms J. M. Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol Microbiol 54, 1287–1294 (2004). [DOI] [PubMed] [Google Scholar]
- Yan K. et al. Biochemical Characterization of the Interactions of the Novel Pleuromutilin Derivative Retapamulin with Bacterial Ribosomes. Antimicrob Agents Chemother 50, 3875–3881 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C. et al. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. P Natl Acad Sci USA 104, 4291–4296 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi T1., Tokiwano T. & Oikawa H. Studies on the later stage of the biosynthesis of pleuromutilin. Biosci. Biotechnol. Biochem. 71, 3116–3121 (2007). [DOI] [PubMed] [Google Scholar]
- Kilaru S., Collins C. M., Hartley A. J., Bailey A. M. & Foster G. D. Establishing molecular tools for genetic manipulation of the pleuromutilin-producing fungus Clitopilus passeckerianus. Appl Environ Microbiol 75, 7196–7204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahirulzaman K. A. K., Williams K. & Lazarus C. M. A toolkit for heterologous expression of metabolic pathways in Aspergillus oryzae. Methods Enzymol 517, 241–260 (2012). [DOI] [PubMed] [Google Scholar]
- Fazakerley N. J., Helm M. D. & Procter D. J. Total Synthesis of (+)-Pleuromutilin. Chem Eur J 19, 6718–6723 (2013). [DOI] [PubMed] [Google Scholar]
- Knauseder F. & Brandl E. Pleuromutilins. Fermentation, Structure and Biosynthesis. J Antibiot 29, 125–131 (1976). [DOI] [PubMed] [Google Scholar]
- Zhang S., Monahan B. J., Tkacz J. S. & Scott B. Indole-Diterpene Gene Cluster from Aspergillus flavus. Appl Environ Microbiol 70, 6875–6883 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T. et al. Cloning of a Gene Cluster Responsible for the Biosynthesis of Diterpene Aphidicolin, a Specific Inhibitor of DNA Polymerase α. Biosci. Biotechnol. Biochem 68, 146–152 (2004). [DOI] [PubMed] [Google Scholar]
- Toyomasu T. et al. Biosynthetic Gene-Based Secondary Metabolite Screening: A New Diterpene, Methyl Phomopsenonate, from the Fungus Phomopsis amygdali. The J Org Chem 74, 1541–1548 (2009). [DOI] [PubMed] [Google Scholar]
- Yao Q. Master of Science in Pharmacy (Oregon State University, 2007). [Google Scholar]
- Wang X. et al. Transgene Induced Co-Suppression during Vegetative Growth in Cryptococcus neoformans. PLos Genet 8, e1002885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurs T. A., Schaeffer E. A. M. & Wessels J. G. H. Homology-Dependent Silencing of the Sc3 Gene in Schizophyllum Commune. Genetics 147, 589–596 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R. et al. Total Biosynthesis of Diterpene Aphidicolin, a Specific Inhibitor of DNA Polymerase α: Heterologous Expression of Four Biosynthetic Genes in Aspergillus oryzae. Biosci. Biotechnol. Biochem 75, 1813–1817 (2011). [DOI] [PubMed] [Google Scholar]
- Heneghan M. N. et al. First heterologous reconstruction of a complete functional fungal biosynthetic multigene cluster. Chem Bio Chem 11, 1508–1512 (2010). [DOI] [PubMed] [Google Scholar]
- Brachmann C. B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998). [DOI] [PubMed] [Google Scholar]
- Jin F. J., Maruyama J., Juvvadi P. R., Arioka M. & Kitamoto K. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett 239, 79–85 (2004). [DOI] [PubMed] [Google Scholar]
- Porebski S., Bailey L. G. & Baum B. R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15 (1997). [Google Scholar]
- Zhang S., Monahan B. J., Tkacz J. S. & Scott B. Indole-Diterpene Gene Cluster from Aspergillus flavus. Appl Environ Microbiol 70, 6875–6883 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. & Russell D. Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor Laboratory Press, 2000). [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N. et al. The PROSITE database. Nucleic Acids Res 34, D227–D230 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28, 1102–1104 (2000). [DOI] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J. & Botstein D. Plasmid construction by homologous recombination in yeast. Gene 58, 201–2016 (1987). [DOI] [PubMed] [Google Scholar]
- Pahirulzaman K. A. K., Williams K. & Lazarus C. M. A toolkit for heterologous expression of metabolic pathways in Aspergillus oryzae. Methods Enzymol 517, 241–260 (2012). [DOI] [PubMed] [Google Scholar]
- Halo L. M. et al. Late Stage Oxidations during the Biosynthesis of the 2-Pyridone Tenellin in the Entomopathogenic Fungus Beauveria bassiana. J. Am. Chem. Soc 130, 17988–17996 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.