Significance
Correlation of expression between genes can offer useful hints regarding their function or underlying regulatory mechanism. Today, large amounts of expression data are publicly available, allowing researchers to estimate expression correlation over thousands of samples. However, extracting information from correlation data is not straightforward, because underlying expression data are generated by different laboratories working on different cell types and under different conditions. Here we present Immuno-Navigator, a database for correlation of expression in cells of the immune system, which addresses these issues. We present examples of ways our database can be used for generating hypotheses for further experimental analysis. We demonstrate how it recapitulates known facts in immunology and successfully predicts key regulators in naturally occurring regulatory T cells.
Keywords: gene expression, network inference, database, immune system, regulatory T cells
Abstract
High-throughput gene expression data are one of the primary resources for exploring complex intracellular dynamics in modern biology. The integration of large amounts of public data may allow us to examine general dynamical relationships between regulators and target genes. However, obstacles for such analyses are study-specific biases or batch effects in the original data. Here we present Immuno-Navigator, a batch-corrected gene expression and coexpression database for 24 cell types of the mouse immune system. We systematically removed batch effects from the underlying gene expression data and showed that this removal considerably improved the consistency between inferred correlations and prior knowledge. The data revealed widespread cell type-specific correlation of expression. Integrated analysis tools allow users to use this correlation of expression for the generation of hypotheses about biological networks and candidate regulators in specific cell types. We show several applications of Immuno-Navigator as examples. In one application we successfully predicted known regulators of importance in naturally occurring Treg cells from their expression correlation with a set of Treg-specific genes. For one high-scoring gene, integrin β8 (Itgb8), we confirmed an association between Itgb8 expression in forkhead box P3 (Foxp3)-positive T cells and Treg-specific epigenetic remodeling. Our results also suggest that the regulation of Treg-specific genes within Treg cells is relatively independent of Foxp3 expression, supporting recent results pointing to a Foxp3-independent component in the development of Treg cells.
High-throughput gene expression data, including microarray and next-generation sequencing data, are widely used in the study of biology. Over time, large amounts of such data have accumulated in public databases such as ArrayExpress and Gene Expression Omnibus (1, 2). In addition to their original purpose, these datasets contain an enormous potential for the study of biological networks, such as signaling pathways and regulatory interactions. For example, correlation of gene expression is widely used for the inference of regulatory networks and signaling pathways (3, 4). Publicly available data could allow researchers to base their predictions on hundreds or even thousands of samples, thus strongly increasing statistical power. Several coexpression databases have been developed, such as ATTED-II (5), COXPRESdb (6), Human Gene Correlation Analysis (HGCA) (7), and STARNET (8), which collect gene expression data and compute a measure of correlation of expression, such as Pearson correlation coefficients (PCCs), between pairs of probes or genes. Other databases and their analysis tools are also accessible (9).
It is reasonable to assume that coexpression networks and regulatory interactions differ significantly among different cell types. In cells of the hematopoietic lineage, for example, cell identities during the progress of differentiation are defined by different combinations of lineage-specific and cell type-specific receptor molecules, signaling pathways, and transcriptional regulators (10, 11). However, most existing coexpression databases do not support the analysis of cell type-specific coexpression. One notable study examined gene coexpression in a several tissues separately and showed that such a tissue-specific approach was more efficient in predicting disease genes (12). Other efforts, such as the Immunological Genome Project (ImmGen) and ImmuNet, offer data-driven approaches for studying the immune system (13, 14). However, low sample counts per cell type in the ImmGen dataset prohibit analysis of cell type-specific correlation of expression. ImmuNet integrates several types of data to infer networks but makes no distinction between cell types and focuses on well-known signaling pathways. At present, no database exists that allows integrative analysis of correlation of gene expression in a cell type-specific manner in cells of the immune system.
An additional weakness of existing coexpression databases is their lack of treatment of batch effects. Batch effects are technical sources of variation in data and are widespread in high-throughput biological data (15–17). Strong laboratory-specific effects, as well as variations associated with data processing (18), have been reported in microarray experiments (19). Batch effects are not removed by normalization (15), making the task of combining data from different studies difficult. Batch effects are expected to affect coexpression databases strongly, because they incorporate gene expression data obtained by different researchers in different laboratories using different experimental protocols and solutions and under different conditions. Nevertheless, the influence of such effects on correlation of gene expression has been scarcely studied, and to the best of our knowledge none of the above databases addresses this problem.
Here, we present Immuno-Navigator (sysimm.ifrec.osaka-u.ac.jp/immuno-navigator/), to our knowledge the first gene expression and coexpression database which addresses the two issues of cell type-specific correlation of expression and the influence of batch effects for cells of the hematopoietic lineage. Immuno-Navigator contains gene expression and expression correlation data for 24 mouse cell types of the immune system, with the use of PCC values to estimate correlation of gene expression in a cell type-specific manner. We first analyzed the influence of batch effects in different studies on estimated correlation of gene expression and attempted to remove these effects. Through genome-wide comparison of data before and after treatment of batch effects, we showed that the batch-effect reduction substantially improved the quality of the expression data and resulting correlation data (see SI Appendix, SI Results for a detailed description). Secondly, our cell type-specific expression data allowed us to find several types of correlation of gene expression, ranging from cell type-specific to widespread correlation. These findings stress the relevance of analyzing coexpression data in a cell type-specific manner.
The combination of cell type-specific correlation data with large-scale analysis functions (SI Appendix, SI Results, Practical Example Analysis Using the Immuno-Navigator Database and Fig. S1) make Immuno-Navigator a valuable resource for network inference and the generation of hypotheses regarding regulatory mechanisms and signaling pathways. We illustrate the usage of the database by a number of examples of applications. In one application we found that expression of the Treg-specific transcription factor forkhead box P3 (Foxp3) within Treg cells is not correlated with the expression of the genes bound by Foxp3. This result supports the existence of additional regulatory mechanisms that are independent of Foxp3 expression levels and that control the expression of Treg-specific genes in Treg cells (20–23). In addition, by using our own data as input to the database functions, we could successfully predict previously unidentified candidate genes of importance in Treg cells. We identified integrin β8 (Itgb8) as one of the genes with high correlation of expression with Treg-specific genes in Treg-derived samples. We experimentally confirmed an association between Itgb8 expression in Foxp3+ T cells and Treg-specific DNA demethylation of the conserved noncoding sequence 2 (CNS2) in the Foxp3 locus.
Results
Gene Expression and Correlation Data in Mouse Hematopoietic Lineage Cells.
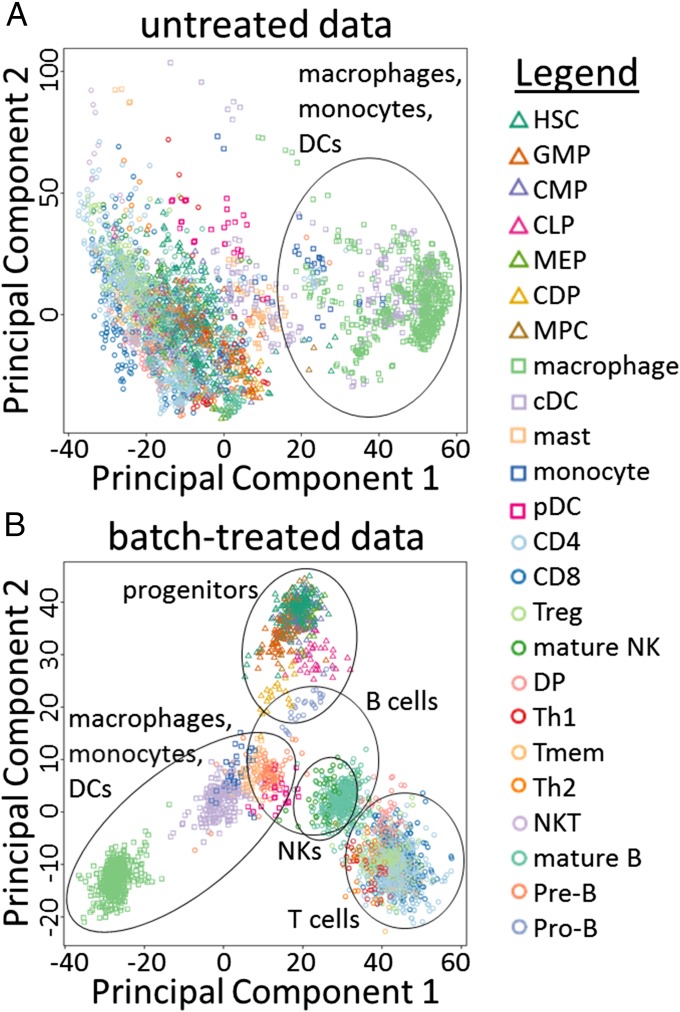
We collected and manually annotated 3,434 microarray samples for 24 mouse hematopoietic lineage cells originating from 261 studies present in ArrayExpress (Materials and Methods and SI Appendix, Fig. S2 and Table S1) (1). These data contain samples from both unstimulated cells and cells treated with various stimuli. Exploratory analysis of this data collection revealed the presence of considerable batch effects (15). One way of illustrating the presence of batch effects is by performing principal component analysis (PCA), followed by the plotting of samples marked by cell type (Fig. 1 and SI Appendix, Fig. S3). Fig. 1A shows the 3,434 samples plotted according to the principal components (PCs) of the data before batch-effect reduction. PC1 (which explains 19.0% of the total variance) is associated with cell types of the myeloid lineage, such as macrophages and dendritic cells (DCs). However, PC2 (explaining 10.8% of the variance) does not seem to be associated with any particular cell type(s). PC2 is likely to reflect some unknown source of variance or batch effect. In addition, hierarchical clustering of these untreated data resulted in strong clustering of samples according to their study of origin, another indicator for the presence of batch effects (SI Appendix, SI Results, Assessment of the Presence of Batch Effects and Fig. S4A).
Fig. 1.
Exploratory analysis of batch effects. Scatter plots are shown for all samples according to the two PCs of the gene expression data before (A) and after (B) treatment of batch effects. Shapes and colors indicate cell types (see legend). A rough indication of cell type clusters is given. △, progenitor cell types; □, myeloid cell types; ○, lymphoid cell types. Cell-type abbreviations are as in SI Appendix, Table S1.
We performed batch-effect reduction on these gene expression data using ComBat (24), treating each study as a batch (Materials and Methods). Inspection of the resulting batch-treated gene expression data suggested that this treatment strongly reduced batch effects. The first two PCs of the batch-treated samples (Fig. 1B) appear to be associated with biologically relevant variables; PC1, which explains 34.0% of the variance in the batch-treated data, divides cell types of the myeloid lineage (negative values), of the lymphoid lineage (positive values), and progenitor cells (intermediate values). PC2, explaining 14.1% of variance, is roughly associated with the degree of maturation of cells, with progenitor cells having high positive values and differentiated cell types having lower values. Similarly, PC3, explaining 8.0% of variance, appears to separate B-cell–derived samples from other samples (SI Appendix, Fig. S3). In addition, compared with the untreated data, hierarchical clustering of the batch-treated data resulted in samples being less clustered by study, another indication that batch effects have been reduced successfully (SI Appendix, Fig. S4B).
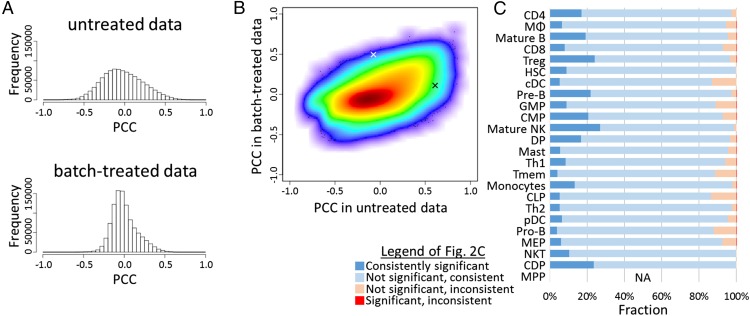
Although the gene expression data describe the behavior of each gene, correlation data reflect the biological networks underlying the relationship between any pair of genes. As a measure for correlation, we used PCCs, which were measured between all pairs of probes, for both untreated and batch-treated expression data. We found that highly (positively or negatively) correlated probe pairs, which could be caused by batch effects, were generally decreased in number after batch effect reduction (Fig. 2A; see also SI Appendix, Fig. S5 for all cell types). Although there was a general tendency for probe pairs with high correlation in the raw data to be correlated in the batch-processed data also (Fig. 2B; see also SI Appendix, SI Results, Evaluation of Batch Effect Reduction and Table S2), only a minority(12.2% on average; roughly 5–25%, depending on the cell type) of significantly correlated probe pairs was also significantly correlated after batch-effect treatment (Fig. 2C), and about 5% (roughly 1–12%) (Fig. 2C) even had a change of sign of the PCC after batch treatment. Most likely, many of these correlations in the untreated data are artifacts caused by the strong batch effects in the original data.
Fig. 2.
Treatment of batch effects strongly changes gene expression correlation. (A) The distribution of PCC values in the set of 601 macrophage-derived gene expression samples before (Upper) and after (Lower) batch-effect reduction. (B) Density scatter plot of the PCCs between all 1.0 × 109 probe pairs in data obtained from macrophages. The x and y axes represent PCC values before and after batch-effect reduction, respectively. The white and black “x” signs mark the two probe pairs shown in Fig. 3 A and B and Fig. 3 C and D, respectively. (C) For probe pairs with significant correlation in the raw data, the fraction of probe pairs that are significantly/not significantly, and consistently/inconsistently (same/different sign) correlated in the batch-treated data are shown. Cell-type abbreviations are as in SI Appendix, Table S1. For MPPs no PCC threshold could be defined.
Two Illustrative Examples of Expression Correlation Affected by Batch Effects.
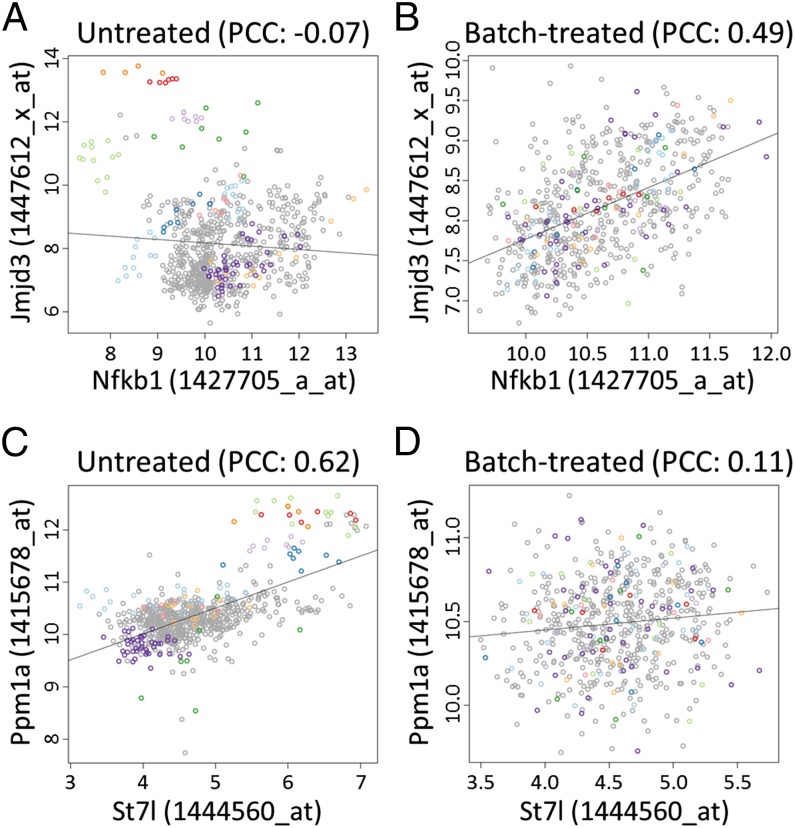
As mentioned above, batch-effect reduction using ComBat had a large impact on the correlation between many probe pairs. We will discuss the large-scale consequences of this treatment in the next section. Here, we present two anecdotal examples (Fig. 3). The histone H3K27 demethylase Jmjd3 (also referred to as “Kdm6b”) is involved in inflammatory control in macrophages (25, 26) and is directly induced by binding of the transcription factor NF-κB to a cluster of three binding sites in the Jmjd3 promoter (25). In the raw, macrophage-derived data (601 samples over 60 batches), no strong correlation was observed between Jmjd3 and Nfkb1 (PCC: −0.07) (Fig. 3A). The scatter plot of the probes for these two genes revealed that, although the samples of many studies showed a clear positive correlation, the overall correlation in the macrophage-derived samples was cancelled out by the samples of a few studies. After batch-effect reduction, the bias caused by these samples was removed, and the PCC between Jmjd3 and Nfkb1 increased to 0.49 (Fig. 3B). Furthermore, the 100 genes with the highest correlation with Jmjd3 in the batch-treated, macrophage-derived data had a clear enrichment of NF-κB–binding sites in their promoter regions (SI Appendix, Table S3). This example is only one illustration of how treatment of batch effects improved the expression correlation estimates and made them more consistent with known biological interactions. Probe pairs for which correlation was reduced after batch treatment were more common; one example is shown in Fig. 3 C and D. In the raw data (Fig. 3C), a strong positive correlation (PCC: 0.62) was observed between suppression of tumorigenicity 7-like (St7l) and magnesium-dependent protein phosphatase 1 alpha (Ppm1a) in macrophage-derived samples. However, this correlation was caused mainly by the samples of a few studies having particularly high values for both probes. Within the samples originating from a single study, no correlation was observed. After batch-effect treatment, the correlation was strongly decreased (PCC: 0.11) (Fig. 3D).
Fig. 3.
Examples of probe pairs with changes in correlation after batch-effect treatment. (A and B) Correlation between Jmjd3 and Nfkb1 in macrophage-derived samples before (A) and after (B) batch-effect reduction. This pair of probes is indicated in Fig. 2B by the white “x.” For a selection of studies, samples are indicated (different studies are represented by different colors). In the raw data, samples of several batches show correlation, but the overall correlation is neutralized by the samples originating from a few studies. After batch-effect treatment, the PCC increases strongly from −0.07 to 0.49. (C and D) Similar plots for Ppm1a and St7l in macrophage-derived samples. This pair of probes is indicated in Fig. 2B by the black “x.” In the raw data, the PCC value is highly influenced by samples originating from only a few studies, although no correlation is observed within the samples of a single study. After batch-effect reduction, the correlation strongly drops.
Treatment of Batch Effects Improves Genome-Wide Gene Correlation Estimates.
Next, we addressed the improvement in the quality of gene correlation data on a larger scale. Because a direct assessment of the accuracy of gene expression correlation is difficult, we used a number of indirect indicators to evaluate the quality of PCC values after batch-effect reduction. Please see SI Appendix, SI Results, Evaluation of Batch Effect Reduction, for a more detailed description of these analyses. In brief, after batch-effect reduction, we found an increased consistency between cell types in terms of the correlated genes that they contained. (i) Between pairs of cell types there was an increase in common correlated gene pairs (SI Appendix, Fig. S6). (ii) The number of gene pairs that had highly correlated expression in multiple cell types increased (SI Appendix, Table S4). (iii) Moreover, similarity in gene correlation between cell types was more consistent with the hematopoietic lineage tree, with relatively high similarity among progenitor cells, among lymphoid cell types, and among myeloid cell types (SI Appendix, Fig. S7). (iv) Gene pairs with similar functional annotations were more often highly correlated (SI Appendix, Fig. S8). (v) Probe pairs assigned to the same gene were more often highly correlated (SI Appendix, Figs. S9 and S10).
Together, these results indicate that treatment of batch effects improved the quality of the expression data and the resulting gene expression correlation measures.
Different Modes of Correlation of Gene Expression.
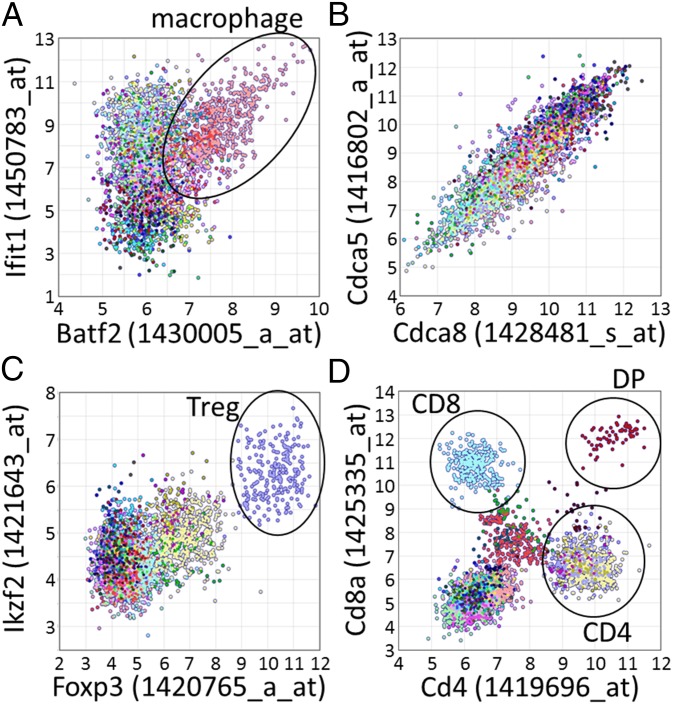
Several coexpression databases exist, but they typically calculate correlation over samples originating from a collection of different tissues and cell types. Our dataset, however, allows the analysis of expression correlation in specific cell types and comparison between cell types and the combined data of all cell types. We found that cell type-specific data contained significantly correlated gene pairs that could not be found in the combined dataset. For example, of the gene pairs that were found to be correlated significantly in one, two, and three cell types, only 2.4, 4.4, and 5.8%, respectively, were also found to be correlated in the combined data of all cell types (SI Appendix, Fig. S11). On the other hand, for 848,675 gene pairs we found significant positive correlation of expression only in the combined data and not in any of the investigated cell types. Visual inspection of such gene pairs revealed that these pairs typically consisted of gene pairs with similar cell type-specific expression. These results illustrate the existence of several modes of correlation of expression, including (i) cell type-specific correlation of expression in which two genes are correlated in only a subset of cell types and not in the combined data (Fig. 4A); (ii) widespread correlation of expression in which two genes are correlated in most cell types and also in the combined data (Fig. 4B); (iii) pairs of genes that have high (or low) expression in the same cell types but whose expression is not correlated within the samples of any individual cell type (Fig. 4C); and (iv) nonlinear relationships between two genes (Fig. 4D).
Fig. 4.
Examples of four types of gene expression correlation observed in our dataset. Each scatter plot shows the values of probes representing two genes, for all 3,434 microarray samples in our database. Different colors represent samples obtained from different cell types. (A) Batf2 (x axis) and Itif1 (y axis) are significantly correlated in a subset of cell types (macrophages, classical DCs, and plasmacytoid DCs) but not in other cell types or in the combined dataset (PCC: 0.32). As example, macrophage samples (pink; PCC: 0.62) are encircled in black. (B) Cdca8 (x axis) and Cdca5 (y axis) are significantly correlated in most cell types (typical PCCs >0.80) and also over the combined dataset (PCC: 0.90). (C) Foxp3 (x axis) and Ikzf2 (y axis) are not correlated in any cell type-specific dataset, but both show high expression in Tregs (purple, encircled in black) and, to a lesser degree, in other CD4+ T cells. As a result of their shared cell type-specific expression, they are significantly correlated in the combined dataset (PCC: 0.69). (D) Cd4 (x axis) and Cd8a (y axis). Both variables differ strongly from a normal distribution, and several distinct clusters can be observed corresponding to cell types that have high/low expression of Cd4 and/or Cd8a, such as CD4+ T cells (high Cd4 but low Cd8a), CD8+ T cells (high Cd8a but low Cd4), double-positive cells (DP, high levels of both Cd4 and Cd8a), and most other cell types (low levels of both Cd4 and Cd8a). This clearly nonlinear relationship between Cd4 and Cd8a results in high mutual information (0.47) but a relatively low PCC value (0.21).
The Immuno-Navigator Database.
We collected all gene expression data and correlation data after the batch-effect treatment in a database and constructed the Immuno-Navigator database. Our database and tools allow users to make a distinction between the modes of correlation described above. This distinction is crucial for the findings we describe further below, such as the lack of correlation between Foxp3 and Foxp3-bound target genes and the prediction of cell type-specific candidate regulators. The use of Immuno-Navigator consists of roughly four parts: (i) basic analysis of single genes; (ii) guilt-by-association analysis; (iii) correlation gene set enrichment analysis (correlation GSEA); (iv) and prediction of genes that are highly connected to a set of genes within the inferred correlation networks. To illustrate the various uses of our database, we briefly explain a few examples here. For more details, see SI Appendix, SI Results, Practical Example Analysis Using the Immuno-Navigator Database and Fig. S1 the online documentation of the Immuno-Navigator database.
Analysis of single genes.
Inspection of the correlation of expression of genes can provide valuable hypotheses regarding the function of a query gene and the regulatory pathways underlying its expression. Here we briefly illustrate the use of our data using Foxp3 as the query gene. Foxp3 has only one probe set, and its highest signals are observed in Treg cell-derived samples (SI Appendix, Fig. S1 A and B), as is consistent with its known function as a key regulator in the development and function of Tregs (27, 28). Genes with high positive or negative correlation with Foxp3 can be easily extracted for each of the cell types in our dataset (SI Appendix, Fig. S1 C–E). Other information, such as links to external databases, functional annotations, and predicted transcription factor-binding sites (TFBSs) are provided also. Visualizing the most highly correlated genes of Foxp3 within Treg cell-derived samples in a correlation network showed that Foxp3 expression was highly correlated with that of interleukin 2 receptor alpha [Il2ra (Cd25)], dystonin (Dst), and IKAROS family zinc finger 4 [Ikzf4 (Eos)] (SI Appendix, Fig. S1F). These genes, in turn, were highly correlated with other Treg markers. On the other hand, Foxp3 also had relatively high correlation with NF-κB subunit 1 (Nfkb1) and B-cell CLL/lymphoma 3 (Bcl3). Nfkb1 encodes a subunit of NF-κB, a key regulator of the response to various immune stimuli, and Bcl3 encodes a transcriptional coactivator of NF-κB. These two genes in turn were connected with Stat3, an important regulator of responses to cytokines and immune tolerance (29). Thus, the inspection of neighboring genes in the correlation network can suggest the function of the query gene and the presence of distinct regulatory modules.
Guilt-by-association analysis.
The prediction of regulatory interactions is one of the key problems in cellular biology. Although the amount of genome-wide transcription factor-binding data is increasing, it is still limited to a small subset of transcription factors in specific cell types or tissues. On the other hand, the scanning of promoter regions using position weight matrices (PWMs) to predict TFBSs remains a widely used method for finding candidate regulators but is well known to have low accuracy. In Immuno-Navigator, TFBS predictions can be further supported by the guilt-by-association principle: We make the assumption that a gene of interest is controlled by the same regulators as genes with similar expression profiles. In particular, Immuno-Navigator allows TFBS predictions for the individual query gene to be supplemented by the enrichment analysis of TFBS motifs in the set of 100 top correlated genes, in a cell type-specific way. SI Appendix, Fig. S1G shows the enriched motifs for genes that are correlated with interferon-induced protein with tetratricopeptide repeats 1 (Ifit1) in macrophage samples. Top motifs here include those of STAT and interferon-regulatory factor (IRF) transcription factor family members, which are well known to regulate expression of IFN-induced genes, including Ifit1 (30). The Ifit1 promoter itself also contains predicted sites for several STAT and IRF transcription factors. In this way, Immuno-Navigator uses motif enrichment in correlated genes to increase the confidence of predicted TFBSs in the query gene’s promoter.
In a similar way, functional annotations of a gene of interest can be inferred from the functions of its correlated genes [Gene Ontology (GO) enrichment].
Correlation GSEA.
It often is interesting to see if a gene of interest has any bias in its correlation with a set of genes that share some particular feature. We implemented correlation GSEA, an approach to detect such biases using a modification of the widely used GSEA approach (31). We applied this method in the analysis of Foxp3 and its targets in Treg cells (see Foxp3-Bound Genes Lack Correlation of Expression with Foxp3 in Treg Cells below) and on a large collection of ChIP-sequencing (ChIP-seq) data (SI Appendix, SI Results, Correlation gene set enrichment analysis).
Correlation network hub prediction.
A typical problem in molecular biology is the prediction of candidate regulators for a set of genes of interest. Under the assumption that the expression of regulators should affect the expression of downstream target genes (i.e., the input set), genes that are more frequently highly correlated with a set of input genes than with noninput genes may include genes that are relevant to their regulation. Such genes might include direct regulators (for example transcription factors) or other genes that indirectly affect the expression of the input set. Thus, genes with high correlation with many of the downstream genes could be potential candidate genes for further investigation. Although mere correlation is not enough for inferring causal relationships, additional analyses (such as TFBS motif enrichment) could reinforce such a hypothesis. Immuno-Navigator offers a tool for predicting such frequently correlated genes. We refer to this methodology as “correlation network hub prediction” (CNHP).
As a proof of concept, we applied CNHP on 345 genes with induction of expression 4 h after LPS stimulation in mouse DCs, a relatively well-studied system for which several regulators of importance are known. For a detailed description see SI Appendix, SI Results, Analysis of LPS-inducible genes in dendritic cells and Fig. S12. In brief, our analysis could successfully predict several known regulators of the response to LPS stimulation, including STAT and IRF family members and NF-κB subunits (SI Appendix, Fig. S12A). Promoter regions of the input genes were strongly enriched for binding sites for several of these transcription factors, further supporting the CNHP result (SI Appendix, Fig. S12B).
These results on a relatively well-known system show that our database and its tools could be used successfully for predicting known regulators of importance in a cell type-specific manner.
Foxp3-Bound Genes Lack Correlation of Expression with Foxp3 in Treg Cells.
Next, we used our data and analysis methods on an unresolved problem in immunology. Tregs are essential for immune homeostasis and can suppress excessive immune reactions harmful to the host. The transcription factor Foxp3 is essential for developing functional Tregs (27, 28), but it has also been shown that its expression alone is not sufficient for Treg function, stability, and lineage establishment (20, 22, 32). Nevertheless, additional necessary and sufficient factors for developing stable Tregs remain unknown.
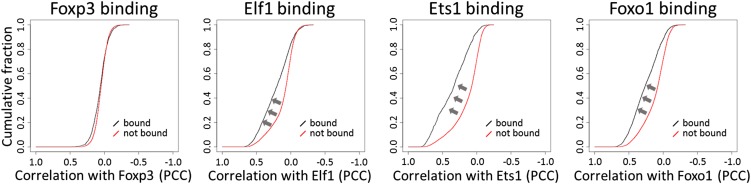
To clarify the extent to which Foxp3 controls gene expression within Treg cells, we analyzed the correlation of expression between Foxp3 and genes that are bound by Foxp3 in Tregs. Using ChIP-seq data for Foxp3 binding in Treg cells, we identified 13,879 genomic regions bound by Foxp3 and selected a set of 1,300 genes with strong Foxp3 binding in proximity of their transcription start sites. Using our correlation GSEA approach, we found that this set of Foxp3-bound genes showed a significant tendency to have correlated expression with Foxp3 over our entire expression dataset (i.e., the data for all cell types combined) (SI Appendix, Fig. S13A). This correlation of expression with Foxp3 reflects the high expression levels of many of these Foxp3-bound genes in Treg cells. However, surprisingly, this tendency was absent within the Treg-derived expression data (Fig. 5): No difference in correlation with Foxp3 expression was observed in Treg-derived expression data between Foxp3-bound and unbound genes. This lack of correlation contrasts strikingly with other transcription factors such as E74-like factor 1 (Elf1), E26 oncogene homolog 1 (Ets1), and forkhead box O1 (Foxo1), which tend to have expression in Treg cells correlated with the genes they bind to (Fig. 5 and SI Appendix, Fig. S13A). To confirm the validity of the lack of correlation between Foxp3 and Foxp3-bound genes further, we analyzed the correlation between Foxp3 and a set of Foxp3-dependent genes as defined by Gavin et al. (21). As expected, these Foxp3-dependent genes indeed showed increased correlation with Foxp3 in Treg cells (SI Appendix, SI Results, Analysis of Foxp3-dependent and -independent genes). Nevertheless, even between Foxp3 and these Foxp3-dependent genes, correlation was in general low (mostly PCC <0.4) (SI Appendix, Fig. S16A). Together, these results indicate that Foxp3 plays a critical role in controlling Treg suppressive function and also that the expression dynamics of most Foxp3-bound genes within Treg cells are relatively independent of changes in Foxp3 expression.
Fig. 5.
Lack of correlation of expression between Foxp3 and Foxp3-bound genes in Tregs. For transcription factors Foxp3, Elf1, Ets1, and Foxo1, the cumulative distribution of PCC values in the Treg-derived data are shown for genes bound by each transcription factor (black line) and genes not bound by the transcription factor (red line). For Elf1, Ets1, and Foxo1 there was a tendency for bound genes to have higher correlation (arrows) with the transcription factor in question; this tendency was not observed for Foxp3.
In a large-scale correlation GSEA of 104 ChIP-seq datasets (SI Appendix, SI Results, Correlation gene set enrichment analysis, Figs. S14 and S15, and Table S5), we made similar observations for several other regulators, including PU.1 (encoded by Sfpi1) and CCAAT/enhancer binding protein beta (C/EBPβ) in DCs. PU.1 and C/EBPβ tend to bind to genes with increased expression in DCs, but correlation between these regulators and the genes to which they bind was very low within the DC-derived samples (SI Appendix, Fig. S13 B and C). These results suggest that this pattern might be common for master regulators that prime the cell fate at the early stage of development. In contrast, Nfkb1 and Stat1 have clearly correlated expression in DCs with the target genes of the transcription factors they encode (SI Appendix, Fig. S13 B and C).
Analysis of Expression Correlation of Treg-Specific Genes Reveals Previously Unidentified Candidate Regulators of Importance in Treg Cells.
Given the lack of correlation between Foxp3 expression and Foxp3-bound genes in Tregs, and even between Foxp3 and Foxp3-dependent genes, it is reasonable to assume that other regulators are required for defining a Treg-specific transcriptome. So far, several studies have attempted to find key regulators in Treg cells by focusing on genes with highly Treg-specific expression (33) or on regulators associated with a Treg cell signature in expression data obtained from various CD4+ T cells (34). The Immuno-Navigator dataset, on the other hand, allows us to find genes that are highly correlated with Treg-specific genes specifically in Treg-derived expression data. Such genes may play a role in the definition of Treg-specific transcriptomes, especially if the correlation with Treg-specific genes is observed only within Treg cell-derived samples and not in those other cell types.
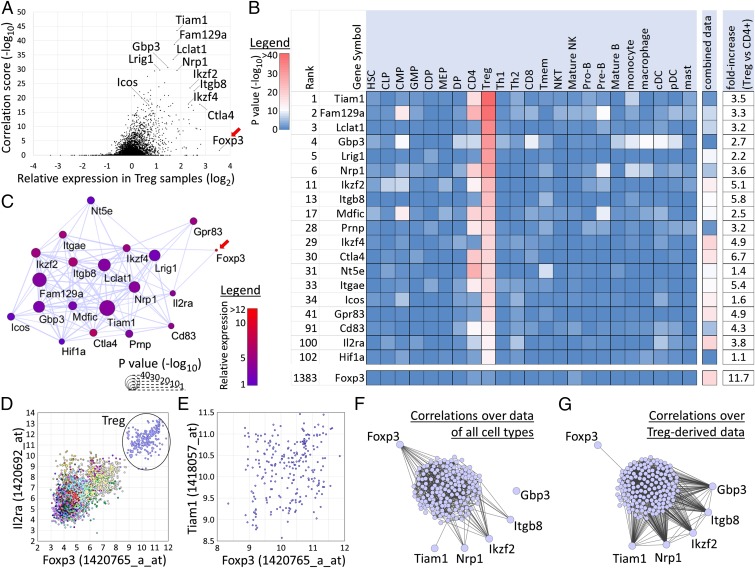
For this purpose, we examined gene expression profiles of Treg cells by RNA sequencing (RNA-seq) and defined a set of 248 Treg-specific genes as genes with higher expression in Treg cells, which are CD25+, than in unstimulated and stimulated CD25− T cells (SI Appendix, Fig. S17). We used CNHP to predict genes that have highly correlated expression specifically with these genes, especially in Treg-derived expression data. From the results we can make several observations (Fig. 6). First, several genes known to play a role in Treg functionality [IKAROS family zinc finger 2 (Ikzf2), Ikzf4, Ctla4, Icos, Il2ra] are among the top-scoring genes (Fig. 6 A and B). This category also includes genes that only recently have been shown to be of importance, such as neuropilin 1 (Nrp1) (ranked sixth out of 22,399 genes) (35) and Itgb8 (ranked 13th). The use of Itgb8 as a marker for thymically derived Tregs and its importance in Treg-mediated immunosuppression was verified experimentally and reported during the preparation of this paper (36, 37). Furthermore, top-scoring genes include multiple genes that have been reported by other studies as having high expression in Tregs, including Mdfic (ranked 17th), Prnp (ranked 28th; see also ref. 38), and Nt5e (ranked 31st) (33). In general, high-scoring genes have a relatively high expression in Treg cells compared with CD4 T cells (Fig. 6 A and B). For most of the top-scoring genes, correlation of expression with the Treg-specific genes is mainly observed only in Treg cell-derived samples, and, for some, to a lesser extent in CD4 T-cell samples, but not in the expression data of other cell types or in the combined data of all cell types (Fig. 6B).
Fig. 6.
Application of CNHP on Treg-specific genes. (A) Plot showing the correlation score in Treg-derived expression data (−log10 of P values; y axis) of all genes vs. their relative expression in Tregs vs. CD4 T cells (x axis). Genes with high correlation and/or Treg-specific expression are indicated. Foxp3 is indicated by a red arrow. (B) Table showing the top five genes with the highest enrichment score (ranked 1–5) and several genes of importance in Tregs and Treg-marker genes. Enrichment scores are shown in 23 cell types and in the combined dataset. At the right, the relative fold-increase in mean expression between Tregs and CD4+ T cells is shown (also see the x axis in A). Genes are sorted by their enrichment score in Treg-derived data (also see the y axis in A). A color code represents the score (−log10 P value): blue indicates no enrichment; red indicates high enrichment. Cell-type abbreviations are as in SI Appendix, Table S1. (C) Correlation network of the genes shown in B within Treg-derived samples. Nodes represent genes, and significantly correlated genes are connected with an edge. The size of nodes reflects their correlation score as shown in A, and their color reflects their relative mean expression in Tregs vs. CD4 T cells as in B. Foxp3 is indicated by a red arrow. (D) Scatter plot for probes representing Foxp3 and Il2ra (Cd25) over all 3,434 samples in our database. Treg-derived samples are indicated. Both genes have high expression in Treg-derived samples, resulting in high correlation (PCC: 0.81). Correlation is high within the Treg-derived samples, also (PCC: 0.49). (E) Scatter plot for probes representing Foxp3 and Tiam1 for Treg-derived samples. No significant correlation is observed (PCC: 0.22). (F) Correlation network of the Treg-signature genes, Foxp3, and five representative genes with high enrichment scores (Tiam1, Nrp1, Ikzf2, Itgb8, and Gbp3). Nodes represent genes, and edges represent significantly high correlation of expression in the combined gene expression data of all cell types. (G) The same network for the same genes as in F, with edges representing significant correlation of expression in the Treg-derived expression data.
More importantly, the top-ranked genes also include several genes for which no clear role in Treg cells has been discovered so far and which could be missed by more traditional approaches. For example, T-lymphoma invasion and metastasis-inducing protein 1 (Tiam1) is highly correlated with 45 of the input genes in Treg cells (P value: 1e-44). Although Tiam1 also has some correlation with Treg-specific genes in CD4 T-cell–derived samples, such correlation is not observed in other cell types. CNHP using as input a set of Foxp3-independent genes (21) also led to Tiam1 being the top-scoring gene (SI Appendix, SI Results, Analysis of Foxp3-dependent and -independent genes and Fig. S16D). Recently, Tiam1 has been shown to be important in the activation of LFA-1 through T-cell receptor (TCR) signaling (39), which is known to be relatively strong in Treg cells (28). Tiam1 has relatively high expression in Treg cells compared with other CD4 T cells (3.5-fold higher).
Surprisingly, Foxp3 was not present among the top genes (ranked 1,383rd) (Fig. 6B). Indeed, Foxp3 had significantly high correlation of expression in Treg cell-derived samples with only one of the Treg-specific input genes: Ikzf4. To illustrate the discrepancy between top-scoring genes and Foxp3, we created a correlation network for these genes in which pairs of genes with significantly high correlation of expression in Treg-derived samples are connected (Fig. 6C). Many of the top-scoring genes have correlated expression in Treg cells, resulting in a tightly interconnected subnetwork with Tiam1 positioned relatively centrally. On the other hand, among these genes, only Ikzf4 and Il2ra are correlated with Foxp3 in the Treg data. In particular, Foxp3 and Il2ra are correlated not only over the entire dataset (PCC: 0.81) but also within the Treg cell-derived samples only (PCC: 0.49) (Fig. 6D). Thus, the expression profile of Foxp3 is not similar to that of most of the high-scoring genes in Fig. 6B. For example, correlation between Foxp3 and Tiam1 is low within Treg cells (PCC: 0.22) (Fig. 6E).
In relation with the above, although correlation between Foxp3 and the 248 Treg-specific genes was almost absent within Treg cell-derived samples, Foxp3 was highly correlated with these genes over the expression data of all cell types combined (Fig. 6B, combined data column). This correlation of expression mainly reflects the similar Treg-specific expression of Foxp3 and many of the input genes. To clarify the different tendencies of the top-scoring genes and Foxp3 further, we compared the correlation networks over the entire dataset (combined over all cell types) (Fig. 6F) and the network over the Treg-derived samples only (Fig. 6G). Foxp3 is highly correlated with many Treg-specific genes over the entire dataset (Fig. 6F) but with only one gene in the Treg-derived data (Fig. 6G). In contrast, five representative high-scoring genes [Tiam1, guanylate-binding protein 3 (Gbp3), Nrp1, Ikzf2, and Itgb8] are frequently correlated with the Treg-specific genes within the Treg-derived data (Fig. 6G).
Taken together, these results suggest that Foxp3 plays a key role in conducting Treg suppressive functions but that the dynamics in expression of these signature genes are regulated by an additional mechanism that is independent of Foxp3 expression. Immuno-Navigator and its tools can be used to predict candidate regulators that might be involved in this regulation in a cell type-specific manner. Many of the top predicted genes are known to play an important role in Treg cells, but they also include genes for which no role in Treg cells is known at present. Such genes represent interesting candidate regulators for future investigations.
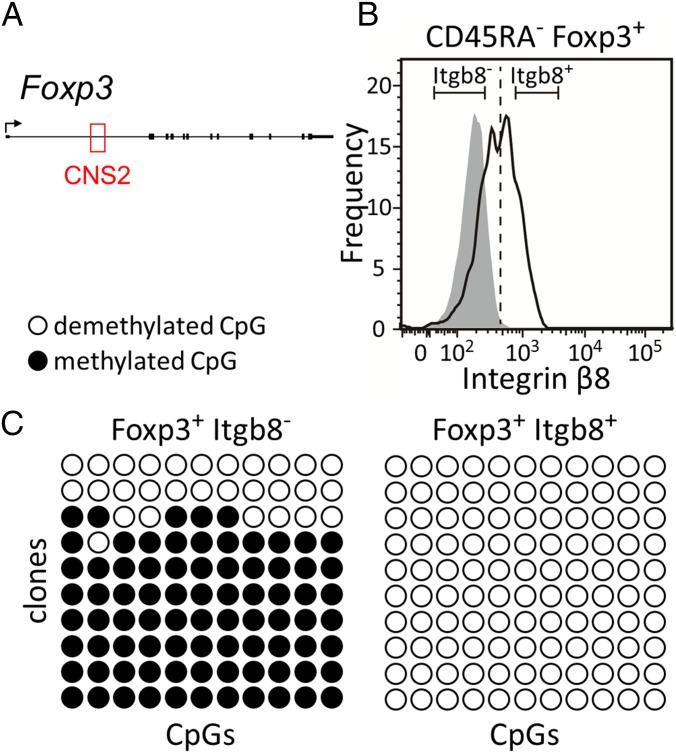
The CNS2 Region Is Demethylated in Foxp3+ Itgb8+ Human T Cells.
Previous studies have shown that functionally stable Treg cells possess a number of characteristic epigenetic features (23, 32). One is the Treg-specific DNA demethylation of the CNS2 within the first intron of the Foxp3 gene (Fig. 7A) (40). Here, for one of the top-scoring genes, Itgb8 (ranked 13th) (Fig. 7), we analyzed the DNA methylation status of CNS2 in CD45RA− Foxp3+ T cells as a function of their Itgb8 levels. Cells were sorted into two fractions, Foxp3+Itgb8− and Foxp3+Itgb8+ (Fig. 7B), and DNA methylation was evaluated using bisulfite sequencing. Although Foxp3+Itgb8− cells had mostly methylated CpG dinucleotides in CNS2, we observed that this region was demethylated in Foxp3+Itgb8+ cells (Fig. 7C). Recent studies showed that Itgb8 expression by Treg cells plays a role in controlling the release of active TGB-β1 and in suppressing inflammation (36, 37). Although the underlying molecular mechanism remains unclear, our result suggests that Itgb8-expressing Treg cells might represent functionally stable Treg cells.
Fig. 7.
Itgb8 and DNA methylation of CNS2. (A) A schematic summary of the human Foxp3 locus, with the CNS2 element indicated in red. (B) Cell sorting of CD45RA− Foxp3+ T cells according to Itgb8 levels. DNA was subsequently extracted from the Itgb8− and Itgb8+ fraction and subjected to bisulfite sequencing. (C) DNA methylation of the CNS2 region by bisulfite sequencing in Foxp3+ Itgb8− (Left) and Foxp3+ Itgb8+ (Right) T cells. Black and white circles indicate methylated and unmethylated CpG residues, respectively. Each column represents one CpG dinucleotide in the CNS2 region. These results are representative of two replicates.
Discussion
In this study, we present Immuno-Navigator, a batch-corrected gene expression and coexpression database for cells of the immune system, its underlying gene expression data and analysis tools, and several examples of applications. An important advantage of Immuno-Navigator is that our database allows us to distinguish between different modes of expression correlation easily (Fig. 3). Users can look up correlated genes and additional supporting information for a gene of interest in a cell type-specific manner. For any query gene, enrichment of regulatory motifs or functional annotation of its top correlated genes can be accessed easily in a cell type-specific manner. Correlation between regulators and sets of genes (e.g., candidate target genes) can be inspected, thus assisting the interpretation of ChIP-seq (or similar) data. Genes or regulators that have highly correlated expression with a set of input genes can be predicted, thus creating hypotheses for further experimental validation. All the expression and correlation data used in this study are available for download at sysimm.ifrec.osaka-u.ac.jp/immuno-navigator/.
In one application, we show how our data can be used to find genes that have highly correlated expression with a set of Treg-specific genes. Top-scoring genes contained not only many known regulators of importance but also several genes that thus far have not been reported to play a role in defining Tregs. An important point is that for many of the top-scoring genes, correlation of expression with the input genes was observed predominantly in Treg-derived samples only and not in other cell types. Indeed, the combination of cell type-specific data and our analysis tools allows users to distinguish easily between highly cell type-restricted correlation (such as Treg-restricted correlation) and more widespread correlation of expression. Genes that show high correlation of expression with Treg-specific genes only in Treg-derived samples may present attractive candidates for identifying key regulators of Treg development. An additional important point is the Foxp3-independence of the Treg-associated genes. In our analysis we found a surprising lack of correlation of expression between Foxp3 and Foxp3-bound genes in Treg-derived data. This finding was supported by similar observations for a number of other regulators. Although Foxp3 has been reported to be a key regulator for Treg development, it also has been shown that Foxp3 overexpression in conventional T cells could not recapitulate the whole gene expression profiles of Tregs (22) and that Foxp3-null Tregs obtained from Foxp3gfpko mice express Treg signature genes (21). In addition, we previously demonstrated that Foxp3-binding genes are correlated with the repressed genes after TCR stimulation in Tregs but not with the Treg signature genes (23). These observations are consistent with our findings and suggest that Foxp3 does not function as an initiator for Treg development itself. We hope that Immuno-Navigator provides genuine candidates for inducing Treg signature gene expression and thus initiating Treg development. As one example, we found that the expression of one of the top-scoring genes in Foxp3+ T cells, Itgb8, was associated with Treg-specific DNA demethylation in the CNS2 region. Although the molecular mechanisms remain unclear, our findings might indicate that Itgb8-dependent signaling plays a role in the establishment of Treg-specific epigenetic modifications during Treg development, before the induction of Foxp3 expression.
The analysis of large-scale biological data can be severely hampered by insufficient consideration of the underlying biological (cell types, stimuli, and other) and experimental (batches, platforms, and other) variables. Batch effects have been reported to be widespread in biological data (15), but such effects are often ignored. The biases caused by batch effects in gene coexpression data have not been studied thoroughly, and thus far, existing gene coexpression databases have paid no attention to them. This lack of attention to batch effects is particularly dangerous, because our results show that batch effects tend to increase the overall correlation observed in a dataset and, naturally, have a large impact on the resulting inferred coexpression networks. On the level of pairs of genes, batch effects can strongly affect correlations. As shown in this study, in extreme cases, negatively correlated genes can become significantly positively correlated after batch-effect treatment, and vice versa. Discrepancies that have been observed among existing databases of coexpression (9) might be explained, at least in part, by such batch effects.
Although the present study focuses on gene expression data originating from cells of the immune system, our approach (SI Appendix, SI Results, General Data Analysis Approach and Fig. S22) is generally applicable, regardless of species or biological system. Cell type-specific regulatory interactions are of key importance in immunology. We therefore explicitly treated data in a cell type-specific manner, used this prior knowledge to guide batch-effect reduction, and implemented analysis and visualization tools allowing easy comparison between cell types. Only this approach allowed us to predict Treg-specific candidate regulators. Nevertheless, this approach could easily be applied to other biological variables, e.g., types of tumor cells. Regarding experimental variables, we presented several genome-wide analyses indicating that batch treatment improves the quality of inferred PCC values. Among these quality indicators, only the consistency with the hematopoietic lineage tree is immune-specific. In applications on other systems, alternative indicators can be used that reflect generally accepted prior knowledge of the system of interest. Furthermore, recent advances in batch-effect treatment methodology are expected to make this general approach more widely applicable, including application on other omics data (16, 41).
Finally, it is widely known that correlation alone does not imply a causal relationship. In addition, coexpression should not be confused with coregulation, and it should be kept in mind that our reported correlations reflect dynamics only on the mRNA level. Gene activity is widely accepted to be regulated at many other levels, including posttranslational modifications, which are independent of mRNA concentration. Conversely, a lack of correlation does not necessarily imply independence. Even small changes in mRNA can be functionally relevant, and unknown biases in the incorporated gene expression data might obscure existing interactions. Nevertheless, when supported by additional analyses and careful interpretation, we believe that our database can help both computational and experimental studies of gene regulation and signaling pathways in the immune system. Furthermore, in the case of Immuno-Navigator, matching the results with the known hematopoietic lineage tree can provide further insights.
Materials and Methods
A complete discussion of all methods, including correlation GSEA, RNA-seq, ChIP-seq, DNA methylation analysis, and supporting computational analysis is included in SI Appendix, SI Materials and Methods. Additional information in Immuno-Navigator includes predicted TFBSs and GO annotation terms for each gene. In addition, for each gene, enriched TFBSs and GO terms for the top 100 most highly correlated genes in each cell type have been precalculated and are available in the database. Please refer to SI Appendix, SI Materials and Methods for a more detailed description of these features.
Microarray Data Collection.
Microarray datasets were downloaded from ArrayExpress (1). The database was searched for samples obtained from several cell types of the hematopoietic lineage. To facilitate downstream analyses, we focused on samples processed by the Affymetrix GeneChip Mouse Genome 430 2.0 platform, the most frequently used platform for mouse samples in ArrayExpress. We manually assigned a cell type to each sample using the annotation data provided in the Sample and Data Relationship Format (SDRF) files in ArrayExpress. The 3,881 samples (for 38 different cell types) obtained by this method were normalized together using the Robust Multiarray Average (RMA) method (42) using the “affy” package in R (43).
Treatment of Batch Effects.
We used the ComBat method in the R package “sva” to reduce batch effects (24, 44). ComBat adjusts for batch effects using parametric and nonparametric empirical Bayes frameworks in datasets in which batch covariates are known. Here, by default, we treated studies and publications as proxies for batches. In practice, we used ArrayExpress accession numbers to designate each study or paper. Six studies contained more than 50 samples; one (ArrayExpress accession number E-TABM-310) contained 216 samples. Annotation data indicated that the samples of the E-TABM-310 study were taken over several years. Therefore we used the dates in the annotation data to subdivide this dataset further into 26 subbatches, 15 of which contained at least five samples. However, to the best of our knowledge, no clear date information was present for other large studies, and therefore each was treated as a single batch. We excluded studies (batches) with fewer than five samples and samples for cell types with fewer than 20 samples. Using the cell-type annotation and batch information of all samples as input for ComBat, we obtained batch-corrected expression data. The final data contained 3,434 samples from 261 studies covering 24 cell types (SI Appendix, Table S1).
Correlation of Gene Expression.
The PCC was used as measure for similarity of expression. For all probe pairs the PCC was calculated over the data obtained for each of the 24 cell types separately and over all combined data. All PCC values (roughly 2.5 × 1010 values for all probe pairs, over 24 cell types and the combined dataset) and associated scatter plots are available in our database. All PCC values are available for download from Immuno-Navigator.
Definition of Significantly Correlated Probe Pairs and Gene Pairs.
Probe pairs with an absolute PCC value exceeding a PCC threshold were defined as being significantly correlated. In the batch-treated gene expression data, the PCC thresholds were decided as follows: all expression data were shuffled, and PCCs for all probe pairs over the shuffled data were calculated for each cell type separately and for all of the combined data. To limit the influence of remaining batch effects, the shuffling was done in a way that would preserve remaining batch effects even in the shuffled data (SI Appendix, SI Materials and Methods, Construction of Shuffled Data and Fig. S18). Next, the distributions of PCC values in the true and shuffled batch-treated data were compared, and false-discovery rates (FDRs) were calculated for each absolute PCC value between 0 and 1 in steps of 1e-6. A PCC threshold then was decided for each cell type and for all combined data, based on the following conditions: (i) the PCC threshold should be at least 0.4; (ii) the corresponding FDR should be at most 0.01; (iii) at least 1 million probe pairs should exceed the PCC threshold in the true data; and (iv) at most 6 million probe pairs should exceed the PCC threshold in the true data. The purpose of these conditions is to set a threshold for each cell type that meets at least a certain level of relevance (in terms of absolute PCC values) and reliability (in terms of FDR) and simultaneously to attempt to obtain similar amounts of significantly correlated probe pairs in each dataset (conditions iii and iv). Using the above conditions, a PCC threshold could be defined for 23 of 24 of the cell types (SI Appendix, Table S6). The one cell type for which no threshold could be set was the dataset for multipotent progenitor cells, which was the smallest dataset in our data (20 samples). In general, the PCC threshold increases with decreasing sample counts.
For each cell type, and for the combined data, we thus obtained a set of significantly correlated probe pairs. Finally, using the gene-to-probe annotation of the microarray platform, we converted these probe pairs to significantly correlated gene pairs. Two genes, A and B, are defined as significantly correlated if at least one probe of gene A is significantly correlated with at least one probe of gene B.
Evaluation of the Influence of Batch-Effect Reduction on Gene Correlation.
To facilitate the comparison of overlap in significantly correlated gene pairs between cell types and between untreated and batch-treated data, we focused only on significantly positively correlated gene pairs. Significantly correlated gene pairs in the batch-treated data were defined using the PCC thresholds described above. For the untreated data, to avoid biases in the comparison with the treated data, the same number of gene pairs with the highest correlation was regarded as significantly correlated for each cell type.
For hierarchical clustering of cell types by similarity in PCC values, we randomly selected 1 million microarray probe pairs and for each cell type collected the corresponding PCC values. These PCC values were used to cluster cell types using hierarchical clustering, using as distance function 1 minus the correlation in PCC values. We performed hierarchical clustering for both for the untreated and the batch-treated data.
For comparison of PCC values among genes with shared functional annotations, GO basic and GO slim (release date January 11, 2015) and mouse annotations (GO Consortium validation January 9, 2015) were downloaded from the Gene Ontology Consortium website (45). More details of the analysis are given in SI Appendix, SI Results, Evaluation of Batch Effect Reduction.
For the comparison between probe sets mapped to the same gene (same-gene probes) and probe sets mapped to different genes (different-gene probes), the PCC was calculated between all pairs of probes representing the same gene (35,164 probe pairs representing 10,556 genes with multiple probes) and between the same number of probe pairs representing randomly selected different genes. Differences in the distribution of PCC values between the same-gene probes and the different-gene probes were measured using the area under the curve of receiver operating characteristic curves.
Prediction of Genes with Frequent High Correlation to an Input Set of Genes.
Given a set S of input genes, we obtain genes that are frequently correlated specifically with the genes in set S as follows. For each gene g, let CS,g be the count of significantly correlated genes in set S and let CG,g be the count of significantly correlated genes in the genome-wide set of genes. The probability p of observing CS,g or a higher number of significant correlations between g and the |S| genes in set S given CG,g can be calculated using the binomial distribution. In case g is also an element of S, we use |S| − 1 to reflect the fact that correlation of g with itself is not considered. Immuno-Navigator allows CNHP on a set of genes of interest over all cell types and the combined data, simultaneously. Typical run times are less than 1 min for all analyses. We describe the application of this analysis on randomly selected sets of genes in SI Appendix, SI Materials and Methods, Application of Correlation Network Hub Prediction on Random Sets of Genes and Fig. S19, and the analysis of its robustness to noise in SI Appendix, SI Materials and Methods, Robustness of Results of Correlation Network Hub Prediction and Figs. S20 and S21.
Supplementary Material
Acknowledgments
We thank the members of the Immunology Frontier Research Center (IFReC) Laboratory of Experimental Immunology, Laboratory of Systems Immunology, and Quantitative Immunology Research Unit for helpful discussions and advice and M. Ogawa for secretarial assistance. Computational time was provided by the computer cluster of the IFReC Laboratory of Systems Immunology. This work was supported by the World Premier International Research Center Initiative of the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), Grant-in-Aid for Scientific Research 26253030 from MEXT, Core Research for Evolutional Science and Technology Grant-in-Aid 15652235 from the Japan Science and Technology Agency (to S.S.), and Grant-in-Aid for Scientific Research B 15H04744 from the Japan Society for the Promotion of Science (to N.O.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the DNA Database of Japan (accession no. DRA004105).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604351113/-/DCSupplemental.
References
- 1.Kolesnikov N, et al. ArrayExpress update--simplifying data submissions. Nucleic Acids Res. 2015;43(Database issue):D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T, et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marbach D, et al. DREAM5 Consortium Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9(8):796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Smet R, Marchal K. Advantages and limitations of current network inference methods. Nat Rev Microbiol. 2010;8(10):717–729. doi: 10.1038/nrmicro2419. [DOI] [PubMed] [Google Scholar]
- 5.Obayashi T, et al. ATTED-II in 2014: Evaluation of gene coexpression in agriculturally important plants. Plant Cell Physiol. 2014;55(1):e6. doi: 10.1093/pcp/pct178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamura Y, et al. COXPRESdb in 2015: Coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic Acids Res. 2015;43(Database issue):D82–D86. doi: 10.1093/nar/gku1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michalopoulos I, et al. Human gene correlation analysis (HGCA): A tool for the identification of transcriptionally co-expressed genes. BMC Res Notes. 2012;5(1):265. doi: 10.1186/1756-0500-5-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jupiter D, Chen H, VanBuren V. STARNET 2: A web-based tool for accelerating discovery of gene regulatory networks using microarray co-expression data. BMC Bioinformatics. 2009;10:332. doi: 10.1186/1471-2105-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Usadel B, et al. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ. 2009;32(12):1633–1651. doi: 10.1111/j.1365-3040.2009.02040.x. [DOI] [PubMed] [Google Scholar]
- 10.Kondo M. Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. Immunol Rev. 2010;238(1):37–46. doi: 10.1111/j.1600-065X.2010.00963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26(6):726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Piro RM, et al. An atlas of tissue-specific conserved coexpression for functional annotation and disease gene prediction. Eur J Hum Genet. 2011;19(11):1173–1180. doi: 10.1038/ejhg.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jojic V, et al. Immunological Genome Project Consortium Identification of transcriptional regulators in the mouse immune system. Nat Immunol. 2013;14(6):633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorenshteyn D, et al. Interactive Big Data Resource to Elucidate Human Immune Pathways and Diseases. Immunity. 2015;43(3):605–614. doi: 10.1016/j.immuni.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11(10):733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leek JT. svaseq: Removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014;42(21):1–9. doi: 10.1093/nar/gku864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilad Y, Mizrahi-Man O. A reanalysis of mouse ENCODE comparative gene expression data. F1000 Res. 2015;4(121):121. doi: 10.12688/f1000research.6536.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherer A. In: Batch Effects and Noise in Microarray Experiments: Sources and Solutions. Scherer A, editor. John Wiley and Sons; Chichester, UK: 2009. [Google Scholar]
- 19.Irizarry RA, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2(5):345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 20.Sugimoto N, et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol. 2006;18(8):1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 21.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445(7129):771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 22.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27(5):786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Morikawa H, et al. FANTOM Consortium Differential roles of epigenetic changes and Foxp3 expression in regulatory T cell-specific transcriptional regulation. Proc Natl Acad Sci USA. 2014;111(14):5289–5294. doi: 10.1073/pnas.1312717110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 25.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 26.De Santa F, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28(21):3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hori S, Nomura T, Sakaguchi S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science. 2003;299(5609):1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 28.Josefowicz SZ, Lu L-F, Rudensky AY. Regulatory T cells: Mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pallandre J-R, et al. Role of STAT3 in CD4+CD25+FOXP3+ regulatory lymphocyte generation: Implications in graft-versus-host disease and antitumor immunity. J Immunol. 2007;179(11):7593–7604. doi: 10.4049/jimmunol.179.11.7593. [DOI] [PubMed] [Google Scholar]
- 30.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 Expression are independent and complementary events required for Treg cell development. Immunity. 2012;37(5):785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: Differentiation, specification, subphenotypes. Nat Immunol. 2009;10(7):689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- 34.Fu W, et al. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat Immunol. 2012;13(10):972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delgoffe GM, et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013;501(7466):252–256. doi: 10.1038/nature12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards JP, Thornton AM, Shevach EM. Release of active TGF-β1 from the latent TGF-β1/GARP complex on T regulatory cells is mediated by integrin β8. J Immunol. 2014;193(6):2843–2849. doi: 10.4049/jimmunol.1401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worthington JJ, et al. Integrin αvβ8-Mediated TGF-β Activation by Effector Regulatory T Cells Is Essential for Suppression of T-Cell-Mediated Inflammation. Immunity. 2015;42(5):903–915. doi: 10.1016/j.immuni.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isaacs JD, et al. The cellular prion protein is preferentially expressed by CD4+ CD25+ Foxp3+ regulatory T cells. Immunology. 2008;125(3):313–319. doi: 10.1111/j.1365-2567.2008.02853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grönholm M, et al. TCR-induced activation of LFA-1 involves signaling through Tiam1. J Immunol. 2011;187(7):3613–3619. doi: 10.4049/jimmunol.1100704. [DOI] [PubMed] [Google Scholar]
- 40.Baron U, et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37(9):2378–2389. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 41.Hicks SC, Teng M, Irizarry RA. On the widespread and critical impact of systematic bias and batch effects in single-cell RNA-Seq data. bioRxiv. 2015 doi: 10.1101/025528. [DOI] [Google Scholar]
- 42.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 43.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy: Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 44.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3(9):1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.