Abstract
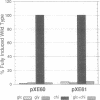
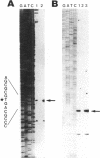
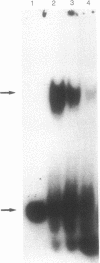
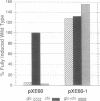
We report the identification and partial characterization of the promoters for two chitinase genes from Streptomyces plicatus. Chitinases are a family of enzymes made by Streptomyces and other soil microbes to digest chitin, an abundant source of carbon and nitrogen in the soil. The promoter regions of two chitinases were defined by using transcriptional fusions to the xylE reporter gene. Transcription was shown to be glucose-sensitive and chitin-dependent. Each promoter contains a putative RNA polymerase binding site with a recognition sequence very similar to that observed in many eubacterial vegetatively expressed genes. In both promoters, a pair of 12-base-pair direct repeat sequences overlap the putative RNA polymerase binding sites. Further analysis of one of the promoters revealed that a single-base change within the direct repeat sequences resulted in glucose-resistant, chitin-independent expression in vivo. In addition, the promoter region that includes the direct repeat sequences was shown to interact with a sequence-specific DNA binding factor in vitro. Similar direct repeat sequences are present in other chitinase genes recently characterized, and we suggest that these repeats may be involved in repression and induction for this entire class of catabolite-controlled genes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brunelle A., Schleif R. Determining residue-base interactions between AraC protein and araI DNA. J Mol Biol. 1989 Oct 20;209(4):607–622. doi: 10.1016/0022-2836(89)90598-6. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Guarente L., Sharp P. A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989 Feb 24;56(4):549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- Buttner M J, Fearnley I M, Bibb M J. The agarase gene (dagA) of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis. Mol Gen Genet. 1987 Aug;209(1):101–109. doi: 10.1007/BF00329843. [DOI] [PubMed] [Google Scholar]
- Buttner M. J. RNA polymerase heterogeneity in Streptomyces coelicolor A3(2). Mol Microbiol. 1989 Nov;3(11):1653–1659. doi: 10.1111/j.1365-2958.1989.tb00151.x. [DOI] [PubMed] [Google Scholar]
- Buttner M. J., Smith A. M., Bibb M. J. At least three different RNA polymerase holoenzymes direct transcription of the agarase gene (dagA) of Streptomyces coelicolor A3(2). Cell. 1988 Feb 26;52(4):599–607. doi: 10.1016/0092-8674(88)90472-2. [DOI] [PubMed] [Google Scholar]
- Fornwald J. A., Schmidt F. J., Adams C. W., Rosenberg M., Brawner M. E. Two promoters, one inducible and one constitutive, control transcription of the Streptomyces lividans galactose operon. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2130–2134. doi: 10.1073/pnas.84.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Seno E. T., Bruton C. J., Chater K. F. Genetic mapping, cloning and physiological aspects of the glucose kinase gene of Streptomyces coelicolor. Mol Gen Genet. 1984;196(3):501–507. doi: 10.1007/BF00436199. [DOI] [PubMed] [Google Scholar]
- Ingram C., Brawner M., Youngman P., Westpheling J. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J Bacteriol. 1989 Dec;171(12):6617–6624. doi: 10.1128/jb.171.12.6617-6624.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E. C. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- Robbins P. W., Albright C., Benfield B. Cloning and expression of a Streptomyces plicatus chitinase (chitinase-63) in Escherichia coli. J Biol Chem. 1988 Jan 5;263(1):443–447. [PubMed] [Google Scholar]
- Smith C. P., Chater K. F. Cloning and transcription analysis of the entire glycerol utilization (gylABX) operon of Streptomyces coelicolor A3(2) and identification of a closely associated transcription unit. Mol Gen Genet. 1988 Jan;211(1):129–137. doi: 10.1007/BF00338403. [DOI] [PubMed] [Google Scholar]
- Vidal-Ingigliardi D., Richet E., Raibaud O. Two MalT binding sites in direct repeat. A structural motif involved in the activation of all the promoters of the maltose regulons in Escherichia coli and Klebsiella pneumoniae. J Mol Biol. 1991 Mar 20;218(2):323–334. doi: 10.1016/0022-2836(91)90715-i. [DOI] [PubMed] [Google Scholar]
- Virolle M. J., Bibb M. J. Cloning, characterization and regulation of an alpha-amylase gene from Streptomyces limosus. Mol Microbiol. 1988 Mar;2(2):197–208. doi: 10.1111/j.1365-2958.1988.tb00021.x. [DOI] [PubMed] [Google Scholar]
- Westpheling J., Ranes M., Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985 Jan 3;313(5997):22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]