Summary
How the genomic landscape of a tumor shapes and is shaped by anti-tumor immunity has not been systematically explored. Using large-scale genomic datasets of solid tissue tumor biopsies, we quantified the cytolytic activity of the local immune infiltrate and identified associated properties across 18 tumor types. The number of predicted MHC Class I-associated neoantigens was correlated with cytolytic activity and was lower than expected in colorectal and other tumors, suggesting immune-mediated elimination. We identified recurrently mutated genes that showed positive association with cytolytic activity, including beta-2-microglobulin (B2M), HLA-A, -B and -C and Caspase 8 (CASP8), highlighting loss of antigen presentation and blockade of extrinsic apoptosis as key strategies of resistance to cytolytic activity. Genetic amplifications were also associated with high cytolytic activity, including immunosuppressive factors such as PDL1/2 and ALOX12B/15B. Our genetic findings thus provide evidence for immunoediting in tumors and uncover mechanisms of tumor-intrinsic resistance to cytolytic activity.
Introduction
With the recent success of checkpoint blockade therapy (against CTLA4 or PD1/PDL1) in inducing durable control of diverse tumors (Hinrichs and Rosenberg, 2014; Sharma et al., 2011), it has become critical to understand why some tumors are not responsive or develop resistance to therapy. Predictors of outcome have been identified in the context of spontaneous tumor immunity (Bindea et al., 2013) and immunotherapy (Herbst et al., 2014; Tumeh et al., 2014; Ji et al., 2012; Snyder et al., 2014) but there remains a need for more extensive mechanistic analyses of human tumor-immune interactions.
High-dimensional datasets – such as The Cancer Genome Atlas (TCGA) that include genome-wide DNA sequencing, RNA sequencing and copy number profiles – have made it possible to dissect the factors driving malignancy with unprecedented depth. Recent analyses of TCGA datasets have linked the genomic landscape of tumors with tumor immunity, implicating neoantigen load in driving T cell responses (Brown et al., 2014) and identifying somatic mutations associated with immune infiltrates (Rutledge et al., 2013). Here, we build on this work and use the extensive TCGA dataset to infer how tumors induce and adapt to immune responses.
To identify and characterize the correlates of anti-tumor immunity, we devised an RNA-based metric of immune cytolytic activity and calculated it for thousands of TCGA solid tumor samples. By correlating this metric with genetic and non-genetic findings from TCGA data, we set out to address several questions, including: does cytolytic activity vary across tumor types? Does the load of predicted neoantigens or viruses in a tumor explain the cytolytic activity? Are particular recurrent mutations in tumors associated with higher cytolytic activity, reflecting mechanisms of tumor escape from cytolytic immune activities?
Results
A metric for immune cytolytic activity based on gene expression in TCGA tumors
To study immune effector activity in solid tumors, we focused on cytotoxic T cells (CTL) and natural killer cells (NK) because of their potent ability to kill tumor cells and numerous studies showing that effector T cells at the tumor site predict favorable outcome across many cancers (Pages et al., 2005; Sato et al., 2005; Schumacher et al., 2001). Using RNA-Seq data from thousands of TCGA solid tumor biopsies, we devised a simple and quantitative measure of immune cytolytic activity (‘CYT’) based on transcript levels of two key cytolytic effectors, granzyme A (GZMA) and perforin (PRF1), which are dramatically upregulated upon CD8+ T cell activation (Johnson et al., 2003) and during productive clinical responses to anti-CTLA-4 and anti-PD-L1 immunotherapies (Ji et al., 2012) (Herbst et al., 2014). Consistent with their coordinated roles, GZMA and PRF1 were tightly co-expressed in TCGA samples (Data S1A, Data S1B) and showed CTL-specific expression in panels of human cell types (Data S1C, Data S1D), thus serving as highly specific markers in heterogeneous tumor samples.
We found that the levels of cytolytic activity were highest in kidney clear cell carcinomas and cervical cancers, lowest in glioma and prostate cancers, and average (albeit skewed to high levels) in melanoma (Figure 1A; Table S1A, B). Most normal tissues (from TCGA or the Genotype-Tissue Expression (GTEx) project (GTEx Consortium, 2013a)) showed definitively lower (6 tissues) or equal (7 tissues) cytolytic activity compared to their corresponding tumors, but two showed definitively higher activity (lung and colon). Of note, CYT in colorectal tumors increased considerably given high microsatellite instability (MSI) (Data S1E) (Schwitalle et al., 2008). The differences in cytolytic activities across tumor types and compared to normal tissues are likely to reflect a combination of tissue- and tumor-specific mechanisms that regulate local immunity.
Figure 1. Immune cytolytic activity (CYT) varies across tumor types and is associated with suppressive factors.
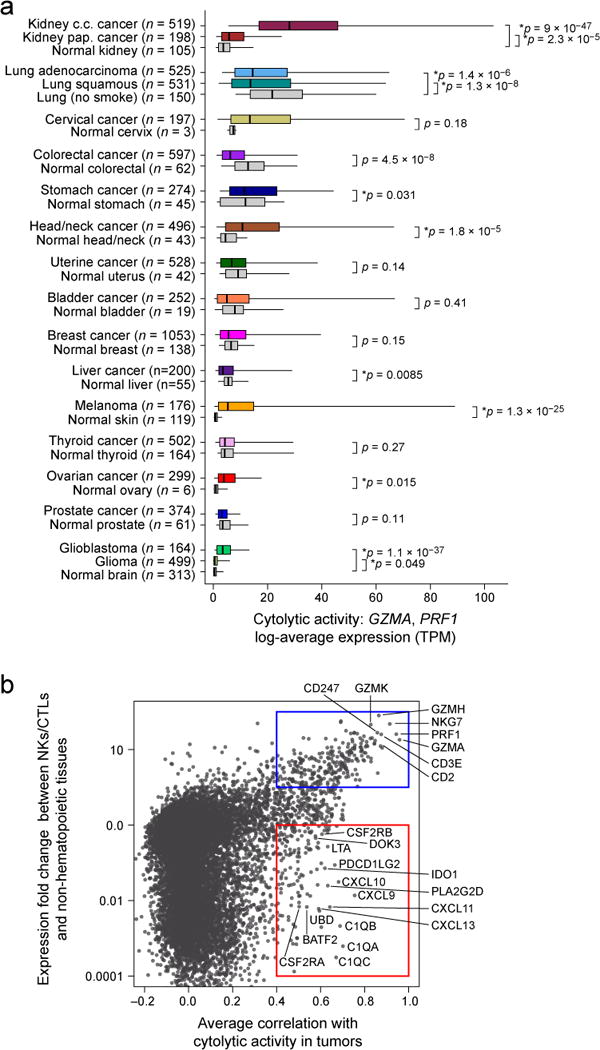
(A) Cytolytic activity (CYT), defined as the log-average (geometric mean) of GZMA and PRF1 expression in transcripts per million (TPM), is shown for each of 18 TCGA tumor types and normal tissues. Normal tissue samples include TCGA controls and GTEx samples, excluding smokers for lung tissues. Boxes in box plot represent interquartile ranges and vertical lines represent 5th–95th percentile ranges, with a notch for the median. P-values are unadjusted and calculated by Wilcoxon rank-sum test (comparison to relevant normal), and asterisks denote events significant at 10% FDR. (B) The correlation of a gene with CYT across all tumor types is shown (X-axis) relative to its relative expression in CTL/NK cells. Top right, genes expressed in CTL/NK cells that are associated with CYT. Bottom right, non-CTL/NK genes associated with CYT. Average Spearman correlation of expression with CYT was calculated across 18 tumor types. Y-axis: for each gene, median expression in NKs and CTLs divided by median expression in non-hematopoietic cells using CAGE data from Fantom5. See also Data S1 and Table S1.
Cytolytic activity is associated with counter-regulatory immune responses and improved prognosis
To determine whether cytolytic activity is associated with other immune cell types and functions, we calculated the enrichment of 15 immune cell type and function gene sets in the same samples (Table S1C; expression data from Fantom5 project (Fantom Consortium et al., 2014)). While CYT showed moderate correlation with B cells and weak correlation with macrophages, it showed strong correlation with: (i) CTL markers, as expected; (ii) plasmacytoid dendritic cells; (iii) counter-regulatory Tregs and known T-cell co-inhibitory receptors, as seen in chronic inflammatory conditions (Data S1F) (Lund et al., 2008). We note that expression of the pre-defined gene sets was similarly enriched in most tumor and normal tissues, with some notable differences (Data S1G), and not typically connected to tumor stage (Data S1H, Data S1I). Finally, when we looked for CYT correlations with any transcript (filtering out CTL and NK genes), we found that CYT was best correlated with immunosuppressive factors (Spranger et al., 2013), such as PDCD1LG2 (PDL2), IDO1/2, DOK3 (Lemay et al., 2000), GMCSF receptor (CSF2RA, CSF2RB) and the C1Q complex (Figure 1B). In addition, it was also associated with interferon-stimulated chemokines (CXCL9, CLCL10, and CXCL11) that attract T cells, as observed previously (Bindea et al., 2013). We conclude that tumors can differ dramatically in their infiltrate levels and composition, and that cytolytic activity is associated with counter-regulatory activities that limit the immune response.
When we used CYT and these other metrics to identify predictors of survival (controlling for tumor histology and stage), we found that high-CYT (and other T cell markers) is associated with a modest but significant pan-cancer survival benefit (Data S1J). While no individual immune cell type metrics were associated with poorer prognosis, higher expression of macrophage markers relative to other markers was consistently linked with poor prognosis, while higher expression of CYT or CTL markers was correlated with improved prognosis (Data S1J).
Tumor cytolytic activity is associated with oncogenic viruses in some tumors
Viruses account for a subset of malignancies and are also known to activate high affinity antigen-specific CTLs against non-self viral antigens. Thus, we tested for correlation of cytolytic activity levels with transcripts from oncogenic viruses – including Epstein Barr virus (EBV), hepatitis B and C (HBV and HCV), human papilloma virus (HPV), Kaposi sarcoma virus (KSV), and polyoma viruses (Table S2A).
Consistent with previous analysis of TCGA data (Tang et al., 2013), HPV infection was most abundant in cervical cancer (91%), but also frequent in head and neck cancer (12%; with more men than women, OR=4.9; p=8.5e-4) and bladder cancer (2%). We also observed occasional cases in colorectal, kidney clear cell, glioma, lung squamous cell carcinoma, and uterine cancer (Figure 2A). Only stomach cancer demonstrated definitive instances of EBV infection (8%; Table S2A), which was associated with high expression of specific EBV genes EBER-1 and RPMS1 (Data S2A). Asian patients, known to exhibit increased rates of stomach cancer (Jemal et al., 2007), were not more likely than other stomach cancer patients to harbor EBV (p=0.63). Consistent with a role for viral infection in the induction of CTLs, >2-fold increases in cytolytic activity were observed in EBV+ vs. EBV− stomach cancers and HPV+ vs. HPV− head and neck cancers, bladder cancers, uterine cancers and possibly cervical cancers (Figure 2B). Strikingly, all the gene sets that were most tightly associated with EBV infection in stomach cancer related to T cell activation (Table S2B).
Figure 2. Viral infection is tumor-specific and associated with higher CYT in a subset of tumor types.
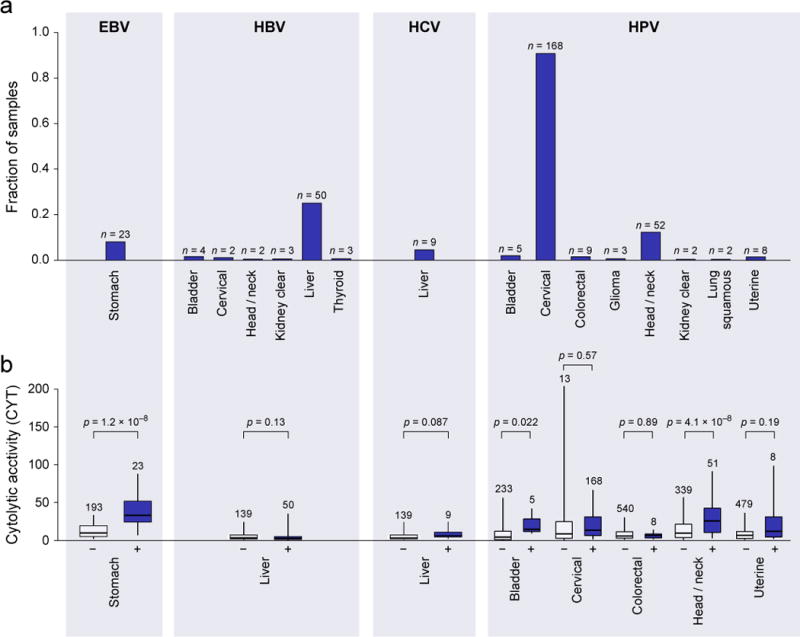
(A) Rates of viral infection, as defined by viral RNA-Seq read counts exceeding those observed in GTEx, for tumor types exhibiting at least one case. Isolated cases of several other viruses were also observed. (B) Distribution of CYT in tumor samples with (+) or without (−) viral infection. In tumor types affected by multiple viruses, “negative” samples include only those negative for all viruses. Box plots as in Figure 1. P-values are according to Wilcoxon rank-sum test. See also Data S2 and Table S2.
HBV and HCV were primarily observed in liver cancer (25% and 5%, respectively), as expected, with occasional instances of HBV infection in diverse tumor types. The extra hepatic cases do not exhibit hepatic gene expression signatures, suggesting that these are not the result of metastases (Data S2B). We also observed singleton cases of Kaposi sarcoma virus (lung squamous cell carcinoma and stomach cancer), BK polyoma (bladder cancer), and Merkel cell polyoma (ovarian cancer). While we did observe type I interferon activation and B cell infiltration for HCV+ liver cancer (Data S2C), these viruses did not show an identifiable association with cytolytic activity.
To probe indirectly for the presence of viruses, we looked for associations between CYT and two other correlates of viral infection, HLA genotype and APOBEC activity. While association with HLA genotype was not observed for a single tumor type (although there was a pan-cancer association with HLA-A31; Data S2D), we did detect association with high APOBEC activity in tumors with viral involvement (head and neck, cervical) and those without known viral involvement (breast, bladder) (Data S2E), suggesting potential for unknown virus infections in some tumors.
Cytolytic cells are likely to be targeting tumor neoantigens
With recent studies from our group and others showing the presence of neoepitope-specific T cells in patients (Fritsch et al., 2014), we tested for CYT association with the overall rate of mutation and the rate of mutations predicted to yield a neoepitopes (i.e., an expressed peptide capable of binding each patient’s imputed HLA alleles) (Data S3A, S3B, Table S3). On average, 50% of non-silent mutations yielded ≥1 predicted neoepitope, and 39% of these impacted a substantially expressed gene (median expression ≥10 TPM in the given tissue type). Despite considerable inter-tumoral heterogeneity (Table S4A), both metrics exhibited significant positive association with CYT in multiple tumor types, most notably uterine cancer, breast cancer, stomach cancer, cervical cancer, and lung adenocarcinoma (Figure 3A, 3B). Consistent with a smoking etiology, lung adenocarcinomas from ever-smokers demonstrated significantly higher CYT than those from never-smokers (p=0.003) (Data S3C). Melanoma mutations exhibited a likely association with CYT. Associations of mutations or neoepitopes with CYT were matched by correlations for other T cell markers, but less so with interferon-responsive genes (Data S3D, S3E). These data are consistent with neoepitopes driving CYT for many tumor types.
Figure 3. Count of predicted antigenic mutations per sample is linked with cytolytic activity and selectively depleted in certain tumor types.
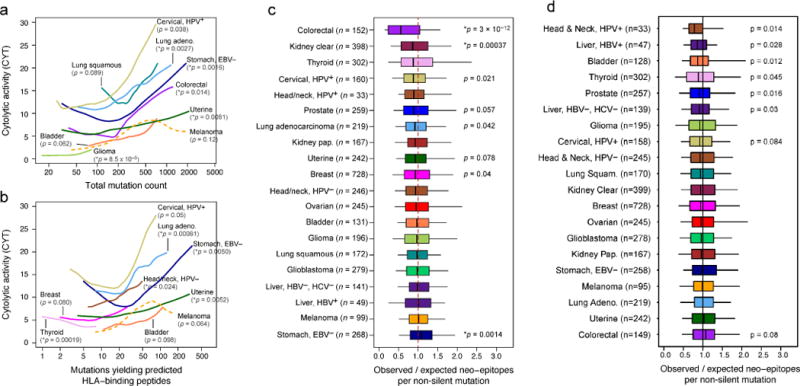
(A) Local regression curves showing significant relationships between CYT and total mutation count in eight tumor types (p<0.1, Spearman rank correlation), plus melanoma (dotted line). Curves span the 5th to 95th percentile of the mutation count variable. Colors correspond to tumor type and are the same as appear in Figure 1. (B) Analogous to (a), but based on the count of point mutations predicted to yield an antigenic neo-epitope. Potential for antigenicity was defined based on gene expression and potential to bind the corresponding patient’s imputed HLA with high affinity. (C) For each tumor, the count of point mutations predicted to generate neo-epitopes was divided by the total count of non-silent point mutations to yield Bobs/Nobs. This observed ratio was compared to an expected ratio, Bpred/Npred, estimated from the mutational spectra of the silent point mutations in the given sample using an empirical model (Methods). The ratio of the observed and predicted ratios represents the relative deviation of the neo-epitope rate from expectation. P-values reflect Wilcoxon rank-sum tests for deviation from 1. (D) Analogous to Figure 3C, but using neo-epitope prediction based on randomly re-permuted HLA genotype assignments (across patients). Asterisks denote trends significant at 10% FDR for all panels. See also Data S3, Table S3, and Table S4.
However, since the per-sample rate of neoepitope yielding mutations closely tracks with the overall rate of mutation (Spearman rho=0.91; Data S3F), CYT may be driven by mutation rate rather than neoepitopes. To test a role for neoepitopes, we reasoned that T cell-mediated immune surveillance would lead to elimination of immunogenic sub-clones expressing neoepitopes. To quantify neoepitope depletion, we determined how the rate of predicted neoepitopes generated per non-silent point mutation deviated from a null model based on the observed mutation rate of silent point mutations. We found that colorectal cancer and kidney clear cell cancer demonstrated dramatic depletions of neoepitopes (Figure 3C; associated gene expression changes, Table S4B). Because neoepitope predictions are dependent on HLA genotypes, we reasoned that random shuffling of HLA genotypes would abrogate the depletion signal (Figure 3D). As expected, depletion was eliminated for colorectal cancer and kidney clear cell cancer (and we note that the residual enrichment for other tumor types may reflect degeneracy of peptide binding across HLA alleles). These findings are consistent with a model in which immune surveillance activities cull subclones expressing immunogenic antigens.
We conclude that neoepitopes are likely to be driving cytolytic activity in a number of tumors, and that the resulting antigen-specific CTLs can eliminate tumor clones harboring these neoepitopes.
Ectopic gene expression, endogenous retroviruses and necrosis associated with CYT
Another potential source of tumor antigens is a unique set of genes, known as cancer testis (CT) antigens, which are not expressed in healthy tissues, except germ cells, but are aberrantly expressed in tumors and associated with antigen-specific responses in patients harboring these tumors. Ectopic expression is likely due to disturbances in genomic methylation and reactivation of stem-like expression programs that may contribute to tumorigenicity (Simpson et al., 2005). Using a set of 276 known CT genes (Almeida et al., 2009), we used GTEx to identify a subset of 60 that are transcriptionally silent in normal non-germline tissues. Ectopic expression was observed for most tumor types, especially melanoma, head and neck, lung, liver, stomach, and ovarian cancer (Data S4A; Table S4A). In no tumor type was there a clear positive association between the CYT and the count of expressed CT antigens (Data S4B). We queried individual CT antigens for correlation with CYT (Table S5A), and observed positive associations for CSAG2 in breast cancer (p=1.2e-15), head and neck cancer (p=1.9e-7), kidney clear cell cancer (p=9.9e-5), and other tumor types. Associations for canonical antigens, such as NY-ESO-1 (CTAG1), were less consistent. We hypothesized that T cell surveillance would lead to CT antigen silencing through chromosomal deletions, but compelling evidence for this was not observed (Data S4C).
Endogenous retroviruses (ERVs) are another class of germline-encoded elements that may be re-activated in tumors, and we considered whether these might also contribute to anti-tumor immunity. TLR7 or RAG knockouts in mice develop uncontrolled ERV expression, ERV infectivity, and ERV insertion-driven tumors (Young et al., 2012; Yu et al., 2012) yet little is known about ERV-immune and ERV-cancer interactions in humans. Given reports that these elements are transcriptionally and sometimes even translationally active in humans (Boller et al., 1997; Schmitt et al., 2013), we considered the possibility that they trigger immune sensing in tumors. Therefore, we mapped TCGA RNA-Seq data to a recently published annotation of 66 expressed ERV family members (Table S5B, Data S4D) and assessed associations with cytolytic activity (Mayer et al., 2011). By comparing GTEx and TCGA tissue controls to TCGA tumor samples, we observed numerous instances of ERVs demonstrating re-activation in tumors, including one instance of an ERVH-2 element exceeding 2,700 reads per million in a stomach adenocarcinoma (Data S4E). From these data we surprisingly discovered a conservative set of three tumor-specific endogenous retroviruses (‘TSERVs’) all with minimal to undetectable expression in normal tissues and elevated expression in tumor tissues (Figure 4A).
Figure 4. Endogenous retroviruses tied to local immunity.
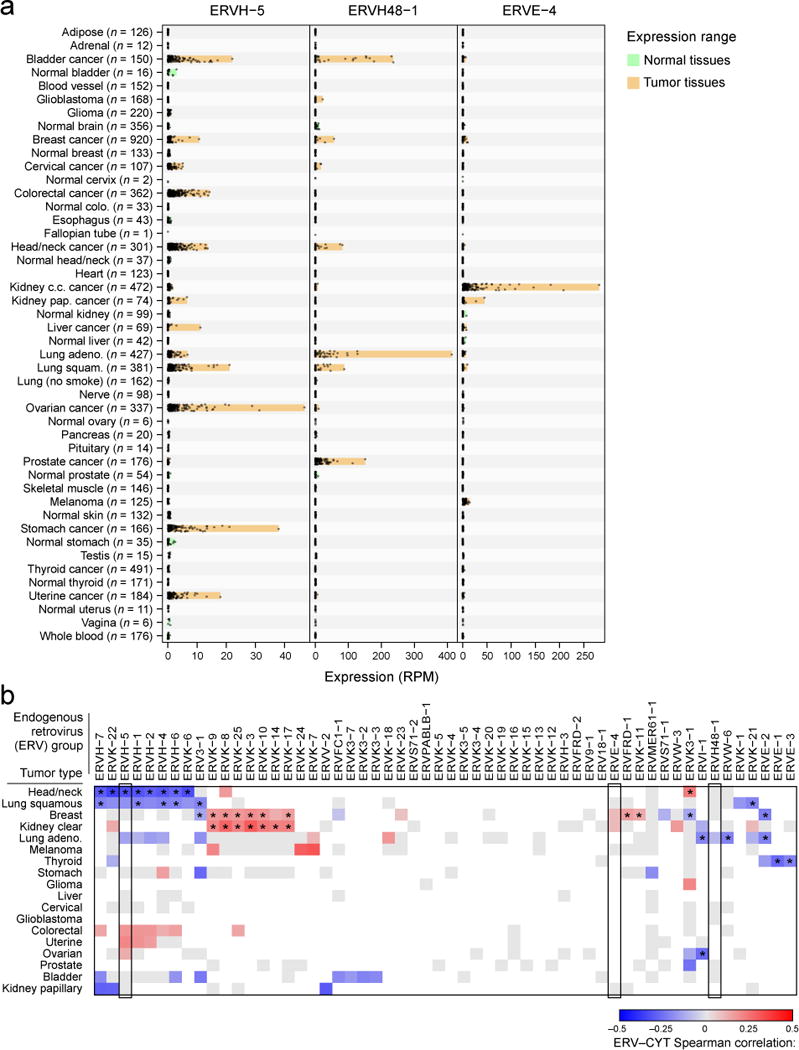
(A) RNA-Seq-derived ERV expression in reads per million (RPM) across 18 TCGA tumor types and 27 non-tumor tissue types (from TCGA and GTEX) for three elements found to be tumor-specific. The expression ranges (minimum value to maximum value) are highlighted in orange (for tumor tissues) or green (for non-tumor tissues). (B) Spearman-rank correlations between CYT and ERV expression. Gray squares indicate non-significant association (unadjusted p>0.05) and blank squares indicate no over-expression of the given ERV in the given tumor type (expression strictly below the normal tissue maximum). Asterisks (*) denote Bonferroni-significant associations (adj. p<0.05). See also Data S4 and Table S5.
Assessing the gene expression correlates of each TSERV in the tumor type exhibiting highest expression, we observed that immune pathways were typically the most significantly enriched (Table S5C). Many ERVs, in addition to the TSERVs, demonstrated association with CYT in multiple tumor types (Figure 4B). While we cannot determine whether ERVs activate immunity or inflammation triggers ERVs (Manghera and Douville, 2013), we conclude that ERVs are highly dysregulated in tumors and speculate that they may yield tumor-specific peptide epitopes (Boller et al., 1997) or act as immunological adjuvants to activate local immunity (Yu et al., 2012).
Another potential source of antigens and immunostimulatory ligands is dying cells. Thus, we explored the potential role for necrosis in driving CYT and immune infiltration in general. Rates of necrosis were highest in glioblastoma (Data S4F) and showed modest positive association (p<0.05) with CYT in glioblastoma, bladder, and ovarian cancer; but notably, association with macrophage markers was consistently stronger (Data S4G).
Mutations in specific driver genes were enriched in tumors with higher cytolytic activity
We hypothesized that high cytolytic activity could select for tumors with somatic mutations that render them resistant to immune attack. We therefore asked whether CYT is associated with mutations in 373 ‘driver’ genes that are frequently mutated in cancer based on analysis of TCGA exome sequencing data (q<0.1 by MutSigCV (Lawrence et al., 2013); Table S6A). Using a regression-based approach to look for pan-cancer association of these mutated genes with CYT, controlling for tumor type and background mutation rate, we found 35 genes (adjusted p<0.1; Figure 5A, Data S5A, Table S6B). In contrast, synonymous somatic mutations were not associated with CYT (adj. pmin=0.09). Of the top 10 CYT-associated mutations, 8 were also associated with an independent marker of CTLs (CD8a; 10% FDR; Data S5B), demonstrating the robustness of our CYT metric. Of the individual tumor types, uterine, stomach and colorectal had the most associations (15, 11, 6 respectively) while kidney clear cell and ovarian, which showed markedly higher CYT compared to normal tissue, had just one each, and lung adenocarcinoma had none. Strikingly, somatic mutations, except TP53, were all positively associated with CYT, consistent with a model in which tumors develop resistance mutations under selection pressure.
Figure 5. Gene mutations associated with high or low immune cytolytic activity.
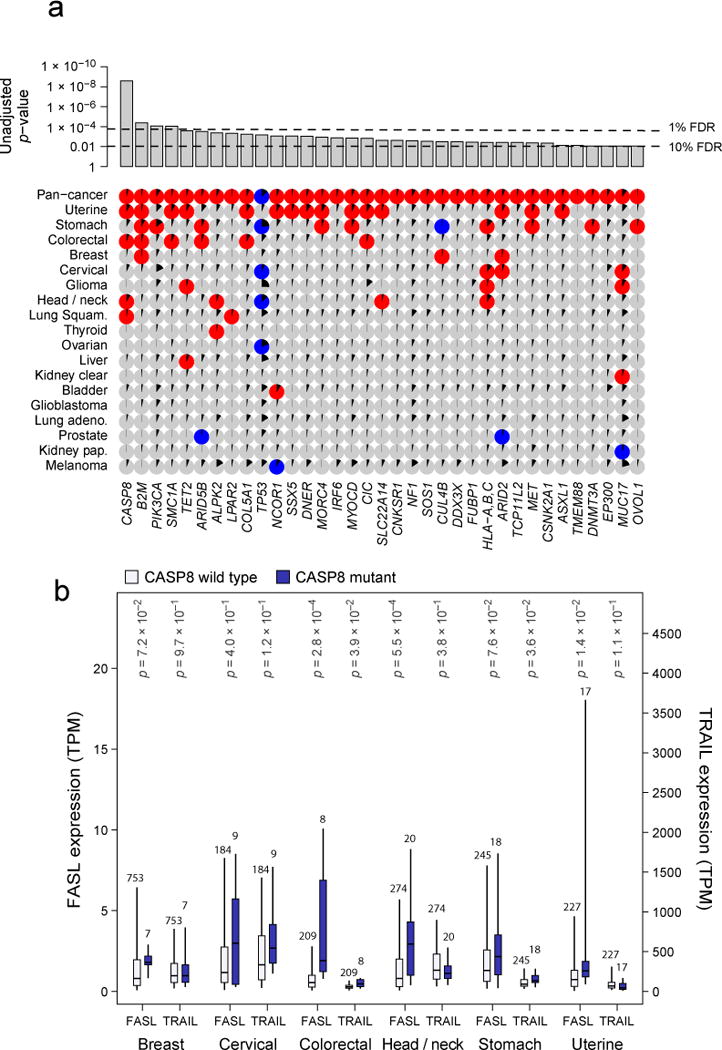
(A) Only genes showing pan-cancer significance (adj. p<0.1, red for positive, blue for negative and grey for non-significant association) for non-silent mutation association with CYT are shown in top row. Additional rows, clustered by similarity, show independent significant (unadjusted p<0.05) enrichment upon sub-analysis. The black wedges represent the share of samples exhibiting mutation. Bar plot indicates unadjusted pan-cancer p-values for mutational association with CYT, dashed lines indicating thresholds yielding 1% and 10% FDRs. (B) Association between CASP8 mutational status and FASLG (left axis) and TRAIL (right axis) gene expression (TPM) for tumor types demonstrating at least 5 instances of nonsynonymous CASP8 mutation. Light and dark bars correspond to wild type and (nonsynonymous) mutant samples, respectively. Box plots as in Figure 1. P-values are calculated by Wilcoxon rank-sum test. See also Data S5 and Table S6.
We note that while we predicted that cytolytic activity would have the strongest impact on the mutation landscape, we also identified gene mutations strongly associated with other immune cell types/functions (adj. p<.01; Data S5B), including STK11 and VHL with reduced macrophage signature, BRAF with increased expression of costimulatory genes, and AXIN2, SNX25 and others with the differential enrichment score of CD8+ T compared to Treg.
Higher CYT was associated with mutations in genes involved in antigen-presentation, extrinsic apoptosis and innate immune sensing
Several themes emerged when we considered the known functions of the identified genes.
First, the most enriched gene, CASP8 (adj. p=8.8e-7), is a critical player in the extrinsic apoptosis pathway and was enriched in head and neck cancer, colorectal cancer, lung squamous cell carcinoma, and uterine cancer (where it showed a maximal mutation frequency of 7.0%). The pattern of mutation was diffuse and suggested loss of function (Data S5C), a potential mechanism by which a tumor cell could evade FasL- or TRAIL-induced apoptosis. Between FasL and TRAIL, FasL is most correlated with CASP8 mutations and thus more consistent with such a hypothesis (Figure 5B). A study in mice indeed demonstrated that blockade of CASP8 results in tumor escape from CTLs (Medema et al., 1999), and our result indicates that this may be a common mechanism in human tumors (that may evade CTLs or NK cells). Interestingly, four additional genes with significant but less definitive statistical enrichment also had well-established roles in regulating extrinsic apoptosis. These include, CNKSR1 (Garimella et al., 2014), MET (Fan et al., 2001; Garofalo et al., 2009), CSNK2A1 (Ravi and Bedi, 2002) (Izeradjene et al., 2005) (Llobet et al., 2008; Wang et al., 2006), and PIK3CA (Saturno et al., 2013; Song et al., 2010). PIK3CA mutations, which were often the well-known activating alterations E545K and H1047R (Samuels and Ericson, 2006), showed their strongest enrichment in stomach cancer, demonstrating a 20% mutation rate and a strong positive association with EBV infection (p=2.9e-10). As in the case of CASP8, mutations in each of these genes were more closely associated with FASL expression than TRAIL expression. We conclude that loss of the extrinsic apoptosis pathway may represent a general mechanism for tumors to escape immune cytolytic activity.
Second, the invariant chain of MHC Class I, B2M, was the next most strongly enriched gene (adj. p=7.1e-3), showing independently significant association in uterine, breast, colorectal cancer, and stomach cancer, which exhibited the highest rate, 5.7%. The most frequent event was the same CT dinucleotide deletion observed previously in melanoma patients relapsing from T cell-based immunotherapy (Chang et al., 2005). The MHC Class I locus itself was also significant (Table S6C; HLA-A, -B, -C mutations considered jointly, adj. p=5.3e-2). HLA-A and HLA-B alleles were mutated about 3 times as frequently as HLA-C alleles. No specific alleles showed strong evidence for being especially frequently mutated. The tumor types with the highest rates of HLA mutation, stomach cancer (14%), cervical cancer (12%), and head and neck cancer (11%), were also among those with frequent viral involvement. However, viral infection was not significantly associated with HLA mutation in any of them (Table S6D). Given the requirement of MHC Class I and B2M in presenting tumor antigens to cytotoxic CD8 T cells, we consider the enrichment of MHC Class I and B2M mutations in high-CYT tumors (Khong and Restifo, 2002) as an independent and strong validation of CYT as a measure of cytolytic activity. While MHC Class II genes were not significantly mutated pan-cancer, class II gene mutations, considered collectively, were positively associated with CYT (unadj. p=0.017) with independent significance in bladder cancer (unadj. p=0.0084).
Other hits included the CT antigens MORC4 (Liggins et al., 2007) and SSX5 (Ayyoub et al., 2004) and genes with roles in innate immune sensing, including DDX3X (Oshiumi et al., 2010) and ARID2 (Yan et al., 2005). We also note that mutant TP53 is negatively correlated with CYT, which may be explained either by a role for p53 in regulating immunity (e.g., loss of p53-regulated stress ligands that induce cytoxicity, (Textor et al., 2011) or from absence of viral infection (consistent with p53 mutations being anti-correlated with viral infection in stomach (p=2.3e-5) and head and neck cancer (p=2.6e-4); Table S6D).
Because MSI-high colorectal tumors are known to be immunogenic (Kloor et al., 2010), we also considered whether MSI-high tumors were enriched for mutations in particular genes with respect to MSI-low and microsatellite stable (MSS) tumors. Mirroring the CYT analysis, CASP8 and MHC Class I mutations were the most enriched mutations in MSI-high tumors (p adj. = 1.5e-5 and 1.4e-12, respectively), with COL5A1, SMC1A, CIC, ARID2, CNKSR1, and DNMT3A also significant (adj. p < 0.05) (Table S6E).
Finally, we note that some candidate genes with well-known immune function (Table S6A) did not show association with CYT. However, enrichment in the expression of immune-related genes were observed in tumors with mutations in some of these genes (TNFRSF14, CLEC4E, CD1D, IL32; Table S6F).
Loci containing known immune regulators show copy number alterations associated with CYT
We also considered the possibility that specific regions of the genome may be preferentially focally amplified or deleted (based on a dataset of TCGA samples profiled with SNP6.0 arrays) in high- or low- CYT tumors. As with the point mutation analysis, we looked for pan-cancer CYT association with copy number alterations (CNAs) using regression and controlling for cancer subtype and background mutation rate (of amplifications and deletions). This approach yielded 13 significantly amplified regions (with 3 adjacent to each other on 6q) and 1 significantly deleted region (FDR=10%) (Figure 6A, Table S7). Although CNAs include variable segments of a chromosomal region and do not typically identify causative genes, many of the identified regions harbored plausible candidates.
Figure 6. Amplifications and deletions are associated with cytolytic activity in tumors.
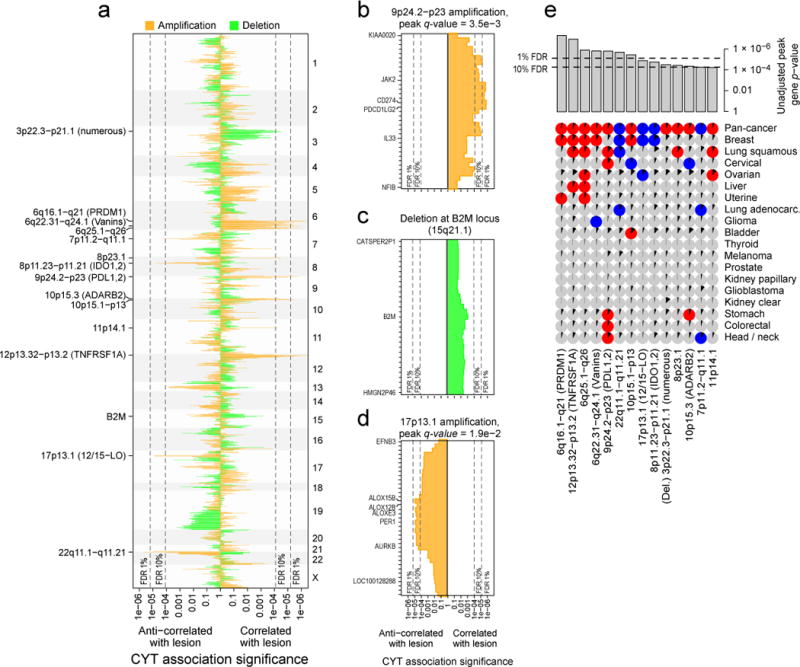
(A) The significance of association between CYT and amplification (orange) and between CYT and deletion (green) for all genic loci. Rightward lines show unadjusted p values for instances in which the lesion was positively associated with CYT, and leftward lines show unadjusted p values for instances in which the lesion was negatively associated with CYT. Dotted lines represent the significance cutoff yielding 1% and 10% FDRs (and also appear in parts B–E). Labels on the right side mark events significant at the 10% FDR, plus B2M. Potential driver genes appear in parentheses. (B) Locus zoom on the 9p24.2-p23 amplification, each bar corresponding to a single gene. Labeled genes include those with driver potential or those on the locus boundary. (C) Locus zoom on the region containing B2M, which was not genome-wide significant. (D) Locus zoom on the 17p13.1 amplification. (E) Significant associations between CNAs and CYT on the pan-cancer and cancer-specific level (as in Figure 5). Pan-cancer significance was defined at a 10% FDR, and significance for individual tumor types was defined at unadjusted p<0.05. Positive association is indicated with red circles, negative with blue circles, and non-association with gray circles. Black wedges indicate the share of samples exhibiting the event (ie. non-zero GISTIC score at the locus). Bar plot indicates unadjusted pan-cancer p-values for CNAs, sorted by significance, with dashed lines indicating thresholds yielding 1% and 10% FDRs. See also Data S6 and Table S7.
On chromosomes 9 and 8, we found two well-known targets of cancer immunotherapy. First, amplification of 9p23-p24.2 (Figure 6B), a region including PDL1 (CD274) and PDL2 (PDCD1LG2),was positively associated with CYT in lung squamous cell carcinoma, head and neck cancer, cervical cancer, stomach cancer, and colorectal cancer (Figure 6E). While tumor cells and tumor infiltrating leukocytes are known to express these ligands, our results suggest that tumor-expressed ligands affect tumor fitness in the presence of cytolytic activity. Second, 8p11.21–8p11.23 (Data S6A) showed increased probability of amplification in low-CYT tumors (pan-cancer and breast) and is adjacent to IDO1 and IDO2, enzymes that degrade extracellular tryptophan and create a potent immunosuppressive microenvironment, which may explain the associated reduction in CYT (Uyttenhove et al., 2003).
In addition, potential new targets were identified. These included 17p13.1, which was preferentially amplified in low-CYT tumors (Figure 6D), including breast and ovarian. The peak genes, ALOX12B/ALOX15B (12/15-LO) regulate immunity in many ways, including blocking the uptake of apoptotic cells by inflammatory monocytes in a manner that decreases antigen presentation to T cells (Uderhardt et al., 2012), which may explain the observed decrease in CYT. Further supporting this model, the amplification was associated with higher necrosis in breast (p=0.002) and kidney clear cell cancer (p=0.0002), though not ovarian cancer. Other peaks included ones near TNFRSF1A and PRDM1 (Data S6A) as well as a suggestive, but not genome-wide significant enrichment at B2M (Figure 6C). In considering how other enrichment signatures might associate with CNAs (Data S6B), we observed a dramatic positive association between increased MHC Class I expression and amplification of the MHC Class II complex (adj. p<5e-4).
Discussion
Based on the notion that effective natural anti-tumor immunity requires a cytolytic immune response (Figure 7A), we quantified cytolytic activity using a simple expression metric of effector molecules that mediate cytolysis. Our analysis was designed to address which genetic and environmental factors drive tumor-associated cytolytic activity, and how this cytolytic activity selects for genetic resistance in tumors. Our results suggest that neoantigens and viruses are likely to drive cytolytic activity, and reveal known and novel mutations that enable tumors to resist immune attack.
Figure 7. Proposed model for evolution of tumor-immune associations.
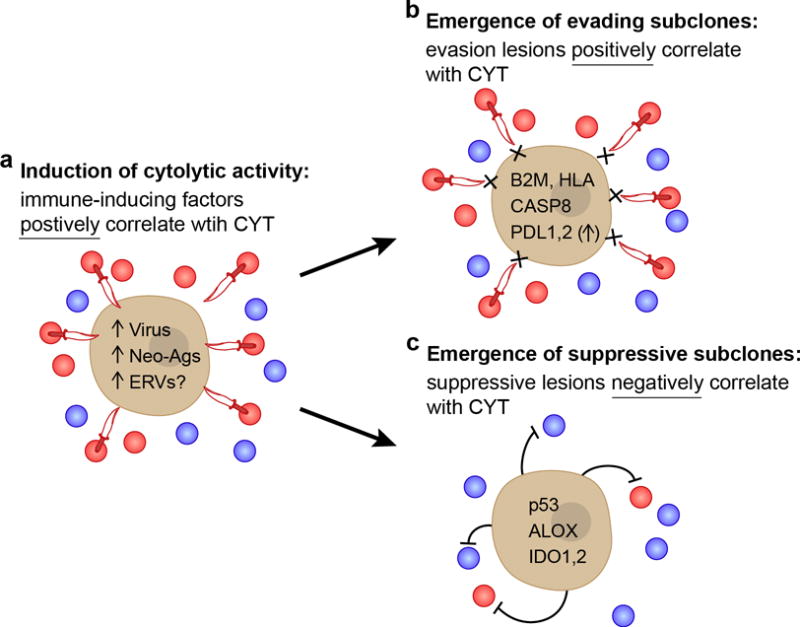
(A) As the tumor develops, we propose that intrinsic tumor factors – such as mutated neoantigens or viruses – induce local immune infiltrates (blue circles) that include cytolytic effector cells (expressing GZMA/PRF1; red circles) that kill tumors (daggers). These factors are expected to be positively correlated with CYT across tumors. (B,C) Under pressure from cytolytic immune cells, subclones with resistance mutations will grow out over time. (B) One subset of these mutations would enable tumors to evade killing, but does not impact the infiltrate, and are positively correlated with CYT (i.e. higher infiltrate samples are enriched for these mutations). (C) Another subset suppresses the immune infiltrate (i.e. lower infiltrate samples are enriched for these mutations), and is negatively correlated with CYT. Notably, p53 mutations and ALOX amplifications were also significantly negatively associated with CD8A, suggesting a reduction in cell numbers and not just activity. See also Data S7 and Table S8.
We considered several explanations for the elevated immune cytolytic activity observed in some tumors (Figure 7A). First, we asked whether neoantigens play a role. These are a compelling set of antigens because of their absence from the thymus and thus lack of central tolerance that would normally delete cognate high-affinity T cells. Indeed, we found that neoantigen load positively associated with cytolytic activity across multiple tumor types, and that neoantigens appear to be depleted in tumors relative to their expected numbers based on the silent mutation rate, consistent with the notion of immunoediting (Schreiber et al., 2011). Second, when we analyzed CT antigens that are expressed selectively in tumors, we could not detect a positive correlation between the number of expressed CT genes and cytolytic activity. In addition, CT antigen genes were not contained within deletions associated with CYT, contrary to what would be expected if there were immune pressure on CT antigens. Although we did not uncover a role for CT antigens in spontaneous immunity (perhaps because our methods were not optimized to detect CT depletion), we did highlight a subset of 60 CT antigens that are highly tumor-specific and may be (or are already) excellent targets for immunotherapy, including vaccines, adoptive T cell transfer or CAR-T therapy. Third, we asked whether viruses could be inducers of immune responses. In some tumors, we observed that cytolytic activity does indeed associate with the presence of exogenous or endogenous viruses, and we expect that some viruses would trigger immunity through RNA and DNA sensors and generate immunogenic antigens for the adaptive immune response.
To learn more about how tumors adapt to attack by cytolytic immune cells, we also searched for enrichment of somatic genetic alterations in tumors with high versus low cytolytic activity. As expected, we observed enrichment of mutations in antigen presentation machinery (thus validating our cytolytic metric), including HLA and B2M, as well as extrinsic apoptosis genes, such as CASP8, that would prevent cytolytic cells from killing tumors via FasL-Fas interactions. In addition, we found cytolytic activity correlating with amplifications in regions containing genes that function in immunosuppression, such as PDL1/2. Most of the identified mutations – including HLA, B2M and CASP8 – were positively correlated with CYT and are likely to represent autonomous escape mechanisms (Figure 7B). In addition, we identified a smaller number of mutations that correlated negatively with cytolytic activity – including IDO1 and IDO2, p53, and the ALOX locus – and may represent non-autonomous mechanisms of suppressing immunity (Figure 7C). Finally, we were surprised that CYT-associated genetic lesions represent ~10% of drivers, and these genes had largely not been studied in the context of immunity. However, given the importance of immune responses in controlling tumor progression (Pages et al., 2005), tumors may have evolved several mechanisms of evasion.
Our approach has allowed us to positively identify the subset of tumor types that are sensitive to spontaneous cytolytic activity (Data S7, Table S8). If we consider positive correlation of HLA, B2M or CASP8 mutations with CYT as a ‘signature’ of selection pressure by the immune system, we find that colorectal, uterine, stomach, head and neck, cervical, lung squamous and breast tumors are most susceptible to immune elimination. If we further consider depletion of neoepitopes as an independent signature of selection, we identify colorectal as well as kidney clear cell cancer as immune-susceptible tumors. For these tumor types, we thus suggest that spontaneous tumor immunity can delete tumor cells.
For several tumor types, we did not find evidence for immunoediting. This could be due to: insufficient power to detect associations in tumors with low rates of spontaneous immunity, non-genetic evasion mechanisms that we cannot detect, or true absence of immune cytolytic activity (perhaps for thyroid and prostate cancers, for example).
Finally, the mutations associated with cytolytic activity reveal potential genetic biomarkers for predicting outcome and candidate targets for immunotherapy. To assess the utility of these markers, one would need to genotype tumors for the 35 identified genes at clonal or subclonal levels, and test if pre-treatment or post-treatment mutations predict refractoriness or relapse in response to cytolytic immunotherapy. We predict that the presence of these mutations (assuming they do not lead to complete loss of susceptibility) indicates that re-activation of CD8 T cells would be therapeutically effective. In addition, we identified new candidates for therapeutic development, including the ALOX enzymes and their products, the PIK3CA protein that is enriched in activating mutations in high-CYT stomach cancers, and FASL which may be useful to upregulate in T cells to enhance the anti-tumor activity of adoptively transferred T cells.
Analysis of TCGA samples has revealed environmental and genetic mechanisms that impact tumor-immune interactions. While we chose to focus on cytolytic activity because of its central role in tumor elimination and the feasibility of monitoring its activity, we did not consider other tumoricidal activities (such as antibody-dependent cell-mediated cytotoxicity) because we are not aware of transcript-based markers for these activities. In addition, the CYT metric we used is transcript-based and thus may not reflect changes in cytolytic activity due to post-transcriptional regulation, and is a snapshot in time that may miss previous activity that impacted tumor growth. We anticipate that improved experimental measurements of anti-tumor immune activity will further reveal the genetic and epigenetic changes that underlie co-evolution of tumor cells and immune cells.
Experimental Procedures
Tumor and normal samples and datasets
Analyzed samples represent untreated primary tumors, except for melanoma, which included metastases. Metastases to lymph nodes were always excluded as were patients that received neo-adjuvant therapy. Gene-level RNA-Seq expression data were accessed from GDAC Firehose (Broad Institute TCGA Genome Data Analysis Center, 2014) (tumors and normals) and from the GTEx web portal (GTEx Consortium, 2013b) (normals only). RNA-Seq-based sequence data from the corresponding projects were accessed through CGHub and the Short Read Archive (SRP012682), respectively, and used to estimate expression of endogenous and exogenous viruses. Additional gene expression data were accessed from the CCLE web portal (http://www.broadinstitute.org/ccle/home) (Barretina et al., 2012) (Affymetrix U133+2 microarrays) and Fantom5 (Fantom Consortium et al., 2014) (cap analysis gene expression) and used to evaluate gene expression markers. Whole exome sequencing-derived point mutation calls were accessed from TumorPortal (Lawrence et al., 2014), Synapse workspace syn1729383 (https://www.synapse.org/#!Synapse:syn1729383; (Kandoth et al., 2013)), TCGA Data Portal (National Institute of Health), GDAC Firehose (Broad Institute TCGA Genome Data Analysis Center, 2014), and the TCGA Research Network stomach adenocarcinoma publication (Cancer Genome Atlas Research Network, 2014). Whole exome sequencing-based sequence data, used to call HLA genotypes and mutations, were accessed through CGHub. GISTIC2 (Mermel et al., 2011) gene-level, zero-centered, focal copy number calls for each patient were accessed from GDAC Firehose (Broad Institute TCGA Genome Data Analysis Center, 2014). Clinical data for each tumor type were accessed from the TCGA public access web portal. Tables S9 and S2 catalog the TCGA and GTEx samples included in the study.
Cytolytic activity and other cell type-specific signatures
Cytolytic activity (CYT) was calculated as the geometric mean of GZMA and PRF1 (as expressed in TPM, 0.01 offset). Marker genes for specific cell types were identified as those with expression at least 2-fold greater than observed in any other cell type (using Fantom5 and DMAP),and enrichment was calculated using ssGSEA (Barbie et al., 2009). CYT-dependent survival analyses via Cox proportional hazards were performed by separating patients into a high-CYT cohort and a low-CYT cohort, each with an identical admixture of histology-stage combinations.
Expression of exogenous and endogenous retroviruses
Viral expression was quantified by mapping unmapped RNA-Seq reads (bowtie2 (Langmead and Salzberg, 2012)) to viral sequence variants deposited in GenBank and normalizing against the count of mapped reads. Positive identification required at least 300nt of unique sequence to map to the viral genome and expression exceeding that observed in GTEx normals. To quantify the expression of endogenous retroviruses, RNA-Seq data (from TCGA and GTEx) was re-mapped (bowtie2 (Langmead and Salzberg, 2012)) to an annotation of known expressed elements (Mayer et al., 2011). For each ERV, the 95th percentile expression value was calculated per tissue type, and if this value was less than <10 TPM in all normal tissues, >10 TPM in a tumor type, and at least 5-fold higher than in all non-tumor tissues, then the ERV was deemed tumor-specific.
Tumor-specific HLA Typing, HLA-binding neoepitope prediction and CT antigen identification
The 4-digit HLA type for each sample was inferred using POLYSOLVER (POLYmorphic loci reSOLVER) which uses a normal tissue .bam file as input and employs a Bayesian classifier to determine genotype (unpublished, SAS, CJW and GG). By comparing to matched tumor .bams, POLYSOLVER also identified HLA mutations. Neo-epitopes were predicted for each patient by defining all novel amino acid 9mers and 10mers resulting from mutation in expressed genes (median >10 TPM in the tumor type) and determining whether the predicted binding affinity to the patient’s germline HLA alleles was <500 nM using NetMHCpan (v2.4) (Nielsen et al., 2007; Rajasagi et al., 2014)). A set of potential cancer testis (CT) antigens was defined by finding known CT antigens (Almeida et al., 2009) with negligible expression in GTEx normal tissues (95th percentile value <1 TPM in all somatic tissue types).
Comparison of expected to observed neoantigen load per tumor
To test whether the count of neo-epitopes was different from expected (ignoring the expression-based filter and excluding indels), the rate at which each mutational spectrum produces neo-epitopes was calculated empirically pan-cancer, and the silent mutations in each patient used to infer the expected ratio of neo-epitopes per non-silent mutation. This was compared to the actual ratio observed in the patient. Random shuffling of HLA genotypes amongst patients served as a control.
Association of CYT with point mutations and amplifications/deletions
Candidate genes were tested for non-silent point mutation association with CYT using a regression-based approach with CYT (rank-transformed) as the dependent variable, mutational status of the gene in question as the independent variable, and cancer histological subtype and the background rate of non-silent point mutations as additional control variables. Hits were defined at q<0.1. Candidate genes were defined by running MutSigCV (Lawrence et al., 2013) on each tumor type separately and all the tumor types collectively (q<0.1) and merging with a previously published result set (Lawrence et al., 2014). To assess for association between CYT and copy number alterations, a regression-based approach was likewise used, using CYT (rank-transformed) as the dependent variable, amplification or deletion signal as the independent variable, and cancer histological subtype and the background rate of copy number alteration as additional control variables. “Peaks” were defined as contiguous regions with p<0.01, and permutation testing was used to determine whether the peak score (based on the most enriched gene in the region) was truly significant (adj. p<0.1).
Supplementary Material
Acknowledgments
We are grateful to the TCGA Research Network (http://cancergenome.nih.gov/), the Genotype-Tissue Expression (GTEx) Project (dbGaP phs000424.v3.p1), the FANTOM research consortium (http://fantom.gsc.riken.jp/), and the Broad-Novartis Cancer Cell Line Encyclopedia (http://www.broadinstitute.org/ccle/home) for providing the data analyzed in this manuscript. We thank Mara Rosenberg, Amaro Taylor-Weiner, Chloe Villani, Arnon Arazi, Pavan Bachireddy, and Dan-Avi Landau for their feedback and help accessing data and analysis tools, and Leslie Gaffney for assistance with artwork. Funding was provided by the Blavatinik Family Foundation (NH), the MGH Research Scholars Program (NH), and the NIH Training Program in Bioinformatics and Integrative Genomics training grant (MSR).
Footnotes
Author Contributions
M.S.R. and N.H. conceived the project, designed analysis strategies, and wrote the manuscript. M.S.R. developed and performed the computational analyses. S.S., C.J.W., and G.G. developed the tools for HLA genotyping, mutation calling, and neo-epitope prediction.
References
- Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, Cohen T, Chen YT, Chua R, Gurung S, Gnjatic S, et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic acids research. 2009;37:D816–819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyoub M, Hesdorffer CS, Montes M, Merlo A, Speiser D, Rimoldi D, Cerottini JC, Ritter G, Scanlan M, Old LJ, et al. An immunodominant SSX-2–derived epitope recognized by CD4+ T cells in association with HLA-DR. Journal of Clinical Investigation. 2004;113:1225–1233. doi: 10.1172/JCI20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Boller K, Janssen O, Schuldes H, Tonjes RR, Kurth R. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J Virol. 1997;71:4581–4588. doi: 10.1128/jvi.71.6.4581-4588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad Institute TCGA Genome Data Analysis Center. Firehose. Clinical Data (Broad Institute of MIT and Harvard) 2014 [Google Scholar]
- Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, Holt RA. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome research. 2014;24:743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Campoli M, Restifo NP, Wang X, Ferrone S. Immune selection of hot-spot beta 2-microglobulin gene mutations, HLA-A2 allospecificity loss, and antigen-processing machinery component down-regulation in melanoma cells derived from recurrent metastases following immunotherapy. Journal of immunology. 2005;174:1462–1471. doi: 10.4049/jimmunol.174.3.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Ma YX, Gao M, Yuan RQ, Meng Q, Goldberg ID, Rosen EM. The multisubstrate adapter Gab1 regulates hepatocyte growth factor (scatter factor)-c-Met signaling for cell survival and DNA repair. Molecular and cellular biology. 2001;21:4968–4984. doi: 10.1128/MCB.21.15.4968-4984.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantom Consortium. Pmi R, Clst. Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Lassmann T, Itoh M, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch EF, Hacohen N, Wu CJ. Personal neoantigen cancer vaccines: The momentum builds. Oncoimmunology. 2014;3:e29311. doi: 10.4161/onci.29311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garimella SV, Gehlhaus K, Dine JL, Pitt JJ, Grandin M, Chakka S, Nau MM, Caplen NJ, Lipkowitz S. Identification of novel molecular regulators of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in breast cancer cells by RNAi screening. Breast cancer research : BCR. 2014;16:R41. doi: 10.1186/bcr3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nature genetics. 2013a;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. GTEx Portal (Broad Institute of MIT and Harvard) 2013b pp. GTEx_Analysis_RNA-seq_RNA-SeQCv1.1.8_gene_reads__Pilot_2013_2001_2031_patch2011.gct.
- Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs CS, Rosenberg SA. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunological reviews. 2014;257:56–71. doi: 10.1111/imr.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeradjene K, Douglas L, Delaney A, Houghton JA. Casein kinase II (CK2) enhances death-inducing signaling complex (DISC) activity in TRAIL-induced apoptosis in human colon carcinoma cell lines. Oncogene. 2005;24:2050–2058. doi: 10.1038/sj.onc.1208397. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA: a cancer journal for clinicians. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer immunology, immunotherapy : CII. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BJ, Costelloe EO, Fitzpatrick DR, Haanen JB, Schumacher TN, Brown LE, Kelso A. Single-cell perforin and granzyme expression reveals the anatomical localization of effector CD8+ T cells in influenza virus-infected mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2657–2662. doi: 10.1073/pnas.0538056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong HT, Restifo NP. Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nature immunology. 2002;3:999–1005. doi: 10.1038/ni1102-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloor M, Michel S, von Knebel Doeberitz M. Immune evasion of microsatellite unstable colorectal cancers. International journal of cancer. 2010;127:1001–1010. doi: 10.1002/ijc.25283. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay S, Davidson D, Latour S, Veillette A. Dok-3, a novel adapter molecule involved in the negative regulation of immunoreceptor signaling. Molecular and cellular biology. 2000;20:2743–2754. doi: 10.1128/mcb.20.8.2743-2754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggins AP, Cooper CD, Lawrie CH, Brown PJ, Collins GP, Hatton CS, Pulford K, Banham AH. MORC4, a novel member of the MORC family, is highly expressed in a subset of diffuse large B-cell lymphomas. British journal of haematology. 2007;138:479–486. doi: 10.1111/j.1365-2141.2007.06680.x. [DOI] [PubMed] [Google Scholar]
- Llobet D, Eritja N, Encinas M, Llecha N, Yeramian A, Pallares J, Sorolla A, Gonzalez-Tallada FJ, Matias-Guiu X, Dolcet X. CK2 controls TRAIL and Fas sensitivity by regulating FLIP levels in endometrial carcinoma cells. Oncogene. 2008;27:2513–2524. doi: 10.1038/sj.onc.1210924. [DOI] [PubMed] [Google Scholar]
- Lund JM, Hsing L, Pham TT, Rudensky AY. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 2008;320:1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghera M, Douville RN. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology. 2013;10:16. doi: 10.1186/1742-4690-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J, Blomberg J, Seal RL. A revised nomenclature for transcribed human endogenous retroviral loci. Mobile DNA. 2011;2:7. doi: 10.1186/1759-8753-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema JP, de Jong J, van Hall T, Melief CJ, Offringa R. Immune escape of tumors in vivo by expression of cellular FLICE-inhibitory protein. The Journal of experimental medicine. 1999;190:1033–1038. doi: 10.1084/jem.190.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome biology. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Health. TCGA Data Portal. [Google Scholar]
- Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Roder G, Peters B, Sette A, Lund O, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PloS one. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Sakai K, Matsumoto M, Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. European journal of immunology. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. The New England journal of medicine. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, Zhang W, Sougnez C, Cibulskis K, Sidney J, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124:453–462. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi R, Bedi A. Sensitization of tumor cells to Apo2 ligand/TRAIL-induced apoptosis by inhibition of casein kinase II. Cancer research. 2002;62:4180–4185. [PubMed] [Google Scholar]
- Rutledge WC, Kong J, Gao J, Gutman DA, Cooper LA, Appin C, Park Y, Scarpace L, Mikkelsen T, Cohen ML, et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:4951–4960. doi: 10.1158/1078-0432.CCR-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Current opinion in oncology. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, Jungbluth AA, Frosina D, Gnjatic S, Ambrosone C, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saturno G, Valenti M, De Haven Brandon A, Thomas GV, Eccles S, Clarke PA, Workman P. Combining trail with PI3 kinase or HSP90 inhibitors enhances apoptosis in colorectal cancer cells via suppression of survival signaling. Oncotarget. 2013;4:1185–1198. doi: 10.18632/oncotarget.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome biology and evolution. 2013;5:307–328. doi: 10.1093/gbe/evt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Haensch W, Roefzaad C, Schlag PM. Prognostic significance of activated CD8(+) T cell infiltrations within esophageal carcinomas. Cancer research. 2001;61:3932–3936. [PubMed] [Google Scholar]
- Schwitalle Y, Kloor M, Eiermann S, Linnebacher M, Kienle P, Knaebel HP, Tariverdian M, Benner A, von Knebel Doeberitz M. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;134:988–997. doi: 10.1053/j.gastro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Sharma P, Wagner K, Wolchok JD, Allison JP. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nature reviews Cancer. 2011;11:805–812. doi: 10.1038/nrc3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nature reviews Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Kim JH, Sun BK, Alcala MA, Jr, Bartlett DL, Lee YJ. c-Cbl acts as a mediator of Src-induced activation of the PI3K-Akt signal transduction pathway during TRAIL treatment. Cellular signalling. 2010;22:377–385. doi: 10.1016/j.cellsig.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and Tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Science translational medicine. 2013;5:200ra116–200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang KW, Alaei-Mahabadi B, Samuelsson T, Lindh M, Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nature communications. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor S, Fiegler N, Arnold A, Porgador A, Hofmann TG, Cerwenka A. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer research. 2011;71:5998–6009. doi: 10.1158/0008-5472.CAN-10-3211. [DOI] [PubMed] [Google Scholar]
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uderhardt S, Herrmann M, Oskolkova OV, Aschermann S, Bicker W, Ipseiz N, Sarter K, Frey B, Rothe T, Voll R, et al. 12/15-lipoxygenase orchestrates the clearance of apoptotic cells and maintains immunologic tolerance. Immunity. 2012;36:834–846. doi: 10.1016/j.immuni.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, Boon T, Van den Eynde BJ. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nature medicine. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- Wang G, Ahmad KA, Ahmed K. Role of protein kinase CK2 in the regulation of tumor necrosis factor-related apoptosis inducing ligand-induced apoptosis in prostate cancer cells. Cancer research. 2006;66:2242–2249. doi: 10.1158/0008-5472.CAN-05-2772. [DOI] [PubMed] [Google Scholar]
- Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes & development. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Lubben W, Slomka H, Gebler J, Konert M, Cai C, Neubrandt L, Prazeres da Costa O, Paul S, Dehnert S, et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 2012;37:867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


